Abstract
When Legionella pneumophila grows in HeLa cells, it alternates between a replicative form and a morphologically distinct “cyst-like” form termed MIF (mature intracellular form). MIFs are also formed in natural amoebic hosts and to a lesser extent in macrophages, but they do not develop in vitro. Since MIFs accumulate at the end of each growth cycle, we investigated the possibility that they are in vivo equivalents of stationary-phase (SP) bacteria, which are enriched for virulence traits. By electron microscopy, MIFs appeared as short, stubby rods with an electron-dense, laminar outer membrane layer and a cytoplasm largely occupied by inclusions of poly-β-hydroxybutyrate and laminations of internal membranes originating from the cytoplasmic membrane. These features may be responsible for the bright red appearance of MIFs by light microscopy following staining with the phenolic Giménez stain. In contrast, SP bacteria appeared as dull red rods after Giménez staining and displayed a typical gram-negative cell wall ultrastructure. Outer membranes from MIFs and SP bacteria were equivalent in terms of the content of the peptidoglycan-bound and disulfide bond cross-linked OmpS porin, although additional proteins, including Hsp60 (which acts as an invasin for HeLa cells), were detected only in preparations from MIFs. Proteomic analysis revealed differences between MIFs and SP forms; in particular, MIFs were enriched for an ∼20-kDa protein, a potential marker of development. Compared with SP bacteria, MIFs were 10-fold more infectious by plaque assay, displayed increased resistance to rifampin (3- to 5-fold) and gentamicin (10- to 1,000-fold), resisted detergent-mediated lysis, and tolerated high pH. Finally, MIFs had a very low respiration rate, consistent with a decreased metabolic activity. Collectively, these results suggest that intracellular L. pneumophila differentiates into a cyst-like, environmentally resilient, highly infectious, post-SP form that is distinct from in vitro SP bacteria. Therefore, MIFs may represent the transmissible environmental forms associated with Legionnaires' disease.
The genus Legionella is one of the most successful of all aquatic bacteria, consisting of over 40 named species, their numerous serogroups (7), and a collection of Legionella-like amoebal pathogens that usually exhibit an obligate intracellular lifestyle requiring a particular protozoan host (3). An obligate requirement for the amino acid cysteine (38), which cannot be substituted for by cystine (the oxidized form most commonly found in aerobic environments), conceivably limits members of the Legionella genus to an intracellular lifestyle (25, 46) or to life in association with other microorganisms (46, 56, 63, 66) that may constitute a source of cysteine. However, in a biofilm coculture model, persistence but not multiplication of legionellae could be strictly demonstrated, suggesting that natural growth may indeed require the intracellular environment of a protozoan host (52). Thus, the natural life cycle of Legionella most likely alternates between periods of intracellular replication in protozoan hosts and planktonic survival, after death and lysis of spent hosts. In environments where suitable hosts are plentiful (e.g., cooling towers, hot tubs, and biofilms) the period between hosts would be shorter than that in environments of low host density, where planktonic survival for extended periods, even years, might be required. Several lines of evidence (reviewed in reference 2) suggest that Legionella pneumophila, the best-studied species of this genus and the one most commonly associated with Legionnaires' disease in humans, must have evolved an exit strategy from protozoan hosts that not only prepares the bacteria for planktonic survival but also ensures a favorable outcome in ensuing encounters with suitable hosts.
In fact, early studies have shown that L. pneumophila is capable of surviving for up to 14 months in water with only a modest loss in viability in the first few months (46, 60, 61). Long-term survival may be associated with the formation of viable but nonculturable forms (55) that remain infectious for amoebae (62). With respect to ensuring a successful infection of new hosts, Cirillo et al. (20, 21) and others (29, 58) have demonstrated that intracellular growth enhances the ability of L. pneumophila to infect amoebae or mammalian cells. This increase in infectivity is accompanied by changes in (i) bacterial morphology, (ii) cell wall composition or structure, as indicated by Giménez staining, and (iii) the route of entry into macrophages, all in relation to those for bacteria grown in vitro (20, 21, 29). Furthermore, studies in which mice were challenged with L. pneumophila, either alone or in various combinations with amoebae, also indicated that growth of L. pneumophila in amoebae always results in a more severe pneumonia (13, 14, 15), as well as increased replication in the lungs of mice (20). Collectively, these findings suggest that L. pneumophila has evolved a developmental program that enables the bacteria to shift from a replicative phase to one of maturation that produces a resilient, highly infectious form (39), perhaps analogous to the shift from reticulate bodies to elementary bodies (EBs) so well characterized in the developmental cycle of Chlamydia spp. (35, 50).
In this respect, we have identified two well-defined, morphologically distinct forms of L. pneumophila in HeLa cells: (i) a replicative form (RF), found inside a specialized cell compartment known as the replicative endosome, and (ii) a cyst-like, mature intracellular form (MIF), observed late in infection of HeLa cells (26, 27, 29). RFs appear as slender rods and display a typical gram-negative cell envelope that is ultrastructurally similar to that of agar-grown bacteria. In contrast, MIFs appear as short, stubby rods containing cytoplasmic inclusions of poly-β-hydroxybutyrate (PHBA) and display a unique electron-dense layer associated with the outer membrane and/or multiple layers of intracytoplasmic membranes (29). We have recently shown that RFs and MIFs alternate in every intracellular growth cycle and have suggested that MIFs are the in vivo equivalent of stationary-phase (SP) bacteria grown in vitro (27). Interestingly, the entry of exponentially growing L. pneumophila into SP in vitro is accompanied by activation of the stringent response regulator RelA (33), which together with the SP sigma factor RpoS coordinates the activation of virulence traits (flagellum synthesis and increased osmotic resistance, cytotoxicity, and infectivity for macrophages) (18, 32, 33, 34). At issue, and the subject of this report, is whether MIFs are distinct from in vitro SP bacteria, which have been proposed to be the transmissible infectious forms associated with disease (34). We took advantage of our ability to purify MIFs (29) to directly compare them with SP forms obtained from broth cultures in vitro. Here we show that MIFs purified from HeLa cells exhibit a lower respiration rate, increased resistance to antibiotics and detergent-mediated lysis, and increased levels of surface-associated Hsp60 and display a unique pattern of proteins compared with SP bacteria. We conclude that the MIF is a unique form with cyst-like properties that enables L. pneumophila to persist as a highly infectious particle in the environment when between hosts.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The L. pneumophila Philadelphia-1 strains Lp1-SVir (streptomycin resistant) and Lp1-Avir (avirulent, salt-tolerant mutant) have been described previously (40). Local clinical isolate 2064 (serogroup 1 Olda) and its avirulent salt-tolerant variant 2064 M (24) were also used in these studies. All strains were kept as frozen stocks at −70°C. Frozen stocks of the virulent strains were made from infected cultures of HeLa cells after intracellular growth had taken place (see below). Frozen stocks were grown on buffered charcoal-yeast extract-agar (BCYE) (54) for 3 to 5 days at 37°C in a humid incubator. BCYE-grown bacteria were then either used immediately for infection of HeLa cells or subcultured in buffered yeast extract broth (BYE) to an optical density at 620 nm (OD620) of 3.5 to obtain SP forms (18). The BYE formulation was based on that of BCYE except that charcoal and agar were omitted.
Cell lines and infection protocols.
HeLa cells were routinely grown in 100-ml spinner bottles in Dulbecco modified Eagle's medium (DMEM; high-glucose formulation) completed with 10% newborn calf serum and an antibiotic-antimycotic mixture (all from GIBCO Laboratories). Before infection with L. pneumophila, HeLa cells were allowed to attach and spread on either tissue culture flasks or multiwell plates (all from Falcon Plastics, Becton Dickinson). To obtain large numbers of MIFs (for isolation purposes), HeLa cells in 75-cm2 cell culture flasks were infected with 2 ml of a bacterial cell suspension with an OD620 of 1 (∼109 bacteria/ml). After an overnight incubation, monolayers were washed twice with Earle's salt solution to eliminate free bacteria and cells, and fresh DMEM (containing 100 μg of gentamicin/ml) was added for 90 min. Monolayers were then washed twice with Earle's salt solution, and fresh DMEM with serum but no antibiotics was added. Monolayers were harvested at day 3 postinfection.
