Abstract
Phot proteins (phototropins and homologs) are blue-light photoreceptors that control mechanical processes like phototropism, chloroplast relocation, or guard-cell opening in plants. Phot receptors consist of two flavin mononucleotide (FMN)-binding light, oxygen, or voltage (LOV) domains and a C-terminal serine/threonine kinase domain. We determined crystal structures of the LOV1 domain of Phot1 from the green alga Chlamydomonas reinhardtii in the dark and illuminated state to 1.9 Å and 2.8 Å resolution, respectively. The structure resembles that of LOV2 from Adiantum (Crosson, S. and K. Moffat. 2001. Proc. Natl. Acad. Sci. USA. 98:2995–3000). In the resting dark state of LOV1, the reactive Cys-57 is present in two conformations. Blue-light absorption causes formation of a proposed active signaling state that is characterized by a covalent bond between the flavin C4a and the thiol of Cys-57. There are differences around the FMN chromophore but no large overall conformational changes. Quantum chemical calculations based on the crystal structures revealed the electronic distribution in the active site during the photocycle. The results suggest trajectories for electrons, protons, and the active site cysteine and offer an interpretation of the reaction mechanism.
INTRODUCTION
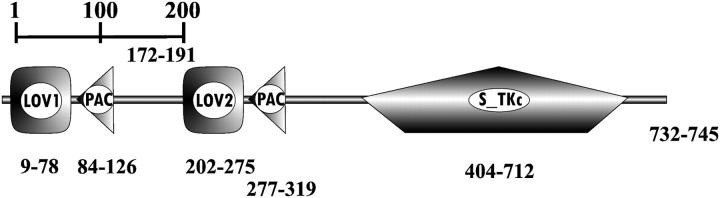
Light is not only essential for plant growth but is also an important signal for plant development. It influences almost all processes in a plant's life cycle from germination to flowering. As the first step in any signaling process is the detection of the signal, plants have several light sensors that cover different spectral ranges and intensities. To date, three widely abundant classes of plant photoreceptors have been identified: the red/far-red light-absorbing phytochromes (Fankhauser, 2001) and two distinct classes of blue-light receptors, the cryptochromes and phototropins (Christie and Briggs, 2001; Briggs et al., 2001). The latter, now dubbed Phot proteins (Briggs et al., 2001) are plasma membrane-associated proteins that function as primary photoreceptor for phototropic plant movement (Huala et al., 1997; Christie et al., 1998; Christie et al., 1999), chloroplast relocation (Jarillo et al., 2001; Kagawa et al., 2001), and stomatal opening in guard cells (Kinoshita et al., 2001). Phot proteins (such as Phot1) contain two functional units: an N-terminal sensory part containing two light, oxygen, and voltage (LOV) sensitive domains (LOV1, LOV2) that bind the chromophore flavin mononucleotide (FMN) (Christie et al., 1999); and a C-terminal serine/threonine kinase domain (Fig. 1, A and B). LOV domains belong to the PAS domain superfamily that is found in a diverse group of sensor proteins (Zhulin et al., 1997). Upon blue-light irradiation, Phot receptors autophosphorylate (Christie et al., 1998), which might be necessary for the in vivo function or for photoreceptor adaptation.
FIGURE 1.
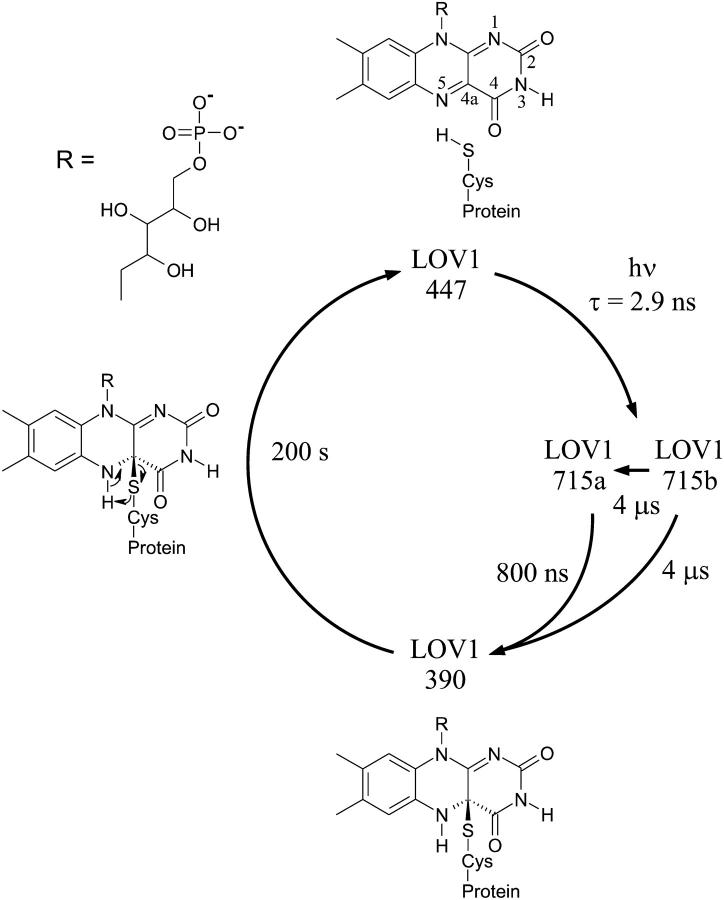
(A) Schematic representation of the modules of the C. reinhardtii phot1 protein, consisting of two PAS-related domains (LOV), two PAS-associated PAC motifs, and a serine/threonin kinase (S-TKc). (B) Amino acid sequence of LOV1 and LOV2 from C. reinhardtii (Cr), Avena sativa (At, oat), and Adiantum capillus-veneris (Ac). Residues identical in C. reinhardtii LOV1 and other LOV sequences are labeled in gray. Reactive Cys is shown in white on black background. *Residues that directly interact with the FMN. (C) Photocycle of LOV1 from C. reinhardtii, adapted from Kottke et al. (2003).
