Abstract
We have investigated the role of the native lipids on bacteriorhodopsin (bR) proton transfer and their connection with the cation-binding role. We observe that both the efficiency of M formation and the kinetics of M rise and decay depend on the lipids and lattice but, as the lipids are removed, the cation binding is a much less important factor for the proton pumping function. Upon 75% delipidation using 3-[(cholamidopropyl)dimethylammonio]-propanesulfonate (CHAPS), the M formation and decay kinetics are much slower than the native, and the efficiency of M formation is ∼30%–40% that of the native. Upon monomerization of bR by Trition X-100, the efficiency of M recovers close to that of the native (depending on pH), M formation is ∼10 times faster, and M decay kinetics are comparable to native at pH 7. The same results on the M intermediate are observed if deionized blue bR (deI bbR) is treated with these detergents (with or without pH buffers present), even though deionized blue bR containing all the lipids has no photocycle. This suggests that the cation(s) has a role in native bR that is different than in delipidated or monomerized bR, even so far as to suggest that the cation(s) becomes unimportant to the function as the lipids are removed.
INTRODUCTION
Bacteriorhodopsin (bR) is an integral membrane protein contained in the purple membrane of the halophilic organism Halobacterium salinarum. It is the only protein in the membrane, and is purple colored due to a covalently bound retinal chromophore at the Lys-216 residue, which has a broad absorption band centered at 570 nm. This chromophore allows the bR molecule to absorb light, which isomerizes the retinal from all-trans to 13-cis initiating a thermal photocycle of events, resulting in the unidirectional transport of a proton across the membrane (Mathies et al., 1991; Lanyi, 1993, 1998). The proton-motive force caused by this photocycle is used by the organism for the ATP synthesis required for its metabolism under nonrespiratory conditions (such as low oxygen). The photosynthetic system of this membrane protein is far simpler than the electron-motive based system of chlorophyll, and has become the model of studying biological ion pumps (Lanyi, 1995), as well as cis-trans based isomerizing retinal proteins such as those used in mammalian vision processes (Birge, 1990a,b). The spectrally distinct intermediates seen throughout the photocycle and the resulting photocurrent have also rendered the system useful in a number of biomolecular electronic and optoelectronic applications (Hampp, 2000; Birge et al., 1999; Hampp et al., 1994).
To have a true, complete understanding of the photocycle, we must discover the effect of the membrane (lipid) environment on the function. Furthermore, it is known that the proton translocation does not proceed if the bR is devoid of cations, which can be imposed by deionization, chelation, or acidification (Mowery et al., 1979; Chang et al., 1985; Kimura et al., 1984). It is postulated that this result is due to the cation affecting the protonation state of the Asp-85 residue, the proton acceptor from the Schiff base (SB) of the retinal, in the ground state bR (Balashov et al., 1996; Subramaniam et al., 1992). However, the exact role of the cations in this process is still not known, as is their exact location. Even the recent high-resolution x-ray structure resolved to 1.55 Å did not elucidate the cation positions (Leucke et al., 1999). The effect of the crystallization procedure on the cations in bR is still unknown and may be a possible cause of this lack of observation, or may simply be due to not high enough spatial resolution of the x rays. We have recently shown that solubilization of the bR lattice into micelles alter the secondary structure of bR more extensive than deionization (Heyes and El-Sayed, 2002). Furthermore, the thermal transitions are found to vary with partial lipid removal and monomerization (Heyes and El-Sayed, 2002). We have also investigated the refolding of bR from the thermally denatured state in both the native and monomerized form and found very different refolding properties (Wang et al., 2002).
The spectral changes in bR throughout the photocycle are identical in the native and Trition X-100 solubilized form, with a small exception in the M intermediate (Varo and Lanyi, 1991a,b). Normally there is only one spectrally resolved M intermediate in the visible region in native bR but, upon solubilization, it was found that there are two M forms, with a very small red-shift in absorption maxima from M1 to M2 (Varo and Lanyi, 1991b). Additionally, the thermodynamic parameters of the photocycle are found to be different in the solubilized form (Varo and Lanyi, 1991c). It is not known if this is due purely to the change in lipid environment, or if there are other indirect effects concerning the cation(s).
Here we present results concerning both partial delipidation and monomerization on the dynamics and efficiency of the photocycle. We investigate these effects for both native and deionized blue bR (deI bbR) in an attempt to discover the connection between lipid, chromophore, protein, and cation.
MATERIALS AND METHODS
Sample preparation
Bacteriorhodopsin was grown and purified by a standard procedure as previously described (Oesterhelt and Stoeckenius, 1974). Deionization was performed by passing bR suspended in 18 MΩ doubly distilled water (DDW) though a cation exchange column in the H+ form (Bio-Rad, AG-50W-X8). The eluted blue bR (bbR) was at pH 4.4–4.7. A solution of 75% delipidated bR was prepared as previously described using 3-[(cholamidopropyl)dimethylammonio]-propanesulfonate (CHAPS) (Szundi and Stoeckenius, 1987). Briefly, 20 mM CHAPS was dissolved in either 5 mM sodium acetate buffer (pH 5.5) or in DDW. Approximately 10 mg bR was centrifuged to a pellet, and 2 ml of the CHAPS solution was added to the pellet. The solution was left to equilibrate overnight. It was spun down again and the supernatant discarded. Then the pellet was resuspended in another 5 ml of detergent solution and left to equilibrate again. The solution was spun down once again, and detergent solution was added to obtain the desired concentration. The relationship between lipid extraction and absorption spectrum has been investigated by Szundi and Stoeckenius (1987). At >8 mM CHAPS (the CMC of CHAPS), the shift in the absorption spectrum and the lipid extraction were shown to be complete and no further changes are observed at higher CHAPS concentrations. A direct relationship between lipid composition and absorption spectrum was shown. This has also been shown by using different concentrations of Trition (Wu et al., 1991) and deoxycholate (Hwang and Stoeckenius, 1977). Thus, we use the absorption spectrum shift as evidence of the lipid removal. The fact that we use 20 mM CHAPS in all sample preparations (more than twice its CMC), and this shifts the λmax in native bR to the published 560 nm, we conclude that the 75% lipid removal is accomplished. bR monomer was prepared by Trition X-100 solubilization as previously described (Huang et al., 1980). This was prepared using 5% Trition in either 0.1 M Tris or DDW, and spun down using an Amicron 5000 MWCO filter cell. The equilibration, resuspension, and concentration control was performed as for the CHAPS treatment. Monomerization was shown to be complete by the shift in absorption energy to 553 nm and the fact that no sediment formed on centrifugation at 19,000 rpm for 45 min. Samples were prepared both by using the normal buffers (5 mM acetate for CHAPS, pH 5.0; 0.1 M Tris for Trition, pH 7.4) and using only detergent in DDW to investigate the effect of the buffers on the samples. The absorption spectrum of each sample was measured on a Shimadzu UV-3101PC spectrometer in plastic cuvettes (the same ones that they were prepared in) that had been rinsed with 10% HNO3 and DDW many times to ensure no cation contamination. Furthermore, the CHAPS in DDW and Trition in DDW solutions were analyzed by inductively coupled plasma-optical emission spectroscopy (ICP-OES) for Ca2+, Mg2+, and Na+ contaminants.
