Abstract
Reverse transcriptase PCR analyses have demonstrated that open reading frames (ORFs) PM0298, PM0299, and PM0300 of the animal pathogen Pasteurella multocida constitute a single transcriptional unit. By cloning and overexpression studies in Escherichia coli cells, the product of ORF PM0300 was shown to bind hemoglobin in vitro; this ORF was therefore designated hgbA. In vitro and in vivo quantitative assays demonstrated that the P. multocida hgbA mutant bound hemoglobin to the same extent as the wild-type strain, although the adsorption kinetics was slightly slower for the hgbA cells. In agreement with this, the virulence of P. multocida hgbA cells was not affected, suggesting that other functional hemoglobin receptor proteins must be present in this organism. On the other hand, P. multocida mutants defective in PM0298 and PM0299 could be isolated only when a plasmid containing an intact copy of the gene was present in the cells, suggesting that these genes are essential for the viability of this bacterial pathogen. By adapting the recombinase-based expression technology in vivo to P. multocida, we also demonstrated that the transcriptional PM0298-PM0299-hgbA unit is iron regulated and that its expression is triggered in the first 2 h following infection in a mouse model. Furthermore, hybridization experiments showed that the hgbA gene is widespread in P. multocida strains regardless of their serotype or the animal from which they were isolated.
Iron is an essential compound for almost all living organisms. Bacterial species have evolved several strategies for the uptake of this element. Some bacteria produce and release into the environment small molecules of various chemical structures, known as siderophores, which are able to chelate free iron (20, 25). Binding of iron-loaded siderophores to specific protein receptors located in the bacterial outer membrane enables iron uptake. Other bacteria have several proteins in the outer membrane which are specific receptors for the different iron-binding host molecules, such as heme, hemoglobin, transferrin, and lactoferrin.
Bacterial transferrin- and lactoferrin-binding proteins have been studied mainly in pathogenic members of the families Pasteurellaceae and Neisseriaceae (9, 31). Typically, they are formed by two different subunits: TbpA and TbpB for transferrin receptors and LbpA and LbpB for lactoferrin receptors (25). TbpB and LbpB are lipoproteins anchored to the outer membrane and are the proteins which more directly interact with the substrate. TbpA and LbpA are inserted into the outer membrane and seem to serve as channels through which iron penetrates to the periplasmic space (25). Moreover, there are two different kinds of hemoglobin receptor proteins (25, 31). Some gram-negative bacteria secrete a hemoglobin-binding protein which extracts the heme group from hemoglobin and delivers it to an outer membrane-specific protein to introduce it into the cell. Other gram-negative bacteria have, anchored in the outer membrane, a specific receptor able to bind hemoglobin directly and afterwards promote internalization of either iron or the heme group. However, the transport of iron or heme across the outer membrane by all of these high-affinity mechanisms requires the products of the exbB, exbD, and tonB genes (1). These TonB-dependent outer membrane proteins have several preserved regions; the most characteristic region is the region which is located at the N-terminal end, which is known as the TonB box and which can be represented as (D/E)TXXVXA(A/S), where X is variable (17). By corollary, the presence of a TonB box in the amino acid sequence of a bacterial protein suggests that it may be involved in the uptake of several nutrients, including iron. The increasing number of bacterial genome sequences, together with comparative genomic analysis, allows identification of gene products which may potentially be TonB-dependent receptors. However, any predictions must be confirmed by experimental data to definitively assign a definite biological function.
Pasteurella multocida is a gram-negative bacterium that is able to cause infectious diseases in a wide spectrum of animals, including several mammal and bird species. As a consequence of this, P. multocida infections result in substantial economic losses worldwide. Nevertheless, and despite its importance, little is known about the iron uptake mechanisms of this pathogen. Determination of the P. multocida genome sequence has revealed the presence in this organism of several genes encoding potential TonB-dependent receptor proteins (18, 24). Nevertheless, there are no experimental data on the function of the product of any of these genes. One example is the hypothetical protein product of the PM0300 open reading frame (ORF), which lies immediately downstream of two other putative ORFs (PM0298 and PM0299) (Fig. 1A).
FIG. 1.
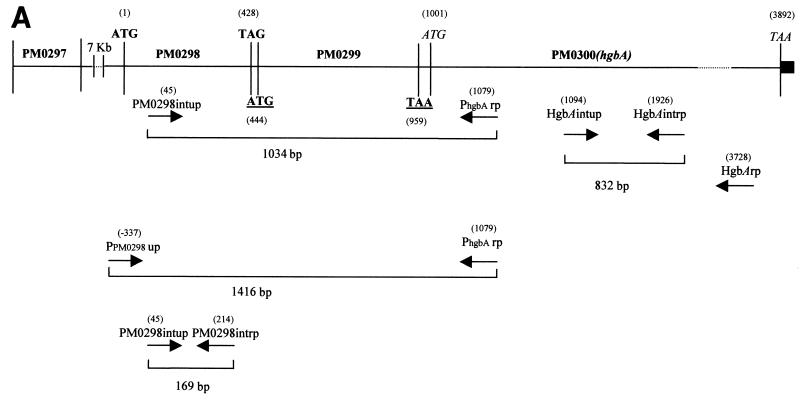
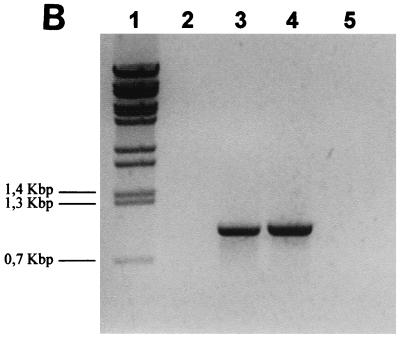
(A) Genetic organization of the P. multocida chromosomal region containing the PM0298-PM0299-PM0300 (hgbA) transcriptional unit. The translational start and stop codons for the PM0298, PM0299, and PM0300 ORFs are indicated by boldface type, by boldface type and underlining, and by italics, respectively. The positions of primers (Table 2) used to show the existence of a single mRNA by RT-PCR, as well as in the several constructions used in this study, are indicated. The numbers in parentheses indicate the positions with reference to the ORF PM0298 translational start point. The location of ORF PM0297 immediately upstream of PM0298 is also shown. The solid box downstream of ORF PM0300 (hgbA) represents the putative rho-independent transcriptional terminator of the operon. (B) RT-PCR analysis of the PM0298-PM0299-PM0300 (hgbA) transcript in P. multocida cells performed with PM0298intup and PhgbArp as the upper and lower primers, respectively, in the presence of RNA (lane 3). As a positive control, the PCR fragment obtained when P. multocida chromosomal DNA was used as the template (lane 4) was also examined. The negative controls were a preparation containing RNA template but no RT (lane 2) and a preparation lacking both RNA and DNA (lane 5). Lane 1 contained BstEII-digested λ DNA employed as the molecular size marker.
