Abstract
Actin-aldolase rafts provide insights into the use of rafts as models for three-dimensional actin bundles. Although aldolase has three twofold axes, filaments in actin-aldolase rafts were not strictly related by a twofold axis. Interfilament angles were on average +15° off the expected 180°, and most rafts appeared handed; that is, rows of cross-bridges were tilted in a clockwise direction off the perpendicular. We can account for both the deviation of the angle from 180° and the handedness of the rafts by a steric constraint due to the lipid layer. We further found that the axial spacings of cross-bridges varied significantly from raft to raft. We suggest that this difference arises from variations in the twist of the filaments that nucleate raft formation; that is, filaments added to a raft adopt the symmetry of those in the raft. We conclude that the organization of filaments in rafts can be modulated by outside factors such as the lipid layer and that the variable twist of filaments in the nucleating core of the raft are imposed on all the filaments in the raft. These results provide a measure of the potential for polymorphism in actin assemblies.
INTRODUCTION
Rafts, or planar arrays of actin filaments cross-linked by actin-bundling proteins, are two-dimensional analogs of three-dimensional actin bundles (Taylor and Taylor, 1992, 1994). Three-dimensional bundles are difficult subjects for structural studies because they are disordered. Although in some bundles the filament axes are arranged on a hexagonal lattice (DeRosier et al., 1977; Matsudaira, 1983; Schmid et al., 1991; Tilney et al., 1987), in others the axes have a liquidlike order (Tilney et al., 1980). Moreover, in bundles with either liquid- or hexagonally packed filaments, actin cross-bridges across the bundle are not in register (DeRosier and Censullo, 1981; DeRosier et al., 1980b; Spudich and Amos, 1979). Rafts offer a tractable alternative to three-dimensional bundles because disorder is constrained to two dimensions (Taylor and Taylor, 1994; Volkmann et al., 2001) so that individual filaments and cross-bridges can be analyzed.
Actin rafts are made on positively-charged lipid layers. In the absence of an actin bundling protein, rafts of pure actin are made. In these rafts, actin filaments are tightly packed, having an interfilament spacing of 70–80 Å and an interfilament rotation of ∼4° (Sukow and DeRosier, 1998; Taylor and Taylor, 1992) or 30–40° (Volkmann et al., 2001). The rafts are largely unipolar (Sukow and DeRosier, 1998; Taylor and Taylor, 1992, 1994; Volkmann et al., 2001). In the presence of an actin bundling protein, the interfilament spacing increases and the interfilament angle changes but the rafts remain unipolar. For example, in actin-fimbrin rafts, the interfilament separation is 115–120 Å with an interfilament rotation of 27° (Volkmann et al., 2001). In actin-aldolase rafts, the interfilament spacing is 126 Å and the interfilament rotation is ∼180° (Taylor and Taylor, 1994). In actin-adducin rafts, the interfilament spacing is 145 Å and the interfilament rotation is 180°. Both of these latter bundling proteins are thought to have twofold axes. In all cases, the actin rafts are monolayers of actin filaments.
The bundling proteins differ in size and the arrangement of their actin binding sites. The fluid lipid layer permits motion of the actin filaments, which can take up positions consistent with the actin binding sites of the bundling proteins. In some cases, for example that of the actin-aldolase rafts, the bundling protein is added to rafts of pure actin. The aldolase is thought to intercalate between the filaments having the same polarity, bind to the specific aldolase-binding sites on each filament, and thereby to drive a reorganization of the filaments from being tightly packed with a short interfilament spacing and small interfilament rotation to a large interfilament spacing and interfilament rotation (Taylor and Taylor, 1999).
In one sense, the cross-linked filaments in a raft are less constrained than those in three-dimensional bundles because the cross-bridging occurs in two dimensions rather than three. This affords an opportunity to view the potential for additional modes of polymorphism. On the other hand, the filaments are differently constrained by their contacts with a planar lipid layer. We set out to determine how the formation of a raft is both governed by and alters the bonding geometries inherent in the component filaments and cross-bridges, and how the lipid layer affects these geometries.
Rafts of actin cross-linked by aldolase are an ideal model system to carry out such a study (Taylor and Taylor, 1994). Aldolase is an actin bundling protein (Clarke and Morton, 1976; Morton et al., 1977; Stewart et al., 1980) and is found associated with the actin in muscle, for example, (Arnold and Pette, 1968; Sigel and Pette, 1969). Aldolase occurs as a tetramer having D2 symmetry (Blom and Sygusch, 1997; Choi et al., 1999; Hester et al., 1991) in which the three twofold axes relate the four actin-binding sites on the tetramer. In order to bundle actin filaments, a cross-bridging protein must have binding sites consistent with a pair of either parallel or antiparallel filaments. Because the actin-binding sites on aldolase are related by a twofold axis, we expect adjacent, cross-linked filaments to be related by a twofold axis. Such rafts give rise to distinctive Fourier transforms and are thus relatively easy to identify. Additionally, aldolase, having a mass of 160,000 Da, is easy to see in micrographs (Taylor and Taylor, 1994). Using the actin-aldolase rafts, we analyzed the organization of filaments and cross-bridges in the rafts and determined the role the lipid layer plays in altering the organization. We further elucidated the nature and degree of disorder in the cross-bridges.
EXPERIMENTAL MATERIALS AND METHODS
Materials used
Except where otherwise noted, reagents and rabbit skeletal muscle aldolase were from Sigma (St. Louis, MO). The lipid DLPC (dilauryl phosphatidylcholine) was from Avanti Polar Lipids (Alabaster, AL), and the surfactant DDDMA (didodecyl dimethyl ammonium bromide) was from Kodak (Rochester, NY). Actin was either purchased from Cytoskeleton (Denver, CO) or prepared from chicken pectoralis muscle using the method of Spudich and Watt (Spudich and Watt, 1971) except that we substituted a centrifugation step after dialysis in place of the column purification step. The chemicals required for making the holey carbon films were obtained from the Ouken Shoji Co. Ltd. (Tokyo, Japan). Uranyl acetate was from Electron Microscopy Sciences (Fort Washington, PA).
Raft-making techniques
The raft-making procedure used was adapted from that developed by Taylor and Taylor (Taylor and Taylor, 1992, 1994). A small humid chamber was assembled in which the rafts were incubated. The apparatus, kept at 4°C, consists of a platform upon which sits a Teflon block containing a number of wells (4mm in diameter and 1 mm deep). The platform and Teflon block are kept inside a petri dish and are surrounded by water to maintain high humidity. A 13 μl droplet of actin polymerization buffer (20 mM PO4, pH 6.0–6.5, 40 mM KCl, 1 mM MgCl2, 1 mM ATP, 1 mM NaN3) was placed into each of the wells, and 0.7 μl of a lipid/surfactant mixture (DLPC 70% and DDDMA 30%, diluted in chloroform to 1 mg/ml) was then layered over the top of each droplet. After 5 min, during which the chloroform evaporated, 2 μl of a G-actin solution (G-actin buffer is 2 mM Tris, pH 8.2, 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP) was injected through the lipid layer on top of the droplet into the polymerization buffer. The G-actin solution was typically 5.0 μM, leading to an actin concentration in the well of ∼0.7 μM.