Flash photolysis
Flash photolysis studies were performed using an Nd:YAG coupled MOPO laser (Spectraphysics, Mountain View, CA) with laser excitation at 570 nm (10-ns pulse width, 10-Hz repetition rate, 17 mJ per pulse). A Xenon arc lamp (PTI, Lawrenceville, NJ) was used for continuous probing combined with a monochrometer set at 412 nm (Acton research 300i, Acton, MA), a photomultiplier tube, and a 500-MHz transient digitizer (LeCroy 9350A, Chestnut Ridge, NY) for which ∼2000 laser shots were averaged. For comparison of the efficiencies of M formation, the OD at 570 nm and the slit widths of the monochrometer were identical for all samples and the laser power was continuously monitored throughout the experiment to ensure equal photon density at the samples. All samples were light adapted for 60 min before measurement.
Data analysis
Efficiency of M formation was obtained by determining the maximal delta absorbance at 412 nm for each sample, and setting the delta absorbance of native at 100% efficiency. Since the OD at the excitation wavelength (570 nm) is controlled to be equal for each sample, the number of bR molecules absorbing a 570-nm photon is equal (assuming that molar absorptivity, ɛ, does not change at 570 nm). Thus, the efficiency of the SB to deprotonate for a given number of absorbing species is compared. In general, kinetic fits were obtained by fitting to a monoexponential decay and inspecting both the fit and the chi-squared values, using initial parameters from the literature when available. These were compared to the fit and chi-squared values obtained by fitting to a biexponential decay. When there was no significant improvement in the fit, or the differences in the fitted lifetimes of two components were small, then the fit was concluded to be monoexponential. If two exponents were required to significantly reduce chi-squared and the lifetimes were separated by more than their errors (generally 1–2 μs due to the excellent signal/noise ratio of the transient signal—omitted in Table 2 for sake of clarity since they are small), then the fit was concluded as biexponential. Since all these curves fit well to one of these two fitting methods (see Figs. 3 and 4), higher exponentials or spread exponentials were not attempted. For M rise and decay kinetics, the origins of these mono- and/or biexponential fits have been extensively discussed in the literature (see later in the text and Hendler et al. (2001); Varo and Lanyi (1991a,b) for more details).
TABLE 2.
| M Rise
|
M Decay