To determine the intracellular growth curve of L. pneumophila, HeLa cell monolayers in six-well plates (∼106 cells per well) were infected with ∼109 plate-grown bacteria per well for 3 h. The wells were then washed twice with phosphate-buffered saline (PBS), treated with gentamicin (100 μg/ml) for 1.5 h, and washed twice with PBS, and cells were incubated for different times in fresh DMEM with serum but no antibiotics. Three wells were sampled at each time point by removing the medium and adding 100 μl of 0.05% Triton X-100 in water (1 min), followed by 900 μl of distilled deionized water (ddH20). One milliliter of lysate was serially diluted in ddH20 and plated on BCYE to obtain the CFU per well. HeLa cell cultures utilized for monitoring the infection by light microscopy and for documenting the Giménez staining (see below) were mounted on glass coverslips (22 by 22 mm; no. 1 thickness) placed in either six-well plates or individual 35-mm-diameter tissue culture dishes as described before (29).
The U937 cell line was grown in suspension in RPMI 1640, supplemented with 5% fetal bovine serum and the antibiotic-antimycotic mixture (GIBCO Laboratories). To obtain adherent macrophage-like cells, U937 cells were treated with 20 ng of phorbol myristoyl acetate (Sigma)/ml and placed in six-well plates at 2 × 105 per well. Following differentiation, adherent cells in the six-well plates were infected with ∼108 L. pneumophila Svir bacteria in accordance with the procedures outlined above for HeLa cells. At 24 and 48 h postinfection, infected cells were gently scraped from the wells, fixed in 0.1 M sodium cacodylate buffer, pH 7, containing 2.5% glutaraldehyde, and subsequently postfixed in 1% OsO4, in preparation for transmission electron microscopy as outlined below.
Purification of MIFs.
L. pneumophila MIFs from HeLa cells were purified in a self-generated, continuous-density gradient of Percoll by a modification of a previously reported method (29). In the modified method, the pellet of infected HeLa cells and the pellet obtained from the HeLa cell culture supernatant were resuspended in 1 ml of ddH2O and passed 10 times through a 27-gauge needle, before high-speed centrifugation in Percoll. Because this process lysed the HeLa cells, the band distribution after centrifugation included only a top band of cell debris and two bottom bands. The upper bottom band (1.061 to 1.074 g/ml) contained cell debris-associated bacteria, and the lower bottom band (>1.074 g/ml) contained free MIFs. MIFs from this lower bottom band were washed twice in ddH2O to remove residual Percoll before use.
Giménez stain.
Giménez staining, originally applied to the identification of rickettsiae in infected yolk sacs (30), was performed exactly as reported by McDade (49). The primary stain for this technique is carbol fuchsin, and the secondary stain is malachite green. While the molecular basis of Giménez staining remains to be determined, it is known that the basic dye carbol fuchsin is retained by acid-fast bacteria, typically mycobacteria, which have a complex envelope composed of glycolipids and glycopeptidolipids (12). A bright red coloration is referred to as a Giménez-positive, or Gim+, phenotype, and a green, blue-green, or gray coloration is referred to as a Giménez-negative, or Gim−, phenotype. Different shades of blue-red colorations are referred to as Giménez-intermediate, or Gimint, phenotypes.
Microscopy.
Light microscopy was performed with a Zeiss microscope equipped with phase contrast, but most observations were made in bright field. The scoring of proportions of Gim+ to Gim− bacteria was performed with the aid of an ocular grid by counting at least 100 bacteria from different randomly chosen fields. Electron microscopy (EM) was performed by using the standard protocols reported previously (26, 29). Briefly, specimens were fixed in glutaraldehyde-OsO4, stained in bloc with aqueous uranyl acetate, dehydrated in acetone, and embedded in epoxy (TAAB 812) resin. Ultrathin sections were poststained with uranyl acetate-lead citrate before observation in a Philips EM300 transmission electron microscope. All materials and reagents for EM were obtained from Marivac Ltd. (Montreal, Canada).
Plaque assay.
Infectivity was determined through a plaque assay, in accordance with the method of Fernandez et al. (24) except for omitting the gentamicin treatment step. Briefly, monolayers of L929 cells in 24-well plates were incubated for 1 h with 200 μl of serial 10-fold dilutions of the bacterial inoculum, washed to eliminate unbound bacteria, and overlaid with cell culture medium containing 0.6% agarose. One hundred-microliter portions of the inoculum dilutions were plated on BCYE to determine the number of CFU added per well. Four days after infection, monolayers were fixed in 4% formaldehyde and stained with crystal violet, and plaques were then enumerated. The percent plaquing efficiency was calculated as (number of plaques/CFU added) × 100.
Preparation of outer membranes, electrophoresis, and immunoblotting.
Outer membranes from SP forms and MIFs were prepared by selectively dissolving the cytoplasmic membrane with Sarkosyl by using a modification of the method of Butler et al. (17). Whole-cell pellets from SP BYE cultures or isolated MIFs from Percoll density gradients were washed twice in ddH2O and frozen until use. A bacterial mass equivalent to ∼1010 bacteria (pelleted from a 10-ml suspension with an OD620 of 1) was washed once in 5 ml of 50 mM Tris, pH 7.2 (OM buffer), and resuspended in 10 ml of 2% (wt/vol) N-lauroylsarcosinate (Sigma) in OM buffer. After 1 h at 37°C, the cleared suspension was thoroughly sonicated in periods of 30 s alternating with cooling on ice for ∼2 min in a Sonifier cell disruptor, model W140 (Heat Systems-Ultrasonics Inc., Plainview, N.Y.), at an output setting of 10. Sonication continued until the suspension had cleared. Unbroken cells were pelleted by centrifugation (5,000 × g, 10 min), and the supernatant was transferred to Beckman polyallomer tubes for ultracentrifugation at 100,000 × g for 30 min to sediment the outer membranes. The outer membrane pellet was treated for 1 h at 37°C with 100 μg of mutanolysin (∼1,000 U) in 100 μl of OM buffer. The mutanolysin-treated outer membranes were then resuspended in 10 ml of 2% (wt/vol) N-lauroylsarcosinate in OM buffer and resedimented by ultracentrifugation. The washed pellet was resuspended in 100 μl of OM buffer, and a 10-μl sample was taken to perform a Lowry method-based protein determination with the DC Protein Assay (Bio-Rad Laboratories Canada, Mississauga, Ontario, Canada). An outer membrane sample equivalent to 10 μg of total protein was solubilized in sample buffer, and the proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 12% acrylamide minigel (Bio-Rad Laboratories). The gel was stained with Coomassie brilliant blue to visualize the separated proteins. Blotting onto nitrocellulose membranes and immunostaining with specific antibodies based on the alkaline phosphatase reaction were performed as reported previously (26).
2-D gel electrophoresis.
Separation of total proteins from whole bacterial cells was performed in an IPGphor system (first dimension) and a Hoefer vertical-slab 14- by 15-cm gel apparatus (second dimension) (Amersham Pharmacia Biotech, Piscataway, N.J.). Sample preparation in buffer containing nonionic detergents, isoelectric focusing in Immobiline DryStrip IPG strips, and SDS-PAGE in 7.5 to 15% acrylamide gradient gels were all performed by closely following protocols and recommendations described previously (T. Berkelman and T. Stenstedt, 2-D electrophoresis using immobilized pH gradients: principles and methods, Amersham Pharmacia Biotech Inc., 1998) and by incorporating sonication, which is required to solubilize MIFs (see “Preparation of outer membranes, electrophoresis, and immunoblotting” above). After electrophoresis, gels were silver stained by the method of Blum et al. (11). Gels were scanned wet in an EPSON ES-1200C scanner equipped with transparency unit EU-13 (Seiko Epson Co., Nagano, Japan) to produce digital images that were analyzed with Z3 software (Compugen, Jamesburg, N.J.). Composites (of three gels each) of SP bacterium and MIF two-dimensional (2-D) gels were made from aligned and registered gel images by using the Raw Master Gel function of Z3. The protein spots of the composites were then matched and compared to produce a color-coded matched-view image and a table of spots differentially or uniquely expressed in MIFs or SP forms.