Injected droplets were incubated for 45–90 min while the actin polymerized and formed rafts on the lipid/surfactant layer. After the incubation period, the rafts were picked up by touching the carbon side of an electron microscopy grid to the top of each droplet. Grids were 400 mesh copper supporting a holey carbon film prepared by the method of Fukami and Adachi (Fukami and Adachi, 1965) and were aged at least one week to render them hydrophobic. If rafts containing only actin were desired, the grids were washed with 2–3 drops of polymerization buffer, blotted with filter paper (Whatman no. 1), and then stained with 1–2% uranyl acetate. To make rafts cross-bridged by aldolase, the grids were washed with the aldolase buffer, blotted, and then two droplets (6–8 μl each) of (5–7 μM) aldolase were applied to the grid. Each drop was allowed to sit for approximately one minute before the grid was washed again with the aldolase buffer, blotted, and stained. The buffer used for aldolase was 10 mM imidazole, pH 6.0–6.8, 10 mM KCl, 1 mM EGTA, 2 mM MgCl2, 1 mM NaN3, and 0.02% β-mercaptoethanol.
Electron microscopy
Micrographs were taken on a Philips 420 microscope (Philips Electronic Instruments, Inc., Mahwah, NJ) operating at 120 kV, equipped with an anticontaminator (model 651, Gatan, Inc. Warrendale, PA). The specimen, held at about −180°C in a Gatan cryoholder (model 626), was recorded at 62,500× using a dose of 20 electrons per Å2. The low dose, low temperature conditions can provide better resolution and is our standard operating condition for negatively-stained preparations. Images were recorded on SO-163 film (Eastman Kodak, Rochester, N.Y.) and developed for 12 min in full-strength D19 developer (Eastman Kodak).
Scanning and initial image processing
Some negatives were scanned on an Eikonix densitometer (Eikonix Corp., Bedford, MA) using an f-stop of 5.6, and a sampling raster of 20–26 μm, for a pixel size of 4–6 Å. Other negatives were scanned on a Zeiss scanner (Carl Zeiss, Oberkochen, Germany) at a raster size of 7μm. Pixels from the latter were binned to give a raster size of 21μm corresponding to a pixel size of ∼4 Å. The scanned rafts were boxed and rotated using the MRC package (Crowther et al., 1996) and Brandeis Helical Package (Owen et al., 1996). Fourier transforms were calculated using the Brandeis Helical Package.
Measuring cross-bridge spacings
The spacings between aldolase tetramers were collected from 10 actin/aldolase rafts. Because the spacings between cross-bridges were consistently multiples of the axial spacing between actin subunits (27.5 Å), cross-bridge spacings were expressed as multiples of the actin subunit spacing.
Determination of rotation and axial shift
We measured the positions of maxima, R, on the first and sixth layer lines. These maxima, which lie at the intersection of the row lines and the layer lines, are then used to determine the axial shift and rotation (Sukow and DeRosier, 1998):
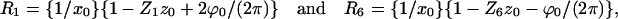 |
where R1 and R6 are the positions of the maxima from the meridian on the first and sixth layer lines, x0 is the separation between filaments, Z1 and Z6 are the layer line heights, and z0 and ϕ0 are the axial shift and rotation.
Simulated filaments and rafts
Simulations were used to predict patterns and distributions of cross-bridge spacings for filaments with different helical symmetries and/or interfilament rotations. These simulations generated ϕij, the azimuthal positions of subunits: ϕij = i × α + j × β, where i denotes the ith subunit in a filament, α is the angle between subunits, j denotes the jth filament in the raft, and β is the angular rotation between neighboring filaments. α is simply 360° divided by the number of units per turn, which we varied between 2.157 and 2.167, the range of values found in rafts. Beta was varied around 180°, the value expected for the approximate twofold relation imposed on filaments by the aldolase. In a simulation, all filaments had the same helical symmetry and adjacent filaments were rotated by a fixed angle relative to their neighbors. No attempt was made to apply z-shifts to the filaments because the sizes of the z-shifts we observed experimentally were small causing a tilt of the cross-bridges rather than a change in the optimal position for cross-bridging.
In the simulation, the perfect cross-bridge is made when two subunits face each other, that is when the one on the left is at 0° and the one on the right is at 180°. Two such subunits are related by an exact twofold axis. Because of the potential of actin to cross-bridge even when the ideal condition is not met, we defined a potential location for cross-bridging to be any position in which the pertinent subunits were both within 40° of their ideal positions (i.e., were angled less than ±40° away from the plane of the raft). In some cases we also noted whether the pair of subunits involved pointed toward or away from a given side of the raft, chosen to represent the side contacting the lipid layer.
χ2 analysis of distribution of cross-bridge spacings in real rafts
The χ2 analysis (Fisher, 1958) was performed as follows: A table was made with entries corresponding to the number of occurrences of the various cross-bridge spacing (i.e., spacings of 11-, 13-, 15-, and 17-subunits). These data are listed by row with one column of such values for each raft analyzed. Under the assumption (to be tested) that there is a single probability distribution that describes the frequency of each spacing, we calculated the expected number of cross-bridges for each according to the formula
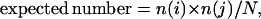 |
where n(i) is the total number of occurrences of a particular spacing (e.g. 11 subunits) for all rafts, n(j) is the total number of spacings for the jth raft, and N is the total number of all observations.
The observed and expected values for each measurement were used to calculate the value of χ2 = ∑{(observed − expected)2/expected}. The number of degrees of freedom is equal to the number of independent entries in the table. For our analysis, this was (number of rafts − 1) × (number of classes of spacings −1) = (10 − 1) × (4 − 1) = 27.
Determination of filament symmetry from the frequencies of intercross-bridge spacings
The relationship between 13- to 15-subunit spacings and units/turn can be expressed in equation form (van der Weide, personal communication). Units/turn customarily refers to the number of monomers in one turn of actin's left-handed, 59 Å helix. We assume that filaments have an average twist between 2.14 and 2.17 units/turn for which the optimal position for a cross-bridge occurs after either 13 subunits or 15 subunits (DeRosier et al., 1977; DeRosier and Censullo, 1981; Spudich and Amos, 1979). A spacing of 13 subunits occurs after 6 turns of the 59 Å helix and the 15-subunit spacings after 7 turns. Thus, the overall helical symmetry of such a filament may be expressed as the average over all the crossovers:
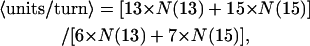 |
where N(13) is the total number of 13-subunit cross-bridges spacings and N(15) is the total number of 15-subunit cross-bridge spacings.
RESULTS
Actin/aldolase rafts are polar; adjacent filaments are rotated by ∼180°
Micrographs showed regions with rafts of pure actin, characterized by the very tight packing of 80 Å seen in rafts of pure actin, and rafts with aldolase, characterized by the rafts having an interfilament spacing of 120 Å and obvious cross-bridges between filaments. In order to determine the organization of the actin and aldolase, we analyzed Fourier transforms of segments of the actin-aldolase rafts. Fig. 1 shows two rafts and their Fourier transforms and Fourier transforms of simulated bipolar and polar rafts for comparison. Images of the sides and ends of rafts (Fig. 2) provide evidence that the actin-aldolase rafts contain a single layer of actin filaments; that is, there are no cases in which one sees one filament on top of another. Moreover, if there were a second layer on top of the first and the filaments were rotated 180° as expected for aldolase cross-bridges (see below), the odd-ordered layer lines (n = −1, 1, 3 etc.) would be absent or very weak. Instead, these layer lines are prominent features of the raft transforms. In the Fourier transforms of the rafts, the absence of reflections corresponding to twice the interfilament spacing (i.e., 240 Å) is evidence that the rafts are polar rather than bipolar (Sukow and DeRosier, 1998); that is, the repeating unit is a single filament rather than a pair of oppositely oriented filaments. Further evidence is provided by the occasional appearance of “interstitial” filaments (Fig. 2). The filaments on either side of the interstitial filament are cross-bridged to it and then to each other after the interstitial filament has ended. This is expected for a polar raft but not for a bipolar raft (Taylor and Taylor, 1994).