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sample | τ1 (A1) | τ2 (A2) | τavg | τ1 (A1) | τ2 (A2) | τavg | Relative % efficiency of M | Measured pH |
| Native bR in DDW | 8.49 μs (0.21) | 75.8 μs (0.79) | 61.7 μs | 8.57 ms | – | 8.57 ms | 100 | 7.0 |
| Native + CHAPS in acetate | 16.4 μs (0.32) | 107 μs (0.68) | 78.0 μs | 25.5 ms | – | 25.5 ms | 36 | 5.6 |
| Native + CHAPS in DDW | 23.5 μs (0.49) | 137 μs (0.51) | 81.4 μs | 27.7 ms | – | 27.7 ms | 45 | 5.0 |
| Native + Trition in Tris | 1.86 μs (0.58) | 13.8 μs (0.42) | 6.87 μs | 2.05 ms (0.54) | 28.3 ms (0.46) | 14.1 ms | 71 | 7.4 |
| Native + Trition in DDW | 1.76 μs (0.42) | 9.71 μs (0.58) | 6.71 μs | 9.47 ms (0.96) | 34.0 ms (0.04) | 10.5 ms | 145 | 5.4 |
| DeI bbR in DDW | – | – | – | – | – | – | <5 | 4.3 |
| DeI + CHAPS in acetate | 29.2 μs (0.61) | 170 μs (0.39) | 84.1 μs | 27.1 ms | – | 27.1 ms | 35 | 5.0 |
| DeI + CHAPS in DDW | 21.0 μs (0.42) | 124 μs (0.58) | 80.7 μs | 27.4 ms | – | 27.4 ms | 33 | 4.2 |
| DeI + Trition in Tris | 1.67 μs (0.57) | 14.3 μs (0.43) | 7.10 μs | 2.76 ms (0.54) | 39.3 ms (0.46) | 19.4 ms | 98 | 7.2 |
| DeI + Trition in DDW | 5.47 μs (0.77) | 69.2 μs (0.23) | 20.1 μs | 7.93 ms (0.58) | 23.2 ms (0.42) | 14.3 ms | 79 | 4.3 |
τavg is calculated by τavg = (A1 × τ1) + (A2 × τ2), where A1 + A2 = 1.
FIGURE 3.
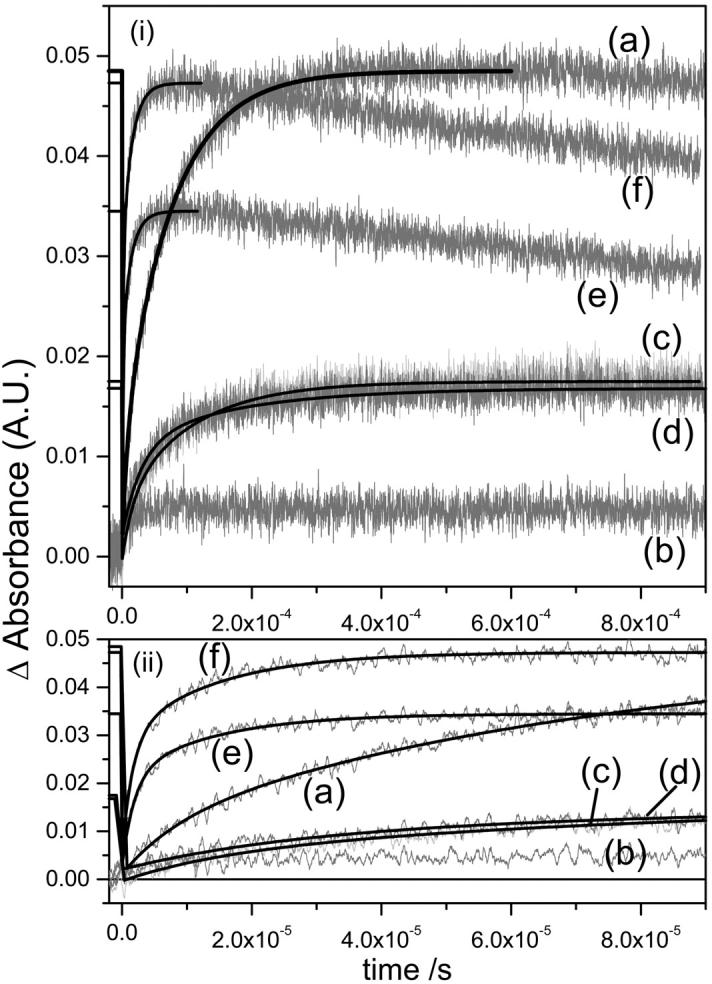
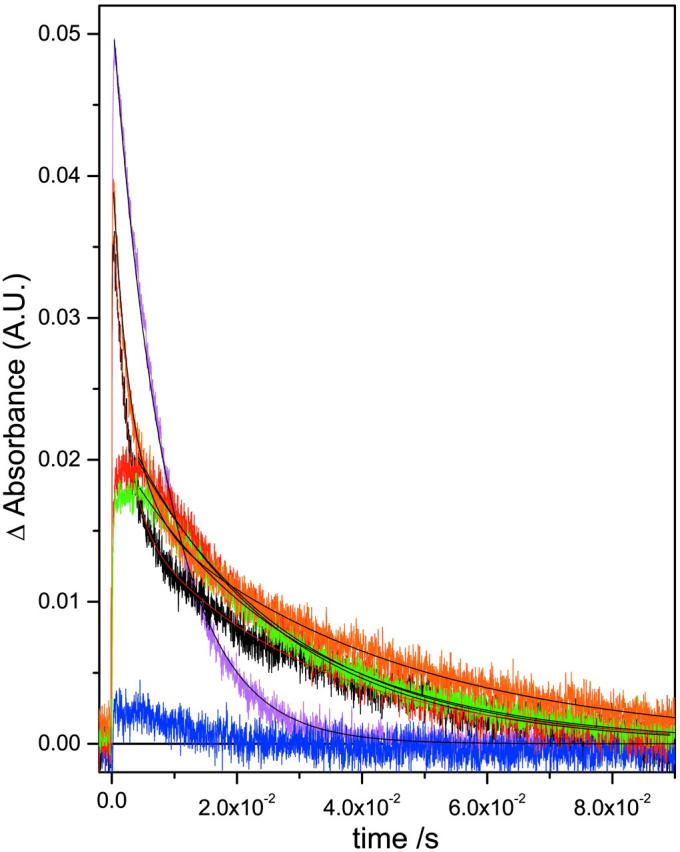
M rise (a) and M decay (b) kinetics for the detergent treated native and deionized bR. For M rise, i displays the whole timescale measured and ii displays the early timescales to aid the reader in comparing the fast component data. The buffers used were sodium acetate for CHAPS (pH 5.5) and Tris for Trition (pH 7.4) as previously described (Szundi and Stoeckenius, 1987; Huang et al., 1980). The legend is: (a) native (magenta), (b) deionized (blue), (c) CHAPS treated native (red), (d) CHAPS treated deI bbR (green), (e) Trition treated native (black), (f) Trition treated deI bbR (orange). Kinetic parameters are shown in Table 2.
FIGURE 4.
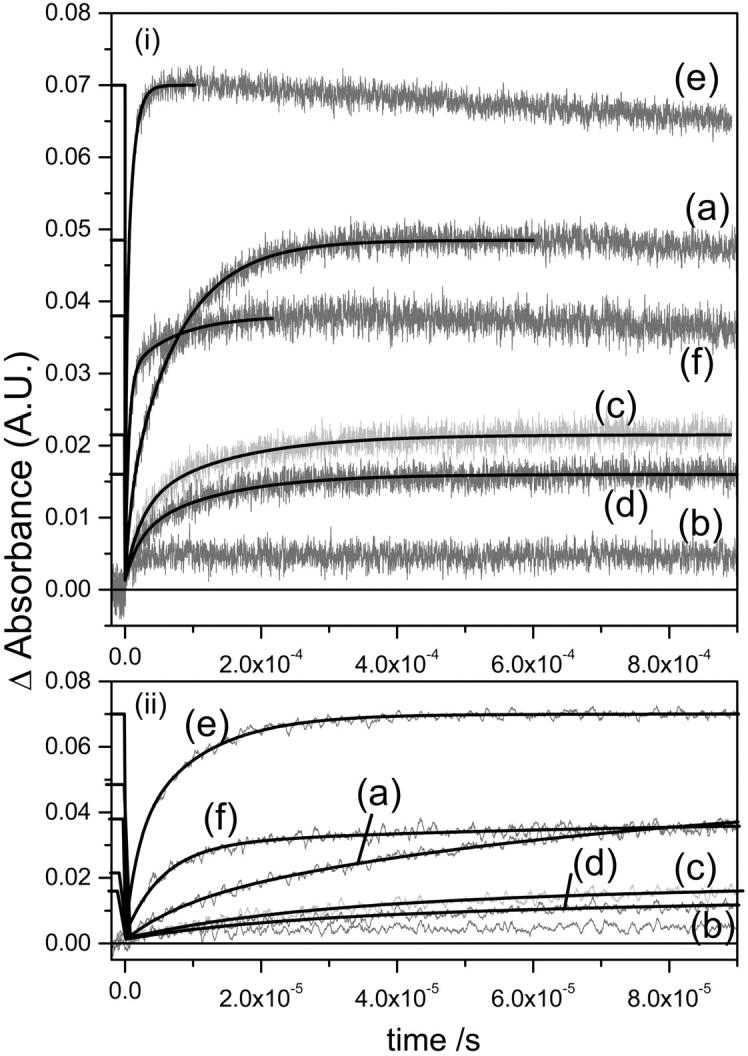
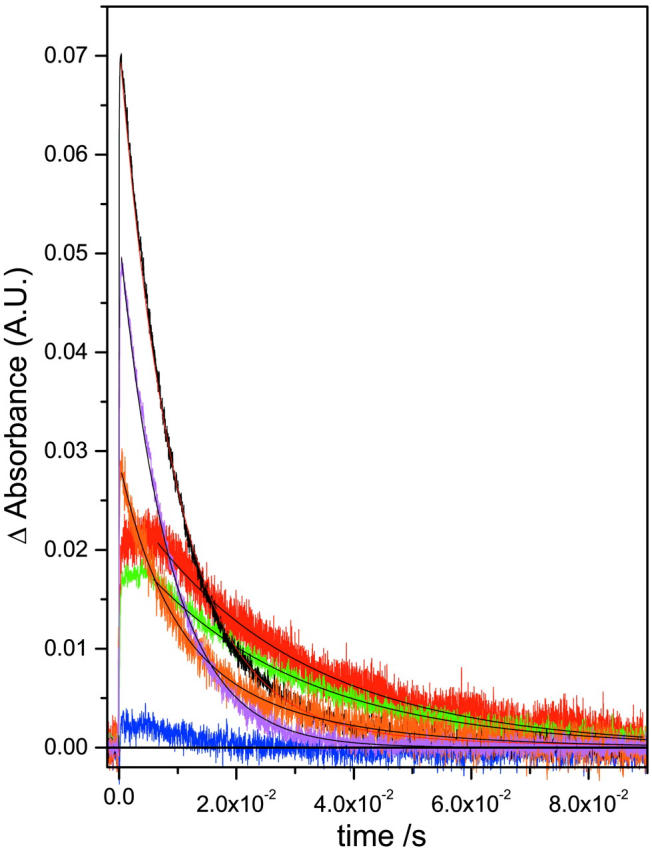
M rise (a) and M decay (b) kinetics for the detergent treated native and deionized bR. For M rise, i displays the whole timescale measured and ii displays the early timescales to aid the reader in comparing the fast component data. No buffers were used in this figure, but the pHs were not able to be controlled (see Table 2 and text). Refer to Fig. 3 for the legend. Kinetic parameters are shown in Table 2.