The crystal structure of the LOV2 domain of the Phytochrome/Phot hybrid photoreceptor Phy3 of the fern Adiantum capillus-veneris constitutes a typical PAS domain fold and reveals how the FMN cofactor is embedded in the protein matrix (Crosson and Moffat, 2001). Moreover, it shows that the conserved Cys-966 is located 4.2 Å from the flavin C4a position. So far, the photocycles have been studied of the LOV2 domain from Avena sativa (oat) (Swartz et al., 2001) and the LOV1 domain from the alga Chlamydomonas reinhardtii (Kottke et al., 2003) (Fig. 1 C): upon absorption of blue light, a short-lived (microseconds) red-shifted species is formed that is consistent with a flavin triplet state. This transient intermediate decays into a metastable form absorbing maximally at 390 nm (LOV-390) that recovers spontaneously but on a timescale of several (oat, Crosson and Moffat (2001)) to many (Chlamydomonas, Kottke et al. (2003)) tens of seconds, to the dark form absorbing maximally at 447 nm. Salomon et al. (2000) suggested that the dominant photoproduct constitutes a FMN-cysteineyl adduct. This would be analogous to other flavoproteins such as mercuric ion reductase (MerA; Miller et al., 1990), lipoamide dehydrogenase, and thioredoxin reductase (Williams, 1992). The prediction that a flavin C4a-thiol adduct is formed (Fig. 1 C) has been confirmed recently by the crystal structure of the photoexcited state of LOV2 from maidenhair fern phy3 (Crosson and Moffat, 2002) and is supported by site-directed mutagenesis studies. Replacing the cysteine residue in the highly conserved sequence GRNCRFLQG by Ala or Ser led to an unbleachable LOV that does not form the intermediate LOV-390 (Salomon et al., 2000; Kottke et al., 2003). NMR studies on the light-induced differences in the chemical shift values (13C, 15N, 31P) of specifically labeled FMN bound to the LOV2 domain of A. sativa (Salomon et al., 2000) are consistent with a flavin-(C4a)-thiol adduct and suggest that conformational changes take place around the FMN chromophore.
The C. reinhardtii Phot1 and Phy3 from Adiantum belong to one subgroup within the phylogram of the Phot family (Briggs et al., 2001). So far, the Phot1 receptor from C. reinhardtii is the only one known of its kind from a lower plant. It is thought to be involved in blue-light-mediated chloroplast development and/or relocation and in the blue-light dependent gametogenesis (Hegemann et al., 2001; Huang and Beck, 2002). Because LOV1 domains of Phot receptors from higher plants are expressed with low yield and the LOV domains from algae and fern seem to be closely related, we set out to determine the spectral (Kottke et al., 2003) and structural characteristics of the LOV1 domain from C. reinhardtii. Having these data from the same protein allows for a one-to-one correlation of the experimental findings, which is important to deduce the structural basis for the functional properties of the protein. We present crystal structures of the dark and illuminated forms of LOV1. The structures are compared with the one of LOV2 from the fern Phy3, interpreted in light of the spectroscopic data, and used as a basis for quantum chemical calculations to obtain insight into the reaction mechanism.
MATERIALS AND METHODS
LOV1 expression and purification
The full-length cDNA clone AV 394090 (Asamizu et al., 1999) of the C. reinhardtii phot1 (formerly named nph1 for nonphototropic hypocotyl-1) was obtained from C. Beck and K. Huang (Freiburg). The gene fragment coding for the FMN-binding LOV1 domain (amino acids 16–133) was inserted into the Escherichia coli expression vector pET16 such that the protein carries 1 Gly, 10 His, and a Factor Xa protease cleavage site derived from the vector sequences at the N-terminus. The protein was expressed in E. coli strain BL21, purified via nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen, Hilden), and stored at a concentration of 10.5 mg/ml in 10 mM sodium phosphate with 10 mM sodium chloride, pH 8.0. Analysis by gel filtration showed that LOV1 exists as a mixture of monomer (80–90%) and tetramer (10–20%). The chromophore in LOV1 was shown to be flavin mononucleotide (FMN, riboflavin-5′-phosphate) by chromophore extraction in 1% TCA, protein precipitation, and electrospray mass spectroscopy (SSQ 7000, Finigan). The molar mass of the apoprotein is 15,609 g/mol, differing by the mass of a methionine from the calculated mass (15,739 g/mol) of the protein and 457.2 g/mol for the chromophore.
Crystallization, spectroscopic characterization, x-ray data collection, and structure determination
Two different crystal forms were grown in the dark at 20°C by vapor diffusion using the hanging-drop geometry. Protein and reservoir solutions (1 μl each) were mixed; the latter contained 100 mM Na+-HEPES buffer at pH 7.3, 1.5–1.8 M ammonium sulfate, and 1–8% PEG 8000. Plate-shaped 200-μm long crystals appeared within days, and much smaller (10 × 10 × 30 μm) pencil-shaped crystals grew after a couple of weeks. Only the latter form gave useable diffraction data. Before flash cooling, crystals were rinsed in a cryoprotection solution consisting of 100 mM Na+-MES at pH 6.0, 1.6 M ammonium sulfate, 20% glycerol. The crystals used for structure determination of the dark state were flash-cooled in the dark with the microscope light bulb shielded with a 2-mm-thick RG 630 filter (ITOS, Mainz, Germany). For the crystals used for determining the photoproduct structure, the crystals were mounted in a loop and placed for 10–20 s in front of a 100-W Xenon lamp shielded with a 2-mm-thick GG 435 filter (ITOS) before flash cooling in liquid nitrogen. Visible absorption spectra were recorded of single crystals mounted in a loop and kept at 100 K using a microspectrophotometer (4Dxray Systems AB, Uppsala, Sweden). Diffraction data were collected at the European Synchrotron Radiation Facility, Grenoble (ESRF; see Table 1 for details) using an ADSC Quantum 4 detector and reduced with the XDS program package (Kabsch, 1993). For structure determination of the photoproduct LOV1-390, the first 15 degrees of data of two randomly oriented crystals were merged (see Results and Discussion). Because LOV1 crystals have differing unit cell lengths (±1.5%), possibly due to an effect of the cryoprotectant, it was difficult to find crystals with diffraction data that could be merged. Due to some nonisomorphism, data and refinement statistics are much poorer for the composite dataset than for the other two datasets. The enantiomorph of the spacegroup (P6522) was determined by molecular replacement (Navaza, 2001) using the structure of the LOV2 domain (Protein Data Base (PDB) code 1G28 (Crosson and Moffat, 2001)) as a search model. The refinement of the molecular replacement solution using the Crystallography and NMR System (CNS; Brunger et al., 1998) included simulated annealing and individual B-factor refinement. The topology and parameter files for the covalent Cys-57–FMN adduct were obtained from quantum chemical calculations (see below). During several cyclic rounds of refinement and manual rebuilding, a sulfate ion and solvent molecules were included in the model. The final models display good stereochemistry (see Table 1). Figures were made with Bobscript (Esnouf, 1997) and Raster3D (Merritt and Murphy, 1994).