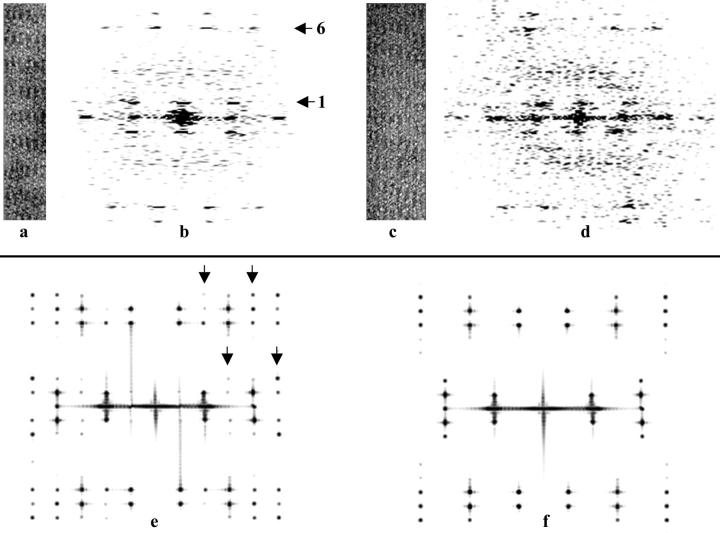
FIGURE 1.
Aldolase rafts and their Fourier transforms. (a) Segment of an actin-aldolase raft. In this raft, successive filaments are rotated by exactly 180°. (b) Fourier transform of the raft in a. The first and sixth layer lines are indicated with arrows. (c) Segment of an actin-aldolase raft in which successive filaments are rotated by ∼195°. (d) Fourier transform of the raft in c. (e). The Fourier transform of a model bipolar raft. Note half row lines (marked by arrowheads), which are not seen in f. (f) The Fourier transform of a model polar raft. Note absence of half row lines seen in e. In this raft successive filaments are rotated by 180° as in a.
FIGURE 2.
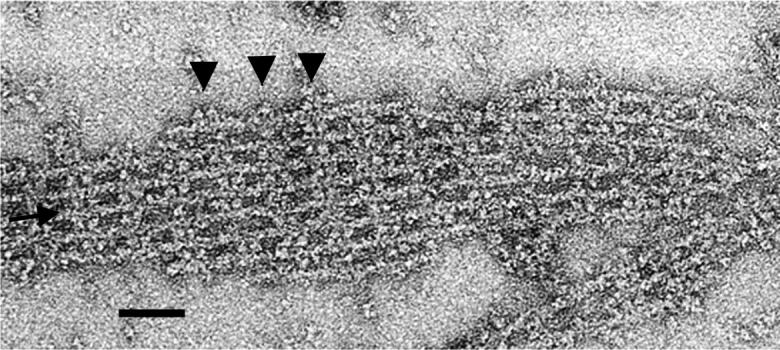
Electron micrograph of a negatively stained actin-aldolase raft. This raft has an interstitial filament (see arrow), which is cross-bridged to its neighbors. To the right of the inserted filament, the two neighbors are cross-bridged to each other. The edges of outer filaments are decorated with aldolase in some places (see arrowheads). Scale bar = 500 Å.
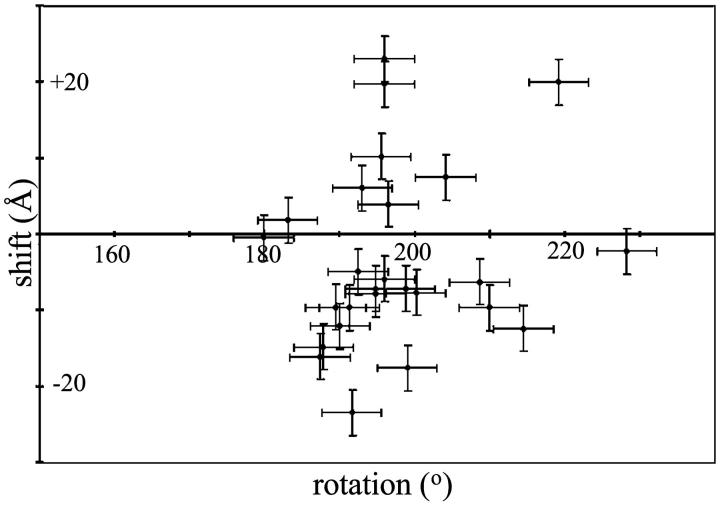
The twofold axis relating pairs of actin-binding sites on aldolase should generate a twofold axis between filaments. A twofold axis requires that adjacent filaments are related by a 180° rotation and that there is no axial shift between filaments. Fig. 1 f shows the transform of a simulated raft in which successive filaments are rotated by 180° and are not shifted axially. Note that the reflections on the sixth layer line are offset from those on the equator whereas on the first layer line, the reflections are in register with the reflections on the equator. The transform in Fig. 1 b exhibits the same pattern. This raft is one in which successive filaments are rotated by 180° and have no axial shift. The pattern shown in Fig. 1 d, however, is one expected for a raft in which successive filaments are rotated by 195° (for a comprehensive discussion see Sukow and DeRosier, 1998). We determined the rotation and shift from the measured positions of row line reflections on the first and sixth layer lines on the Fourier transforms of each raft (see Materials and Methods). Fig. 3 is a plot of axial shift versus rotation. Successive filaments in most rafts are rotated by angles greater than 180° (the average being ∼195° relative to the filament immediately to the left), and have an axial shift of −15 to + 15 Å. The error bars represent the uncertainty of one pixel in selecting the peak positions on the first and sixth layer lines. The deviation from 180° is not due simply to errors in measurement.
FIGURE 3.
Scatter plot of axial shift against interfilament rotation for actin-aldolase rafts.
Unusual cross-bridge spacings of 11 and 17 actin subunits are observed; the frequencies of spacings differ among rafts
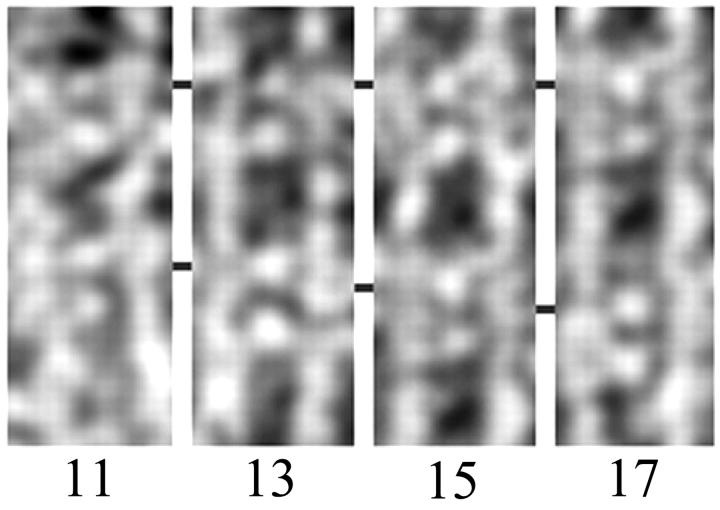
Within most actin crossovers, we observed a pair of aldolase cross-bridges consistently spaced by four actin subunits, or ∼110 Å, suggesting that there are at least two positions with conditions favorable for aldolase binding in each crossover. The spacings between adjacent pairs of cross-bridges, however, are variable. We observed 13-subunit spacings and 15-subunit spacings as expected from earlier works (DeRosier and Censullo, 1981; Spudich and Amos, 1979), but we also observed 11-subunit spacings and 17-subunit spacings (Fig. 4).
FIGURE 4.
