Abstract
A novel interarm interaction of DNA cruciform forming at inverted repeat sequence was characterized using an S1 nuclease digestion, permanganate oxidation, and microscopic imaging. An inverted repeat consisting of 17 bp complementary sequences was isolated from the bluegill sunfish Lepomis macrochirus (Perciformes) and subcloned into the pUC19 plasmid, after which the supercoiled recombinant plasmid was subjected to enzymatic and chemical modification. In high salt conditions (200 mM NaCl, or 100–200 mM KCl), S1 nuclease cut supercoiled DNA at the center of palindromic symmetry, suggesting the formation of DNA cruciform. On the other hand, S1 nuclease in the presence of 150 mM NaCl or less cleaved mainly the 3′-half of the repeat, thereby forming an unusual structure in which the 3′-half of the inverted repeat, but not the 5′-half, was retained as an unpaired strand. Permanganate oxidation profiles also supported the presence of single-stranded part in the 3′-half of the inverted repeat in addition to the center of the symmetry. Both electron microscopy and atomic force microscopy have detected a thick protrusion on the supercoiled DNA harboring the inverted repeat. We hypothesize that the cruciform hairpins at conditions favoring triplex formation adopt a parallel side-by-side orientation of the arms allowing the interaction between them supposedly stabilized by hydrogen bonding of base triads.
INTRODUCTION
Inverted repeat sequences are known to form cruciform structures in negatively supercoiled DNA (for review, Wells, 1988; Sinden, 1994). They are widespread in the genomes of both eukaryotes and prokaryotes, occurring more often than expected among random sequence (Schroth and Ho, 1995). In some bacteria, extrusion of cruciform DNA is required for initiation of replication and transcription (Jin et al., 1997; Glucksmann et al., 1992; Kim et al., 1998), though certain inverted repeat sequences are unstable when subcloned into bacteria (Lockson and Galloway, 1986; Schaaper et al., 1986). Because such structures might have an impedimentary effect on the fidelity of DNA replication, the role of inverted repeats in mutagenesis and in human diseases has been widely investigated (Bissler, 1998, for review).
Atomic force microscopy (AFM) has revealed DNA cruciform structures to be highly dynamic. The DNA cruciforms exhibit a high degree of variability with respect to the interarm angle and occur in two conformations: planar and X-type (Shlyakhtenko et al., 1998). The X-type cruciforms are localized consistently at loops of plectonemic superhelix, suggesting that the conformational transition between planar and X-type serves as a molecular trigger for the slithering of DNA chains (Shlyakhtenko et al., 2000). Ohta et al. (1996) has detected an unusual structure at the inverted repeat of the Staphylococcus aureus HSP70 promoter using AFM technique, suggesting a structural quadruplet model consisting of a pair of stem-loops (Stem-Loop-Loop-Stem; SL2S model). The inverted repeats could change the conformation among B-form duplex, planar cruciform, X-type cruciform, and SL2S conformation, depending upon the nucleotide sequence and environment.
We previously identified a satellite DNA that contains short inverted repeats (11 bp complementary stretches) in the saltwater fish Sillago japonica (Perciformes), and assumed an unusual conformation different from the typical DNA cruciform based on the results of S1 nuclease assay (Kato et al., 1998). It showed a dominant accessibility of S1 nuclease to 3′-half of the inverted repeat and another half was protected from cutting. These profiles of S1 nuclease cutting may propose an involvement of certain triplex-related structure in the formation of alternative DNA structure at the inverted repeat. More recently, we found an A/T-rich DNA fragment that contains an inverted repeat with 17 bp complementary sequences from the bluegill sunfish Lepomis macrochirus (Perciformes) (Takahashi et al., 2001). Here we apply the enzymatic and chemical modification assays as well as visualization with electron and atomic force microscopes to characterize the unusual structures formed in the inverted repeat in the supercoiled DNA of L. macrochirus; we propose a novel model “hairpin triplex” for this unusual DNA conformation.
EXPERIMENTAL PROCEDURES
Plasmid DNA
A DNA fragment containing the inverted repeat (clone name: pBG4) was isolated from bluegill sunfish L. macrochirus and deposited in the GenBank/EMBL/DDBJ international databases under the accession number AB046703 (Takahashi et al., 2001). The AluI-Sau3AI fragment containing the inverted repeat was subcloned into the HincII-BamHI site of plasmid pUC19. The subcloned sequence was as follows:
CTTTTAATCTATGATTAACTGGTGTTGTGTCATCATATGTGTATTTATACACATATGATGACATATGATC (palindromic region is underlined). The recombinant plasmid was termed pBan1.