In the present work, the transcriptional organization of the DNA region containing these three ORFs and the role of the product of the PM0300 gene in P. multocida hemoglobin uptake in vivo and in vitro were investigated. Furthermore, after the recombinase-based in vivo expression technology (RIVET) was adapted to P. multocida characteristics, expression of these three P. multocida ORFs in vitro and during animal infection in a mouse model was studied.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA and RNA techniques.
Bacteria used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium (19). P. multocida was cultured in either buffered peptone water (BPW) or brain heart infusion (BHI) liquid medium or on BHI or sheep blood agar plates. The antibiotic concentrations used have been described previously (3). The DNA method used was also described previously (30). Oligonucleotide primers used in this work are listed in Table 2. The entire nucleotide sequence of P. multocida ORFs PM0298, PM0299, and PM0300 was determined for both DNA strands by the dideoxy method with an ALF sequencer (Pharmacia Biotech). Total cellular RNA extraction and reverse transcriptase PCR (RT-PCR) analysis were performed as reported by Jordan et al. (12).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F′ supE4 lacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Clontech |
| HB101 | supE4 hsdS20 recA13 ara-1 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Clontech |
| MC1061 (λpir) | hsdR mcrB araD139 Δ(araABC-leu) 7679 ΔlacX74 galI galK rpsL thi lysogenized with λpir bacteriophage | Our laboratory |
| BL21 (λDE3) | F−ompT hsdSB (rB− rB+) dcm gal λ(DE3) | 38 |
| P. multocida strains | ||
| PM25 | Wild type | Our laboratory |
| PM1002 | Same as PM25, but Rifr Spcr | Our laboratory |
| PM1077 | Same as PM1002, but carrying pUA961 | This study |
| PM1078 | Same as PM1002, but hgbA::pUA967 | This study |
| PM1079 | Same as PM1002, but PM0298 defective and carrying pUA964 | This study |
| Plasmids | ||
| pGEM-T | PCR cloning vector, Apr | Promega |
| pRK2013 | rep (ColE1), Mob+ Tra+ Kmr | 5 |
| pHRP309 | Broad-host-range cloning vector for transcriptional lacZ fusion construction, Gmr Mob+ | 23 |
| pUA826 | Mob+, R6K replicon, Apr Strr Spcr | 2 |
| pET-22b | Overexpression plasmid, Apr | Novagen |
| pGH436 | Same as pBR322, but carrying the resl-tet-resl construction | 35 |
| pIVET5mut | R6K replicon containing tnpR and lacZY promotorless genes | 15 |
| pUA958 | Same as pUA826, but carrying the promotorless tnpR gene | This study |
| pUA959 | Same as pGH436, but carrying the Cm cassette | This study |
| pUA960 | Same as pUA958, but carrying the res1-Cm-res1 construction | This study |
| pUA961 | Same as pUA960, but carrying a fusion between the P. multocida hgbA 5′ end and the tnpR gene | This study |
| pUA962 | Same as pGEM-T, but carrying the P. multocida PM0298-PM0299-hgbA operon | This study |
| pUA963 | Same as pET-22b, but carrying a fusion between the P. multocida hgbA promoterless gene and the T7 promoter | This study |
| pUA964 | Same as pHRP309, but carrying P. multocida ORFs PM0298 and PM0299 | This study |
| pUA965 | Same as pUA826, but carrying a 169-bp internal fragment of P. multocida ORF PM0298 | This study |
| pUA966 | Same as pUA826, but carrying a 258-bp internal fragment of P. multocida ORF PM0299 | This study |
| pUA967 | Same as pUA826, but carrying a 832-bp internal fragment of P. multocida hgbA gene | This study |
TABLE 2.