Segments of rafts in which pairs of aldolase cross-bridges are separated by 11, 13, 15, and 17 actin subunits. Each panel shows a pair of actin filaments running vertically and two pairs of aldolase cross-bridges. The top pair of aldolase cross-bridges are lined up from one panel to the next. The position of the lower pair changes stepwise from 11 to 17 actin subunits.
Table 1 presents the number of occurrences for each of the four spacings observed. Alongside are the numbers expected if the probability of occurrence of each spacing is the same for each raft. To test whether these frequencies are the same within experimental error, we used a χ2 test (see Materials and Methods). The value of χ2 of 47.8 on (10 rafts − 1) × (4 types of spacings − 1) = 27 degrees of freedom means that there is a >99% probability that the frequencies of spacings are not drawn from the same population. We think this is a result of the nucleation process (see Discussion).
TABLE 1.
Observed and expected frequencies of cross-bridge spacings
| 11 subunits
|
13 subunits
|
15 subunits
|
17 subunits
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Spacing Raft | Observed | Expected | Observed | Expected | Observed | Expected | Observed | Expected | Totals |
| 6758 | 2 | 3.68 | 21 | 17.48 | 7 | 6.03 | 1 | 3.82 | 31 |
| 6765 | 1 | 6.06 | 42 | 28.75 | 8 | 9.91 | 0 | 6.28 | 51 |
| 6820a | 1 | 4.87 | 24 | 23.11 | 8 | 7.97 | 8 | 5.05 | 41 |
| 6848 | 6 | 5.46 | 24 | 25.93 | 10 | 8.94 | 6 | 5.66 | 46 |
| 6834 | 7 | 4.4 | 18 | 20.86 | 7 | 7.19 | 5 | 4.56 | 37 |
| 7594a | 1 | 3.21 | 21 | 15.22 | 3 | 5.25 | 2 | 3.32 | 27 |
| 7594b | 3 | 3.21 | 15 | 15.22 | 7 | 5.25 | 2 | 3.32 | 27 |
| 7604a | 4 | 3.21 | 17 | 15.22 | 4 | 5.25 | 2 | 3.32 | 27 |
| 7606a | 9 | 5.82 | 22 | 27.62 | 10 | 9.52 | 8 | 6.03 | 49 |
| 7606c | 21 | 15.1 | 57 | 71.59 | 26 | 24.7 | 23 | 15.64 | 127 |
| Totals | 55 | 261 | 90 | 57 | 463 | ||||
Differences in cross-bridge frequencies provide estimates of filament symmetries
The layer lines generated from images of rafts are not sufficiently sharp to produce an accurate estimate of the number of units per turn for the actin filaments. We used the frequencies of 13- and 15-subunit cross-bridge spacings, which are set by the helical symmetry of the filaments (DeRosier et al., 1980a; DeRosier and Censullo, 1981; Spudich and Amos, 1979) to determine filament symmetry (see Materials and Methods). The result is approximate because cross-bridge spacings of 11 and 17 subunits are ignored (see Discussion). Most rafts have units/turn values of 2.159 (Table 2), a value commonly observed by other workers (Egelman et al., 1982; Spudich and Amos, 1979). There are also rafts with higher units/turn values, in the range 2.161–2.163, which are values observed for isolated filaments (Orlova and Egelman, 2000; Orlova et al., 2001).
TABLE 2.
Ratio of crossover spacings and derived actin symmetries for rafts
| Raft identification | Ratio N(13)/N(15) | Units/turn |
|---|---|---|
| 7594b | 2.1 | 2.158 |
| 7606a | 2.2 | 2.158 |
| 7606c | 2.2 | 2.158 |
| 6848 | 2.4 | 2.159 |
| 6834 | 2.6 | 2.159 |
| 6820 | 3.1 | 2.160 |
| 6758 | 3.1 | 2.160 |
| 7604a | 4.3 | 2.162 |
| 6765 | 5.3 | 2.162 |
| 7594a | 7.0 | 2.163 |
Some actin/aldolase rafts present handed images; the handedness is correlated with interfilament rotation
In some rafts rows of cross-bridges are essentially perpendicular to the filament axes. An example of such a “nonhanded” raft is shown in Fig. 5 a. This raft, which has an interfilament rotation of 180°, has cross-bridges that alternately are one subunit up and then one subunit down. There is no upward or downward trend in cross-bridge positions across this raft. Other rafts appear handed: the axial positions of cross-bridges between pairs of filaments have a downward trend from left to right across the raft (Fig. 5 b). Of the ten rafts examined, the nonhanded rafts have interfilament rotations of ∼180°. The handed rafts, in contrast, have interfilament rotations of 187°–200°, with an average of 195°. Rafts having the other hand (cross-bridges shifted up instead of down, and having rotations <180°) were not observed.
FIGURE 5.
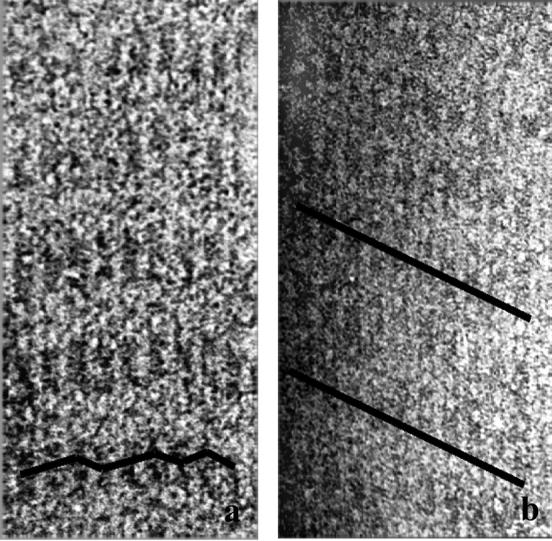
(a) Actin-aldolase raft in which there is no consistent downward shift of cross-bridges from left to right. A line is drawn indicating the path of the cross-bridges. (b) Raft in which there is a consistent downward shift of cross-bridges from left to right.
DISCUSSION
Aldolase interacts with actin in a site-specific manner
The evidence for site-specific interaction between actin and aldolase is threefold. First, site-directed mutants in aldolase alter actin-binding activity while leaving enzyme activity intact (Kusakabe et al., 1997). Because the actin-binding and active sites are in the same domain, the mutational change affects the actin-binding site locally (specifically) rather than causing a global change to the domain. Second, aldolase alters the relationship between actin filaments seen in rafts of pure actin. This is significant because the change in interfilament rotation is accomplished by adding aldolase to already formed plain actin rafts. In rafts cross-bridged by aldolase, the interfilament rotation is ∼180° whereas in rafts of pure actin, the rotation is ∼0° (Sukow and DeRosier, 1998). Hence an equivalent pair of sites on the aldolase tetramer interact with an equivalent pair of sites on adjacent actin filaments. Third, aldolase binds to actin at spacings equal to integral multiples of the actin subunit spacing. Thus the twofold arrangement of sites on aldolase dictates the twofold arrangement of adjacent actin filaments and the spacing of subunits along the actin helix dictates the spacings between aldolase cross-bridges.
Cross-bridge spacings can be accounted for by actin's variable twist
The symmetry (number of units per turn) of actin filaments in the rafts fall within the range found in isolated filaments. Since the helical symmetry of actin is not a crystallographic screw symmetry, the actin-actin and/or the actin-aldolase interactions in different parts of the raft must not all be equivalent. Previously, cross-bridges in actin-fascin bundles (DeRosier and Censullo, 1981) were found at helical positions that most closely approximated the ideal orientation for cross-bridge formation. Sometimes two subunits were approximately equally well (or poorly, depending on your point of view) positioned, in which case a cross-bridge could be found at either position. In electron micrographs of hexagonally packed, actin-fascin bundles, cross-bridges, which were seen in projection, appeared as transverse bands. The variations in cross-bridge position caused a variation in the spacings and widths of these bands as expected from the incompatibility of the actin symmetry and the hexagonal arrangement of filaments. Cross-bridge spacings of 13 and 15 subunits were seen, but not spacings of 11 and 17 subunits, which could have been present but hidden in the confusion of the projection.