S1 nuclease digestion
The pUC19 derivative, pBan1, was propagated in Escherichia coli JM107, after which a Sephaglas FlexiPrep Kit (Amersham Pharmacia Biotech, Piscataway, NJ) was used to prepare supercoiled DNA. Samples of the DNA (10 μg) were treated with S1 nuclease in 200 μl of reaction mixture containing 50 mM sodium-acetate (pH4.6), 1 mM ZnSO4, 0∼200 mM KCl or NaCl, and 100 units of S1 nuclease. Before addition of the S1 nuclease, the reaction mixtures were preincubated on ice for 5 min. After addition of the S1 nuclease, the reaction mixtures were incubated at 25°C for an additional period (1, 2, or 5 min), after which the reaction was stopped by adding 10 μl of 0.5 M EDTA and then chilling the mixture on ice. The S1 nuclease-treated DNA was isolated by phenol extraction and ethanol precipitation and purified using the Sephaglas FlexiPrep kit.
Permanganate oxidation in vitro
Samples of DNA (1 μg) were treated with 0.16 mM potassium permanganate at 37°C for 1, 2, or 4 min in 100 μl of the reaction mixtures (10 mM sodium phosphate (pH 7.0)/100 mM NaCl, or 30 mM sodium acetate (pH 4.6)/100 mM NaCl). After the incubation, an aliquot (1 μl) of 150 mM sodium bisulfite was added to each reaction mixture. The oxidated DNA was recovered by Sephaglas BandPrep kit (Pharmacia), treated with piperidine, and subjected to primer extension assay.
In situ oxidation of DNA with potassium permanganate
Overnight culture (0.3 ml) of E. coli cells harboring pBan1 DNA was inoculated to 10 ml of Luria-Bertani's broth, and incubated with shaking at 37°C. Aliquots (1 ml) of the culture were withdrawn after 5 h and 8 h of incubation. The cells were harvested, washed with 10 mM sodium phosphate (pH 7.0)/100 mM NaCl, and suspended in 100 μl of 10 mM sodium phosphate (pH 7.0)/100 mM NaCl/1.6 mM potassium permanganate. Permanganate oxidation in situ was performed at 20°C for 60 min. The plasmid DNA was isolated from the permanganate-treated cells by FlexiPrep kit, treated with piperidine, and subjected to primer extension assay. Because the intracellular materials (proteins and nucleic acids) and the cell membrane consume permanganate, higher concentration of potassium permanganate and longer incubation time are required for in situ mapping of sensitive sites. The reaction conditions were defined experimentally.
Primer extension assay
Primer extension was carried out using rhodamine-labeled sequencing primers (Takara, Ohtsu, Japan). The reaction mixtures (17 μl) contained 500 ng of template DNA (enzymatically or chemically modified DNA), 3 pmol of primer DNA (M13 forward or reverse), 0.2 mM each of dNTP, 1X Tth DNA polymerase buffer (Toyobo, Osaka, Japan), and 2 units of Tth DNA polymerase (Toyobo). Initially the mixtures were preincubated at 96°C for 5 min, after which three cycles of incubation at 96°C for 36 s, 50°C for 36 s, and 74°C for 84 s were followed by incubation at 74°C for 5 min. Aliquots (5 μl) of the reaction mixtures were then loaded onto a 6% denaturing polyacrylamide gel in parallel with control sequence ladders obtained from a Tth DNA Polymerase AutoSequencer core kit (Toyobo). After the electrophoresis, the bands were visualized using FMBIO-100 or FMBIO-II image analyzer (Takara-Bio, Kusatsu, Japan).
Topoisomer analysis by two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis was performed to examine supercoiling-dependent topological change of pBan1 DNA similarly as described (Peck and Wang, 1983; Hanai and Roca, 1999). Briefly, the aliquot supercoiled pBan1 DNA was fractionated on 1.2% agarose gel in TAE buffer (40 mM Tris-acetate/20 mM Na-acetate/1 mM EDTA, pH7.8) for the first dimension, and in same buffer containing 6 μg/ml chloroquine for the second dimension. Electrophoresis was performed under 0.64V/cm at 6°C for 40 h in the first dimension, and under 1.2V/cm at 6°C for 20 h in the second dimension. After the electrophoresis, the DNA was transferred to Hybond-N membrane and visualized by DIG Nucleic Acid detection kit (Roche Diagnostics, Mannheim, Germany).
Electron microscopy
Supercoiled DNA molecules were dissolved in 40 mM sodium acetate buffer (pH4.6) at the concentration of 2 μg/ml. An aliquot of DNA solution was deposited onto a carbon-coated grid that had been freshly activated by glow-discharging at room temperature, and stained with 2% uranyl acetate, similarly as described by Delain et al. (1992). Electron microscopic imaging was performed with a model JEM 1200 EX (JEOL, Akishima, Japan), operating at 100 kV in dark field mode realized by tilted beam. Images were photographed, digitized by scanning negatives, and handled in TIFF files. Size measurements of the DNA structure were performed with NIH Image software (version 1.62).