RESULTS AND DISCUSSION
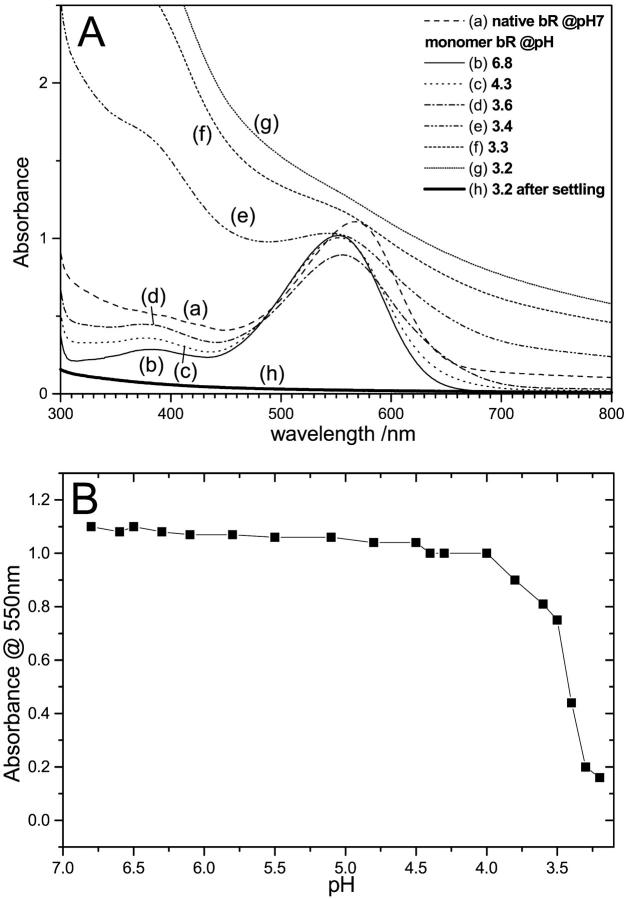
Fig. 1 shows the absorption spectra of the native and deionized bR after treatment by each of the detergents. Spectra are shown for samples in the buffered detergent (Fig. 1 a) as well as the detergent in unbuffered DDW only (Fig. 1 b). Deionization is known to shift the absorption maximum of native bR to 603 nm (Mowery et al., 1979; Chang et al., 1985; Kimura et al., 1984). Monomerization of native bR blue shifts the absorption maximum to 550 nm (Dencher and Heyn, 1978, 1982), whereas 75% delipidation by CHAPS shifts it to 560 nm (Szundi and Stoeckenius, 1987). Table 1 summarizes the specific peak positions and shifts. Our results agree with these reported previously for the native bR. Unsurprisingly, this is similar if the samples are buffered or not, since none of the samples fall below the pKa of the Asp-85, ∼3.3. Upon 75% delipidation of deionized blue bR in the buffer-free detergent, the shift is from 603 to 567 nm—the same blue-shifted direction as the CHAPS treated native, but not to the same wavelength. In the presence of the 5 mM sodium acetate buffer, this blue shift does completely shift to 560 nm, showing that there are some effects of cations (from the buffer) on the final λmax. Monomerization of the deionized bR shifts the maximum to the same as that of the monomerized native, 553 nm, whether buffer is present or not. This suggests that the chromophore is affected similarly whether the cation is present before monomerization or not, which shows that there are no effects of (buffer) cations on the final λmax. This may imply movement of the native cation(s) to an inactive location (from the color-controlling perspective), or removal altogether. This led to the question of effect of acidification on the monomerized native bR. This was seen by the eye and is shown spectrally in Fig. 2. The most striking observation from this titration is that there is denaturation of the protein at pH lower than ∼3.4, unlike native bR, which retains the bound chromophore at pH down to 0 or even less (Mowery et al., 1979). The protein aggregates and loses the opsin-shifted color. This is shown by the higher baseline at nonabsorbing energies, typical of light scattering from aggregated samples. In addition, there is a peak rising roughly concurrent with this aggregation at 380 nm, the λmax of unbound retinal. Interestingly, as the aggregates settle out of the beam path, so does the unbound retinal peak. This suggests that retinal is within this aggregate, and not free in the solution. The Schiff base hydrolyses, but retinal does not leave the aggregated protein.
FIGURE 1.
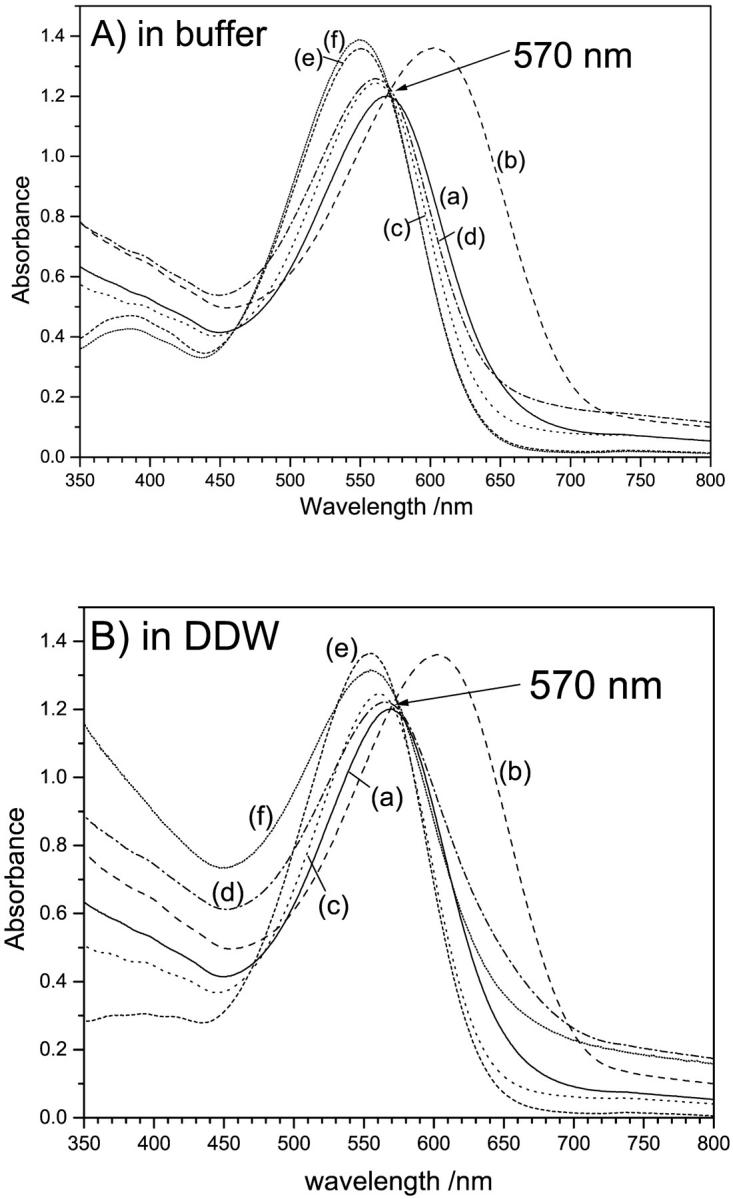
Absorption spectra of CHAPS (75% delipidation) and Trition (monomerized) treated native and deionized bR (A) in the presence and (B) absence of buffers. The legend is as follows: (a) native, (b) deionized, (c) CHAPS treated native, (d) CHAPS treated deI bbR, (e) Trition treated native, (f) Trition treated deI bbR. In general, deionization causes a red shift, whereas delipidation causes a blue shift. The specific peak positions and shifts are summarized in Table 1. The buffer has no effect on the bR monomer, and the λmax is the same whether the deionized bR or native bR is monomerized, suggesting these species are the same. The buffer does have an effect on CHAPS treated bR. The λmax of deionized, delipidated bR is the same as native delipidated bR in the presence of buffers, 560 nm, but is 567 nm for unbuffered CHAPS treated bR, suggesting some cation involvement in color regulation (but not as much as in native).