Oligonucleotide primers used in this work
| Primer | Sequencea | Position | Aplication |
|---|---|---|---|
| PM0298intup | 5′-TTACCTGACGAGTTTGTTCG-3′ | +45b | Upper primer to detect the present of the single transcrit in RT-PCR experiment; also used as upper primer to obtain the 169-bp internal fragment of P. multocida ORF PM0298 |
| PhgbArp | 5′-TCTAGAACTATCCGCCAAAATGGCC-3′ | +1079b | Lower primer to detect the present of the single transcrit in RT-PCR experiment; also used as lower primer to obtain the 360-bp P. multocida hgbA 5′ end |
| PPM0298up | 5′-GAGCATCAAATTAGGTCTG-3′ | −337b | Upper primer to obtain a fragment containing the P. multocida PM0298-PM0299-hgbA operon |
| HgbAextrp | 5′-CGCTAGCCGATCTCTAATCC-3′ | +4026b | Lower primer to obtain a fragment containing the P. multocida PM0298-PM0299-hgbA operon |
| NdehgbA | 5′-CATATGCGTACACAACAACAATAAAAATTTCTGC-3′ | +1001b | Upper primer to obtain the 3-kb fragment used to overexpress the P. multocida hgbA gene |
| NothgbA | 5′-GCGGCCGCCGCTAGCCGATCTCTAATCC-3′ | +4026b | Lower primer to obtain the 3-kb fragment used to overexpress the P. multocida hgbA gene |
| HgbAintup | 5′-CTGAACTTGATACGATTACC-3′ | +1094b | Upper primer to obtain the 832-bp internal fragment of the P. multocida hgbA gene |
| HgbAintrp | 5′-CCAAAATGGCGTAACAG-3′ | +1926b | Lower primer to obtain the 832-bp internal fragment of the P. multocida hgbA gene |
| HgbArp | 5′-CGGTTTAATATAACCAACC-3′ | +3728b | Primer to confirm disruption of the P. multocida hgbA gene by insertion of the pUA967 plasmid |
| PM0298intrp | 5′-GTTCTGGAAATAGTTGACGC-3′ | +214b | Lower primer to obtain the 169-bp internal fragment of P. multocida ORF PM0298 |
| PM0299intup | 5′-TCGGTGAAGATGGTAATCCC-3′ | +538b | Upper primer to obtain the 258-bp internal fragment of P. multocida ORF PM0299 |
| PM0299intrp | 5′-TTGCTTTGAGTGCCTCAACG-3′ | +796b | Lower primer to obtain the 258-bp internal fragment of P. multocida ORF PM0299 |
| PhgbAup | 5′-TCTAGAATCTTTTGATGCGGTTGCGCG-3′ | +719b | Upper primer to obtain the 360-bp containing the P. multocida hgbA 5′ end |
| Aad | 5′-CGGCGATCACCGCTTCCC-3′ | +2c | Plasmid primer to confirm disruption of P. multocida ORFs PM0298 and hgbA by insertion of the pUA965 and pUA967 plasmids, respectively |
| RTHgbAup | 5′-TATTGTGGGGTCATTTTGGCG-3′ | +1052b | Upper primer to detect hgbA transcript |
| RTHgbArp | 5′-TGAATATCTTCTTTCACTGGG-3′ | +2343b | Lower primer to detect hgbA transcript |
| RecAup | 5′-GCTCTATTATGAAATTGGGCG-3′ | +100d | Upper primer to test PM1078 RNA integrity |
| RecAdw | 5′-CTAACCATTTCATCGCG-3′ | +957d | Lower primer to test PM1078 RNA integrity |
When present, added restriction sites are underlined.
Position of the 5′ end of the oligonucleotide with respect the translational start point of P. multocida ORF PM0298.
Position of the 5′ end of the oligonucleotide with respect the translational start point of the aad gene of the pUA826 plasmid.
Position of the 5′ end of the oligonucleotide with respect the translational start point of the P. multocida recA gene.
Genetic methods.
P. multocida mutants were obtained by insertional mutagenesis by using the strategy described previously (3). Briefly, an internal fragment of the gene to be inactivated was inserted into the suicide plasmid pUA826, which is unable to replicate in host strains lacking the R6K-specified Π product of the pir gene (3). pUA826 derivatives harboring the internal P. multocida gene fragments were then introduced by triparental mating, using pRK2013 as the mobilizing plasmid, into P. multocida wild-type cells, and ampicillin-resistant exconjugants were selected. The presence of the desired mutations in these clones was confirmed by PCR analysis by using primers shown in Table 2. When done, biparental mating was performed by using the E. coli S-17 mobilizing system (32).
Virulence assays.
Female Swiss mice that were 3 to 8 weeks old, were obtained from Harlan Iberica Inc. (Barcelona, Spain), and were housed under specific-pathogen-free conditions were used for studies of virulence. Bacteria were grown for 16 h in BHI medium prior to infection. The 50% lethal doses of the strains were determined in triplicate as previously reported (8). Basically, groups of three mice were injected intraperitoneally with 0.1-ml portions of serial 10-fold dilutions of bacteria in BPW. The concentrations of the original bacterial suspensions were determined by the plate count method. The numbers of animals which were alive at 24, 48, and 72 h postinoculation were recorded, and the 50% lethal dose was calculated as described previously (26). The stability of the hgbA mutation in vivo was analyzed in cells recovered from the hearts of dead animals. Heart samples were homogenized in 1 ml of BPW, and 0.1-ml portions of diluted homogenates were plated on BHI medium plates. Following this, the presence of the mutation in 100 recovered colonies was determined by both PCR analysis and replica plating on BHI medium plates in the presence and absence of ampicillin (50 μg/ml).
In vitro and in vivo expression assays of the PM0298-PM0299-hgbA operon.
An overnight culture of the P. multocida PM1077 strain carrying the pUA961 plasmid in its chromosome harboring both the PM0298-PM0299-hgbA::tnpR transcriptional fusion and the res1-Cm-res1 construction and grown in BHI liquid medium containing streptomycin (75 μg/ml) and chloramphenicol (50 μg/ml) was diluted 1:100 into BHI medium containing streptomycin and supplemented or not supplemented with either 2,2-dypiridyl (DPD), DPD plus FeSO4, or DPD plus hemoglobin. Samples were removed periodically, and serial dilutions of the samples were plated on BHI agar containing 75 μg of streptomycin per ml. The percentage of Cms colonies of the resultant Strr colonies was then calculated by replica plating colonies onto BHI agar supplemented with 50 μg of chloramphenicol per ml and incubating the preparations at 37°C for 24 h.
To analyze the in vivo expression, a sample of an overnight culture of P. multocida PM1077 in BHI medium containing chloramphenicol and streptomycin was diluted in BHI medium to obtain a concentration of approximately 103 CFU/ml. Portions (100 μl) of this suspension were injected intraperitoneally into 3- to 8-week-old female Swiss mice. To recover the viable bacteria from the peritoneum after different postinoculation periods, 2 ml of BPW was inoculated into the peritoneal cavity of each Swiss mouse used in the trials. Before the bacteria were recovered, the animals were sacrificed by intrapulmonary inoculation of 0.5 ml of T-61 (Hoechst Roussel Vet). The maximum amount of BPW was collected after the abdominal compartment of each mouse was massaged. Serial dilutions of the cleaning fluid, which was maintained at 4°C until examination, as well as serial dilutions of the overnight culture used for the inoculum, were plated on BHI agar containing streptomycin. The percentage of Cms clones was calculated for both the inocula and the peritoneally grown bacteria by replica plating as described above. Loss of Cmr was due to TnpR-mediated excision of the res1-Cm-res1 construction of the P. multocida PM1077 chromosome.
Digoxigenin labeling of hemoglobin.