Atomic force microscopy
Imaging with atomic force microscope was performed as reported (Kato et al., 2002). Briefly, an aliquot (5 μl) of DNA solution (0.3 μg/ml in 50 mM sodium acetate, pH 4.35) was deposited on the mica functionalized with aminopropyl silatrane (APS-mica) as described (Shlyakhtenko et al., 2000). Images were acquired by MM SPM NanoScope III system (Veeco Instruments, Santa Barbara, CA) operating in Tapping Mode in air at ambient conditions using silicon probes from MikroMash (Tallinn, Estonia), and converted to the TIFF files. Size measurements of the DNA structure were performed with NIH Image software (version 1.62).
RESULTS
S1 nuclease mapping and permanganate oxidation
The inverted repeat sequence from L. macrochirus was subcloned into pUC19 plasmid, after which the recombinant plasmid was digested with S1 nuclease. Most of the supercoiled molecules have been cut under the assay conditions employed (5 min of S1 nuclease treatment), as no superhelix and only a few relaxed molecules were detected on agarose gel electrophoresis (data not shown). Majority of the sites cut by the enzyme was localized within the DNA inserted in the recombinant plasmid (Fig. 1).
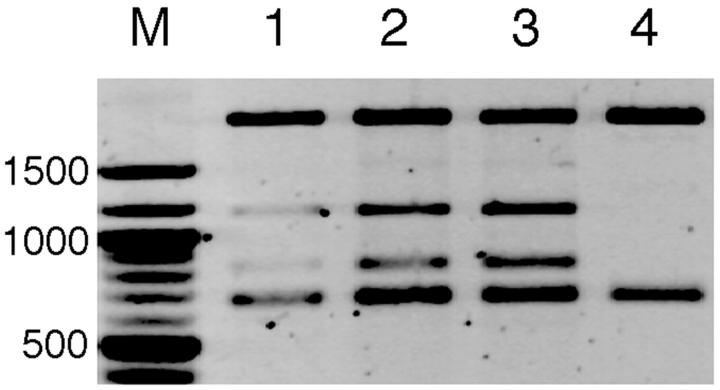
FIGURE 1.
S1 nuclease digestion of supercoiled DNA. Supercoiled recombinant plasmid containing the inverted repeat was treated with S1 nuclease in a reaction buffer containing 200 mM NaCl for 1 min (lane 1), 2 min (lane 2), or 5 min (lane 3) at 25°C followed by DraI digestion. DraI-digested, intact DNA (S1-untreated) was loaded onto lane 4. DNA size markers (New England BioLabs, Beverly, MA) were loaded onto lane M. The efficiency of specific S1-cutting was evaluated by comparing the intensity of the top and bottom DNA bands; ∼40% of the plasmid molecules were specifically cut at the inserted DNA region after 5 min of incubation.
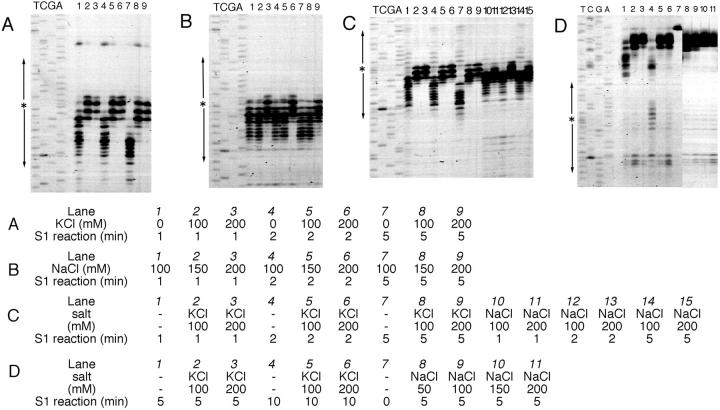
The sites cut by S1 nuclease were mapped using a primer extension assay, where the synthesis of complementary DNA terminated (Fig. 2). In the presence of KCl (100–200 mM) or high concentration of NaCl (200 mM), S1 nuclease cut the supercoiled DNA at the center of palindromic symmetry, whereas in low salt conditions (150 mM or lower NaCl concentration) mainly the 3′-half of the repeat was cleaved (Fig. 2, A–C). The potassium ion seems to stabilize the formation of DNA cruciform, and also inhibits formation of the alternative DNA structure that is distinct from the cruciform, as does a higher concentration of sodium ion (200 mM NaCl). Potassium cation is known to inhibit intermolecular triplex formation by a G-rich oligonucleotide and a Pur/Pyr-biased duplex even at low concentrations (10–100 mM) in a manner that cannot be explained solely on the basis of potassium-induced aggregation of G-rich oligonucleotide (Cheng and Dyke, 1993). The present results may suggest the involvement of triple-stranded DNA in the alternative DNA structure forming at the short inverted repeat that is sensitive to potassium. S1 nuclease cut the linearized molecules around the center of palindromic symmetry in the presence of either NaCl or KCl, though the efficiency of the specific cleavage was much lower (Fig. 2 D).