In the actin-aldolase rafts, however, the precise locations of all cross-bridges can be determined. If cross-bridges were found only at those subunits closest to the ideal position, and if actin were a perfectly regular helix, we would expect to find a regular pattern of spacings of 13 and 15 subunits (DeRosier and Censullo, 1981). In the rafts we find mainly spacing of 13 and 15 subunits but we also we find spacings of 11 and 17 subunits.
Recently, Galkin et al. (Galkin et al., 2001) showed that actin subunits in filaments exist in more than one conformation. In the most common conformation, the angle between subunits is 166°, giving rise to a filament with 2.17 units/turn; there are several other conformations in the range 162°–169° as well. There is also a conformation in which the angle is 158° or 2.28 units/turn. The average twist of all filament segments in the population they examined is 2.17 units/turn. Galkin et al. propose that the variable twist of actin filaments results from a mixture of two or probably more such conformations. If the spacings of the cross-bridges we have observed reveal the underlying local twist of the actin filaments, then a spacing of 11 subunits (in 5 turns) would indicate an intersubunit angle of 5 × 360/11 = 164° and a spacing of 17 subunits (in 8 turns) would correspond to an angle of 8 × 360/17 = 169°. The difference in intersubunit angles we observed is thus 5°, which is comparable to the range seen by Galkin et al. Thus, the variation in subunit spacing can be explained by actin's variable twist. In such a case, the actin subunits take up positions offset from those predicted for an ideal helix in order to make an ideal cross-bridge.
Are such large deviations from equivalence observed in nature? The actin bundle found in the sperm of the horseshoe crab may be just such a case (DeRosier and Tilney, 1984). The bundle in its coiled state is not continuously bent, but rather contains straight segments interrupted periodically by bends having an angle of 126°. The bends are locked into the bundle by a slippage of the cross-bridges by two actin subunits, that is, subunit n on one filament is cross-bridged to subunit n + 2 instead of subunit n on a neighboring filament. Such a slippage could convert spacings of 13 and 15 subunits into spacings of 11 and 17 in a bend. The ability of filaments to accommodate such variability may be important in bundle form and function.
The frequencies of cross-bridge spacings vary significantly among rafts, suggesting a seeding mechanism for raft formation
The χ2 test indicates that, in a statistical sense, the frequencies of cross-bridge spacings in rafts are not all drawn by chance from a uniform population; that is, the probabilities of cross-bridge spacings of 11, 13, 15, and 17 actin units in different rafts differ significantly. Because the relative frequencies of the four classes of cross-bridge spacings depend on the average number of units per turn of the filaments in a raft, the average number of units per turn for the filaments in these rafts must also differ significantly. Since the filaments in a given raft are drawn from the same population of actin filaments as any other raft, why should the filament symmetries in one raft differ significantly from those in another?
The average filament symmetries found in rafts fall within the symmetries observed for isolated filaments (Egelman et al., 1982; Orlova and Egelman, 2000; Orlova et al., 2001). What makes the rafts differ significantly is that the distribution (of cross-bridge spacings) within one raft is much tighter than one would predict if the filaments in one raft had symmetries as variable as those in the general population of isolated filaments. The enforced symmetry among filaments in a raft is a result of the cross-bridging. For cross-bridges to form along a pair of filaments, subunits on one filament must be related to those on the other filament by a twofold rotation (at least approximately so). Thus, the subunits in both sets of filaments must have the same intersubunit angles. For example, consider a pair of filaments such that the intersubunit angle in one filament is 164° and in the adjacent filament is 169°. Align the two filaments so that a cross-bridge is made between subunit 1 (orientation = 0°) on the left filament and subunit 1 (orientation = 180°) on the right filament. On the left filament, the next potential cross-bridging position is 11 subunits up (orientation = 4°), whereas on the right filament, it is 17 subunits up (orientation = 173°). Because these two subunits are not opposite one another, no cross-bridge can be made for this crossover; and in fact very few cross-bridges can be made at all between such a pair of filaments. Thus the symmetries of any pair of filaments must be the same (at least approximately so) for cross-bridging to occur, and it follows that the symmetries of all filaments in a raft must be essentially the same.
If the process of assembly were one of arriving at a consensus (average) symmetry prior to or during assembly, then we would expect all rafts to have the essentially the same symmetry (e.g., approximately the average symmetry within the actin population). As this is not observed, we propose that raft assembly begins with the local cross-bridging of two or a few filaments. A new filament added to an edge of the growing raft must have or must adopt the same local symmetry of the filament to which it is being attached. In this assembly mechanism, the initial seed raft enforces its symmetry on the filaments that subsequently join the raft. Thus the filaments within a raft are more similar to one another than they are to filaments in a different raft.
Most rafts appear handed, and the lipid layer produces the hand
One of the three twofold axes that relate the actin-binding sites in aldolase should be manifest in the rafts. Although this is true (i.e., the interfilament angle is 180°) in some cases (Fig. 3), more often than not adjacent filaments are related by somewhat larger angles, on average 195°. In such rafts, the cross-bridges are displaced (axially) downwards as one moves left to right across the raft. No case has been seen in which the cross-bridges move up as one goes from left to right nor in which the interfilament angle is less than 180°.
The appearance of only one hand must be a consequence of the lipid layer. To see why this is so, imagine lifting the handed raft free of the lipid layer, flipping it over like a pancake, and placing it back on the lipid layer. The image will be of a raft with the opposite hand; the cross-bridges are displaced upwards going from left to right. In addition, the filament orientations will all change by 180°. In raft with 195° interfilament rotations, going from right to left: filament 1 at 0° and filament 2 at 0° + 195° = 195°. When we add 180° to each filament and reverse the order on the left is filament 2 at 195° + 180° = 375° = 15° and filament 1 is at 0° + 180° = 180°. The interfilament angle is 180° − 15° = 165°. Thus, we are merely viewing the same raft from the opposite side, but in micrographs we only see one side of the raft. The fact that only one side is seen means that the presence of the lipid layer, which is always on the side away from the viewer, is determining the hand we see.
A similar situation was seen in micrographs of the complex of nine adenovirus hexons (van Oostrum et al., 1986). In the electron micrographs, each complex appeared with the same hand. The conclusion was that they were all stuck onto the carbon film in the same orientation, that is all face up or all face down. The groups of nine hexons have two different sides, one that faces outward toward the environment and the other that faces inwards toward the nucleic acid. These two different faces are chemically different, and it is easy to imagine that one face has a strong preference for the carbon foil so that all groups of nine sit down in the same way.
Can this same explanation work for actin aldolase rafts? We think it unlikely. First, the actin filament, no matter how oriented parallel to the lipid layer, presents subunits in the same way to the lipid because subunits point radially outward in all possible orientations. Thus, the interaction between an actin filament and the lipid layer is independent of rotation of the filament about its axis. Second, in the rafts, aldolase has a twofold axis parallel to the lipid layer so that when flipped over about this axis, it presents an equivalent face to the lipid. Thus it must be that some other aspect of the interaction of the lipid with the assembled components is producing the handedness.