Digoxigenin was purchased from Roche, and the labeling procedure used was the procedure recommended by the supplier. Briefly, bovine hemoglobin (5 mg/ml; Sigma, St. Louis, Mo.) was dissolved in phosphate-buffered saline (pH 7.4), and digoxigenin-3-O-succinyl-ɛ-aminocaproic acid-N-hydroxysuccinamide ester was dissolved at a concentration of 2 mg/ml in dimethyl sulfoxide. To 1 ml of the hemoglobin solution 215 μl of the digoxigenin solution was added, and after incubation for 2 h at room temperature, unbound digoxigenin was removed by passage through a Sephadex G-25 column (Roche Laboratories). The amount of labeled hemoglobin was determined by measuring the absorption at 280 nm (1 U of A280 was equivalent to 0.55 mg of protein/ml).
Expression of HgbA in E. coli.
The pUA962 plasmid containing ORFs PM0298, PM0299, and PM0300 was used as a template in PCR amplifications with primers NdehgbA and NothgbA (Table 2), and the 3-kb fragment obtained was cloned in pGEM-T. The ATG translational start codon of PM0300 is part of the NdeI restriction site of the NdehgbA primer, which allows isopropyl-β-d-thiogalactopyranoside (IPTG)-mediated overexpression of the PM0300 product. After digestion with NdeI and NotI, the PM0300 ORF was cloned into the pET22b expression vector, and the ligation mixture was transformed into DH5α competent cells, resulting in pUA963. After confirmation by sequencing that no mutation had been introduced into the PM0300 ORF contained in pUA963, this plasmid was transformed into E. coli BL21(λDE3) for overexpression of the PM0300 protein. Hemoglobin binding to either whole or sonicated E. coli BL21(λDE3)/pUA963 cells was tested by a dot blot assay. These cells, grown to the mid-exponential phase in Luria-Bertani medium supplemented with appropriate antibiotics, were treated with 1 mM IPTG for 3 h and then harvested and resuspended in phosphate-buffered saline to a concentration of about 2 × 106 CFU/ml. In a dot blot manifold (Bio-Rad), 150 μg of cell suspension was filtered through nitrocellulose membranes. The membranes were then air dried for 20 min and blocked for 1 h in Tris-buffered saline (TBS) containing 5% skim milk and 0.025% Tween 20. Then 50 nM digoxigenin-labeled hemoglobin was added. After 1 h of incubation, the membranes were washed three times (10 min each) with TBS and incubated for 1 h in TBS containing 5% skim milk, 0.025% Tween 20, and 7.5 U of antidigoxigenin Fab fragments (Roche). Following this, the membranes were washed three times with TBS, and the contents were revealed in alkaline phosphatase buffer (100 mM NaCl, 50 mM Tris-HCl, 5 mM MnCl2) containing 4-nitroblue tetrazolium chloride and X-phosphate-5-bromo-4-chloro-3-indolylphosphate (BCIP, 4-toluidine salt), as recommended by the supplier (Roche). All procedures were carried out at room temperature. When necessary, sonication was performed in an NaCl-ice bath for a total of 3 min in 30-s bursts with 30 s between bursts. Sonicates (750 μg/ml) were filtered onto nitrocellulose membranes in a dot blot manifold, and the membranes were probed and developed as described above. In vitro hemoglobin binding to P. multocida was analyzed like in vitro hemoglobin binding to E. coli.
In vivo binding of hemoglobin by P. multocida.
Quantitative analysis of hemoglobin adsorption by P. multocida cells was performed basically as described previously (22). Ten milliliters of an overnight culture of P. multocida was harvested by centrifugation, washed with 50 mM acetate buffer (pH 6.0), and resuspended in the original volume of the same buffer. The optical density at 650 nm of the cell suspension was adjusted to 0.7. Then 7.3 ml of the cell suspension was mixed with 2.7 ml of bovine hemoglobin (at a concentration of 1 mg/ml in the same buffer). The mixture was incubated at 37°C, and at the desired times samples were removed and centrifuged at 10,000 × g for 15 min. The absorbance of the supernatant at 410 nm was then measured. Adsorbed hemoglobin was evaluated by the decrease in absorbance of the supernatant.
RESULTS
Cloning and transcriptional analysis of ORFs PM0298, PM0299, and PM0300.
ORFs PM0298, PM0299, and PM0300 were amplified from total DNA of the P. multocida PM25 strain by using the oligonucleotide primers shown in Table 2 and designed by using the P. multocida genome sequence (18). The 4.2-kb PCR fragment obtained was cloned into the pGEM-T vector and sequenced to confirm that no mutation was introduced during the amplification reaction. To confirm that the close proximity of PM0298, PM0299, and PM0300 implies that they are transcriptionally linked, RT-PCRs were performed with total RNA by using primers PM0298intup and PhgbArp (Fig. 1A), which amplified a 1-kb DNA fragment if an mRNA spanning the three ORFs was present. Figure 1B shows that the expected band was obtained, indicating that, effectively, the three ORFs constitute a single transcriptional unit.
Hemoglobin binding by the product of PM0300 in E. coli.
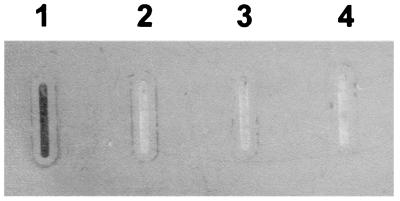
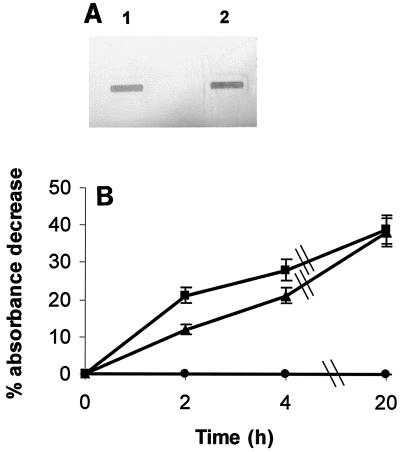
As described above, PM0300 contains the consensus sequence which is characteristic of the TonB-dependent receptor proteins, including hemoglobin-binding proteins. A dot blot assay was therefore used to determine whether E. coli cells carrying a plasmid containing ORF PM0300 were able to bind hemoglobin. Sonicated E. coli BL21(λDE3) cells containing the pUA963 plasmid, a derivative of the pET22b expression vector in which ORF PM0300 had been cloned, bound digoxigenin-labeled bovine hemoglobin after IPTG induction (Fig. 2, lane 1). However, whole cells of IPTG-induced E. coli BL21(λDE3)/pUA963 did not bind hemoglobin (Fig. 2, lane 2). This negative result was probably due to the fact that the recombinant protein cannot be exported to the surface of E. coli cells. Moreover, sonicated E. coli BL21(λDE3) cells containing the pET22b vector alone were unable to bind hemoglobin in either the presence or the absence of IPTG (Fig. 2, lanes 3 and 4). These data clearly indicate that the product of ORF PM0300 is able to bind hemoglobin. For this reason and since to our knowledge this is the first protein of P. multocida in which hemoglobin-binding ability has been demonstrated, the gene was designated hgbA (for hemoglobin-binding protein).