FIGURE 2.
Primer extension assay of S1 nuclease-treated DNA. The products of primer extension were electrophoresed with control sequence ladders. Supercoiled (panels A, B, and C) and linearized (panel D; EcoRI-digested) DNA was treated with S1 nuclease and subjected to primer extension assay. The reaction conditions for S1 nuclease digestion are listed in the table under the panels. (A and B) The results with M13 forward primer; (C and D) M13 reverse primer. Asterisks and arrows at the left of the panels indicate the positions of the inverted repeat symmetry.
Potassium permanganate is an effective probe to detect the conformation of nucleic acids (Hayatsu, 1996, for review). It attacks 5,6-double bond of the thymines in single-stranded regions more efficiently than in double-stranded DNA. The results of in vitro oxidation of pBan1 DNA were similar to the S1 nuclease-cutting profiles although the center of the inverted repeat was the most susceptive to permanganate; the 5′-half of the inverted repeat was protected from the attack of the probe (Fig. 3, A and B). Essentially identical patterns of permanganate oxidation were shown in the E. coli cells (Fig. 3, C and D), meaning that the alternative structure on pBan1 DNA existed in the cell and it was not generated during the process of plasmid preparation in which alkaline-denaturing step was involved. Although the plasmid DNA isolated from mid-log phase was more supercoiled than that from late-log phase (1–2 helical turns per pUC18 plasmid DNA (Kato and Furuno, 1992), no significant difference was observed in the profiles of permanganate oxidation between two growth stages of E. coli. No oxidation has occurred in the linearized pBan1 DNA by permanganate as similar to the S1 nuclease assay (data not shown).
FIGURE 3.
Primer extension assay of permanganate-treated DNA. The products of primer extension were electrophoresed with control sequence ladders. Permanganate oxidation was performed in vitro (A and B) and in situ (C and D). (A and C) The results with M13 forward primer; (B and D) M13 reverse primer. Asterisks and arrows at the left of the panels indicate the positions of the inverted repeat symmetry. Summary of the susceptive sites (indicated by arrowheads) for permanganate oxidation is shown at the bottom. (A and B) Lanes 1 and 4, 1 min permanganate reaction; lanes 2 and 5, 2 min permanganate reaction; lanes 3 and 6, 4 min permanganate reaction. Reactions in the neutral buffer are lanes 1, 2, and 3; reactions in acidic buffer are lanes 4, 5, and 6. Lanes T, C, G, and A are control sequence ladders. Although the permanganate reacted with DNA more efficiently in the acidic conditions (lanes 4, 5, and 6), the profile of permanganate oxidation was essentially identical in both neutral and acidic conditions. (C and D) Lane 1, permanganate reaction with the mid-log growth phase (5 h culture) of E. coli cells; lane 2, permanganate reaction with the late-log growth phase (8 h culture) of E. coli cells. Lanes T, C, G, and A are control sequence ladders.