Survival assays.
Survival following exposure to ddH2O, antibiotics, and a series of environmental challenges was determined through viable-bacterial-cell counts performed before and after exposure to the condition or agent. Viable-bacterial-cell counts were performed by a standard dilution-plating method using ddH2O as the dilution medium and BCYE as the plating medium. The number of colonies was scored after incubation for 3 to 5 days at 37°C. Results are reported as percent survival in relation to the initial number of CFU per milliliter, obtained before challenges.
Bacterial suspensions of ∼108 bacteria/ml were made in ddH2O (and incubated for up to 2 weeks) or in 1-ml aqueous solutions of 100 mM glycine, pH 2; 100 mM Tris base, pH 11 (incubated overnight, ∼16 h); 30 and 70% ethanol (incubated for 15 min at room temperature); or proteinase K (1 mg/ml; incubated for 1 h at 37°C). For antibiotic challenges, bacterial suspensions in minimal essential medium adjusted to ∼108 bacteria/ml were challenged with 5 or 100 μg of gentamicin per ml or 6 μg of rifampin per ml for 3 h. After incubations, samples were diluted and plated. Similar 1-ml bacterial suspensions (108/ml) were made in ddH2O and were incubated at 50°C in a water bath for 20 min or were frozen at −20°C and thawed before dilution plating. Desiccation was tested in 6-mm-diameter disks of filter paper onto which ∼106 bacteria were absorbed. Disks were placed in a sterile petri dish and left at room temperature for different periods. At each sampling time a paper disk was placed on a BCYE plate and 10 μl of BYE was added to the disk. Plates were incubated at 37°C, and bacterial growth was scored daily. Susceptibility to lysis by detergent was tested by making suspensions of ∼109 bacteria/ml in N-lauroylsarcosinate, SDS, sodium desoxycholate, or Triton X-100 (all at 10 mg/ml in 10 mM Tris, pH 7.5) and measuring OD at regular intervals. A graph of OD versus time was made, and the time in minutes required to reach one-half of the initial OD620 was interpolated and reported as the detergent lysis index.
Respiration rates.
The consumption of oxygen by whole viable bacteria was measured in a 1.5-ml Gilson water-jacketed chamber with the oxygen electrode of a Yellow Springs Instruments (Yellow Springs, Ohio) oxygen monitor, model 53, as previously described (41). Approximately 109 bacteria in aerated ddH2O were placed in the chamber and equilibrated to 37°C for about 5 min to determine the basal respiration rate in water. Oxygen levels were traced by a chart recorder, and the oxygen consumption rate was obtained as the slope from the trace. Oxygen saturation in the aerated chamber with no bacteria was determined to be 220 μM (41, 64). Once basal respiration reached a constant slope, substrates were added with a Hamilton syringe through a glass capillary bore stopper (one at a time and at a final concentration of 6.67 mM). Then, oxygen consumption was recorded until the trace in the recorder stayed at a fairly constant slope for 3 to 5 min. Respiration rates for each substrate were calculated in nanomoles of oxygen consumed per minute per 109 CFU. CFU were determined by dilution plating in duplicate on BCYE agar medium.
RESULTS
MIFs as intracellular SP forms.
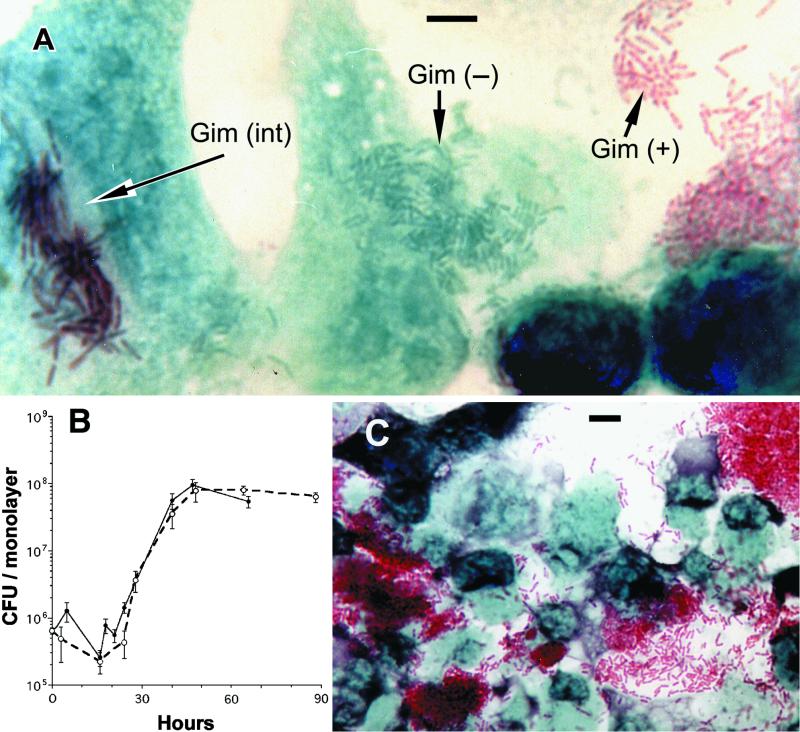
In agreement with our previous observations (27, 29), coverslip cultures of HeLa cells infected with purified Gim+ MIFs developed foci of numerous bright green Gim− bacteria or of long rods stained with a blue-red Gimint shade (Fig. 1A). The appearance of these foci coincided with the onset of the replicative phase of the intracellular growth curve shown in Fig. 1B (15 to 24 h postinfection). Twenty-four hours also marked the onset of the previously reported expansion in the percentage of infected HeLa cells in coverslip cultures (29). Only Gim+ forms accumulated in large numbers between 48 and 72 h postinfection (Fig. 1C), coinciding with the end of the replicative phase (Fig. 1B) and the peak (at ∼70%) in the proportion of infected HeLa cells (29). As reported before, no accumulation of Gim− or Gimint forms was observed (27), strongly suggesting that MIFs are the in vivo equivalent to in vitro SP forms.
FIG. 1.
Giménez phenotypes observed in HeLa cell monolayers infected with purified MIFs. (A) Micrograph showing three different Giménez phenotypes (+, −, and int) in a single microscopical field from a specimen fixed and stained 24 h after infection. Foci of green bacteria (such as the ones shown in the center) were easier to find 15 to 24 h after infection. (B) Growth curves of Lp1-SVir in HeLa cells, defining establishment of the in vivo SP at 45 to 50 h postinfection. Each curve represents an independent experiment run in triplicate (○, experiment 1; •, experiment 2), and error bars represent standard deviations from the means (n = 3). (C) Massive accumulation of bright red MIFs 48 h after infection. Bars (A and C), 5 μm.
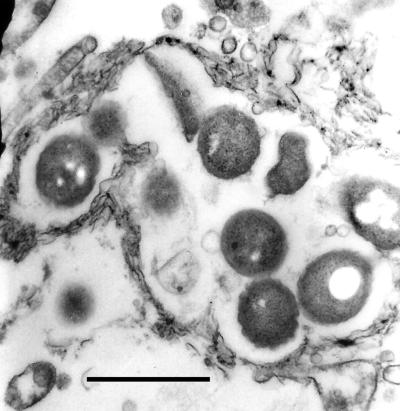
MIF formation in macrophages was investigated in U937 cells infected with L. pneumophila SVir. By 48 h, the majority of infected U937 cells had detached and signs of lysis were evident in the form of abundant free cell debris. By EM, most U937 cells were highly vesiculated and displayed a very electron-transparent cytoplasm. Intracellular bacteria were observed inside membrane-bound compartments with characteristics typical of replicative endosomes (i.e., associated with the endoplasmic reticulum and/or following the contour of the contained bacteria). While some signs of maturation were evident (i.e., appearance of prominent inclusions and lack of a wavy outer membrane around a visible periplasmic space), most intracellular bacteria still lacked the envelope complexity typical of MIFs (Fig. 2). The fact that replicating or immature bacteria were contained in already-dying macrophages led us to conclude that the early disintegration of macrophages may be a determining factor in whether or not replicating bacteria fully differentiate into MIFs.