FIG. 2.
Solid-phase assay for binding of bovine hemoglobin to sonicated and intact E. coli BL21(λDE3)/pUA963 cells (lanes 1 and 2, respectively). Sonicated E. coli BL21(λDE3)/pET22b cells in the presence and absence of IPTG were the negative controls (lanes 3 and 4, respectively).
Construction of P. multocida PM0298, PM0299, and hgbA mutants.
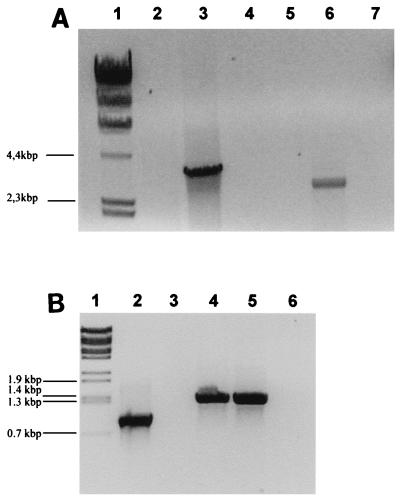
To further characterize the role of the products encoded by the PM0298-PM0299-hgbA transcriptional unit, we attempted to obtain mutants with mutations in each of the three ORFs by insertion of a derivative of the pUA826 suicide plasmid containing an internal region of the corresponding ORF. An hgbA mutant was obtained, and its structure was confirmed by PCR amplification of the junction segment of the vector with the chromosomal DNA by using an internal primer for hgbA and the Aad plasmid primer (Fig. 3A, lane 3). Likewise, an RT-PCR analysis performed with oligonucleotides flanking the insertion point of the pUA967 plasmid (RTHgbAup and RTHgbArp as the upper and lower primers, respectively) clearly demonstrated the absence of the hgbA mRNA in the P. multocida hgbA mutant (Fig. 3B). In contrast to the hgbA results, attempts to isolate either PM0298 or PM0299 insertional mutants were unsuccessful despite the fact that four independent experiments were carried out. This result could indicate that these two ORFs are essential for normal cell growth of P. multocida. To test this possibility, a P. multocida merodiploid strain containing the pUA964 plasmid, a derivative of the broad-host-range pHRP309 plasmid which is stable in P. multocida (23), and carrying a 1.4-kb fragment that included ORFs PM0298 and PM0299 was used as a recipient in new insertional mutagenesis experiments. In this case, strains with an insertional mutation in the chromosomal copy of ORF PM0298 were obtained (Fig. 3A, lane 6). These data strongly suggest that mutations in either ORF PM0298 or ORF PM0299 are lethal. However, the level of hemoglobin binding of the P. multocida hgbA mutant seemed to be the same as that of the wild-type strain (Fig. 4A), although in vivo quantitative assays revealed that the adsorption kinetics of hgbA cells were slightly slower than those of wild-type cells (Fig. 4B). Furthermore, the growth rate and virulence of the P. multocida hgbA mutant were not affected compared to the growth rate and virulence of the wild-type strain (data not shown).
FIG. 3.
(A) PCR analysis of P. multocida mutants. Chromosomal DNA from hgbA (lane 3) and PM0298 (lane 6) mutants were subjected to PCR analysis with the Aad oligonucleotide (Table 2) as the upper primer and the HgbArp and HgbAintrp oligonucleotides (Fig. 1A), respectively, as the lower primers. PCRs performed with chromosomal DNA from the wild-type strain (lanes 2 and 5) and these primer pairs and PCRs performed with preparations lacking DNA template (lanes 4 and 7) were the negative controls. Lane 1 contained HindIII-digested λ DNA as the molecular size marker. (B) RT-PCR study of the P. multocida hgbA mutant. RNA from hgbA (lane 3) and wild-type (lane 4) cells were subjected to RT-PCR analysis with the RTHgbAup and RTHgbArp oligonucleotides (Table 2). The control for RNA integrity was an RT-PCR amplification performed with RNA from hgbA cells and oligonucleotides RecAup and RecAdw belonging to the internal sequence of the P. multocida recA gene (lane 2). PCR performed with chromosomal DNA from the wild-type strain and oligonucleotides RTHgbAup and RTHgbArp (lane 5) and RT-PCR amplification of preparations containing the same primers but lacking both DNA and RNA templates (lane 6) were the product size and negative controls, respectively. Lane 1 contained BstEII-digested λ DNA as the molecular size marker.
FIG. 4.
(A) Solid-phase assay for in vitro binding of bovine hemoglobin to whole P. multocida wild-type (lane 1) or hgbA mutant (lane 2) cells. (B) In vivo quantitative analysis of hemoglobin adsorption to P. multocida wild-type (▪) and hgbA (▴) cells. The behavior of E. coli cells (•) was used as a negative control. All values are the means of three experiments (each performed in triplicate), and the standard error of any value was never greater than 10%.
Expression of the hgbA transcriptional unit in vitro and during infection.