TABLE 1.
Crystal parameters, data collection, and refinement statistics
| Illuminated state
|
|||
|---|---|---|---|
| Complex | Dark state LOV1-447 | Dataset of a single crystal | Composite dataset of LOV1-390 |
| PDB code | 1N9L | 1N9N | 1N9O |
| Crystal parameters: P6522 unit cell (a = b, c), Å | 122.1, 46.1 | 120.9, 46.0 | 121.9, 45.5 |
| Data collection | |||
| Beamline (ESRF, Grenoble) | ID14-4 | ID29 | ID29, ID14-4 |
| Wavelength, Å | 0.9393 | 0.969 | 0.969/0.939 |
| Data statistics | |||
| Resolution, Å | 15.0–1.9 | 15.0–2.3 | 15.0–2.8 |
| No. of observations/unique reflections | 77,830/16142 | 69,773/9184 | 16,983/5009 |
| Completeness (total/high) [%]* | 98.0/96.6 | 99.5/99.9 | 95.2/82.5 |
| I/σ(I) (total/high)* | 12.8/3.5 | 12.6/4.6 | 7.5/2.6 |
| Rsym,(total/high)*† | 6.8/35.1 | 13.2/43.1 | 12.9/32.2 |
| Refinement statistics | |||
| Resolution range, Å | 15.0–1.9 | 15.0–2.3 | 15.0–2.8 |
| Included amino acids | 17–125 | 18–125 | 18–125 |
| No. of protein atoms | 844 | 834 | 834 |
| No. of waters | 108 | 114 | 26 |
| No. of SO42− ions | 1 | 0 | 1 |
| Rwork, Rfree, %‡ | 23.2/24.8 | 21.2/26.5 | 25.0/29.8 |
| Root-mean square deviation for bonds/angles, Å/degrees | 0.008/1.28 | 0.008/1.34 | 0.009/1.32 |
Completeness, Rsym, and I/σ(I) are given for all data and for the highest resolution shell: dark, 1.9–2.0 Å; illuminated 1, 2.3–2.4 Å; composite dataset, 2.8–2.9 Å.
Rsym = ∑|I − 〈I〉|/∑I.
Rwork = ∑|Fobs| − k|Fcalc|/∑|Fobs|.
5% of randomly chosen reflections were used.
Quantum chemical calculations
Quantum chemical calculations were used to analyze the electronic properties of van der Waals complexes of FMN and Cys-57 (Cys-57°FMN) in the dark state and upon photon absorption and to create model, topology, and parameters for the covalent Cys-57–FMN adduct. Cys-57°FMN includes two conformations of Cys-57, Cys-57(1) and Cys-57(2), and the flavin fragment of FMN in ground singlet (FMN0) and first excited triplet (FMN1) states, corresponding to the LOV1-447 and LOV1-715 states of the LOV1 photocycle, respectively (Fig. 1 C).
The Cys-57°FMN models were constructed on the basis of the crystal structures. Geometry optimization of the models and electronic property calculations were performed with ab initio complete active space self-consistent field method (CASSCF; Roos, 1987). The energies of the optimized structures were refined using second-order multireference quasi-degenerate perturbation theory corrections with Møller-Plesset zero-order Hamiltonian (MCQDPT2; Nakano, 1993). All ab initio calculations were performed using the PC GAMESS version (Alex A. Granovsky, http://www.classic.chem.msu.su/gran/gamess/index.html) of the GAMESS (US) Quantum Chemistry package (Schmidt et al., 1993) in 6-31G* basis set of atomic orbitals. The position of the Cys-57 Cα atom was restrained during geometry optimization to ensure the effect of the protein framework. The optimized geometries of the Cys-57°FMN models were very close to the experimental ones with root-mean square (r.m.s.) deviation of all nonhydrogen model atoms < 0.05 Å.
Highest occupied and lowest unoccupied molecular π−orbitals (HOMO and LUMO) for the closed-shell electronic configurations of the Cys-57°FMN complexes belong to a flavin subsystem and have compositions similar to those of isolated FMN. These orbitals were used to generate CASSCF wave functions. The energy differences between the calculated low-lying electronic states are in good agreement with the experimental absorption spectrum (e.g., the experimental band at 447 nm is reproduced with a precision of 10 nm). The extension of the active space or basis set of atomic orbitals led to an overall lowering of the absolute energy but did not change the energy differences significantly. Thus the computational approach was accurate and ensured good agreement with experimental data. To estimate more precisely the energies of Cys-57°FMN complexes, MCQDPT2 corrections were made for CASSCF wave functions. All occupied valence-shell molecular orbitals and all virtual orbitals were included in the perturbation calculation. The MCQDPT2 energies of the Cys-57°FMN complexes are used for the comparisons described below (see Results and Discussion).