Topoisomer analysis by two-dimensional agarose gel electrophoresis
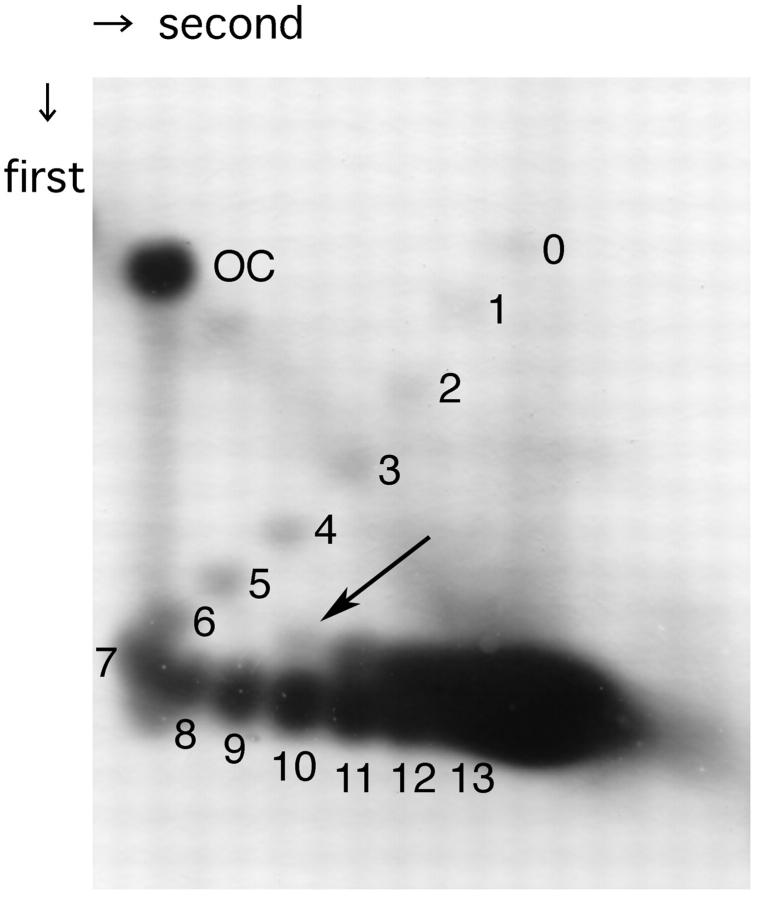
Supercoiling-dependent structural transition was examined by two-dimensional agarose gel electrophoresis. As shown in Fig. 4, structural transition that absorbs ∼3.5 helical turns has been observed in pBan1 DNA molecules having 10 or more negative supercoiling turns. This relaxation effect is consistent with the length of inverted repeat of pBan1 DNA (17 + 2 + 17 = 36 bp; 36/10.5 = 3.4 turns). Although the formation of H-DNA and Z-DNA in the supercoiled plasmids had occurred in all topoisomers with superhelical density above the threshold value (Lyamichev et al., 1985; Peck and Wang, 1983), the pBan1 showed structural transition in a small population of molecules with 10 superhelical turns (superhelical density is ∼−0.04) and in ∼50% of population of more supercoiled molecules. Therefore, 2D gel electrophoresis data as well as the results on chemical and enzymatic probing suggest that under negative supercoiling stress, the plasmid pBan1 undergoes local structural transition, presumably cruciform at the inverted repeat.
FIGURE 4.
Two-dimensional gel electrophoresis of pBan1 DNA. The plasmid pBan1 with bacterial native superhelicity was fractionated on 1.2% agarose gel two-dimensionally. In the first dimension, standard TAE buffer was used and 6 μg/ml chloroquine was added in the second dimension. OC represents open circular pBan1, and the numbers of negative supercoils present in sample DNA are given at respective spots. Structural transition has occurred in the DNA molecules having 10 negative supercoils (indicated by arrow) or more, and it absorbs ∼3.5 helical turns (the spot marked with arrow migrated between the spot 6 and spot 7 in the first dimension, and the upper spot of number 11 migrated between spot 7 and spot 8, and so on).
Visualization of pBan1 DNA with electron and atomic force microscopes (EM and AFM)
The possibility of the alternative DNA structure formation in pBan1 DNA was tested by electron (Fig. 5) and atomic force microscopies (Fig. 6). We have observed thick protrusions at the sharp turn region of pBan1 DNA molecules, in both EM and AFM images, that were similar as the intramolecular triplex (H-DNA) reported previously (Stokrova et al., 1989; Tiner et al., 2001; Kato et al., 2002). We have measured the lengths of thick stems forming in two different DNA molecules, pBan1 (inverted repeat) and pTIR10 (polypurine/polypyrimidine) to evaluate the relationship between the stem size observed in microscopic imaging and the length of potential DNA region. Distribution of the size of the stem part in pBan1 DNA was listed in Table 1 along with the data for pTIR10 DNA. The data show that stems forming at pBan1 DNA are significantly shorter than those at pTIR10 in both EM and AFM analyses (examined by Kolmogorov-Smirnov test, p < 0.01). This is consistent with the lengths of the potential DNA sequences. The short inverted repeat in pBan1 consists of 17 bp complementary stretches (17 + 2 + 17 = 36 bp in total) that is consistent with the formation of the cruciform, whereas the 230 bp polypurine/polypyrimidine stretch in pTIR10 is able to adopt longer intramolecular triplex (H-DNA). These AFM data are in line with the length measurements for the H-DNA stem formed by 46 bp purine/pyrimidine mirror repeat; average stem length obtained with AFM performed at similar sample preparation and imaging conditions was 12 nm (Tiner et al., 2001). Therefore, the results obtained by both two different microscopic methods (EM and AFM) suggest that the stem structure observed in pBan1 DNA is originated from the short inverted repeat sequence. The microscopic analyses have shown clearly the presence of thick stem structure on pBan1 that is distinct from simple DNA cruciform.
FIGURE 5.