To study the iron starvation effect on transcription of the hgbA operon, as well as the pattern of temporal expression in a mouse model, the RIVET method was used. In this technique, a suicide plasmid containing a fusion between the promoter to be analyzed and the region encoding the resolvase gene (tnpR) from Tnγδ is constructed (2). This plasmid is then introduced into a bacterial strain in which a copy of the res1-tet-res1 construction is present in the chromosome. In this way, after integration of the plasmid harboring the promoter-tnpR fusion, expression of the promoter produces an increase in the intracellular level of resolvase, which facilitates specific recombination between the two res1 sequences, resulting in a loss of tetracycline resistance (2). Alternatively, the res1-tet-res1 construction may also be introduced into the same plasmid which contains the promoter-resolvase fusion (34).
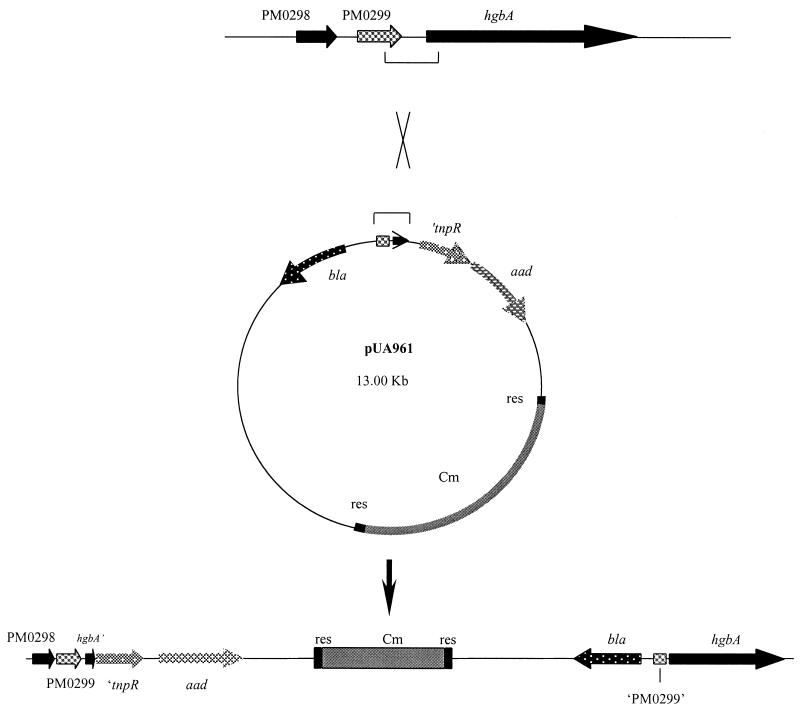
To develop a RIVET system for P. multocida, the tnpR-encoding region was amplified by PCR from the pIVET5mut plasmid (15) and ligated into the pUA826 suicide plasmid to obtain pUA958. Since P. multocida isolates are often tetracycline resistant (13) and because the RIVET system could be generally used with any strain of this organism, we decided to replace the tet marker of the res1-tet-res1 construction with the Cm cassette of plasmid pHP45ΩCm. Accordingly, the Cm cassette was isolated from the pHP45ΩCm plasmid by HindIII digestion and inserted into plasmid pGH436 which had previously been digested with AccI and SphI to delete the tet region. Cm cassette and pGH436 plasmid ends were previously made blunt by Klenow fragment treatment. This yielded plasmid pUA959, from which the res1-Cm-res1 fragment was recovered by NdeI-ScaI digestion and, after end filling, ligated into the SmaI-digested pUA958 plasmid to obtain pUA960 containing both the tnpR-encoding region and the target region (res1-Cm-res1) of the resolvase. Finally, a 360-bp PCR fragment which contained 71 bp of the 5′ end of the hgbA gene and 241 bp of the 3′ end of PM0299 and was obtained by amplification with the PhgbAup and PhgbArp primers was digested with XbaI and cloned into the XbaI restriction site present upstream of the tnpR region of the pUA960 plasmid, giving rise to pUA961 (Fig. 5). The structure of the desired construction was then confirmed by PCR and restriction analysis (data not shown). This 360-bp fragment belonging to the last gene of the transcriptional unit, hgbA, was used rather than another fragment containing the immediate upstream region of PM0298 in order to be certain that transcription of the whole operon was really being measured.
FIG. 5.
Schematic representation of specific single recombination between the pUA961 plasmid and the P. multocida chromosome through the 370-bp internal fragment of the PM0298-PM0299-hgbA transcriptional unit cloned upstream of the tnpR gene-encoding region.
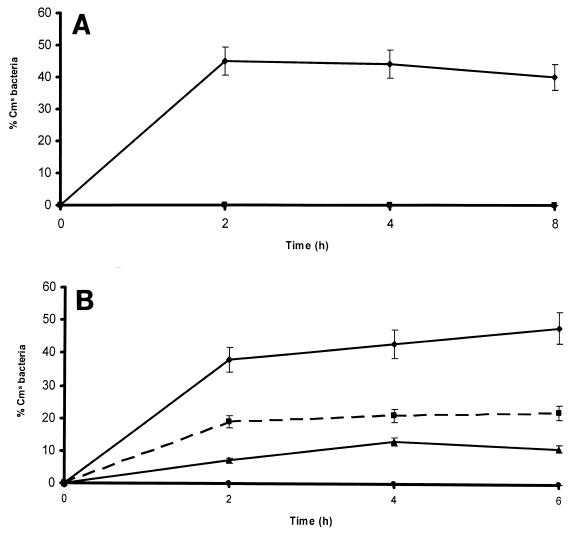
The pUA961 plasmid was then introduced into P. multocida by mating, streptomycin- and chloramphenicol-resistant clones were selected, and insertion of the plasmid into the expected point between the PM0299 and hgbA genes (Fig. 5) was confirmed by PCR and Southern analysis (data not shown). One of the clones, PM1077, was kept and used for subsequent experiments. Analysis of the development of a number of chloramphenicol-sensitive cells after mouse infection with the PM1077 strain indicated that the hgbA operon is rapidly induced after intraperitoneal inoculation. Thus, 35% of the cells recovered from the peritoneal cavity lost chloramphenicol resistance within 2 h after challenge (Fig. 6A). A similar loss of chloramphenicol resistance was also observed in vitro in the presence of the iron chelator DPD (Fig. 6B). Moreover, this induction could be strongly suppressed by addition of FeSO4 or hemoglobin (Fig. 6B).