TABLE 1.
λmax and spectral shifts from the native or deionized blue bR upon treatment of the bR sample with the detergents
| Sample | λmax/nm | Δλmax upon detergent treatment/nm |
|---|---|---|
| Native | 570 | – |
| CHAPS treated native (in buffer) | 560 | −10 from native |
| Trition treated native (in buffer) | 553 | −17 from native |
| CHAPS treated native (in DDW) | 560 | −10 from native |
| Trition treated native (in DDW) | 553 | −17 from native |
| DeI bbR | 603 | – |
| CHAPS treated deI bbR (in buffer) | 560 | −43 from deI bbR |
| Trition treated deI bbR (in buffer) | 552 | −51 from deI bbR |
| CHAPS treated deI bbR (in DDW) | 567 | −36 from deI bbR |
| Trition treated deI bbR (in DDW) | 552 | −51 from deI bbR |
CHAPS removes 75% of the lipids, and Trition monomerizes the bR samples.
FIGURE 2.
(A) absorption spectra and (B) titration curve of absorbance at 550 nm of monomeric bR with microliter additions of HCl to the cuvette. There is no red shift associated with Asp-85 protonation as is seen in native bR. The protein denatures and the SB hydrolyses at pH ∼3.4.
From Fig. 1 and Table 1, the effects of lipid removal on the chromophore absorption energy follow the general trend of red shifting upon cation removal and blue shifting on lipid removal. Even if the cation(s) is removed first, and the λmax is at 603 nm, 75% lipid removal blue shifts it to 567 nm—a larger blue shift than 75% delipidation of the native. An even larger effect is seen upon monomerization, 570–550 nm for native bR compared to 603–550 nm for deionized bR. This suggests that the monomer is identical whether deionized first or not. This may be explained by the fact that the cation(s) is no longer in the (color-controlling) active site upon monomerization, or its electrostatic and binding effect is completely negated by the more hydrophilic environment caused by exposure to water. It is possible that the cation is necessary only to increase the electrostatics when the lipid environment is present to counter the effect of the hydrophobic lipid environment. This is no longer necessary once this lipid hydrophobicity is reduced or eliminated. This is further supported by the fact that even though the lipid-depleted bR and monomer bR pump protons, and therefore have their proton acceptor (Asp-85) initially deprotonated, there is no deprotonation pKa observed for this acceptor group before denaturation of the monomer (Fig. 2). Normally the cation is thought to keep the Proton acceptor deprotonated in the hydrophobic environment. This may not be necessary once the lipid environment is reduced or eliminated, since the charge density is increased already, as evidenced from the absorption spectra (Fig. 1).
When performing these detergent treatments, it is possible to either control the pH by adding buffers, or to control the added cation by using only the detergent in unbuffered DDW. Previously reported data on the detergent treatment of bR (Szundi and Stoeckenius, 1987; Dencher and Heyn, 1982; Lozier, 1982; Jang and El-Sayed, 1988) has had the treatment performed in the presence of pH buffers. In native bR, this is usually not a problem in data interpretation since the monovalent cations present in the buffers have very low affinity for the native bR and displacing native cations requires very high concentrations (>1 M) of monovalent ions. Fig. 3, a and b shows the flash photolysis M rise and decay at 412 nm in the presence of the buffered detergents (acetate for CHAPS, and Tris for Triton X-100) and is summarized in Table 2. Native bR that has been 75% delipidated shows a lowered efficiency of M formation of ∼35% compared to the untreated native, whereas monomerized native bR has an efficiency of ∼70% of the native bR. Also, it is well known that deionized blue bR has no M intermediate due to the lack of a proton acceptor group (deprotonated Asp-85). From the figure, there is in fact a very small signal from the deionized bR. This has been observed before, and is likely due to very small amounts of remaining cation-bound bR in the sample due to <100% deionization efficiency (Chang et al., 1986). However, when the deionized bR is delipidated to 75%, there is an increase in the photocycle efficiency. In fact, the efficiency is comparable to that of 75% delipidated native bR. Also, deionized bR in Triton (monomerized deionized bR) shows further increased efficiency, and is actually even slightly more efficient than the native monomerized bR. These results seem to suggest that upon delipidation, the photocycle is not affected by the presence of the native cation(s) as it is in the native membrane.
To investigate if the presence of buffer cations affects the recovery of the M intermediate upon delipidation of deI bbR, the experiments were also done in the absence of buffers—just the detergent in DDW. DeI bbR can be regenerated with monovalent cations such as Na+, but are usually required to be in large concentrations (∼100–500:1 cation:bbR ratios). However, since our aim is to separate the cation and lipid effects on the proton transfer steps, we have examined the possibility of buffer cation interference. The results are shown in Fig. 4, a and b, and Table 2. The CHAPS treated native bR in the absence of buffer is almost identical to the buffered CHAPS treated native bR—in fact, the efficiency is slightly higher. This is also true for the native monomer in the absence of buffers. It is actually more efficient without the buffer and even more efficient than the native untreated bR. The reason for this increased efficiency is not clear at the present time, but the stock solution used for the monomerization is identical to the measured native bR, so bR sample variation is excluded. This increased M efficiency of the bR monomer in the absence of any other cations is consistent with the increased M formation time as previously reported, and discussed later in this paper. Most likely, the fact that bR monomers have much less interaction within micelles and therefore more flexibility could explain the increased efficiency. For the deionized CHAPS bR, the percent efficiency is the same whether buffers are present or not. The deionized monomer without buffer has the percent efficiency reduced from ∼100% to ∼80% of the native compared to the deionized monomer in buffer. The most likely explanation for deionized monomer efficiency variation is the pH. Since the CHAPS buffer (acetate) is ∼pH 5, and unbuffered CHAPS is about the same pH, there is little change, whereas the Triton buffer (Tris) is ∼pH 7.4 and without the buffer, the pH of the deionized monomer is ∼4.3. The efficiency of M of the solubilized native bR in buffered solutions has been found to vary with pH so this is not surprising (Drachev et al., 1993). The assignment made in the previous paragraph of the cation being less involved in the photocycle upon delipidation still seems to hold true from these results on unbuffered samples. This is principally based on the recovery of the M intermediate efficiency upon delipidation or monomerization even in the absence of buffers, and the fact that the Na+ ions in the CHAPS buffer have no effect at all. To ensure that the detergent solutions alone do not introduce contaminant cations, we analyzed them by using inductively coupled plasma-optical emission spectroscopy for Ca2+, Mg2+, and Na+. It was shown that the concentrations of cations in the detergent solutions are close to the detection limit of the instrument, and therefore the ratio of cation:bR is less than 0.5:1 for Ca:bR, 0.01:1 for Mg:bR, and 3:1 for Na:bR. All of these ratios are known not to convert blue bR to purple bR, and can be excluded as a factor in these observations.