Electron microscopic images of alternative DNA structure forming at supercoiled DNA containing short inverted repeat (pBan1) and polypurine/polypyrimidine sequence (pTIR10). Representative images of pBan1 (A) and pTIR10 (B) are shown. Stem part is indicated by arrow in each panel. Scale bar (100 nm) is given at the bottom and common to both panels. The plasmid pBan1 retains the inverted repeat of 17 bp complementary stretches (17 + 2 + 17 = 36 bp in total); the length of potential stem-loop may be around 6 nm (0.34 nm/bp). The plasmid pTIR10 retains the polypurine/polypyrimidine sequence (∼230 bp) that may conform to intramolecular triplex (Kato et al., 2002).
FIGURE 6.

Atomic force microscopic images of pBan1 DNA. Large-scale scanning image is shown in panel A, and enlarged-rescanned images are shown in panels B and C. The molecules having the stem part at sharp turn region are numbered from 1 to 5.
TABLE 1.
Size distribution of thick protrusion visualized by microscopic techniques
| Number of molecules observed
|
||||
|---|---|---|---|---|
| Class (size of triplex-like stem: nm)
|
Measurement of EM images
|
Measurement of AFM images
|
||
| pTIR10 | pBan1 | pTIR10* | pBan1 | |
| 4–6 | 0 | 1 | 0 | 1 |
| 6–8 | 0 | 2 | 1 | 4 |
| 8–10 | 0 | 2 | 3 | 6 |
| 10–12 | 0 | 2 | 7 | 2 |
| 12–14 | 1 | 1 | 11 | 2 |
| 14–16 | 0 | 0 | 5 | 1 |
| 16–18 | 0 | 0 | 4 | 0 |
| 18–20 | 3 | 0 | 2 | 0 |
| 20–22 | 0 | 1 | 2 | 0 |
| 22–24 | 4 | 0 | 1 | 0 |
| 24–26 | 0 | 0 | 1 | 0 |
| 26–28 | 0 | 0 | 1 | 0 |
| 28–30 | 3 | 0 | 0 | 0 |
| 30–32 | 1 | 0 | 0 | 0 |
| 32–34 | 2 | 0 | 0 | 0 |
| 34–36 | 4 | 0 | 0 | 0 |
| 36–38 | 0 | 0 | 0 | 0 |
| 38–40 | 1 | 0 | 0 | 0 |
| 40–42 | 0 | 0 | 0 | 0 |
| Over 42 nm | 1 | 1 | 0 | 0 |
| Total | 20 | 10 | 38 | 16 |
Results from Kato et al. (2002).
DISCUSSION
The results of enzymatic and chemical modification of pBan1 DNA together with the microscopic analyses propose a structural model for the alternative DNA structure forming at the inverted repeat, termed hairpin triplex, which consists of a triple-stranded stem with a single stranded extension (Fig. 7). Two stem-loops of the cruciform arrange in parallel allowing them to interact with each other, and one stem-loop melts to donate the third strand to another stem-loop. In this model, the length of the stem may be critical; longer stem is too stable to donate the third strand, and the triple-stranded structure is unstable with shorter stem. Loop-loop interaction may be also possible as proposed for slipped stranded DNA in triplet repeat sequences (Sinden et al., 2002).
FIGURE 7.
Hairpin triplex model formed at the inverted repeat sequence. Two stem-loops of the cruciform DNA are arranged in parallel, with one stem-loop melting to yield a third strand. DNA cruciform (center) and two isomers of hairpin triplex (right and left) may be interchangeable.
Ohta et al. (1996) proposed a quadruplet model consisting of a pair of stem-loops for the alternative DNA structure at short inverted repeat based on the results of atomic force microscopy; our model is consistent with their results. Shlyakhtenko et al. (2000) have proposed that the structural transition of cruciforms can be a molecular trigger for the slithering of DNA chains. Since the formation of a hairpin triplex causes sharp bending at the center of a palindromic symmetry, it probably restricts DNA slithering more effectively than the X-type cruciform. The hairpin triplex may be formed in a short palindromic sequence when the X-type cruciform faces the stress of diminishing the radius of the loop at the edge of plectonemic superhelix; for example, when there is an increase of negative superhelicity. As observed in the 2D gel electrophoresis (Fig. 4), two discrete topological states exist in the molecules above the threshold superhelical density (10 or more negative supercoils). Slower-migrating populations (upper spots) are expected to be forming the cruciform. The populations of faster migration (lower spots) would not be in B-form, because the supercoiling-induced formation of cruciforms occurs similarly to Z-DNA and H-DNA; all of the molecules conform to alternative DNA structure with superhelical density above the threshold value (Sinden, 1994). We hypothesize that tight folding of the cruciform (formation of the hairpin triplex) compacts the molecules in such a way that their electrophoretic mobility increases. The coexistence of two conformations leads to the 2D gel electrophoresis pattern observed in the present paper. Structural transition between the cruciform and hairpin triplex may readily occur in living cell. For instance, interarm interaction involving triplex part and single-stranded region occurring at four-way Holliday junction may affect the branch migration.