Steric hindrance can explain both the deviations of the interfilament angles from 180° and the selection of the hand of the rafts seen in the images
Table 3 (left side) lists the angles between binding sites in a raft of three filaments in which alternate filaments are rotated by 180°. Let us assume the cross-bridge between the left-side and middle filaments at subunit 0 is the ideal cross-bridge, that is one subunit on the left-side filament is facing right (angle = 0°) while subunit 0 on the middle filaments is facing left (angle = 180°). These two binding sites are related by an exact twofold axis and will perfectly accommodate an aldolase cross-bridge. If we look in the same region between middle and right-side filaments, we see that the best choices for a cross-bridge is either at subunit −1 or subunit +1. In either case the binding sites are rotated 14° off their ideal positions. One subunit is rotated 14° toward the lipid layer and the other 14° away from the lipid layer. What happens when successive filaments are rotated by 195° instead of 180°?
TABLE 3.
Angular orientations of subunits in rafts
| Interfilament angle = 180°
|
Interfilament angle = 195° = −165°
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fil.1 | Fil.2 | Fil.3 | Fil.4 | Subunit | Fil.1 | Fil.2 | Fil.3 | Fil.4 | ||||||
| 0 | → ← | −180 | 0 | → ← | −180 | 1 | 0 | →↙ | −165 | 30 | −135 | |||
| 166 | −14 | ↘↖ | 166 | −14 | 2 | 166 | 1 | →↙ | −164 | 31 | ||||
| −28 | 152 | −28 | 152 | 3 | −28 | 167 | 2 | →↙ | −163 | |||||
| 138 | −42 | 138 | −42 | 4 | 138 | −27 | 168 | 3 | ||||||
| −55 | 125 | −55 | 125 | 5 | −55 | 140 | −25 | 170 | ||||||
| 111 | −69 | 111 | −69 | 6 | 111 | −54 | 141 | −24 | ||||||
| −83 | 97 | −83 | 97 | 7 | −83 | 112 | −53 | 142 | ||||||
| 83 | −97 | 83 | −97 | 8 | 83 | −82 | 113 | −52 | ||||||
| −111 | 69 | −111 | 69 | 9 | −111 | 84 | −81 | 114 | ||||||
| 55 | −125 | 55 | −125 | 10 | 55 | −110 | 85 | −80 | ||||||
| −138 | 42 | −138 | 42 | 11 | −138 | 57 | −109 | 87 | ||||||
| 28 | −152 | 28 | −152 | 12 | 28 | −137 | 58 | −107 | ||||||
| −166 | 14 | ↗↙ | −166 | 14 | 13 | −166 | 29 | −136 | 59 | |||||
| 0 | → ← | 180 | 0 | → ← | 180 | 14 | 0 | →↙ | −165 | 30 | −135 | |||
| 166 | −14 | ↘↖ | 166 | −14 | 15 | 166 | 1 | →↙ | −164 | 31 | ||||
| −28 | 152 | −28 | 152 | 16 | −28 | 167 | 2 | →↙ | −163 | |||||
| 138 | −42 | 138 | −42 | 17 | 138 | −27 | 168 | 3 | ||||||
In the raft represented on the left side of the table, the filaments are alternately rotated by 180°. The pairs of arrows in the columns between the four filaments mark positions where one expects to find aldolase cross-bridges. Arrows that angle upwards indicate that the aldolase binding site on actin points into the lipid layer. Horizontal arrows indicate binding sites that lie parallel to the plane of the lipid layer. Arrows that angle downwards represent sites that point away from the lipid layer. Note that the rows of arrows are essentially horizontal. In the raft represented by the right side of the table, successive filaments are rotated by 195° instead of 180°. With this additional +15° of rotation, no aldolase binding sites point into the lipid layer. Note that rows of arrows slope down and to the right.
A rotation of successive filaments, going from left to right, of 195° instead of 180° (see Table 3, right side) moves the binding sites away from the lipid layer, which we take to be at the top of the cells with arrows in Table 3. Although subunit 1 in the left-side filament is at 0°, the same subunit on the middle filament is rotated 15° away from the lipid layer. Subunit 2 on the right side of the middle filament is now at 1° toward the lipid instead of 14° away from the lipid but the corresponding subunit on the right-side filament is now pointing 16° away from the lipid instead of 14° into the lipid. Between the middle and the right-side filament, a cross-bridge can be made at subunit 3. At that position, the subunit on the middle filament is pointed 2° into the lipid whereas that on the right-side filament points 17° away from the lipid layer. Subunits in bonding position now point at most 2° toward the lipid layer instead of the 14° found in the 180° raft. Thus, potential steric hindrances caused by the lipid layer are minimized by this rotation. Rotations that are much larger or much smaller either do not relieve the steric hindrance or they increase the angular deviation from the ideal angle for cross-bridge formation (simulations not shown). Thus a rotation of ∼195° is about ideal. What we suggest is that assembly of the raft on the lipid layer favors rotations of succeeding filaments by angles ∼15° greater than 180°.
In actin-fimbrin rafts, a similar handedness was seen, but in this case there were two distinct classes of rafts. In one class, actin filaments were in register and the bands of fimbrin cross-bridges were perpendicular to the axes of the filaments. In the other class, successive filaments were rotated by ∼27° and the cross-bridges were sloped always from upper left to lower right. The authors suggest that, in the second class of rafts, the fimbrin is rotated by 180° relative to that in the first. The two actin binding domains are thus interchanged in the second class relative those in the first. In the actin-aldolase rafts, such an interchange is a symmetry operation and could not result in a different class of raft.
In rafts of actin-α actinin, the long cross-bridges always slope from upper left to lower right (Taylor and Taylor, 1994). Moreover, there are handed spirals in which a single actin filament is cross-bridged to itself. Since the opposite hand would be observed if the raft or spiral were flipped over, it seems inescapable that the lipid layer is forcing the handedness although no mechanism has been suggested. It is not clear that the mechanism operating in actin-aldolase rafts operates on actin-α actinin rafts. In the cases where the rafts have been analyzed in some depth, however, the lipid layer exerts an influence on the rafts either by selecting a handedness or, as we suggest here, by altering the rotational relationship between adjacent filaments. How the lipid effects the changes may well depend on the particular properties (e.g., size, shape, and distribution of charges) of the cross-bridging protein.
The lipid layer may also explain the presence of the second cross-bridge per crossover
In the simulations above, we have used the simplifying assumption of one cross-bridging aldolase tetramer per actin crossover. In actin/aldolase rafts, we observed that most crossovers in fact had two cross-bridges and that the spacing between a pair of tetramers was 4 actin subunits measured along the genetic helix. This presents a puzzle. If the ideal cross-bridge is made at position 1, then at position 4, the subunits are rotated by over 50° off the ideal and in opposite directions. The situation is no better in the 195° rafts. In order for both cross-bridges to make identical, stereospecific contacts with actin, a much larger amount of angular distortion would be required than has previously been observed in actin cross-bridges. Such distortions might be possible but it motivated us to look at other possibilities.
As an alternative, it is possible that the two cross-bridges are not equivalent. The specific cross-bridging contacts may instead be made by only one of the two tetramers, and the second tetramer may make the correct specific contact with only one filament, while being nonspecifically stabilized by the presence of the lipid. This second possibility is supported by the observations that aldolase tetramers decorate the edges of a raft in a periodic manner, and that when filaments in a raft are separated beyond the distance bridgeable by aldolase, the tetramers remain bound to one filament (Fig. 2). These observations demonstrate that aldolase binding does not require two actin filaments (no edge decoration would then be observed), nor is one actin filament alone, without the stabilization by the lipid layer, sufficient for binding aldolase (in which case all accessible actin subunits would be decorated by aldolase). The stabilization of a second aldolase by the lipid is a bit puzzling because both aldolase and the lipid layer are positively charged. On the other hand, the lipid layer is only doped with a positive surfactant; it may be that the negatively charged actin filaments are adequate to offset the charge or it may be that one of the surfaces that does not bear a significant net positive charge may be the one adjacent to the lipid layer.