FIG. 2.
Electron micrograph of a U937 cell, taken 48 h after infection with L. pneumophila Lp1-SVir. Shown is a group of legionellae contained in an endosome, the membrane of which follows the contour of the contained bacteria. Notice the electron transparency of the cytoplasmic material surrounding the endosome, as well as the signs of early maturation of the contained bacteria (i.e., presence of cytoplasmic inclusions and the lack of a visible periplasmic space). Bar, 1.0 μm.
Giménez staining and ultrastructure of SP L. pneumophila.
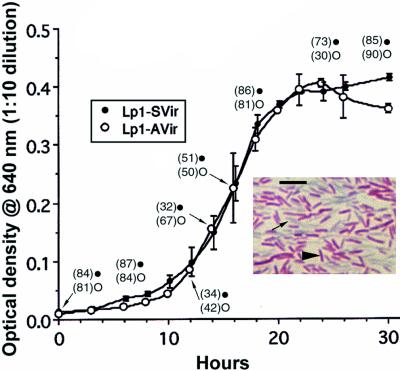
We monitored the growth and changes in Giménez staining of both virulent and avirulent L. pneumophila strains in vitro and found that both entered SP at the same time (Fig. 3). As reported before (27) no correlation between growth phase in BYE and Giménez staining could be established (Fig. 3), except for late SP bacteria, which showed a predominant Gim+ phenotype (Fig. 3, inset). The Giménez phenotype did not correlate with virulence status, since Gim+ avirulent SP bacteria remained noninfectious for HeLa and L929 cells.
FIG. 3.
Growth curves in BYE of virulent Lp1-Svir (•) and nonvirulent Lp1-Avir (○), showing the percentages of Gim+ forms (numbers in parentheses) at selected points along the growth curve. Notice that both strains grew similarly and entered SP at the same time. (Inset) Light micrograph of Giménez-stained Lp1-SVir in SP (30 h). SP Lp1-AVir had the same appearance. Arrowhead, typical Gim+ bacterium; arrow, Gim− bacteria in the background. Bar (inset), 5 μm.
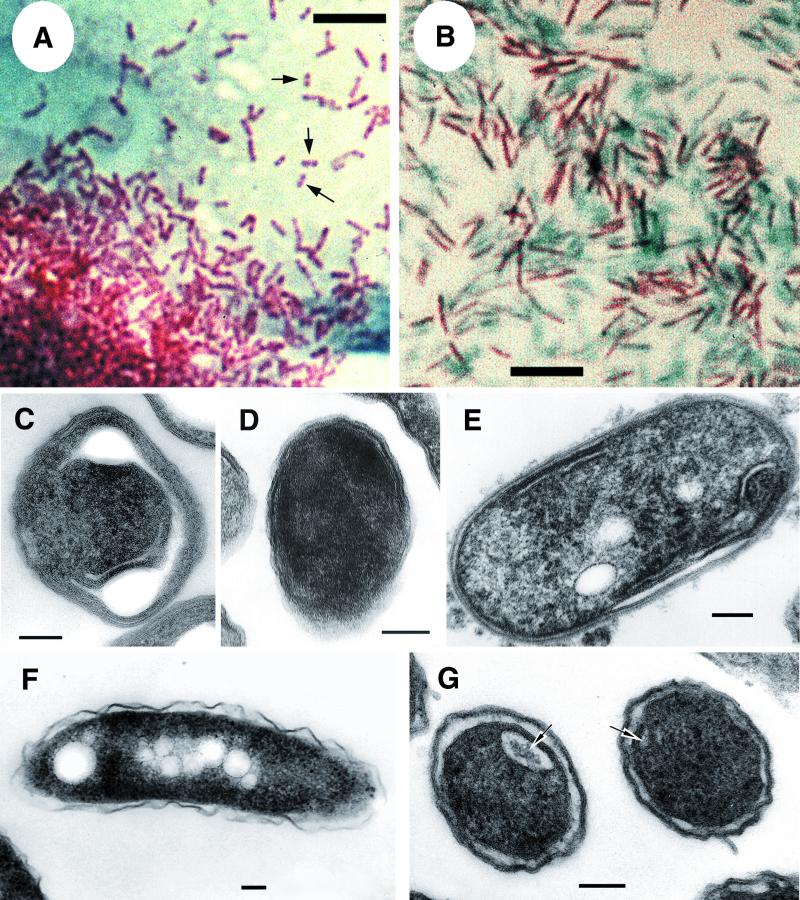
By light microscopy after Giménez staining, SP bacteria and MIFs were readily distinguishable; the short, stubby, rod-shaped MIFs typically displayed a bright shade of red that was often bipolar (Fig. 4A), whereas the slender, rod-shaped SP forms displayed a homogeneous dull red staining (Fig. 3, inset, and 4B). In agreement with previous reports (29), transmission electron microscopy showed that the envelope ultrastructure of MIFs was defined by long invaginations of the cytoplasmic membrane forming putative compartments (Fig. 4C), the presence of an electron-dense laminated outer membrane structure that no longer resembled the typical gram-negative cell wall ultrastructure (Fig. 4D), or prominent stacks (laminations) of intracytoplasmic membranes (Fig. 4E). In contrast, most sections of SP legionellae displayed the typical cell wall ultrastructure of gram-negative bacteria (Fig. 4F), but approximately 30% of the sections showed some signs of maturation in SP bacteria, such as the presence of cytoplasmic inclusions (Fig. 4F), darkened outer membranes, and internal membranes (Fig. 4G). The dark thick layer associated with the outer membranes of MIFs was not apparent in sections of SP bacteria.
FIG. 4.
Tinctorial and morphological features of MIFs and SP forms of L. pneumophila strain 2064. Light microscopy of Giménez-stained smears of MIFs (A) and SP bacteria (B) show differences in size, shade of red, and general appearance. Notice the bipolar staining of some MIFs (arrows in panel A). EM (C to G) showed differences in cell envelope ultrastructure and internal inclusions. MIFs displayed long invaginations of the cytoplasmic membrane (C) and a darkened cytoplasm and periplasm, which made the definition of membranes very difficult, so that the envelope appeared as a thick laminated layer (D) or stacks of internal membranes at both parallel sides (E). Most bacterial sections of SP forms (like the ones shown in panel F) displayed the envelope ultrastructure typical of gram-negative bacteria, with a wavy outer membrane. Others, however, showed darkening of the outer membrane and the presence of internal membranes defining pseudocompartments (arrows in panel G). Notice also the presence of inclusions in SP bacteria. Bars, 5 (A and B) and 0.1 μm (C and G).
Differences in protein profiles of MIFs and SP forms.