The effect of removing the cations from bR on the photocycle has long been studied, as has the effect of adding the native Ca2+ and Mg2+, or other (nonnative), cations to the deionized bR (Ariki and Lanyi, 1986; Jonas et al., 1990; El-Sayed et al., 1995). Since it was discovered that cations were necessary for the proton pumping, many efforts have tried to explain their binding to bR. Two models were proposed. The first is the specific binding model, which describes the cation binding directly to specific sites in the protein, with at least one site affecting the retinal absorption energy (i.e., binding directly to the protein) (Ariki and Lanyi, 1986; Jonas et al., 1990; El-Sayed et al., 1995). The second model assumes that the negatively charged membrane surface randomly binds cations (and protons). This binding of cations and protons affects the surface pH and regulates the protonation state of Asp-85 indirectly according to the Guoy-Chapman theory (Szundi and Stoeckenius, 1987, 1988, 1989; Varo et al., 1999). All resolved structures of bR to date have found no bound cations (Leucke et al., 1999; Grigorieff et al., 1996), either due to too low resolution or to cation removal on sample preparation. Effects of removing and adding cations to bR structure and stability have been intensively studied by our group and others (Heyes and El-Sayed, 2001; Heyes et al., 2002; Krescheck et al., 1990; Dunach et al., 1989; Sanz et al., 2001). It has been found that the cation has an important contribution to both secondary structure at physiological temperature and thermal stability. More recently, we have investigated the temperature dependence of the FTIR spectrum on delipidation and monomerization of bR (Heyes and El-Sayed, 2002), and found that even at physiological temperatures protein structural changes more extensive than deionization occur upon monomerization. This was not evident on 75% delipidation, suggesting that the lipids that hold the lattice structure together are the important contributors to structure (Heyes and El-Sayed, 2002). In addition, the thermal transitions are different in the lipid-depleted bR than in native suggesting different thermodynamic effects upon lipid removal. One of the major dilemmas is the observation that cation removal causes the proton pumping function to cease, whereas lipid removal does not. We have shown here that the proton pumping is restored upon lipid removal and upon monomerization from deionized bR, suggesting that the cation that was so important in the native bR is not so important in the lipid depleted bR.
The effects of lipid removal and monomerization on the M kinetics are both interesting and complex (Table 2). In general, 75% lipid removal using CHAPS increases both the M formation and decay times and is independent of the buffer ions. Monomerization causes a decrease in the M formation time, slightly increases the M lifetime, and is dependent on pH and buffer. This general trend is consistent with previously reported observations (Lozier, 1982; Jang and El-Sayed, 1988). We have shown here that this occurs whether native or deionized bbR is delipidated/monomerized. However, the details of these changes are more in depth than this. It is well known that M rise in native bR fits to a biexponential curve (Hanamoto et al., 1984; Varo and Lanyi, 1990, 1991b). The origin of this biexponential behavior has been the subject of much debate in the literature over a number of years. Two primary models have been put forward to explain the M kinetics. The first is one of parallel photocycles, starting with two ground state bR species, which go through their own photocycle kinetics but have the same spectral intermediates (Balashov et al., 1991; Eisfeld et al., 1995; Hendler et al., 2001). The second model assumes that there is a transition between two M states that is spectrally silent in the visible region (Varo and Lanyi, 1990, 1991b; Perkins et al., 1992; Oka et al., 2002). These two states have been given the terms M1 and M2, Mearly and Mlate, or M and MN. All the samples measured here show biexponential fits for M rise (Figs. 3 a and 4 a, and Table 2). The fast component in native bR has a rise time of 8.49 μs with a 21% weighting. This time increases to 16–23 μs upon 75% delipidation of native bR with ∼30%–50% weighting, depending on the presence of the acetate buffer. Upon monomerization of native BR, the fast component decreases to 1.8 μs and 50% amplitude. Similar values are present in detergent treated deI bbR, with the exception of unbuffered deI monomer. Again, this inconsistency is most likely due to low pH as discussed earlier rendering proton transfer from the Schiff base to Asp-85, and the appearance of the surface proton, slower. The slow component in native bR is 75.8 μs, which increases to 100–130 μs upon 75% delipidation and decreases upon monomerization to ∼10–15 μs. Again, the exception is the unbuffered deI monomer. The fast Schiff base deprotonation in the monomer samples could suggest that the pKa difference between the SB and the Asp-85 proton acceptor is larger during the M intermediate, and/or that there is a more direct connection from the SB to Asp-85. This may be a result of a greater degree of exposure to water, which reduces the hydrophobicity in this vicinity. This could be the same effect brought on by the cation in native bR, but is now unnecessary in the lipid-depleted environment. Whether or not a cation is actually present in lipid-depleted bR is still under investigation, but the results presented here suggest that if there is, it is not important to the function once at least 75% of the lipids are removed. An interesting observation concerns the longer rise time (and decay time—see later) of the 75% delipidated bR. The proton transfer from the SB to Asp-85 is inhibited slightly upon 75% lipid removal. Since the lattice is still in tact in these samples, it is likely that the extra lipids present in native bR facilitate this proton transfer. Perhaps the further apart the bR molecules are, the larger the pKa difference in the SB and Asp-85, and hence the monomer has the fastest M formation time.
M decay has been described in the literature as fitting a monoexponential decay in some reports (Jang and El-Sayed, 1988), or biexponetial in others (Hanamoto et al., 1984; Varo and Lanyi, 1990). The data presented here fits more to a single exponent in native or 75% delipidated bR, and have biexponential fits for the monomerized bR samples. To explain monoexponential decay of the M intermediate, either this would mean that the two M intermediates of parallel photocycles have different rise times, but the same decay time, or that there is an irreversible step of M1-M2, and M2 is the only species that decays at 412 nm. In the monomeric bR of either native or deionized origin, two decay components are present. Furthermore, the relative amplitudes of each component are the same as the M rise components in the presence of the Tris buffer, but different in the DDW solutions. It is not possible to conclude if this inconsistency is due solely to pH or to the Tris ions. However, considering the fact that the acetate buffer does not seem to affect the M kinetics of the CHAPS-treated samples, and that the large bulky Tris molecule should interact with the bR less than the sodium ions of the acetate buffer, it seems more likely that the pH of the solutions is the major contributing factor to the complicated M decay kinetics. The result is a destabilizing of the deprotonated Schiff base at lower pH compared to the neutral pH monomer. However, as we mentioned earlier, we cannot control both the pH systematically and exclude cations from the system simultaneously, and thus this pH effect is only inferred from this data, and cannot be measured directly. The exact reason that the M lifetime is longer in the 75% delipidated bR than in the monomer or the native is also not clear. The most likely reason is that removal of 75% of the lipids, but maintaining the lattice, lowers the unit cell dimensions of the membrane (Szundi and Stoeckenius, 1987). The closer bR molecules in the 75% delipidated bR are likely to inhibit the protein conformational changes necessary to provide access of the Schiff base to the proton donor in the cytoplasmic region (Asp-96). In contrast, monomerization would have no such restraints on the conformational changes. The decay time of M is slightly longer than the native, and may imply that the micelle causes some inhibitory effect on protein conformational changes, but the effect is relatively small.