According to the hairpin triplex model, one 5′-half of the inverted repeat becomes the third strand and should be arranged in parallel with the 5′-half of the stem-loop structure, as shown in Fig. 7. Four types of base triad are involved in the hairpin triplex: G*G-C, A*A-T, T*T-C, and C*C-G (where asterisks indicate the interaction between the third strand and the stem, and hyphens indicate the Watson-Crick type baseparing of the stem). Rao and Radding (1994) have proposed a possible arrangement of these base triads in parallel orientation. The G*G-C triad is stable enough to form a triplex having two hydrogen bonds between the two guanines (Soyfer and Potaman, 1996). The other three triads are less stable than G*G-C, but may form two hydrogen bonds. In that regard, Shchyolkina et al. (1995, 2001) have also provided evidence of the existence of a parallel purine*purine-pyrimidine triplex. More recently, Walter et al. (2001) have reported stable A*A-T triplex in parallel orientation. Furthermore, the parallel G*G-C triplex appears to be stabilized by the presence of zinc ion (Khomyakova et al., 2000). In the present study, 1 mM ZnSO4 was added to the reaction mixture to optimize the activity of S1 nuclease. The presence of zinc ions can stabilize the hairpin triplex structure, although the addition of ZnSO4 did not affect the profiles of permanganate probing (data not shown) and ZnSO4 was not used in the sample preparation for microscopic analysis.
The inverted repeat sequences of the guanine-rich 5′-half are expected to have a higher potential of forming the hairpin triplex. Using a NCBI BLAST search, we have found several DNA sequences in the GenBank/EMBL/DDBJ international databases that are similar or nearly identical to the inverted repeat tested here (data not shown). They may have a potential of forming hairpin triplexes and may be involved in the gene regulation and chromosome construction, given the effect of the structural transition on the global topology of chromosomal DNA.
Acknowledgments
The authors thank Mr. Masanori Yoshida for preparation of bluegill genomic DNA and DNA sequencing and Takara-Bio for the use of FMBIO-II image analyzer.
This work was supported in part by funds from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.K. and N.S.), Japan Society for the Promotion of Science (to N.S.), and the National Institutes of Health (GM 62235 to Y.L.L.).
References
- Bissler, J. J. 1998. DNA inverted repeats and human disease. Front. Biosci. 3:d408–d418. [DOI] [PubMed] [Google Scholar]
- Cheng, A.-J., and W. V. Dyke. 1993. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 21:5630–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delain, E., A. Fourcade, B. Revet, and C. Mory. 1992. New possibilities in the observation of nucleic acids by electron spectroscopic imaging. Microsc. Microanal. Microstruc. 3:175–186. [Google Scholar]
- Glucksmann, M. A., P. Markiewicz, C. Malone, and L. B. Rothman-Denes. 1992. Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell. 70:491–500. [DOI] [PubMed] [Google Scholar]
- Hanai, R., and J. Roca. 1999. Two-dimensional agarose-gel electrophoresis of DNA topoisomers. Methods Mol. Biol. 94:19–27. [DOI] [PubMed] [Google Scholar]
- Hayatsu, H. 1996. The 5, 6-double bond of pyrimidine nucleosides, a fragile site in nucleic acids. J. Biochem. 119:391–395. [DOI] [PubMed] [Google Scholar]
- Jin, R., M. E. Fernandez-Beros, and R. P. Novick. 1997. Why is the initiation nick site of an AT-rich rolling circle plasmid at the tip of a GC-rich cruciform? EMBO J. 16:4456–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., and A. Furuno. 1992. Unusual structure of plasmid DNA formed in transformants of Escherichia coli. Res. Microbiol. 143:665–669. [DOI] [PubMed] [Google Scholar]
- Kato, M., K. Matsunaga, and N. Shimizu. 1998. A novel unusual DNA structure formed in an inverted repeat sequence. Biochem. Biophys. Res. Commun. 246:532–534. [DOI] [PubMed] [Google Scholar]
- Kato, M., C. J. McAllister, S. Hokabe, M. Shimizu, and Y. L. Lyubchenko. 2002. Structural heterogeneity of pyrimidine/purine-biased DNA sequence detected by atomic force microscopy. Eur. J. Biochem. 269:3632–3636. [DOI] [PubMed] [Google Scholar]