The model of the covalent Cys-57–FMN adduct was obtained by combining quantum mechanics/molecular mechanics (QM/MM) semiempirical quantum chemical geometry optimization using parametric method 3 (PM3; Stewart, 1989) and x-ray crystallographic refinement using experimental diffraction data. Model construction and QM/MM PM3 calculations were performed with the program HyperChem (evaluation version 6.03, Hypercube, Inc., 2001), and the crystallographic refinement was done using CNS and O (see above). At the beginning a preliminary model of FMN with hydrogen-saturated N5 and C4a atoms was created and optimized using PM3. This model was inserted into the protein structure, and the C4a–hydrogen bond was replaced with a C4a–S(Cys-57) bond. Subsequently, the QM/MM PM3 geometry optimization was performed, where the Cys-57–FMN adduct was treated quantum mechanically, and the rest of the protein molecule was considered as producing a fixed potential field. The PM3-optimized structure was refined manually, fit into the experimental electron-density map, and subjected to automatic crystallographic refinement by simulated annealing of the protein part containing residues Asn-56, Cys-57, and Arg-58. This structure was used as an initial approximation for the next QM/MM PM3 geometry optimization. The cycle was repeated until self-consistency was reached. The final Cys-57–FMN geometry was used to generate topology and parameters for the crystallographic refinement.
RESULTS AND DISCUSSION
The overall organization of the C. reinhardtii Phot1 protein resembles that of higher plant Phot1 (Fig. 1, A and B). It consists of two LOV domains (Taylor and Zhulin, 1999) that are composed of PAS-A and -B domains, each of which is linked to a conserved PAS-associated PAC domain. The spacing between the two LOV domains is short compared with higher plant Phot proteins. Of the 104 amino acids in the LOV1 core (Fig. 1 B), 32 are fully conserved in the two LOV domains from C. reinhardtii and the four domains from oat and fern, which demonstrates the low divergence between LOV1 and LOV2 and between lower and higher plants, respectively.
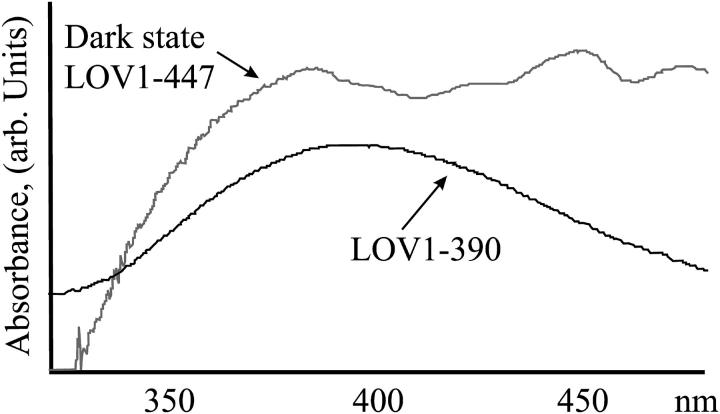
The LOV1 domain of the algal phot1 cDNA was expressed in E. coli. LOV1 was purified via affinity chromatography, characterized spectroscopically (Holzer et al., 2002; Kottke et al., 2003), and crystallized. To ensure the structure determination of well-defined states of the photosensitive LOV1 crystals, light exposure of the crystals was controlled carefully. For the structure determination of the dark state, crystals were mounted under orange lighting, and for the photoproduct structure, crystals were exposed to light with λ > 345 nm to minimize a possible blue-light-induced back-reaction (Kottke et al., 2003). Before x-ray data collection, the state of the cryocooled crystals was verified by microspectroscopy (see Fig. 3 A). The spectra are less fine structured than might be expected due to the low transmittance of the crystals.
FIGURE 3.
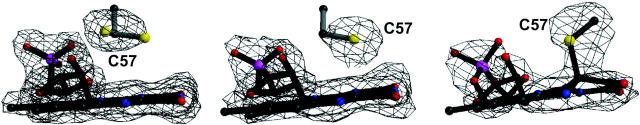
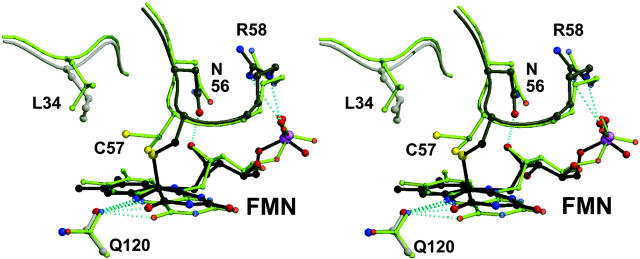
(A) Optical spectra of LOV1 crystals used for the determination of the dark state (upper trace) and the photoproduct LOV1-390 (lower trace). (B) Simulated annealing omits electron-density maps of FMN and Cys-57. Left, dark state; middle, data obtained of a single illuminated crystal (the bond between C4a and Cys-57 has broken due to x-ray radiolysis); right, composite dataset of two illuminated crystals. See text for further explanation. Maps are contoured at 3.0 σ, and the final model is overlaid. (C) Stereoview of the superposition of the dark state of LOV1 and the photoproduct LOV1-390. Atom coloring is the same as in Fig. 2; model of the dark state is shown in gray. Structural differences are restricted to the neighborhood of the FMN chromophore.