FIG. 6.
(A) Kinetics of transcription of the PM0298-PM0299-hgbA operon in mice inoculated intraperitoneally with the P. multocida PM1077 strain, measured as the proportion of Cms bacteria recovered from the peritoneal cavity (♦). The absence of Cms bacteria in cultures of the P. multocida PM1077 strain growing in BPW medium without chloramphenicol demonstrates the genetic stability of the construction (▪). Three animals were used for each point, and each Cms percentage is the average of three determinations for each of the inoculated mice. In all cases, the standard error of any value was never greater than 10%. (B) Kinetics of transcription of the PM0298-PM0299-hgbA operon in the P. multocida PM1077 strain measured in vitro as the proportion of Cms bacteria recovered from BPW medium (•) or BPW medium supplemented with either DPD (50 μg/ml) (♦), DPD and FeSO4 (1 mM) (▴), or DPD and hemoglobin (40 mM) (▪). All values are the means of at least three experiments (each performed in triplicate), and the standard error of any value was never greater than 10%.
Presence of the hgbA gene in several P. multocida serotypes.
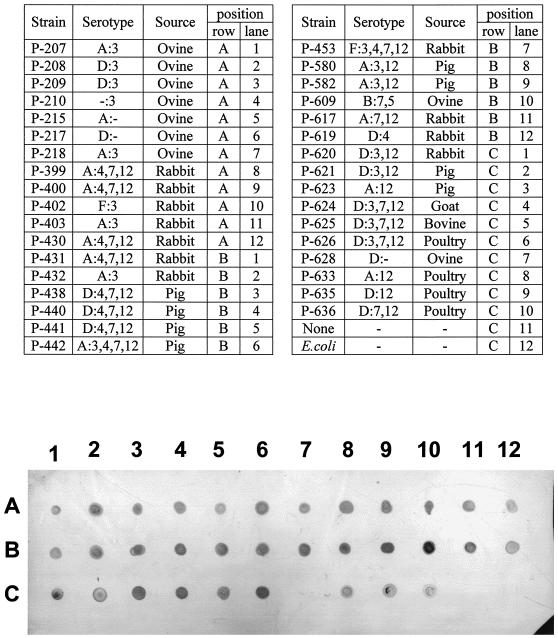
Genomic DNA was extracted from 34 isolates belonging to several P. multocida serotypes and obtained from different animal sources, and a hybridization dot blot analysis was performed to investigate the presence of the hgbA gene in these strains with a digoxigenin-labeled 989-bp internal fragment of hgbA as the probe. As Fig. 7 shows, DNA from most of the P. multocida strains strongly hybridized with the hgbA probe; the only exception was the nontypeable P-628 strain. No hybridization was detected when chromosomal DNA from E. coli (Fig. 7) or Mannhemia haemolytica (data not shown) was used. These results indicate that the hgbA gene may be widely present in P. multocida strains regardless of the specific serotype.
FIG. 7.
Presence of the hgbA gene in P. multocida strains: dot blot hybridization of chromosomal DNA from 34 P. multocida strains obtained from different animal sources and belonging to several serotypes with a digoxigenin-labeled 989-bp internal fragment of the hgbA gene. The DNA position corresponding to each of the strains is indicated above the nitrocellulose membrane. Equal amounts of chromosomal DNA (1 μg) of all strains were applied to the nitrocellulose membrane, and hybridization was performed under high-stringency conditions.
DISCUSSION
In this paper we report that ORFs PM0298, PM0299, and PM0300 of P. multocida constitute a single transcriptional unit. Mutation analysis by plasmid insertion showed that strains containing inactivated ORF PM0298 or PM0299 could be obtained only when another copy of the inactivated ORF is present. This fact suggests that PM0298 and PM0299 are essential for P. multocida cell viability. The biological role of PM0298 and PM0299 is unknown, although these ORFs show 49 and 61% identity with products of the Plesiomonas shigelloides hugX and hugZ genes, respectively. These P. shigelloides genes seem to be necessary to prevent heme toxicity, as demonstrated by complementation assays performed with E. coli strains growing in media with heme as the only iron source (10). Nevertheless, there are no data on the importance of these two genes in P. shigelloides since construction of derivative mutants of this bacterial species has not been reported. Our data concerning the lack of viability of P. multocida PM0298 and PM0299 mutants are consistent with a detoxifying role under certain growth conditions. In contrast, our results showed that a P. multocida PM0300 mutant was viable and did not show any significant defect in the ability to grow either in vitro or during animal infection.
Cloning and overexpression in E. coli of ORF PM0300 demonstrated that it is a hemoglobin receptor, and PM0300 was therefore designated hgbA. However, the extent of hemoglobin binding by the P. multocida hgbA mutant was similar to that of the wild-type strain, although the hemoglobin adsorption kinetics of hgbA cells were a little slower than those of the wild-type strain. This fact suggests that other functional hemoglobin receptor proteins must be present in P. multocida cells, which can compensate for the loss of HgbA. In fact, when other P. multocida ORFs, such as PM0741, PM0745, and PM1282 present a TonB box, are used as a query in a TBLASTN analysis performed with the GenBank database, the results obtained predict the presence of putative hemoglobin-binding motifs in them (data not shown). Additionally, the presence of several hemoglobin receptor proteins able to support cell growth when one of them is inactivated has also been demonstrated for Haemophilus influenzae (27). Thus, this pathogen possesses three hemoglobin receptor proteins encoded by the hgpA, hgpB, and hgpC genes (21). Mutants of H. influenzae defective in either one or two of these three genes show the same behavior as the wild-type strain, whereas triple mutants show a reduced ability to use hemoglobin (21, 27). The presence of more than one gene encoding hemoglobin receptors may be useful to bacterial cells either because it increases the level of hemoglobin binding to the cell or because it prevents the negative effects of mutations. It is worth noting that H. influenzae hgpA, hgpB, and hgpC contain several tandem repeats of the CCAA tetranucleotide, which can introduce frameshifts due to the loss or gain of one or more of these repeats, giving rise to a high frequency of phase variation in these genes (28). This changing ability has been proposed to be related to mechanisms that enhance evasion of the host immune response (11). If there are several genes encoding the same biological function (in this case a hemoglobin receptor), variability is strongly increased. However, the P. multocida hgbA gene does not show any of the several tandem repeats which are implicated in phase variation (28), suggesting that its expression is not under this kind of regulation. Moreover, the virulence of some bacteria, such as Neisseria meningitidis (37) and Porphyromonas gingivalis (33), was significantly reduced when a single gene encoding a hemoglobin receptor protein was mutagenized. These data suggest that the presence of more than one gene for hemoglobin receptors is not a general feature of all bacterial species.