- Khomyakova, E. B., H. Gousset, J. Liquier, T. Huynh-Dinh, C. Gouyette, M. Takahashi, V. L. Florentiev, and E. Taillandier. 2000. Parallel intramolecular DNA triple helix with G and T bases in the third strand stabilized by Zn2+ ions. Nucleic Acids Res. 28:3511–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. L., H. Peng, F. M. Esparza, S. Z. Maltchenko, and M. K. Stachowiak. 1998. Cruciform-extruding regulatory element controls cell-specific activity of the tyrosine hydroxylase gene promoter. Nucleic Acids Res. 26:1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockson, D., and D. A. Galloway. 1986. Cloning and characterization of oriL2, a large palindromic DNA replication origin of herpes simplex virus type 2. J. Virol. 58:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev, V. I., S. M. Mirkin, and M. D. Frank-Kamenetskii. 1985. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J. Biomol. Struct. Dyn. 3:327–338. [DOI] [PubMed] [Google Scholar]
- Ohta, T., S. Nettikadan, F. Tokumasu, H. Ideno, Y. Abe, M. Kuroda, H. Hayashi, and K. Takeyasu. 1996. Atomic force microscopy proposes a novel model for stem-loop structure that binds a heat shock protein in the Staphylococcus aureus HSP70 operon. Biochem. Biophys. Res. Commun. 226:730–734. [DOI] [PubMed] [Google Scholar]
- Peck, L. J., and J. C. Wang. 1983. Energetics of B-to-Z transition in DNA. Proc. Natl. Acad. Sci. USA. 80:6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, B. J., and C. M. Radding. 1994. Formation of base triplets by non-Watson-Crick bonds mediates homologous recognition in RecA recombination filaments. Proc. Natl. Acad. Sci. USA. 91:6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper, R. M., B. N. Danforth, and B. W. Glickman. 1986. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J. Mol. Biol. 189:273–284. [DOI] [PubMed] [Google Scholar]
- Schroth, G. P., and P. S. Ho. 1995. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 23:1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchyolkina, A. K., O. F. Borisova, E. E. Minyat, E. N. Timofeev, I. A. Il'icheva, E. B. Khomyakova, and V. L. Florentiev. 1995. Parallel purine-pyrimidine-purine triplex: experimental evidence for existence. FEBS Lett. 367:81–84. [DOI] [PubMed] [Google Scholar]
- Shchyolkina, A. K., E. N. Timofeev, Y. P. Lysov, V. L. Florentiev, T. M. Jovin, and D. J. Arndt-Jovin. 2001. Protein-free parallel triple-stranded DNA complex formation. Nucleic Acids Res. 29:986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko, L. S., P. Hsieh, M. Grigoriev, V. N. Potaman, R. R. Sinden, and Y. L. Lyubchenko. 2000. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J. Mol. Biol. 296:1169–1173. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko, L. S., V. N. Potaman, R. R. Sinden, and Y. L. Lyubchenko. 1998. Structure and dynamics of supercoil-stabilized DNA cruciforms. J. Mol. Biol. 280:61–72. [DOI] [PubMed] [Google Scholar]
- Sinden, R. R. 1994. DNA Structure and Function. Academic Press, New York.
- Sinden, R. R., V. N. Potaman, E. A. Oussatcheva, C. E. Pearson, Y. L. Lyubchenko, and L. S. Shlyakhtenko. 2002. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 27(Suppl. 1):53–65. [DOI] [PubMed] [Google Scholar]
- Soyfer, V. N., and V. N. Potaman. 1996. Triple-Helical Nucleic Acids. Springer, New York.
- Stokrova, J., M. Vojtiskova, and E. Palecek. 1989. Electron microscopy of supercoiled pEJ4 DNA containing homopurine.homopyrimidine sequences. J. Biomol. Struct. Dyn. 6:891–898. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Y. Kawamura, N. Sakata, G. E. Elmesiry, Y. Takemon, K. Tanida, S. Minoshima, N. Shimizu, and M. Kato. 2001. Nucleotide sequence of BamHI family satellite DNA and its unit length polymorphism in bluegill sunfish Lepomis macrochirus. Mol. Biol. Rep. 28:119–122. [DOI] [PubMed] [Google Scholar]
- Tiner, W. J., Sr., V. N. Potaman, R. R. Sinden, and Y. L. Lybchenko. 2001. The structure of intramolecular triplex DNA: atomic microscopy study. J. Mol. Biol. 314:353–357. [DOI] [PubMed] [Google Scholar]
- Walter, A., H. Schutz, H. Simon, and E. Birch-Hirschfeld. 2001. Evidence for a DNA triplex in a recombination-like motif: I. Recognition of Watson-Crick base pairs by natural bases in a high-stability triplex. J. Mol. Recognit. 14:122–139. [DOI] [PubMed] [Google Scholar]
- Wells, R. D. 1988. Unusual DNA structures. J. Biol. Chem. 263:1095–1098. [PubMed] [Google Scholar]