Abstract
Members of the antigen I/II family of cell surface proteins are highly conserved, multifunctional adhesins that mediate interactions of oral streptococci with other oral bacteria, with cell matrix proteins (e.g., type I collagen), and with salivary glycoproteins, e.g., gp340. The interaction of gp340 (formerly designated salivary agglutinin) with Streptococcus mutans requires an alanine-rich repetitive domain (A region) of antigen I/II that is highly conserved in all members of this family of proteins. In this report, we show that the A regions from the two Streptococcus gordonii M5 antigen I/II proteins (SspA and SspB) interact differently with the salivary gp340 glycoprotein and appear to be structurally distinct. Recombinant polypeptides encompassing the A region of SspA or from a highly related S. mutans antigen I/II protein (SpaP) competitively inhibited the interaction of gp340 with intact S. gordonii and S. mutans cells, respectively. In contrast, an A region polypeptide from SspB was inactive, and furthermore, it did not bind to purified gp340 in vitro. Circular dichroism spectra suggested that all three polypeptides were highly α-helical and may form coiled-coil structures. However, the A region of SspB underwent a conformational change and exhibited reduced α-helical structure at pH 8.5, whereas the A region polypeptides from SspA and SpaP were relatively stable under these conditions. Melt curves also indicated that at physiological pH, the A region of SspB lost α-helical structure more rapidly than that of SspA or SpaP when the temperature was increased from 10 to 40°C. Furthermore, the SspB A region polypeptide denatured completely at a temperature that was 7 to 9°C lower than that required for the A region polypeptide of SspA or SpaP. The full-length SspB protein and the three A region peptides migrated in native gel electrophoresis and column chromatography with apparent molecular masses that were approximately 2- to 2.5-fold greater than their predicted molecular masses. However, sedimentation equilibrium ultracentrifugation data showed that the A region peptides sedimented as monomers, suggesting that the peptides may form nonglobular intramolecular coiled-coil structures under the experimental conditions used. Taken together, our results suggest that the A region of SspB is less stable than the corresponding A regions of SspA and SpaP and that this structural difference may explain, at least in part, the functional variation observed in their interactions with salivary gp340.
The antigen I/II proteins are a family of related major surface polypeptides that are expressed by virtually all species of oral streptococci (37), but they have been most extensively characterized from several strains of Streptococcus mutans and Streptococcus gordonii (12, 14, 19, 25, 26, 32, 42, 44, 53). These studies have shown that the overall structural organization of individual members of the antigen I/II family of proteins is highly conserved and that they exhibit approximately 65 to 70% primary sequence identity (24).
Each of the polypeptides possesses seven structural regions, comprising a signal sequence, a highly charged N-terminal region, an alanine-rich repetitive domain (A region), a divergent central region, a proline-rich repetitive region, a C-terminal domain, and cell wall-anchoring sequences (24). The A region of antigen I/II comprises three to five copies of an 82-amino-acid repeat and is predicted to assume a highly α-helical structure (14, 26, 31). This region also exhibits a seven-residue periodicity of hydrophobic amino acids, suggesting that it may form a coiled coil. Similarly, structural predictions suggest that the proline-rich repeats assume a highly extended structure. A truncated antigen I/II protein that lacks the proline-rich region is not secreted by S. mutans, suggesting that the recombinant polypeptide does not fold properly in the cell (4). However, very little experimental data exist to support these structural predictions, and little is known about the secondary and tertiary structures of antigen I/II.
Antigen I/II proteins are multifunctional adhesins that contribute to the initiation and development of the oral biofilm by mediating interactions with salivary constituents, host cell matrix proteins, and other oral bacteria. Several studies have shown that antigen I/II mediates interbacterial interactions with other oral organisms, including actinomyces (11, 13, 18, 25), Porphyromonas gingivalis (7, 8,15), and Candida albicans (23). These interactions suggest that antigen I/II proteins may play an important role in the maturation of the oral biofilm by promoting the colonization of an existing streptococcal community by other bacteria. Antigen I/II proteins also have been shown to interact with human and rat collagen (35, 46, 48) and with human fibronectin and laminin (48). These interactions are thought to contribute to the development of streptococcal abscesses and infective endocarditis and may also be important in the invasion of dentinal tubules by streptococci (35, 36).