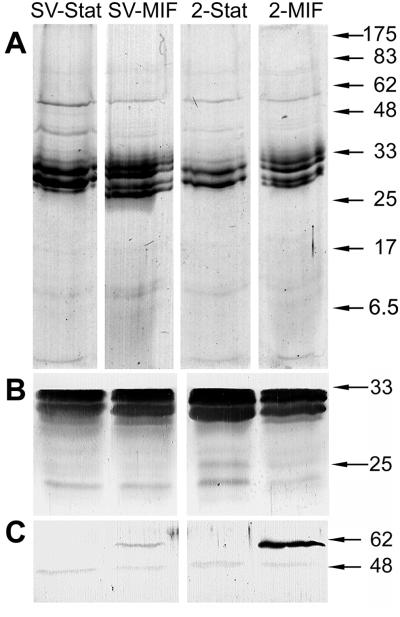
The morphological differences between MIFs and SP forms, particularly at the cell envelope level, were expected to reflect differences in the compositions of outer membranes. In Fig. 5A, outer membrane preparations of MIFs showed four protein bands whereas SP forms showed only two: those expected for the major outer membrane protein OmpS (43). Immunoblots with anti-OmpS rabbit hyperimmune serum (16) revealed a doublet in each of the samples (Fig. 5B). To confirm the identity of such extra bands, an immunoblot was run by using an anti-OmpS rabbit hyperimmune serum (16). As shown in Fig. 5B, only the typical OmpS doublet was similarly revealed in all samples. This was indicative of a good degradation of the peptidoglycan mesh and the consequent release of the 31-kDa, peptidoglycan-bound OmpS subunit, in addition to the 28-kDa peptidoglycan-free subunit (16, 43). The multiple protein bands observed in MIFs most likely represent posttranslational modifications to OmpS subunits that are not observed in SP bacteria. Previous immunolabeling studies had indicated that MIFs apparently display increased amounts of envelope- and surface-associated Hsp60 (28). We ran an immunoblot stained with an Hsp60-specific monoclonal antibody (Fig. 5C) and confirmed the exclusive presence of Hsp60 in the outer membranes of MIFs. We also noticed that MIFs of the strain 2064 had much more outer membrane-associated Hsp60 than MIFs of Lp1-SVir.
FIG. 5.
Outer membrane proteins of SP forms and MIFs. (A) Coomassie blue-stained gel of outer membrane proteins. (B) Immunoblot stained with a major outer membrane protein (OmpS)-specific rabbit antiserum and alkaline phosphatase-conjugated second antibody (goat anti-rabbit immunoglobulin [Ig]). (C) Immunoblot stained with an Hsp60-specific monoclonal antibody and alkaline phosphatase-conjugated second antibody (rabbit anti-mouse Ig). The positions of reference molecular size markers (in kilodaltons) are indicated on the right. Stat, SP forms; SV, Lp1-Svir strain; 2, 2064 strain.
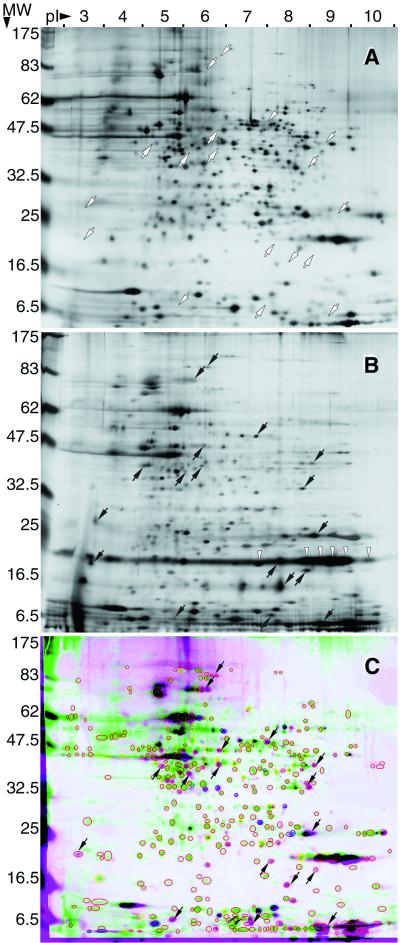
2-D gel electrophoresis indicated numerous differences between the protein profiles of SP bacteria (Fig. 6A) and MIFs (Fig. 6B). MIFs showed 43% (i.e., only 254 spots) of the 591 more noticeable spots (filtered to have an area of more than 75 pixels and a contrast higher than 16) identified in 2-D gels of SP bacteria, indicating that MIFs have a limited complement of proteins. The proteins exclusively present in SP bacteria (i.e., not present in gels of MIFs) are shown in the matched-view image presented in Fig. 6C as green spots surrounded by a red oval. As a result of the much lower number of protein spots in the MIF gels, those spots present appeared slightly overloaded, as equivalent amounts of protein were run in both gels. In spite of this expected unbalance, MIFs truly overexpressed (2.5-fold more than SP bacteria) the previously identified ∼20-kDa protein MagA (27, 37a). Interestingly, MagA appeared as a series of spots spread over a range of pI values (Fig. 6B). From its DNA sequence, MagA has a predicted pI of 7.73, but it is typically identified as a doublet or a smear in immunoblots from one-dimensional SDS-PAGE protein gels (27, 37a), collectively suggesting that this MIF-associated protein may be posttranslationally modified. Finally, the proteins exclusively present in MIFs are shown in the matched-view image (Fig. 6C) as magenta spots surrounded by a red oval. These protein spots are shown in Fig. 6B and C, and their equivalent positions are also shown in the gel of SP forms (Fig. 6A). It is possible that some of these MIF-associated spots represent host-derived proteins or peptides, as the NH3-terminal amino acid sequence of the abundant, very-low-molecular-weight streak with a high pI showed significant homology to mouse and other eukaryotic histones (unpublished results). The identification of the complete set of spots exclusively expressed in MIFs should provide invaluable information on the gene expression pattern of these differentiated forms.
FIG. 6.
2-D protein gels of SP forms and MIFs. Shown is the best-matched pair of 2-D gels of SP bacteria (A) and MIFs (B). (C) Color-coded matched-view image obtained by overlaying and comparing three in-register gels of SP bacteria (green) and three in-register gels of MIFs (magenta). Only differentially expressed spots are shown, and the colors of circles and ovals around spots indicate spots overexpressed (>2-fold; yellow) and spots underexpressed (<0.5-fold; blue) in SP bacteria. Red circles and ovals, unmatched spots expressed only in either SP bacteria (green) or MIFs (magenta). The latter (or their corresponding areas) are marked with small arrows in all panels. Spots of ∼20 kDa and pIs of 7 to 10 (white arrowheads in panel B) correspond to MagA, a protein known to be overexpressed in MIFs (27, 37). Parameters of 2-D separation, molecular weight (MW; in thousands) and pI, are indicated on the left and top, respectively.
Environmental resistance and infectivity.
MIFs displayed higher survival in the presence of rifampin and gentamicin than SP forms (Table 1). In addition, only MIFs tolerated exposure to alkaline pH, and, notably, MIFs were extremely stable against detergent-mediated lysis. Not shown in Table 1 are the survival results for exposure to ethanol, acid pH, and desiccation (because both forms were quite susceptible to these) or to high temperature, osmotic shock, freezing, proteinase K, or starvation in ddH2O (as both forms were similarly resistant to them). In general, the survival of the two different forms against environmental challenges was quite variable among experiments, something we have as yet been unable to explain.
TABLE 1.
Environmental fitness and infectivity of SP forms and MIFs of two virulent L. pneumophila strainse
| Strain and form | Environmental resistance (% survival) in the presence of:
|
Lysis (T50 index [min]) mediated by detergent:
|
Infectivity (% efficiency)f | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Rifampin (6 μg/ml) | Gentamicin at:
|
pH 11 | T-100 | SDS | Sarkosyl | DOC | |||
| 5 μg/ml | 100 μg/ml | ||||||||
| Lp1-SVir | |||||||||
| SP | 4.4 ± 1.3 | 2.58 ± 1.82 | 0.02 ± 0.001 | <0.001a | 60 | 0.33 | 1.6 | 4.4 | 0.89 ± 0.22 |
| MIF | 21.4 ± 4.80 | 28.6 ± 19.4 | 30.0 ± 14.0 | 0.18 ± 0.15 | ∞b | ∞ | ∞ | ∞ | 10.1 ± 1.9 |
| 2064 | |||||||||
| SP | 37c | 0.038 ± 0.005 | 0.007 ± 0.003 | <0.001 | 640 | 0.6 | 2.0 | 3.4 | 1.9 ± 1.6 |
| MIF | 108c | 21.8 ± 21.6 | 10.7 ± 4.8 | 1.0 ± 1.35 | ∞ | ∞ | ∞ | ∞ | 12.1 ± 4.2 |
Indicates that survival was very low since no CFU were detected at the lower dilution plated (normally 1:100).
∞, apparently infinite index since the lysis curve became asymptotic at a value above 50% of the original OD.