By using RIVET, we were able to show that expression of the PM0298-PM0299-hgbA transcriptional unit is triggered within 2 h following intraperitoneal inoculation of mice. In addition, this operon is regulated by iron, as shown by the decrease in its transcription when FeSO4 is added to cultures growing in the presence of DPD (Fig. 6B). Likewise, these results are consistent with preliminary data obtained in microarray analyses for the pattern of P. multocida whole-genome expression when preparations are treated with chelating agents (24). Nevertheless, the rapid induction of these genes in infected mice is consistent with the low concentrations of free iron present in mammalian tissues. However, to our knowledge, this is the first case in which temporal expression of iron-regulated genes has been analyzed in vivo in a nonenteric bacterial species (14, 15). It is known that iron regulates bacterial gene expression through the Fur protein, which in the presence of this element specifically recognizes and binds a given sequence (Fur box), blocking transcription (36). The Fur box is composed of at least three contiguous NAT(A/T)AT-like hexamers in either direct or inverse orientations (7). One putative copy of this motif, which responds to the TATTATCAATATTGATAAT sequence, is found 114 bp upstream of the PM0298 translational start codon. Similar iron-dependent regulation of the transcription of genes encoding hemoglobin receptor proteins has been described for other pathogenic bacteria, such as H. influenzae, P. gingivalis, and N. meningitidis (27, 33, 37). Finally, our data also indicate that the hgbA gene is widespread in P. multocida strains regardless of the serotype analyzed or the source of the isolate. This suggests that the hgbA gene may be useful as a specific probe to detect P. multocida in field samples. It has been demonstrated that some TonB-dependent iron receptors, such as transferrin- and lactoferrin-binding proteins, are able to induce protection when they are used as prophylactic immunogens (4, 6, 16, 29). Further work is required to determine whether the widely distributed HgbA protein may also be useful as an antigen that provides protection against virulent P. multocida strains.
Acknowledgments
This work was funded by grant BIO99-0779 from the Ministerio de Ciencia y Tecnología de España and by grant 2001SGR-206 from the Comissionat per Universitats i Recerca de la Generalitat de Catalunya. M. Elena Garrido was a recipient of a predoctoral fellowship from the Direcció General d'Universitats de la Generalitat de Catalunya.
We are deeply indebted to Ben Adler for his critical reading of the manuscript and his suggestions. We also acknowledge A. Camilli for his generous gift of bacterial strains and plasmids. We thank Joan Ruiz and Susana Escribano for their excellent technical assistance.
Editor: B. B. Finlay
REFERENCES
- 1.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 2.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 91:2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardenas, M., A. R. Fernández de Henestrosa, S. Campoy, A. Perez de Rozas, J. Barbé, I. Badiola, and M. Llagostera. 2001. Virulence of Pasteurella multocida recA mutants. Vet. Microbiol. 80:53-61. [DOI] [PubMed] [Google Scholar]
- 4.Danve, B., L. Lissolo, M. Mignon, P. Dumas, S. Colombani, A. B. Schryvers, and M. J. Quentin-Millet. 1993. Transferrin-binding proteins isolated from Neisseria meningitidis elicit protective and bactericidal antibodies in laboratory animals. Vaccine 11:1214-1220. [DOI] [PubMed] [Google Scholar]
- 5.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 6.Du, R. P., Q. Wang, Y. P. Yang, A. B. Schryvers, P. Chong, M. H. Klein, and S. M. Loosmore. 1998. Cloning and expression of the Moraxella catarrhalis lactoferrin receptor. Infect. Immun. 66:3656-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escolar, L., J. P. Martín, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez de Henestrosa, A. R., I. Badiola, M. Saco, A. M. Perez de Rozas, S. Campoy, and J. Barbé. 1997. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol. Lett. 154:311-316. [DOI] [PubMed] [Google Scholar]
- 9.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan, A., E. Aragall, I. Gibert, and J. Barbé. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol. Microbiol. 19:777-790. [DOI] [PubMed] [Google Scholar]
- 13.Kehrenberg, C., S. A. Salmon, J. L. Watts, and S. Schwarz. 2001. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMT1. J. Antimicrob. Chemother. 48:631-640. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 180:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, S. H., L. D. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 16.Loosmore, S. M., Y. P. Yang, D. C. Coleman, J. M. Shortreed, D. M. England, R. E. Harkness, P. S. Chong, and M. H. Klein. 1996. Cloning and expression of the Haemophilus influenzae transferrin receptor genes. Mol. Microbiol. 19:575-586. [DOI] [PubMed] [Google Scholar]
- 17.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 18.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 21.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 23.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram negative bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 24.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 26.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 27.Ren, Z., H. Jin, D. J. Morton, and T. L. Stull. 1998. HgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin- binding protein. Infect. Immun. 66:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren, Z., H. Jin, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin- haptoglobin binding protein genes of Haemophilus influenzae. J. Bacteriol. 181:5865-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi-Campos, A., C. Anderson, G. F. Gerlach, S. Klashinsky, A. A. Potter, and P. J. Wilson. 1992. Immunization of pigs against Actinobacillus pleuropneumoniae with two recombinant protein preparations. Vaccine 10:512-518. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 33.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182:5737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slauch, J. M., and A. Camilli. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 326:73-96. [DOI] [PubMed] [Google Scholar]
- 35.Stark, W. M., N. D. F. Grindley, G. F. Hatfull, and M. R. Boocock. 1991. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 10:3541-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojilykovic, I., A. Baumler, and K. Hantke. 1994. Fur regulation in Gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 37.Stojilykovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 38.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorf. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]