If aldolase has the potential to bind to one filament and stabilize this interaction by association with the lipid but without the need to bind specifically to the other filament (as we propose for the decorating aldolase tetramer), can both aldolase tetramers be so bound and the raft simply be the result of physical forces rather than specific cross-bridging by the cross-bridging aldolase tetramer? For example, might aldolase tetramers be bound to only one actin filament and simply forced into a groove on the neighboring filament as a result of crowding? In such a case, the filaments in a raft would exhibit screw disorder which is easily recognized in the Fourier transforms (Sukow and DeRosier, 1998) by sharp sampling on one family of layer lines (e.g., layer lines 1 and 2) but continuous sampling on others (e.g., layer lines 6 and 7). This is not seen. The appearance of sharp sampling on all layer lines means that filaments are arranged with equivalent sites in a particular geometry in each raft as though fixed by specific cross-bridges. Another possibility is that one of each pair of aldolase tetramers in a crossover is bound to the left side filament and the other to the right side filament and that the decorated but not cross-bridged filaments pack together as tightly as possible. In this model, a filament would be identical to its neighbor but merely shifted down ∼4 subunits or ∼100 Å. Fourier transforms of the raft images, however, reveal shifts of at most 15 Å. Thus the most straightforward interpretation of the images is that the rafts consist of filaments held together by specific interfilament cross-bridges of aldolase tetramers.
Observed sequences of cross-bridge spacings may be explained by assuming that cross-bridges in a pair are not equivalent
Some cross-bridges are apparently spaced by 11 actin subunits, others by 17. It is possible that these spacings reflect local variations in the twist of actin (Galkin et al., 2001). However, recall that we measured spacings between pairs of closely spaced cross-bridges. We suggested that one aldolase tetramer of each pair makes specific cross-bridging contacts to both filaments, and the second makes specific contacts to only one filament. The cross-bridging tetramer is assumed to be placed in a favorable cross-bridging position, and the decorating tetramer will be found four subunits up or down the filament from the first. Fig. 6 illustrates how shifting the decorating cross-bridge could produce spacings of 11 or 17 subunits. In Fig. 6, a–d, two pairs of cross-bridged filaments are shown. The aldolase cross-bridges are represented by ovals with the shaded oval indicating the cross-bridging tetramer and the unshaded oval indicating the decorating tetramer. The pair of filaments on the left (the same for all panels) has the cross-bridging tetramer placed at the position in which the actin subunits are in closest proximity. The decorating tetramer is then placed either four subunits up or down. The pair of filaments on the right in each panel shows how shifting both the cross-bridging tetramer and the decorating tetramer four subunits could produce different spacings. Note that the spacing between the cross-bridging tetramer is always either 13 or 15 but that the shifting of the decorating cross-bridge can produce spacings between the pairs of 11 or 17 subunits.
FIGURE 6.
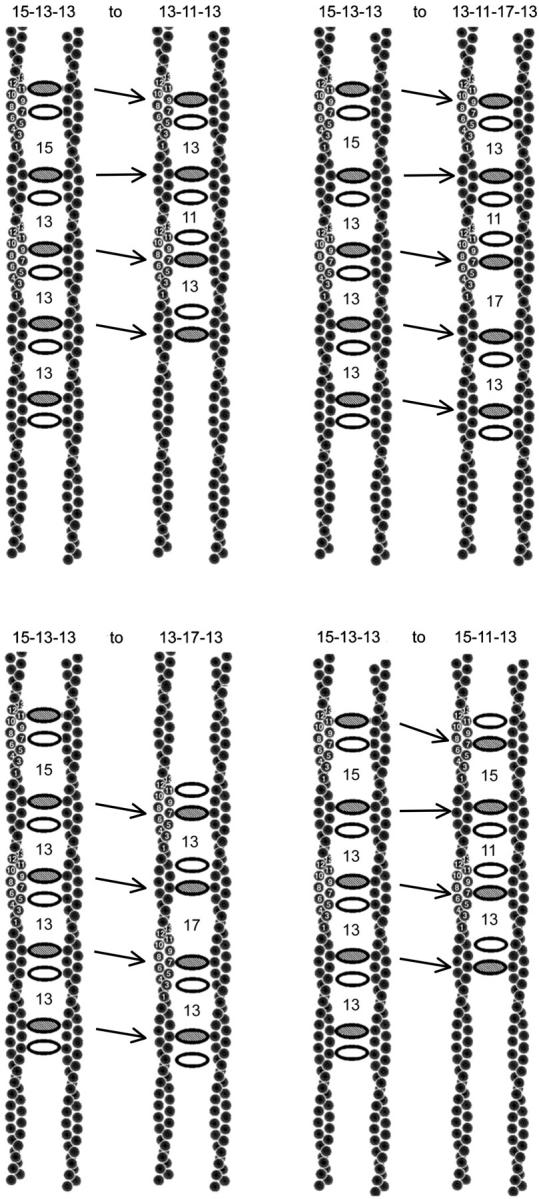
Cartoon showing how the swapping and shifting of cross-bridging and decorating aldolase tetramers results in a change of spacings between pairs of aldolase tetramers. The decorating aldolase is shown as shaded; the cross-bridging aldolase is unshaded. The figures on the left contain the same set of starting positions of each pair as expected set given the symmetry of actin. The one on the right shows the change in spacing of the cross-bridges that accompanies moving one or both of the aldolase tetramers. The restricting feature is that all cross-bridging aldolase tetramers are either 13 or 15 subunits from a neighboring cross-bridging aldolase. Note that the patterns 13-11-13 and 13-17-13 can be generated under these restrictions.
In micrographs of rafts, we are unable to tell which is the cross-bridging and which the decorating tetramer. Furthermore, there is no a priori reason to expect that they would appear in the same order in every tetramer pair. Therefore, we can only measure the cross-bridge spacings as the difference between tetramer pairs. The spacings in the simulated rafts were thus measured as the number of subunits from the first tetramer of one pair to the first tetramer of the next. These spacings are indicated between each tetramer pair in Fig. 6. The observed sequence 13-17-15, seen just once in ten rafts, was the only one that could not be generated by switching around cross-bridging and decorating tetramers while maintaining spacings of only 13 or 15 between cross-bridging tetramers. Thus we have two mechanisms by which most of the observed patterns of spacing could be generated, namely the shifting of the decorating tetramer relative to the cross-bridging tetramer, or the variability of twist in the actin filament. We cannot tell them apart. This is the reason that in using cross-bridge spacings to estimate filament symmetry we had to ignore spacings of 11 and 17.
How are rafts relevant to bundles?
In vivo, most bundles contain more than one kind of cross-bridging protein; how do such proteins work in combination? For example, the bundles found in the brush border of the intestinal epithelium have villin and fimbrin as their two major bundling proteins. We would like to know how each of these proteins taken singly interacts with actin and how they interact when combined. Unfortunately, ordered actin bundles have not been made from fimbrin or villin, so little structural information can be gleaned from them. A study of rafts offers a more tractable approach. We therefore need to know how and to what extent the lipid layer can influence the organization of actin in bundles. We find that the lipid layer can cause a 15° bias in the interfilament rotation and can stabilize the binding of additional actin bundling proteins. While this at first may seem to invalidate the use of the raft as an analog, the polymorphism actually provides insight into properties of cross-bridged filaments. When more than one actin-bundling protein is involved in bundle formation, we want to know to what extent the bonding rules for the two proteins can differ and still be compatible for bundle formation. We know from the results with aldolase, that angular incompatibilities of on average 15° can be tolerated and that axial shifts of ∼15 Å are acceptable. Rafts thus provide a system in which one can accurately determine the positions of cross-bridges and the rotation, axial shift and lateral separation of filaments. Because one can potentially overcome the effects of disorder in rafts, one can potentially get higher resolution structural information than one can extract from disordered bundles. Rafts can provide the background information needed to interpret images of bundles.