No duplicate experiment was run for these results, so that no mean and standard deviation are indicated.
Abbreviations used: T50, lysis index, defined as the time that it takes to reduce the original OD by 50%; T-100, Triton X-100; DOC, sodium desoxycholate.
Only those environmental challenges for which a difference between forms was observed are shown. In general, results were taken from at least two independent experiments (and as many as four) and are shown as means ± standard deviations.
A measure of plaquing efficiency.
It should be noted that others and we have determined MIFs to be more infectious to cultured host cells than bacteria grown on agar plates (21, 29). However, the infectivity tests reported previously relied on a gentamicin treatment to eliminate extracellular bacteria. Because we determined here that MIFs survive a gentamicin treatment much better than SP forms (up to 1,000-fold) (Table 1), we turned to a plaque assay that did not include a gentamicin treatment to confirm the high infectivity of MIFs. In addition, because previous experiments had compared MIFs with agar-grown legionellae, we also wanted to compare MIFs head to head with SP bacteria grown in broth to meet the definitions outlined by Byrne and Swanson for SP forms (18). MIFs were confirmed to be highly infectious as they consistently had approximately a 10-fold-higher percent plaquing efficiency than SP forms (Table 1).
Differences in respiration rate between MIFs and SP forms.
MIFs had a 2-fold-lower basal rate of respiration than SP forms ([1.83 ± 0.32]-fold, n = 2, for SVir, and 2.5-fold, n = 1, for 2064), but, most importantly, MIFs did not respond to the addition of respirable substrates. In contrast, SP forms of both strains increased their respiration rates (1.5 ± 0.33)-fold (n = 6) in response to pyruvate and α-ketoglutarate. It is important that the numbers of viable bacteria in these assays were confirmed to be very similar for all forms (in average, [0.92 ± 0.27] × 109/ml for SP bacteria and [0.97 ± 0.33] × 109/ml for MIFs).
DISCUSSION
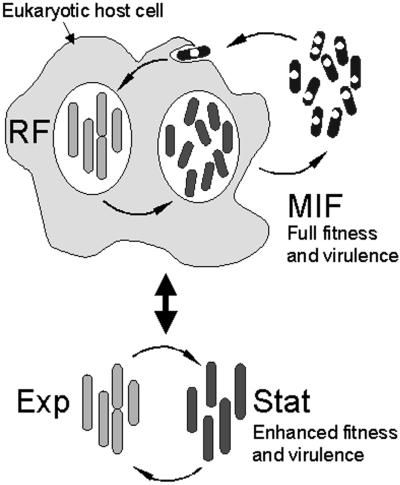
In natural environments, L. pneumophila alternates between an intracellular replicative phase in protozoa and an extracellular nonreplicative (planktonic) phase in which the bacteria remain highly infectious. In vitro, L. pneumophila progresses through the stages of a typical bacterial growth cycle, alternating between an exponentially replicative phase and a postreplicative SP, in which virulence traits are acquired (18, 33, 34) (refer to model in Fig. 7). We have tested the hypothesis that MIFs, observed in great numbers late in infection of HeLa cells, represent a developmental form distinct from in vitro SP bacteria. The results of our studies showed that MIFs indeed constituted a unique form, different from SP bacteria, as they (i) were exclusive to the intracellular milieu (did not develop in vitro), (ii) were metabolically dormant (did not show substrate-inducible respiration), (iii) exhibited a unique cell wall ultrastructure, (iv) were resistant to antibiotics and detergent-mediated lysis, (v) were enriched for surface-associated Hsp60, (vi) displayed a unique protein profile, and (vii) were ∼10-fold more infectious than SP bacteria by plaque assay of L929 cells. On the other hand, we established some similarities between MIFs and SP forms including survival in ddH2O, resistance to osmotic shock and proteinase K treatments, thermal tolerance, retention of Giménez stain (albeit intensity differences were noted), and development of cytoplasmic inclusions and invaginations of the cytoplasmic membrane. Moreover, others have reported that SP forms induce flagellum synthesis and expression of cytotoxic factors associated with egress from infected hosts and exhibit increased infectivity and thermal tolerance (4, 18), indicating their differentiated state beyond the exponentially RF (18, 34). Taken together, these results establish SP bacteria and MIFs as two distinct post-RFs that have reached different levels of differentiation.
FIG. 7.
Simplified model illustrating the intracellular and extracellular growth cycles of L. pneumophila. Only the intracellular growth cycle produces the fully differentiated MIF, which is more infectious and environmentally fit than the in vitro-grown SP bacteria (Stat). Presumably, host-derived signals (inducers) are required for post-stationary-phase differentiation of MIFs. Intracellular RFs would be equivalent to exponentially growing bacteria (Exp) in vitro, as they display similar morphological traits, but it remains to be determined whether or not they are physiologically different. The central double arrow indicates that L. pneumophila can move from one cycle to the other when the proper conditions are met. Notice that, to initiate a new intracellular cycle, L. pneumophila has to exit into the aquatic environment, where it is possible for MIFs to continue their maturation in response to extracellular signals (inducers).
One of the key questions raised by our studies is why MIFs are not formed in vitro. The obvious corollary to this question is whether SP forms exist in natural environments, where L. pneumophila replication takes place in protozoan hosts. For Chlamydia spp., development and the growth cycle cannot possibly be separated, as the bacteria are incapable of in vitro growth. In contrast, the free-living, nitrogen-fixing and cyst-forming bacterium Azotobacter vinelandii, which displays a typical bacterial growth cycle in nutrient-rich media (with exponential-phase and SP forms) (42), can enter SP without differentiating into cysts. However, when treated with PHBA (inducer) it proceeds to encystment (59). Clearly, then, the highly differentiated cyst does not constitute an SP form. By analogy, we suggest that SP forms of L. pneumophila initiate the developmental program inferred for MIFs but, in the absence of additional signals (inducers) conceivably unique to the host milieu, become arrested in maturation (Fig. 7). It is also reasonable to hypothesize that in MIFs those regulatory factors associated with in vitro SP transitions in other bacteria (e.g., RelA and RpoS) are likely adapted to the tasks of coordinating the morphogenesis of infectious forms and inducing the transmission phenotype (34), both in anticipation of host cell demise and in response to host-derived signals (inducers), not available in vitro.
While clear morphological attributes supporting the existence of distinct developmental forms have been observed for Legionella spp. grown in mammalian cells (31, 45, 53) or even in vitro (31), these were never considered to be part of a differentiation process or a developmental cycle. Retrospectively, differentiated MIFs have been observed (but unrecognized) as ultrastructurally distinct forms in amoebae (9, 57, 65) and as Gim+ forms in clinical specimens (10), yolk sac cultures (49), and infected Vero cells (47). It is also likely that MIFs may have accounted for the positive acid-fast staining (retention of carbol fuchsin after an acidic wash) of Legionella spp. in sputum samples (8, 37). While the MIF morphology is readily observed in protozoan hosts (9, 57, 65), it has not been conspicuous in macrophages. This does not imply that MIFs cannot develop in macrophages, as others (44) have published electron micrographs of infected macrophages where MIFs can be recognized. L. pneumophila grows rapidly in macrophages, induces early apoptosis (51), and produces lytic cytotoxins (4), which contribute to an early demise of this host. We have confirmed here that the morphology of intracellular L. pneumophila in already-disintegrating macrophages is not that typical of HeLa cell- or protozoon-derived MIFs. Furthermore, historically, most studies with macrophages were terminated at 24 h postinfection (insufficient time for MIF formation) (44). It seems, then, that HeLa and other nonlymphoid mammalian cell lines (29, 53), as well as natural protozoan hosts, appear to be more tolerant than macrophages to legionella infection and thus favor the full differentiation of L. pneumophila into MIFs.