The best-characterized functional property of antigen I/II polypeptides is their interaction with salivary proteins. Numerous studies have suggested that antigen I/II is important for the initial streptococcal colonization of human teeth through its interaction with various proteins that constitute the salivary pellicle, including a mucin-like salivary glycoprotein that has been designated salivary agglutinin (6, 12, 25, 30, 39, 43). Recently, the salivary agglutinin has been shown to be identical to the lung surfactant protein D scavenger protein gp340 (2, 34, 47). The interaction of antigen I/II with gp340 requires carbohydrate constituents of the glycoprotein and is calcium dependent (14, 33). Indeed, Duan et al. (17) showed that the S. gordonii antigen I/II polypeptide SspB is a high-affinity calcium-binding protein that binds a single calcium ion, suggesting that antigen I/II may require calcium for activity.
Furthermore, specific regions of antigen I/II polypeptides have been identified as being important for interacting with gp340. Monoclonal anti-antigen I/II antibodies that react with epitopes in the C-terminal region of the protein inhibit binding of intact S. mutans cells to gp340-coated hydroxyapatite (5, 6). Consistent with this, Munro et al. (40) showed that a recombinant peptide comprising residues 816 to 1161 of the S. mutans antigen I/II (SpaP) bound to gp340. Subsequently, two specific sequences within this region of SpaP, designated Ad1 and Ad2, were shown to interact with gp340 (27). However, antibodies directed against the A region of antigen I/II also inhibit adhesion of streptococci to gp340-coated hydroxyapatite surfaces, and peptides encompassing the A region of S. mutans antigen I/II have been shown to bind salivary gp340 (10, 39, 41). The relative contributions of the A region and C-terminal sequences in the interaction of antigen I/II with gp340 are not clear, and both of these regions of the S. mutans antigen I/II protein have been targeted as potential subunit anticaries vaccine candidates (28, 49-52).
In spite of the high conservation of antigen I/II structure and primary sequence, the functional properties of individual members of this family of proteins differ. For example, Egland et al. (18) showed that SspA and SspB from S. gordonii DL1 differed in their coaggregation with Actinomyces naeslundii strains. Holmes et al. (22) also showed that SspA and SspB differ in their interactions with type 1 collagen and with Candida albicans. Other functions of antigen I/II are species specific. S. gordonii SspB interacts with the minor fimbrial protein of P. gingivalis, but the related SpaP protein of S. mutans does not (7, 8). Furthermore, specific amino acid residues of SspB that are not conserved in SpaP are essential for this interaction and may confer a discrete structural motif in SspB that is recognized by P. gingivalis (15). The interactions of SspB and SpaP with gp340 also differ. SspB is sensitive to inhibition by sialic acid-containing sugars and by neuraminidase treatment of gp340, whereas SpaP is not (14).
In this report, we show that the structural and functional properties of the A region of S. gordonii SspB differ from those of the related SspA and SpaP proteins. Polypeptides representing the A regions of SspA and SpaP interacted with salivary gp340, whereas the A region peptide from SspB did not. Structural analyses of each of these polypeptides suggested that the A region of SspB is less stable than that of SspA and SpaP. Circular dichroism spectra and sedimentation equilibrium ultracentrifugation showed that each peptide contained approximately 60% α-helix at pH 7.5 and existed in solution as nonglobular monomers. However, SspB but not SspA or SpaP exhibited a loss of α-helix and a concomitant increase in β-sheet at pH 8.5. In addition, melt curves at physiological pH showed that the A region of SspB lost α-helical content more rapidly as a function of temperature than did the corresponding peptides from SspA and SpaP. Furthermore, SspB denatured at temperatures that were 7 to 9°C lower than that required to denature SspA and SpaP. Together, these results suggest that the structure and function of the A region of SspB differs from that of SspA or SpaP.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptococcus gordonii M5 and Streptococcus mutans KPSK2 were grown in Todd-Hewitt broth at 37°C without aeration. Escherichia coli strains DH5α and BL21 were used for propagation of pGEX plasmids and expression of glutathione S-transferase-antigen I/II fusion proteins. These strains were grown at 37°C with aeration in Luria-Bertani broth (LB) supplemented where necessary with 100 μg of ampicillin per ml. E. coli pEB5 is a recombinant clone that carries pUC19 with a 4.8-kbp streptococcal insert containing sspB (12) and encodes the full-length S. gordonii M5 SspB protein. E. coli pPAc7 contains pUC19 and spaP of S. mutans KPSK2 serotype c, encoding the full-length SpaP polypeptide. E. coli pPAc7 is a subclone of the previously described lambda phage strain MSL-1 (16). Strains pEB5 and pPAc7 were cultured in LB supplemented with 100 μg of ampicillin per ml.