Acknowledgments
We thank Karen Allen and Dean Tolan for generously supplying samples and for helpful discussions. We thank Chris Mercogliano for helpful suggestions on the manuscript.
This work was supported by a research grant from NIGMS R01-GM26357 to D.J.D. and by a training grant from NIGMS T32-GM07596 (C.S.).
References
- Arnold, H., and D. Pette. 1968. Binding of glycolytic enzymes to structure proteins of the muscle. Eur. J. Biochem. 6:163–171. [DOI] [PubMed] [Google Scholar]
- Blom, N., and J. Sygusch. 1997. Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase. Nat. Struct. Biol. 4:36–39. [DOI] [PubMed] [Google Scholar]
- Choi, K. H., A. S. Mazurkie, A. J. Morris, D. Utheza, D. R. Tolan, and K. N. Allen. 1999. Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 Å. Biochemistry. 38:12655–12664. [DOI] [PubMed] [Google Scholar]
- Clarke, F. M., and D. J. Morton. 1976. Aldolase binding to actin-containing filaments. Formation of paracrystals. Biochem. J. 159:797–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9–16. [DOI] [PubMed] [Google Scholar]
- DeRosier, D. J., and R. Censullo. 1981. Structure of F-actin needles from extracts of sea urchin oocytes. J. Mol. Biol. 146:77–99. [DOI] [PubMed] [Google Scholar]
- DeRosier, D., E. Mandelkow, A. Silliman, L. Tilney, and R. Kane. 1977. Structure of actin-containing filaments from two types of non-muscle cells. J. Mol. Biol. 113:679–695. [DOI] [PubMed] [Google Scholar]
- DeRosier, D. J., and L. G. Tilney. 1984. How to build a bend into an actin bundle. J. Mol. Biol. 175:57–73. [DOI] [PubMed] [Google Scholar]
- DeRosier, D., L. Tilney, and P. Flicker. 1980a. A change in the twist of the actin-containing filaments occurs during the extension of the acrosomal process in Limulus sperm. J. Mol. Biol. 137:375–389. [DOI] [PubMed] [Google Scholar]
- DeRosier, D. J., L. G. Tilney, and E. Egelman. 1980b. Actin in the inner ear: the remarkable structure of the stereocilium. Nature. 287:291–296. [DOI] [PubMed] [Google Scholar]
- Egelman, E. H., N. Francis, and D. J. DeRosier. 1982. F-actin is a helix with a random variable twist. Nature. 298:131–135. [DOI] [PubMed] [Google Scholar]
- Fisher, R. 1958. Statistical Methods for Research Workers. New York: Hafner Publishing Company Inc. 356 p.
- Fukami, A., and K. Adachi. 1965. A new method of preparation of a self-perforated micro plastic grid and its application. J Electron Microsc. (Tokyo). 14:112–118. [PubMed] [Google Scholar]
- Galkin, V. E., A. Orlova, N. Lukoyanova, W. Wriggers, and E. H. Egelman. 2001. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 153:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester, G., O. Brenner-Holzach, F. A. Rossi, M. Struck-Donatz, K. H. Winterhalter, J. D. Smit, and K. Piontek. 1991. The crystal structure of fructose-1,6-bisphosphate aldolase from Drosophila melanogaster at 2.5 A resolution. FEBS Lett. 292:237–242. [DOI] [PubMed] [Google Scholar]
- Kusakabe, T., K. Motoki, and K. Hori. 1997. Mode of interactions of human aldolase isozymes with cytoskeletons. Arch. Biochem. Biophys. 344:184–193. [DOI] [PubMed] [Google Scholar]
- Matsudaira, P. T. 1983. Structural and functional relationship between the membrane and the cytoskeleton in brush border microvilli. Ciba Found. Symp. 95:233–252. [DOI] [PubMed] [Google Scholar]
- Morton, D. J., F. M. Clarke, and C. J. Masters. 1977. An electron microscope study of the interaction between fructose diphosphate aldolase and actin-containing filaments. J. Cell Biol. 74:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, A., and E. H. Egelman. 2000. F-actin retains a memory of angular order. Biophys. J. 78:2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, A., V. E. Galkin, M. S. VanLoock, E. Kim, A. Shvetsov, E. Reisler, and E. H. Egelman. 2001. Probing the structure of F-actin: cross-links constrain atomic models and modify actin dynamics. J. Mol. Biol. 312:95–106. [DOI] [PubMed] [Google Scholar]
- Owen, C. H., D. G. Morgan, and D. J. DeRosier. 1996. Image analysis of helical objects: The Brandeis helical package. J. Struct. Biol. 116:167–175. [DOI] [PubMed] [Google Scholar]
- Schmid, M. F., P. Matsudaira, T. W. Jeng, J. Jakana, E. Towns-Andrews, J. Bordas, and W. Chiu. 1991. Crystallographic analysis of acrosomal bundle from Limulus sperm. J. Mol. Biol. 221:711–725. [DOI] [PubMed] [Google Scholar]
- Sigel, P., and D. Pette. 1969. Intracellular localization of glycogenolytic and glycolytic enzymes in white and red rabbit skeletal muscle: a gel film method for coupled enzyme reactions in histochemistry. J. Histochem. Cytochem. 17:225–237. [DOI] [PubMed] [Google Scholar]
- Spudich, J. A., and L. A. Amos. 1979. Structure of actin filament bundles from microvilli of sea urchin eggs. J. Mol. Biol. 129:319–331. [DOI] [PubMed] [Google Scholar]
- Spudich, J. A., and S. Watt. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- Stewart, M., D. J. Morton, and F. M. Clarke. 1980. Interaction of aldolase with actin-containing filaments. Structural studies. Biochem. J. 186:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukow, C., and D. DeRosier. 1998. How to analyze electron micrographs of rafts of actin filaments crosslinked by actin-binding proteins. J. Mol. Biol. 284:1039–1050. [DOI] [PubMed] [Google Scholar]
- Taylor, K. A., and D. W. Taylor. 1992. Formation of 2-D paracrystals of F-actin on phospholipid layers mixed with quaternary ammonium surfactants. J. Struct. Biol. 108:140–147. [DOI] [PubMed] [Google Scholar]
- Taylor, K. A., and D. W. Taylor. 1994. Formation of two-dimensional complexes of F-actin and crosslinking proteins on lipid monolayers: demonstration of unipolar alpha-actinin-F- actin crosslinking. Biophys. J. 67:1976–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, K. A., and D. W. Taylor. 1999. Structural studies of cytoskeletal arrays formed on lipid monolayers. J. Struct. Biol. 128:75–81. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., D. J. Derosier, and M. J. Mulroy. 1980. The organization of actin filaments in the stereocilia of cochlear hair cells. J. Cell Biol. 86:244–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L. G., Y. Fukui, and D. J. DeRosier. 1987. Movement of the actin filament bundle in Mytilus sperm: a new mechanism is proposed. J. Cell Biol. 104:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oostrum, J., P. R. Smith, M. Mohraz, and R. M. Burnett. 1986. Interpretation of electron micrographs of adenovirus hexon arrays using a crystallographic molecular model. J. Ultrastruct. Mol. Struct. Res. 96:77–90. [DOI] [PubMed] [Google Scholar]
- Volkmann, N., D. DeRosier, P. Matsudaira, and D. Hanein. 2001. An atomic model of actin filaments cross-linked by fimbrin and its implications for bundle assembly and function. J. Cell Biol. 153:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]