Crystal structure of the LOV1 dark state
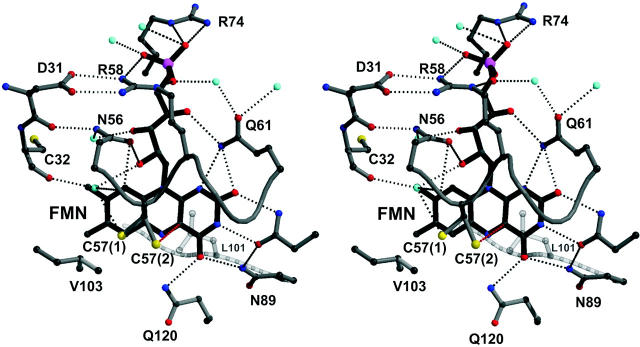
The LOV1 structure was determined by molecular replacement using the structure of fern LOV2 as a search model (Crosson and Moffat, 2001). The structures are very similar with a root-mean square deviation of 0.98 Å between 104 Cα atoms. The similarity extends to the FMN-binding pocket that is mainly polar on the pyrimidine side of the isoalloxazine ring and nonpolar around the dimethylbenzene moiety (Fig. 2). As in the case of oat LOV2, the phosphate interacts with the guanidinium group of Arg-58 (Arg-58-NE–O1P, 2.8 Å; Arg-58-NH2–O2P, 3.0 Å), whereas Arg-74 has weak electron density and seems to be mobile. The hydroxyl groups of the ribityl chain form the following interactions: O4′–Gln-61–NE2, 3.2 Å; O2′–Asn-56-OD1, 2.7 Å; and O2′–Wat-515, 2.9 Å. The heteroatoms of the isoalloxazine ring interact with the following side chains: N1–Gln-61-NE2, 3.3 Å; O2–Gln-61-NE2, 3.1 Å; O2–Asn-89-ND2, 3.0 Å; N3–Asn-89-OD1, 2.9 Å; O4–Asn-99-ND2, 3.0 Å; O4–Gln-120-NE2, 2.9 Å; and N5–Gln-120-NE2, 3.3 Å. Differences close to the bound FMN exist between algal LOV1 and fern LOV2, respectively, at the residue stacking below the isoalloxazine ring (Leu-101/Phe-1010), at Leu-87/Val-996, Ile-77/Val-986, Cys-32/Asn-941, Ala-25/Thr-934, and Leu-34/Ile-943. The latter is located next to Cys-57/Cys-966 involved in formation of the C4a-flavin photoproduct. The conformation of Cys-57 is therefore of particular interest. The structure of the dark state of LOV1 was determined to 1.9 Å resolution (Table 1). The thiol of Cys-57 has two conformations. The higher occupied one (70 ± 10%) is located 3.8 Å from C5a and 4.4 Å from C4a and points toward Leu-60. In the lower occupied conformation, the thiol is located 3.5 Å from C4a. The finding of two conformations of the active site cysteine has not been reported for LOV2, possibly due to the lower resolution (2.73 Å) of the structure (Crosson and Moffat, 2001). There are no charged residues or water molecules close to C4a or N5 of the flavin and the thiol group of Cys-57.
FIGURE 2.
Stereoview of the FMN binding site of Chlamydomonas LOV1. To facilitate comparison with fern LOV2, an orientation has been chosen that resembles the one in Fig. 2 B in Crosson and Moffat (2001). Hydrogen bonds are indicated by dotted lines, water molecules by cyan spheres, atoms are colored by elements: carbon, gray; oxygen, red; nitrogen, blue; phosphorus, magenta; and sulfur, yellow. As described in detail in the text, the Cys57 S-H group has two orientations.
Structure of the photoproduct LOV1-390
Microspectrophotometric analysis of illuminated cryocooled crystals showed an almost complete conversion of LOV1-447 into LOV1-390 (Fig. 3 A). A dataset was collected to 2.3 Å resolution of a single illuminated LOV1 crystal (see Table 1). Surprisingly, although the side chain of Cys-57 rotated to fully occupy the position atop the flavin C4a atom, there was no electron density indicative of a bond between C4a and the thiol (Fig. 3 B). The distance is 3.1 Å, which is too short for a van der Waals contact but too long for a covalent bond. Because the adduct between the flavin C4a and the cysteinyl would have to be somewhat unstable to ensure reversibility of the photocycle, we suspected that the bond might have broken during data collection due to x-ray radiolysis. Photoreduction of cysteine disulfide bonds (Ravelli and McSweeney, 2000; Burmeister, 2000) and other radiation-induced effects (Ravelli and McSweeney, 2000; Burmeister, 2000; Schlichting et al., 2000; Berglund et al., 2002) have been observed before. To reduce the detrimental effect of the x rays, we merged the first 15 degrees of data of two crystals in a composite dataset. Due to nonisomorphism of LOV1 crystals, it was not possible to merge data of more crystals, which would have allowed use of fewer degrees of data per crystal to obtain a full dataset. As can be seen from Fig. 3 B, there is clear electron density for a covalent bond between the C4a atom of the isoalloxazine ring and the thiol group of Cys-57. The parameters for the refinement of the adduct were obtained as described (see Materials and Methods). There are no gross changes between the structures of dark and illuminated states of LOV1. As can be seen from Fig. 3, B and C, the flavin is no longer planar since the hybridization of C4a is sp3. The bond length between C4a and the Cys-75 thiol is 1.89 Å. Because Cys-57 has to move to “reach” C4a, Leu-34 follows to fill the void. As shown in Fig. 3 C, the flavin ring tilts, and larger conformational changes take place at the residues surrounding Cys-57, Asn-56, and Arg-58. In the photoproduct LOV1-390 N5 is protonated (see Fig. 1 C). The side chain of Gln-120 can interact with both the lone pair of N5 and a protonated N5; in LOV1-390 the carbonyl of Gln-120 is within 3.3 Å (Figs. 2 and 3 C). The structural changes upon photoexitation seem to be similar in LOV1 (this study) and LOV2 (Crosson and Moffat, 2002). Because the latter coordinates are not in the Protein Data Bank, a detailed comparison is not possible.