The intracellular dimorphic life cycle of L. pneumophila (which alternates between the RF and the differentiated MIF) thus resembles the stage-specific developmental cycle of Chlamydia spp., in which reticulate bodies differentiate late in infection into a nearly homogeneous population of EBs, which are metabolically dormant, resilient, and highly infectious (35, 50). However, MIFs differ from EBs in ultrastructure, as the latter exhibit a distinguishable inner and outer membrane wall structure, as well as an absence of both cytoplasmic inclusions (PHBA) and laminations of intracytoplasmic membranes (19). Laminations of intracytoplasmic membranes, however, are observed in the resilient small-cell variants (SCV) of Coxiella burnetii (48), but SCVs differ from MIFs in that they exhibit condensed chromatin (nucleoid material) and cytoplasm and lack cytoplasmic inclusions of PHBA. Perhaps most notably, SCVs show evidence of cell division (36), which has never been observed for MIFs or EBs by EM. Conceivably, it is also possible for MIFs to be equivalent to the differentiated cysts of A. vinelandii (59). Like the cysts of A. vinelandii, MIFs have lost the typical gram-negative cell wall architecture, but, unlike those cysts, which produce a thick laminated exine coat, MIFs produce a much thinner electron-dense structure and there is no evidence for a capsule. In addition, it is not known whether MIFs use PHBA as an energy source during maturation, as has been noted for A. vinelandii cysts (59).
Of interest was the resistance of MIFs to antibiotics. Whereas Barker et al. (5, 6) previously noted the increased antibiotic resistance of intracellular L. pneumophila, its emergence was not correlated with a developmental cycle. In their dormant state, MIFs may be resistant to antibiotics that rely on active protein synthesis, DNA replication, transcription, or cell wall biosynthesis. Alternatively, such resistance could be related to putative characteristics of the MIF envelope, such as altered permeability. In fact, proteome comparisons suggest that MIFs display both a unique, perhaps limited, set of proteins, which reflects a state of terminal differentiation and dormancy, and an unusual complement of outer membrane-associated proteins. Proteome changes are also consistent with previous analyses that reported major protein differences between legionellae grown in vitro and in vivo (1, 21, 27). One of the proteins enriched in MIFs was an ∼20-kDa protein that we have named MagA (27, 37) and have subsequently determined to be a marker of MIF differentiation (unpublished data). Finally, the selective expression of a set of proteins by MIFs provides evidence for stage-specific gene expression.
Cirillo et al. (21, 22) initially reported that L. pneumophila grown in amoebae were 100-fold more infectious to epithelial cells and 10-fold more infectious to macrophages than agar-grown bacteria. We also previously compared the infectivities of HeLa cell-passaged and agar-passaged L. pneumophila (29) and obtained results similar to those of Cirillo et al., but in both cases it was assumed (albeit not specifically determined) that the infecting agar-grown bacteria were in SP and that the intracellular bacteria were sensitive to gentamicin. To be consistent with the definitions of Byrne and Swanson (18) and to rule out an artifactual effect of gentamicin, we adopted both the liquid culture protocol to obtain SP forms (18) and a gentamicin-less method of infectivity. After this, we still demonstrated a reproducible 10-fold increase in MIF infectivity in L929 cells over that for broth-grown SP bacteria. The increased infectivity of MIFs may be attributable in part to their enrichment with surface-associated Hsp60, previously shown to mediate invasion of HeLa cells (28). While other putative invasins have been identified, mostly through genetic analyses (22, 23), only Hsp60 has been demonstrated to directly promote invasion and uptake of inert latex beads by HeLa cells (28). In addition to Hsp60, DotO and DotH proteins, which optimize the infectivity of L. pneumophila for new hosts, become localized to the bacterial surface during the late phases of intracellular growth (corresponding to MIF maturation) but not during SP in vitro (67). Thus, the increased association of Hsp60 and other proteins with the surfaces of MIFs (in relation to SP bacteria) might contribute to the increased invasiveness observed in this and previous studies (21, 29).
In summary, we have characterized a highly differentiated form of L. pneumophila (MIF) that appears in great numbers in infected HeLa cells and to a lesser extent in infected macrophages. The MIF is highly infectious, is arrayed with surface-exposed Hsp60 (an invasin in L. pneumophila), and differs from SP bacteria in protein expression patterns, morphology, and some environmental fitness traits. As depicted in the model presented in Fig. 7, we propose that intracellular vegetative bacteria (RFs) differentiate late in infection into cyst-like MIFs. We further suggest that in nature MIFs are the predominant extracellular planktonic forms that survive in a highly infectious mode for long periods while between hosts. SP forms, while capable of developing the transmission phenotype (34), seem arrested in their developmental program and therefore possess a decreased environmental fitness and infectivity. Clinically, L. pneumophila-infected amoebae or free L. pneumophila-laden vesicles, which are proven sources of MIFs, might constitute the basic infectious unit commonly aerosolized into the lungs of susceptible humans (9, 13, 58). Interestingly, the noncommunicable nature of Legionnaires' disease among humans might be attributed (at least in part) to the less permissive nature of the alveolar macrophage for promoting full differentiation of the infectious MIF. Further study of MIF biology could be of paramount importance in understanding L. pneumophila pathogenesis, the transmission of Legionnaires' disease, and, in particular, the regulatory factors controlling its stage-specific development.
Acknowledgments
The technical help and EM expertise of Mary Anne Trevors is gratefully acknowledged, as is the assistance of Kimberly Jefferies in running the growth curves of L. pneumophila in vivo. We sincerely thank Ken Chisholm for his assistance with the 2-D gel work and analysis.
Funding for this work was provided by a CIHR grant (MT14443) to P.S.H. and R.A.G.
Editor: B. B. Finlay
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., L.-Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. Fields. 1996. Legionella-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alli, O. A. T., L.-Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, J., and M. R. Brown. 1995. Speculations on the influence of infecting phenotype on virulence and antibiotic susceptibility of Legionella pneumophila. J. Antimicrob. Chemother. 36:7-21. [DOI] [PubMed] [Google Scholar]
- 6.Barker, J., H. Scaife, and M. R. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 8.Bentz, J. S., K. Carroll, J. H. Ward, M. Elstad, and C. J. Marshall. 2000. Acid-fast positive Legionella pneumophila: a possible pitfall in the cytologic diagnosis of mycobacterial infection in pulmonary specimens. Diagn. Cytopathol. 22:45-48. [DOI] [PubMed] [Google Scholar]
- 9.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackmon, J. A., F. W. Chandler, and M. D. Hicklin. 1979. Pathologic features of Legionnaires' disease, p. 10-12. In G. L. Jones and G. A. Hébert (ed.), “Legionnaires'” the disease, the bacterium and methodology. Center for Disease Control, Bureau of Laboratories, U.S. Public Health Service, U.S. Department of Health, Education, and Welfare, Atlanta, Ga.
- 11.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 12.Brennan, P. J., S. W. Hunter, M. McNeil, D. Chatterjee, and M. Daffe. 1990. Reappraisal of the chemistry of mycobacterial cell walls, with a view to understanding the roles of individual entities in disease processes, p. 55-75. In E. M. Ayoub, G. H. Cassell, W. C. Branche, Jr., and T. J. Henry (ed.), Microbial determinants of virulence and host response. American Society for Microbiology, Washington, D.C.