Recently, Padros and coworkers described a series of experiments (Lazarova et al., 2000; Sanz et al., 1999, 2001), which focused on the importance of the extracellular Glu residues to bR structure and function. They described a complex role of Glu-194, Glu-204, Glu-9, and water molecules in the pKa regulation of Asp-85, which were proposed to involve specific cation binding sites—one cation coordinated to Glu-194 and Glu-204, and another acting as a ligand to Glu-9, bridged by water molecules (Sanz et al., 2001). Eliash et al. (2001) used electron spin resonance (ESR) to also determine a specific binding site located less than 9.8 Å from Glu-74 and Tuzi et al. (1999) used 13C NMR to observe a binding site near Ala-196 (and hence Glu-194). These extracellular binding sites are thought to control the Asp-85 protonation state and, therefore, the color and M intermediate by a hydrogen-bonded chain in the extracellular proton channel. These sites would indeed be very sensitive to the lipid environment. Removal of the lipids would undoubtedly change the pKa and binding constants of extracellular acidic residues, possibly rendering these sites unnecessary for occupation to keep Asp-85 deprotonated upon delipidation or monomerization. This coupling of lipidic charges to the pKa of acidic groups of the protein has been the major complicating factor in interpretation of cation binding to bR, and has led to the two proposed roles of cation binding. Our previous results on the effect of different cations on the thermal stability (Heyes et al., 2002), together with the results presented here, support the existence of lipid exposed, specific binding sites in bR such as those proposed by Sanz et al. (2001). Without the direct observation of cations in the high-resolution bR structure, we must rely on spectroscopic experiments such as these to elucidate their role in the structure, function, and stability of ion pumps.
Acknowledgments
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Science, Office of Sciences, U.S. Department of Energy (under grant DE-FG02-97ER14799).
References
- Ariki, M., and J. K. Lanyi. 1986. Characterization of metal ion-binding sites in bacteriorhodopsin. J. Biol. Chem. 261:8167–8174. [PubMed] [Google Scholar]
- Balashov, S. P., R. Govindjee, and T. G. Ebrey. 1991. Red shift of the purple membrane absorption band and the deprotonation of tyrosine residues at high pH. Origin of the parallel photocycles of trans- bacteriorhodopsin. Biophys. J. 60:475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov, S. P., E. S. Imasheva, R. Govindjee, and T. G. Ebrey. 1996. Titration of aspartate-85 in bacteriorhodopsin: what it says about chromophore isomerization and proton release. Biophys. J. 70:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge, R. R. 1990a. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta. 1016:293–327. [DOI] [PubMed] [Google Scholar]
- Birge, R. R. 1990b. Photophysics and molecular electronic applications of the rhodopsins. Annu. Rev. Phys. Chem. 41:683–733. [DOI] [PubMed] [Google Scholar]
- Birge, R. R., N. B. Gillespie, E. W. Izaguirre, A. Kusnetzow, A. F. Lawrence, D. Singh, Q. W. Song, E. Schmidt, J. A. Stuart, S. Seetharaman, and K. J. Wise. 1999. Biomolecular electronics: protein-based associative processors and volumetric memories. J. Phys. Chem. B. 103:10746–10766. [Google Scholar]
- Chang, C. H., J. G. Chen, R. Govindjee, and T. Ebrey. 1985. Cation binding by bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 82:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. H., R. Jonas, S. Melchiore, R. Govindjee, and T. G. Ebrey. 1986. Mechanism and role of divalent cation binding of bacteriorhodopsin. Biophys. J. 49:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencher, N. A., and M. P. Heyn. 1978. Formation and properties of bacteriorhodopsin monomers in the non-ionic detergents octyl-beta-D-glucoside and Triton X-100. FEBS Lett. 96:322–326. [DOI] [PubMed] [Google Scholar]
- Dencher, N. A., and M. P. Heyn. 1982. Preparation and properties of monomeric bacteriorhodopsin. Methods Enzymol. 88:5–10. [Google Scholar]
- Drachev, L. A., S. V. Dracheva, and A. D. Kaulen. 1993. pH dependence of the formation of an M-type intermediate in the photocycle of 13-cis-bacteriorhodopsin. FEBS Lett. 332:67–70. [DOI] [PubMed] [Google Scholar]
- Dunach, M., E. Padros, A. Muga, and J. L. R. Arrondo. 1989. Fourier-transform infrared studies on cation binding to native and modified purple membranes. Biochemistry. 28:8940–8945. [DOI] [PubMed] [Google Scholar]
- Eisfeld, W., T. Althaus, and M. Stockburger. 1995. Evidence for parallel photocycles and implications for the proton pump in bacteriorhodopsin. Biophys. Chem. 56:105–112. [DOI] [PubMed] [Google Scholar]
- Eliash, T., L. Weiner, M. Ottolenghi, and M. Sheves. 2001. Specific binding sites for cations in bacteriorhodopsin. Biophys. J. 81:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed, M. A., D. Yang, S.-K. Yoo, and N. Zhang. 1995. The effect of different metal cation binding on the proton pumping in bacteriorhodopsin. Isr. J. Chem. 35:465–474. [Google Scholar]
- Grigorieff, N., T. A. Ceska, K. H. Downing, J. M. Baldwin, and R. Henderson. 1996. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259:393–421. [DOI] [PubMed] [Google Scholar]
- Hampp, N. 2000. Bacteriorhodopsin as a photochromic retinal protein for optical memories. Chem. Rev. 100:1755–1776. [DOI] [PubMed] [Google Scholar]
- Hampp, N., R. Schmid, and D. Zeisel. 1994. Genetic modified bacteriorhodopsin for holographic interferometry. Mater. Res. Soc. Symp. Proc. 330:269–274. [Google Scholar]
- Hanamoto, J. H., P. Dupuis, and M. A. El-Sayed. 1984. On the protein (tyrosine)-chromophore (protonated Schiff base) coupling in bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 81:7083–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendler, R. W., R. I. Shrager, and S. Bose. 2001. Theory and procedures for finding a correct kinetic model for the bacteriorhodopsin photocycle. J. Phys. Chem. B. 105:3319–3328. [DOI] [PubMed] [Google Scholar]
- Heyes, C. D., and M. A. El-Sayed. 2001. Effect of temperature, pH, and metal ion binding on the secondary structure of bacteriorhodopsin: FT-IR study of the melting and premelting transition temperatures. Biochemistry. 40:11819–11827. [DOI] [PubMed] [Google Scholar]
- Heyes, C. D., and M. A. El-Sayed. 2002. The role of the native lipids and lattice structure in bacteriorhodopsin protein conformation and stability as studied by temperature-dependent Fourier transform-infrared spectroscopy. J. Biol. Chem. 277:29437–29443. [DOI] [PubMed] [Google Scholar]