Crystal structures and spectroscopic findings
The crystal structures suggest a structural basis for the finding that the back-reaction, i.e., the decay of the photoproduct LOV1-390, is strongly pH dependent with an apparent pKa value between 5 and 6 (Kottke et al., 2003). There are no basic residues close to the active site that could modulate, for example, the abstraction of a proton at the N5 position in the long-lived intermediate LOV1-390. As described above, if one compares the crystal structures of LOV1-447 and LOV1-390, the largest changes occur at and around Cys-57, the residues move closer to the isoalloxazine ring to allow bond formation with C4a. Obviously, for the back reaction this process has to be reversed. Asn-56 and Arg-58 interact with the 2′-hydroxyl group of the ribityl chain and the phosphate group of FMN, respectively. If the phosphate becomes protonated, interactions with Arg-58 become unfavorable and Arg-58 stays closer to the flavin ring thereby stabilizing the bond between Cys-57 and C4a. Thus, we propose that the residue with a pKa between 5 and 6 that governs the pH dependence of the back-reaction is the FMN phosphate.
The flavoenzyme MerA is believed to catalyze the reduction of Hg(II) to Hg(0) via formation of a flavin C4a thiol adduct intermediate. Because the structure of MerA has been known for quite some time (Schiering et al., 1991), and a wealth of mutational and spectroscopic data exists (Miller et al., 1990), MerA has served as a paradigm for other flavoproteins including LOVs. In this context, it is interesting to note that there are differences in the active-site geometry. In MerA, the reactive Cys-140 is located atop the dimethylbenzene moiety of the isoalloxazine ring. In contrast, in LOV1 and LOV2 (Crosson and Moffat, 2001), the reactive cysteine is located not above the flavin ring but instead protrudes “over” the ring and would sit between N5 and C6 when projected. Moreover, MerA undergoes ground-state reactions, whereas all LOVs react from an excited triplet state.
There has been some debate on the protonation state of the reactive thiol in LOV domains (Kottke et al., 2003; Swartz et al., 2001; Crosson and Moffat, 2001). On the basis of the reduced fluorescence in wt-LOV2 compared to the C39S and nonlinear pH dependence between 4 and 10, Swartz et al. (2001) interpreted the reactive Cys as a thiolate. However, the reduced fluorescence is readily explained by the heavy-atom effect of the sulfur. Moreover, the weaker interaction between sulfur and FMN makes the Cys-57 less acidic in LOV1, leaving it protonated over the whole pH range between 4 and 10 (see below), which is supported by spectroscopic findings (Kottke et al., 2003; Iwata et al., 2002; Ataka et al., 2002).
LOV1 photocycle
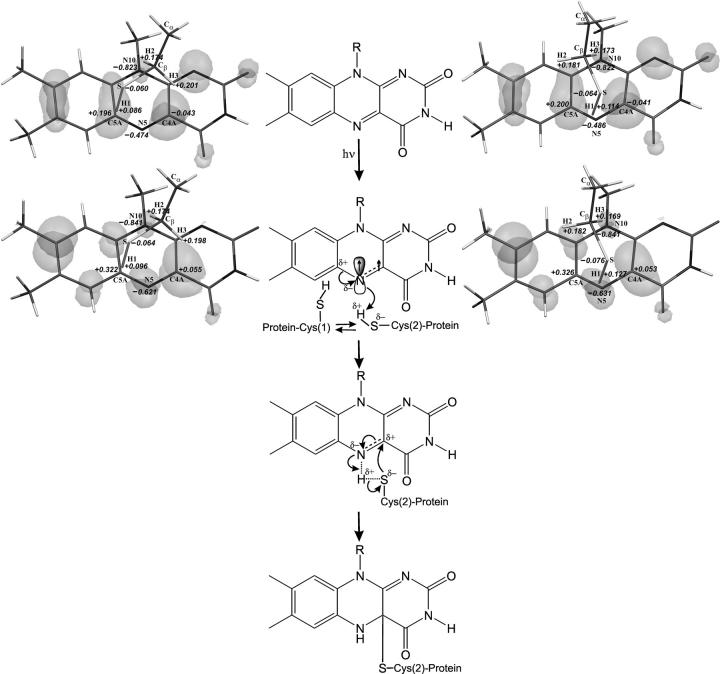
The formation of the covalent Cys-57–FMN adduct is a key step in the LOV1 photocycle. To elucidate the molecular reaction mechanism, we calculated the electronic properties of the ground and first excited triplet state of the isoalloxazine Cys-57 model system. In the crystal structure of the dark resting state (Figs. 2 and 3 B), Cys-57 has two conformations: one close to C5a (conformation 1, Cys-57(1)) and another closer to C4a (conformation 2, Cys-57(2)). Quantum chemical calculations on the ground-state van der Waals complexes Cys-57(1)°FMN0 and Cys-57(2)°FMN0 (Fig. 4) show that the two conformations are stabilized by different interactions of the Cys-57–Cβ hydrogen atoms H2 and H3 with the flavin N10 nitrogen. H2 and H3 carry noticeable partial positive charges, whereas N10 possesses significant partial negative charge. There is no orbital overlap between the H2 or H3 and N10 atoms (the distances DH2-N10 = 2.91 Å and DH3-N10 = 2.88 Å), but the HOMO and next occupied π-orbitals localized on N10 protrude far toward H2 and H3, which adds a significant polarization component to the coulombic interaction. The energies of the two Cys-57°FMN0 complexes differ by 0.4 kcal/mol with Cys-57(2)°FMN0 having the lower energy. In both cases, the N5–C5a bond is polarized with N5 carrying a partial negative charge and C5a carrying a partial positive charge. The C4a atom is almost neutral in that it has negligible negative charge. It is interesting to note that the S–H1 bond is more polarized in Cys-57(2) than in Cys-57(1), rendering H1 more acidic in Cys-57(2). Molecular orbital analysis and electron-density calculations show that there is no orbital overlap between FMN in the ground state and Cys-57, explaining why the reaction does not occur in the dark.
FIGURE 4.
Mechanism of the covalent Cys-57–FMN adduct formation proposed on the basis of experimental and theoretical studies (middle column). Ground (Cys-57(1)°FMN0, upper lane) and excited triplet states (Cys57°FMN1, lower lane) are shown for the two van der Waals complexes of the flavin and Cys-57 in conformation 1 (Cys-57(1), left) and conformation 2 (Cys-57(2), right) as described in the text. HOMO orbitals are shown in gray. Atomic charges were calculated on the basis of Mulliken population analysis (Mulliken, 1955).