- 13.Brieland, J., J. C. Fantone, D. G. Remick, M. LeGendre, M. McClain, and C. Engleberg. 1997. The role of Legionella pneumophila-infected Hartmanella vermiformis as an infectious particle in a murine model of Legionnaires' disease. Infect. Immun. 65:5330-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brieland, J., M. McClain, L. Heath, C. Chrisp, G. Huffnagle, M. LeGendre, M. Hurley, J. Fantone, and C. Engleberg. 1996. Coinoculation with Hartmanella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect. Immun. 64:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brieland, J., M. McClain, M. LeGendre, and C. Engleberg. 1997. Intrapulmonary Hartmanella vermiformis: a potential niche for Legionella pneumophila replication in a murine model of legionellosis. Infect. Immun. 65:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler, C. A., and P. S. Hoffman. 1990. Characterization of a major 31-kilodalton peptidoglycan-bound protein of Legionella pneumophila. J. Bacteriol. 172:2401-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler, C. A., E. D. Street, T. P. Hatch, and P. S. Hoffman. 1985. Disulfide-bonded outer membrane proteins in the genus Legionella. Infect. Immun. 48:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi, E. Y., C.-C. Kuo, and J. T. Grayston. 1987. Unique ultrastructure in the elementary body of Chlamydia sp. strain TWAR. J. Bacteriol. 169:3757-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellani affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellani enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirillo, S. L. G., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 23.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez, R. C., S. H. S. Lee, D. Haldane, R. Sumarah, and K. R. Rozee. 1989. Plaque assay for virulent Legionella pneumophila. J. Clin. Microbiol. 27:1961-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 26.Garduño, R. A., G. Faulkner, M. A. Trevors, N. Vats, and P. S. Hoffman. 1998. Immunolocalization of Hsp60 in Legionella pneumophila. J. Bacteriol. 180:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garduño, R. A., E. Garduño, M. Hiltz, D. Allan, and P. S. Hoffman. 2001. Morphological and physiological evidence for a developmental cycle in Legionella pneumophila, p. 82-85. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Lück (ed.), Legionella. ASM Press, Washington, D.C.
- 28.Garduño, R. A., E. Garduño, and P. S. Hoffman. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garduño, R. A., F. D. Quinn, and P. S. Hoffman. 1998. HeLa cells as a model to study the invasiveness and biology of Legionella pneumophila. Can. J. Microbiol. 44:430-440. [PubMed] [Google Scholar]
- 30.Giménez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 31.Gress, F. M., R. L. Myerowitz, A. W. Pascule, C. R. Rinaldo, Jr., and J. N. Dowling. 1980. The ultrastructural morphologic features of Pittsburgh pneumonia agent. Am. J. Pathol. 101:63-78. [PMC free article] [PubMed] [Google Scholar]
- 32.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer, B. K., and M. S. Swanson. 1999. Coordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 34.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 35.Hatch, T. P., I. Allan, and J. H. Pearce. 1984. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J. Bacteriol. 157:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinzen, R. A., T. Hackstadt, and J. E. Samuel. 1999. Developmental biology of Coxiella burnetii. Trends Microbiol. 7:149-154. [DOI] [PubMed] [Google Scholar]
- 37.Hilton, E., R. A. Freedman, F. Cintron, H. D. Isenberg, and C. Singer. 1986. Acid-fast bacilli in sputum: a case of Legionella micdadei pneumonia. J. Clin. Microbiol. 24:1102-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Hiltz, M. 2000. Characterization of magA, a developmentally regulated gene of Legionella pneumophila demonstrating strain variability. M.Sc. thesis. Dalhousie University, Halifax, Nova Scotia, Canada.
- 38.Hoffman, P. S. 1984. Bacterial physiology, p. 61-67. In C. Thornsberry, A. Balows, J. C. Feeley, and W. Jakubowski (ed.), Legionella, Proceedings of the 2nd International Symposium. American Society for Microbiology, Washington, D.C.
- 39.Hoffman, P. S. 1997. Invasion of eukaryotic cells by Legionella pneumophila: a common strategy for all hosts? Can. J. Infect. Dis. 8:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman, P. S., C. A. Butler, and F. D. Quinn. 1989. Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Infect. Immun. 57:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman, P. S., and T. G. Goodman. 1982. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman, P. S., T. V. Morgan, and D. V. DerVartanian. 1980. Respiratory properties of cytochrome-c-deficient mutants of Azotobacter vinelandii. Eur. J. Biochem. 110:349-354. [DOI] [PubMed] [Google Scholar]
- 43.Hoffman, P. S., J. H. Seyer, and C. A. Butler. 1992. Molecular characterization of the 28- and 31-kilodalton subunits of the Legionella pneumophila major outer membrane protein. J. Bacteriol. 174:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz, S. M., and P. Nash. 1978. The morphology of the Legionnaires' disease organism. Am. J. Pathol. 90:701-722. [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, J. V., and A. A. West. 1991. Survival and growth of Legionella species in the environment. Soc. Appl. Bacteriol. Symp. Ser. 20:121S-129S. [PubMed] [Google Scholar]
- 47.Maruta, K., M. Ogawa, H. Miyamoto, K. Izu, and S.-I. Yoshida. 1998. Entry and intracellular localization of Legionella dumoffii in Vero cells. Microb. Pathog. 24:65-73. [DOI] [PubMed] [Google Scholar]
- 48.McCaul, T. F., and J. C. Williams. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDade, J. E. 1979. Primary isolation using guinea pigs and embryonated eggs, p. 70-76. In G. L. Jones and G. A. Hébert (ed.), “Legionnaires'” the disease, the bacterium and methodology. Center for Disease Control, Bureau of Laboratories, U.S. Public Health Service, U.S. Department of Health, Education, and Welfare, Atlanta, Ga.
- 50.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller, A., J. Hacker, and B. Brand. 1996. Evidence for apoptosis of human macrophage-like HL-60 cells by Legionella pneumophila infection. Infect. Immun. 64:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121-3126. [DOI] [PubMed] [Google Scholar]
- 53.Oldham, L. J., and F. G. Rodgers. 1985. Adhesion, penetration and intracellular replication of Legionella pneumophila: an in vitro model of pathogenesis. J. Gen. Microbiol. 131:697-706. [DOI] [PubMed] [Google Scholar]
- 54.Pascule, A. W., J. C. Feeley, R. J. Gibson, L. G. Cordes, R. L. Meyerowitz, C. M. Patton, G. W. Gorman, C. L. Carmack, J. W. Ezzell, and J. N. Dowling. 1980. Pittsburgh pneumonia agent: direct isolation from human lung tissue. J. Infect. Dis. 141:727-732. [DOI] [PubMed] [Google Scholar]
- 55.Paszko-Kolva, C. M. Shahamat, and R. R. Colwell. 1992. Long term survival of Legionella pneumophila serogroup 1 under low-nutrient conditions and associated morphological changes. FEMS Microbiol. Ecol. 102:45-55. [Google Scholar]
- 56.Pope, D. H., R. J. Soracco, H. K. Gill, and C. B. Fliermans. 1982. Growth of Legionella pneumophila in two-membered cultures with green algae and cyanobacteria. Curr. Microbiol. 7:319-322. [Google Scholar]
- 57.Pruckler, J. M., R. F. Benson, M. Moyenuddin, W. T. Martin, and B. S. Fields. 1995. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect. Immun. 63:4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 59.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schofield, G. M. 1985. A note on the survival of Legionella pneumophila in stagnant tap water. J. Appl. Bacteriol. 59:333-335. [DOI] [PubMed] [Google Scholar]
- 61.Skaliy, P., and H. V. McEachern. 1979. Survival of the Legionnaires' disease bacterium in water. Ann. Intern. Med. 90:662-663. [DOI] [PubMed] [Google Scholar]
- 62.Steinert, M., L. Emödy, R. Amann, and J. Hacker. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellani. Appl. Environ. Microbiol. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tison, D. L., D. H. Pope, W. B. Cherry, and C. B. Fliermans. 1980. Growth of Legionella pneumophila in association with blue-green algae (cyanobacteria). Appl. Environ. Microbiol. 39:456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umbreit, W. W., R. H. Burris, and J. F. Stauffer. 1974. Manometric techniques. Burgess Publishing Co., Minneapolis, Minn.
- 65.Wadowsky, R. M., A. Fleisher, N. J. Kapp, M. El-Moufti, J. N. Dowling, R. M. Agostini, and R. B. Yee. 1993. Multiplication of virulent and avirulent strains of Legionella pneumophila in cultures of Hartmannella vermiformis, p. 145-147. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella, current status and emerging perspectives. ASM Press, Washington, D.C.
- 66.Wadowsky, R. M., and R. B. Yee. 1985. Effect of non-Legionellaceae bacteria on the multiplication of Legionella pneumophila in potable water. Appl. Environ. Microbiol. 49:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watarai, M., H. L. Andrews, and R. R. Isberg. 2001. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39:313-329. [DOI] [PubMed] [Google Scholar]