- Heyes, C. D., J. Wang, L. S. Sanii, and M. A. El-Sayed. 2002. Fourier transform infrared study of the effect of different cations on bacteriorhodopsin protein thermal stability. Biophys. J. 82:1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K.-S., H. Bayley, and H. G. Khorana. 1980. Delipidation of bacteriorhodopsin and reconstitution with exogenous phospholipid. Proc. Natl. Acad. Sci. USA. 77:323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, S. B., and W. Stoeckenius. 1977. Purple membrane vesicles: morphology and proton translocation. J. Membr. Biol. 33:325–350. [DOI] [PubMed] [Google Scholar]
- Jang, D. J., and M. A. El-Sayed. 1988. Deprotonation of lipid-depleted bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 85:5918–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas, R., Y. Koutalos, and T. G. Ebrey. 1990. Purple membrane: surface charge density and the multiple effect of pH and cations. Photochem. Photobiol. 52:1163–1177. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., A. Ikegami, and W. Stoeckenius. 1984. Salt and pH-dependent changes of the purple membrane absorption spectrum. Photochem. Photobiol. 40:641–646. [DOI] [PubMed] [Google Scholar]
- Krescheck, G. C., C. T. Lin, L. N. Williamson, W. R. Mason, D. J. Jang, and M. A. El-Sayed. 1990. The thermal stability of native, delipidated, deionized and regenerated bacteriorhodopsin. J. Photochem. Photobiol. B. 7:289–302. [Google Scholar]
- Lanyi, J. K. 1993. Proton translocation mechanism and energetics in the light-driven pump bacteriorhodopsin. Biochim. Biophys. Acta. 1183:241–261. [DOI] [PubMed] [Google Scholar]
- Lanyi, J. K. 1995. Bacteriorhodopsin as a model for proton pumps. Nature. 375:461–463. [DOI] [PubMed] [Google Scholar]
- Lanyi, J. K. 1998. Understanding the structure and function in the light-driven proton pump bacteriorhodopsin. J. Struct. Biol. 124:164–178. [DOI] [PubMed] [Google Scholar]
- Lazarova, T., C. Sanz, E. Querol, and E. Padrós. 2000. Fourier transform infrared evidence for early deprotonation of Asp85 at alkaline pH in the photocycle of bacteriorhodopsin mutants containing E194Q. Biophys. J. 78:2022–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucke, H., B. Schobert, H. T. Richter, J. P. Cartailler, and J. K. Lanyi. 1999. Structure of bacterierhodopsin at 1.55 angstrom resolution. J. Mol. Biol. 291:899–911. [DOI] [PubMed] [Google Scholar]
- Lozier, R. H. 1982. Rapid kinetic optical absorption spectroscopy of bacteriorhodopsin photocycles. Methods Enzymol. 88:133–162. [Google Scholar]
- Mathies, R. A., S. W. Lin, J. B. Ames, and W. T. Pollard. 1991. From femtoseconds to biology: mechanism of bacteriorhodopsin's light-driven proton pump. Annu. Rev. Biophys. Biophys. Chem. 20:491–518. [DOI] [PubMed] [Google Scholar]
- Mowery, P. C., R. H. Lozier, Q. Chae, Y.-W. Tseng, M. Taylor, and W. Stoeckenius. 1979. Effect of acid pH on the absorption spectra and photoreactions of bacteriorhodopsin. Biochemistry. 18:4100–4107. [DOI] [PubMed] [Google Scholar]
- Oesterhelt, D., and W. Stoeckenius. 1974. Isolation of the cell membrane of halobacterium halobium and its fractionnation into red and purple membrane. Methods Enzymol. 31:667–678. [DOI] [PubMed] [Google Scholar]
- Oka, T., N. Yagi, F. Tokunaga, and M. Kataoka. 2002. Time-resolved x-ray diffraction reveals movement of F helix of D96N bacteriorhodopsin during M-MN transition at neutral pH. Biophys. J. 82:2610–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, G. A., E. Liu, F. Burkard, E. A. Berry, and R. M. Glaeser. 1992. Characterization of the conformational change in the M1 and M2 substates of bacteriorhodopsin by the combined use of visible and infrared spectroscopy. J. Struct. Biol. 109:142–151. [DOI] [PubMed] [Google Scholar]
- Sanz, C., T. Lazarova, F. Sepulcre, R. González-Moreno, J.-L. Bourdelande, E. Querol, and E. Padrós. 1999. Opening the Schiff base moiety of bacteriorhodopsin by mutation of the four extracellular Glu side chains. FEBS Lett. 456:191–195. [DOI] [PubMed] [Google Scholar]
- Sanz, C., M. Marquez, A. Peralvarez, S. Elouatik, F. Sepulcre, E. Querol, T. Lazarova, and E. Padros. 2001. Contribution of extracellular Glu residues to the structure and function of bacteriorhodopsin. Presence of specific cation-binding sites. J. Biol. Chem. 276:40788–40794. [DOI] [PubMed] [Google Scholar]
- Subramaniam, S., D. A. Greenhalgh, and H. G. Khorana. 1992. Aspartic acid 85 in bacteriorhodopsin functions both as proton acceptor and negative counterion to the Schiff base. J. Biol. Chem. 267:25730–25733. [PubMed] [Google Scholar]
- Szundi, I., and W. Stoeckenius. 1987. Effect of lipid surface charges on the purple-to-blue transition of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 84:3681–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szundi, I., and W. Stoeckenius. 1988. Purple-to-blue transition of bacteriorhodopsin in a neutral lipid environment. Biophys. J. 54:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szundi, I., and W. Stoeckenius. 1989. Surface pH controls purple-to-blue transition of bacteriorhodopsin. A theoretical model of purple membrane surface. Biophys. J. 56:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzi, S., S. Yamaguchi, M. Tanio, H. Konishi, S. Inoue, A. Naito, R. Needleman, J. K. Lanyi, and H. Saitô. 1999. Location of a cation-binding site in the loop between helices F and G of bacteriorhodopsin as studied by 13C NMR. Biophys. J. 76:1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varo, G., L. S. Brown, R. Needleman, and J. K. Lanyi. 1999. Binding of calcium ions to bacteriorhodopsin. Biophys. J. 76:3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varo, G., and J. K. Lanyi. 1990. Pathways of the rise and decay of the M photointermediate(s) of bacteriorhodopsin. Biochemistry. 29:2241–2250. [DOI] [PubMed] [Google Scholar]
- Varo, G., and J. K. Lanyi. 1991a. Effects of the crystalline structure of purple membrane on the kinetics and energetics of the bacteriorhodopsin photocycle. Biochemistry. 30:7165–7171. [DOI] [PubMed] [Google Scholar]
- Varo, G., and J. K. Lanyi. 1991b. Kinetic and spectroscopic evidence for an irreversible step between deprotonation and reprotonation of the Schiff base in the bacteriorhodopsin photocycle. Biochemistry. 30:5008–5015. [DOI] [PubMed] [Google Scholar]
- Varo, G., and J. K. Lanyi. 1991c. Thermodynamics and energy coupling in the bacteriorhodopsin photocycle. Biochemistry. 30:5016–5022. [DOI] [PubMed] [Google Scholar]
- Wang, J., C. D. Heyes, and M. A. El-Sayed. 2002. Refolding of thermally denatured bacteriorhodopsin in purple membrane. J. Phys. Chem. B. 106:723–729. [Google Scholar]
- Wu, S., E. S. Awad, and M. A. El-Sayed. 1991. Circular dichroism and photocycle kinetics of partially detergent solubilized and partially retinal regenerated bacteriorhodopsin. Biophys. J. 59:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]