Upon photon absorption, the electron distributions of the Cys-57°FMN complexes change. The most important difference is that the HOMO component appears to be localized on the N5 atom of the flavin (which has no HOMO component in the ground-state complexes), resulting in orbital overlap between N5 and the H1 atom of Cys-57(2) (but not of Cys-57(1)). The HOMO component on the C4a atom is less obvious. It is too far from the sulfur of Cys-57(2) to allow orbital overlap. The analysis of the spin-density distribution shows that the unpaired electrons are localized on the N5 and C4a atoms of the flavin, which accompanies the loosening of the N5–C4a double bond. The negative charge on the N5 atom and the positive charge on the C5a atom increase significantly. However, in contrast to published schemes (Swartz et al., 2001; Crosson and Moffat, 2001), C4a remains almost neutral. The positive charge on the H1 atom of Cys-57(2) increases slightly, rendering H1 more acidic. The excitation does not seem to influence the interaction of N10 with H2 or H3 in either conformation.
The correlation of experimental structures and molecular orbitals, partial charges, and spin-density distribution of the ground and excited states of both Cys-57 conformations allows us to suggest the following reaction mechanism for the covalent Cys-57–FMN adduct formation (Fig. 4, middle column). In the dark state, the two conformations of Cys-57 exist in equilibrium with a 70% preference for Cys-57(1). Upon photon absorption, FMN is excited to the triplet state, which results in localization of the highest occupied molecular π-orbital on the N5 atom and ensues interaction of this HOMO with the hydrogen atom H1 bound to the sulfur of Cys-57(2). Preliminary calculations of the reaction pathway suggest that the H1 proton will move toward the N5 atom to be accepted by the HOMO, and during this process, the cysteine sulfur will gain more negative charge. To compensate for the positive charge of the H1 proton, the N5 atom will have to pull electron density from the neighboring C4a and C5a atoms, and because the positive charge on the C5a atom is already high, the process of H1 acceptance will lead to further loosening (and finally, breaking) of the N5–C4a double bond and polarization of the C4a atom so that it will gain additional positive charge. Concomitant with the movement of H1 to N5, the interaction between the sulfur of Cys-57 and C4a increases until the orbitals on the sulfur start overlapping with the ones on C4a, and eventually the covalent adduct is formed. Thus our experimental data and theoretical analysis clearly indicate that the primary interaction between Cys-57 and FMN after absorption of light occurs via cysteine hydrogen atom H1 and N5 atom of FMN. The role of light in this process is to facilitate the orbital overlap between H1 and N5 and to ensure the increase of the basic properties of N5.
The functional role of conformation 1 of Cys-57 is not clear. Upon photon absorption, the equilibrium between the two conformations should shift toward conformation 2. Correspondingly, at some point of the back-reaction, conformation 2 should start passing into conformation 1 and, thus, the equilibrium should be restored. From this point of view conformation 1, remote from the reaction center of FMN, may render the back-reaction easier. The presence of two conformations with apparently different reactivity may be related to the spectroscopic finding of two triplet states that decay with two time constants (800 ns and 4 μs) into the LOV1-390 photoproduct (Kottke et al., 2003).
CONCLUSION
We determined the crystal structures of the dark state of LOV1 (LOV1-447), of a state where the thiol of Cys-57 is close to the planar FMN, and of the metastable covalent intermediate LOV1-390. The structures form the basis for quantum chemical calculations that allowed us to propose an atomic model for the formation of the photoproduct. Although we cannot exclude an effect of the crystal lattice, our study suggests that the conformations of the dark state and the photoproduct of LOV1 are very similar. This is in line with a crystallographic study on the light-induced structural changes in maidenhair fern LOV2 (Crosson and Moffat, 2002) and is supported by infrared-difference spectroscopy (Ataka et al., 2002). Although the protein, the crystal forms and packing, and the experimental strategy (freeze-trap versus continuous illumination at room temperature) differ, the results are similar and show that the light-induced structural changes take place around the chromophore. These observations raise the question how does LOV1 function as a light-induced signaling switch that promotes activation of the Phot1 kinase domain? The structures of the isolated LOV1 domain do not give an answer to this question. It is conceivable that illumination triggers a release of the LOV1/LOV2 interaction and of the kinase domain. Studies on and structures of larger constructs of Phot1 are needed to investigate this possibility.
Acknowledgments
Data deposition: Coordinates and structure factor amplitudes have been deposited with the Protein Data Bank (accession codes 1N9L, 1N9N, and 1N9O).
The authors thank Tina Schireis for excellent technical assistance; E. Hochmuth for taking the MS-spectra; the staff at the ESRF, Grenoble for help during data collection; and Roger S. Goody for continuous encouragement and support. The authors also thank Emil F. Pai for sending them the coordinates of mercuric ion reductase.
This work was supported by the Deutsche Forschungsgemeinschaft (GK640 and SFB521). I.S. is very grateful to the “Richard und Anne-Liese Gielen-Leyendecker Stiftung.” R.F. was supported by the Alexander von Humboldt Foundation.
Roman Fedorov and Ilme Schlichting contributed equally to this work.
References
- Asamizu, E., Y. Nakamura, S. Sato, H. Fukuzawa, and S. Tabata. 1999. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6:369–373. [DOI] [PubMed] [Google Scholar]
- Ataka, K., P. Hegemann, and J. Heberle. 2002. Vibrational spectroscopy of an algal Phot-LOV1 domain probes the molecular changes associated with blue-light reception. Biophys. J. 84:466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, G. I., G. H. Carlsson, A. T. Smith, H. Szoke, A. Henriksen, and J. Hajdu. 2002. The catalytic pathway of horseradish peroxidase at high resolution. Nature 417:463–468. [DOI] [PubMed] [Google Scholar]
- Briggs, W. R., J. M. Christie, and M. Salomon. 2001. Phototropins: a new family of flavin-binding blue light receptors in plants. Antioxid. Redox. Signal. 3:775–788. [DOI] [PubMed] [Google Scholar]
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. [DOI] [PubMed] [Google Scholar]
- Burmeister, W. P. 2000. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. D Biol. Crystallogr. 56:328–341. [DOI] [PubMed] [Google Scholar]
- Christie, J. M. and W. R. Briggs. 2001. Blue light sensing in higher plants. J. Biol. Chem. 276:11457–11460. [DOI] [PubMed] [Google Scholar]
- Christie, J. M., P. Reymond, G. K. Powell, P. Bernasconi, A. A. Raibekas, E. Liscum, and W. R. Briggs. 1998. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282:1698–1701. [DOI] [PubMed] [Google Scholar]
- Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson, S. and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98:2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson, S. and K. Moffat. 2002. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132–133. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C. 2001. The phytochromes, a family of red/far-red absorbing photoreceptors. J. Biol. Chem. 276:11453–11456. [DOI] [PubMed] [Google Scholar]
- Hegemann, P., M. Fuhrmann, and S. Kateriya. 2001. Algal sensory photoreceptors. J. Phycol. 37:668–676. [Google Scholar]
- Holzer, W., A. Penzkofer, M. Fuhrmann, and P. Hegemann. 2002. Spectroscopic characterization of flavin mononucleotide bound to the LOV1 domain of phototropin from Chlamydomonas reinhardtii. Photochem. Photobiol. 75:479–487. [DOI] [PubMed] [Google Scholar]
- Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen, and W. R. Briggs.1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278:2120–2123. [DOI] [PubMed] [Google Scholar]
- Huang, K. and C. F. Beck. 2002. 10th International Conference on the Cell and Molecular Biology of Chlamydomonas, Vancouver, BC. 57a.
- Iwata, T., S. Tokutomi, and H. Kandori. 2002. Photoreaction of the cysteine S-H group in the LOV2 domain of adiantum phytochrome3. J. Am. Chem. Soc. 124:11840–11841. [DOI] [PubMed] [Google Scholar]
- Jarillo, J. A., H. Gabrys, J. Capel, J. M. Alonso, J. R. Ecker, and A. R. Cashmore. 2001. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410:952–954. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800. [Google Scholar]
- Kagawa, T., T. Sakai, N. Suetsugu, K. Oikawa, S. Ishiguro, T. Kato, S. Tabata, K. Okada, and M. Wada. 2001. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291:2138–2141. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., M. Doi, N. Suetsugu, T. Kagawa, M. Wada, and K. Shimazaki. 2001. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414:656–660. [DOI] [PubMed] [Google Scholar]
- Kottke, T., J. Heberle, D. Hehn, B. Dick, and P. Hegemann. 2003. Phot LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys. J. 1192–1201. [DOI] [PMC free article] [PubMed]
- Merritt, E. A. and M. E. P. Murphy. 1994. Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50:869–873. [DOI] [PubMed] [Google Scholar]
- Miller, S. M., V. Massey, D. Ballou, C. H. Williams, Jr., M. D. Distefano, M. J. Moore, and C. T. Walsh. 1990. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry 29:2831–2841. [DOI] [PubMed] [Google Scholar]
- Mulliken, R. S. 1955. Electron population analysis on LCAO-MO molecular wave functions. IV. Bonding and antibonding in LCAO and valence-bond theories. J. Chem. Phys. 23:2343–2346. [Google Scholar]
- Nakano, H. 1993. Quasidegenerate perturbation theory with multiconfigurational self-consistent-field reference functions. J. Chem. Phys. 99:7983–7992. [Google Scholar]
- Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57:1367–1372. [DOI] [PubMed] [Google Scholar]
- Ravelli, R. B. and S. M. McSweeney. 2000. The “fingerprint” that x-rays can leave on structures. Struct. Fold. Des. 8:315–328. [DOI] [PubMed] [Google Scholar]
- Roos, B. O. 1987. The CASSCF method and its application in electronic structure calculations. In Advances in Chemical Physics, vol. 69. K. P. Lawley, editor. Wiley Interscience, New York. 339–445.
- Salomon, M., J. M. Christie, E. Knieb, U. Lempert, and W. R. Briggs. 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39:9401–9410. [DOI] [PubMed] [Google Scholar]
- Schiering, N., W. Kabsch, M. J. Moore, M. D. Distefano, C. T. Walsh, and E. F. Pai. 1991. Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature 352:168–172. [DOI] [PubMed] [Google Scholar]
- Schlichting, I., J. Berendzen, K. Chu, A. M. Stock, S. A. Maves, D. E. Benson, R. M. Sweet, D. Ringe, G. A. Petsko, and S. G. Sligar. 2000. The catalytic pathway of cytochrome p450cam at atomic resolution. Science 287:1615–1622. [DOI] [PubMed] [Google Scholar]
- Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. J. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su, T. L. Windus, M. Dupuis, and J. A. Montgomery. 1993. The general atomic and molecular electronic structure system. J. Comput. Chem. 14:1347–1363. [Google Scholar]
- Stewart, J. J. P. 1989. Optimization of parameters for semiempirical methods. I. Method. J. Comput. Chem. 10:209–220. [Google Scholar]
- Swartz, T. E., S. B. Corchnoy, J. M. Christie, J. W. Lewis, I. Szundi, W. R. Briggs, and R. A. Bogomolni. 2001. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276:36493–36500. [DOI] [PubMed] [Google Scholar]
- Taylor, B. L. and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C. H., Jr. 1992. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase—a family of flavoenzyme transhydrogenases. In Chemistry and Biochemistry of Flavoenzymes, vol. III. F. Mueller, editor. CRC Press, Boca Raton, FL. 121–211.
- Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331–333. [DOI] [PubMed] [Google Scholar]