Cloning and expression of DNA fragments encoding AR peptides.
DNA fragments encoding the N-terminal repetitive domains of SspA, SspB, and SpaP were amplified by PCR from the following templates: S. gordonii M5 genomic DNA for SspA, plasmid pEB5 for SspB, and plasmid pPAc7 for SpaP. Each fragment encoded a polypeptide of 248 amino acids starting at Tyr202 of SspB (or the corresponding Tyr residue in SspA or SpaP) and encompassing 3 of the 82 residue A region repeat units. The primers used for amplification reactions are shown in Table 1 and contained restriction sites to facilitate cloning.
TABLE 1.
PCR Primers
| Primer sequencea | Restriction site | Target gene |
|---|---|---|
| 5′-CCGGATCCCCGTATGAAGCGAAGCTAGC-3′ | BamHI | sspA |
| 5′-TCAGAATTCATAGTCAGTTTTAGC-3′ | EcoRI | sspA |
| 5′-CCGGATCCCCGTATGAGGCTAAGCTAGC-3′ | BamHI | sspB |
| 5′-TCAGAATTCATAATCTGTTTTAGC-3′ | EcoRI | sspB |
| 5′-CCGGATCCCCGTATGAAGCTAAATTGGC-3′ | BamHI | spaP |
| 5′-TCAGAATTCGTAATCAGCTTTAGC-3′ | EcoRI | spaP |
Restriction sites are indicated in boldface.
PCR conditions were as follows: 94°C for 1 min; 55°C for 1.5 min; and 72°C for 2 min for 30 cycles. Reaction products were analyzed by agarose gel electrophoresis in 0.8% gels, and the desired DNA fragments were excised and purified with the Concert rapid gel extraction system (Gibco-BRL). Purified fragments were ligated with pGEM-T Easy (Promega) and transformed into subcloning-grade competent E. coli DH5α (Gibco-BRL). Plasmid from recombinant clones was isolated with the Wizard Mini-Prep DNA purification system (Promega) and analyzed for incorporation of the desired insert by digestion with EcoRI.
Insert fragments from the plasmids selected above were then excised by digestion with EcoRI and BamHI, purified as described above, and cloned into pGEX6-2P. Ligation mixtures were transformed into competent E. coli BL21. Purified plasmid from the recombinant clones was digested with EcoRI and BamHI to confirm incorporation of the correct insert. Finally, to confirm the identity of the insert fragments and to show that no errors were introduced by the amplification reaction, the sequence of the inserts from selected clones was determined by the DNA Sequencing Facility at the University of Pennsylvania. Clones containing the desired sequences were designated E. coli AR-A, E. coli AR-B, and E. coli AR-P and encoded fusion proteins containing the N-terminal repetitive domains of SspA, SspB, and SpaP, respectively.
Expression of glutathione S-transferase-antigen I/II fusion proteins was carried out by inoculating 2 liters of LB broth with 20 ml of an overnight culture of each recombinant E. coli clone and incubating with vigorous aeration for 3 h at 37°C (optical density at 600 nm [OD600], ≈0.5). Isopropylthiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM, and incubation was continued for an additional 2 to 3 h (final OD600 ≈1.3). Cells were pelleted by centrifugation at 10,000 × g, washed three times with phosphate-buffered saline (PBS; 50 mM sodium phosphate, 150 mM NaCl, pH 7.4), and suspended in 20 ml of B-PERII bacterial protein extraction reagent (Pierce). The cell suspension was mixed to achieve a homogenous suspension, and then an additional 20 ml of B-PERII lysis solution was added. The resulting suspension was shaken gently at 25°C for 10 min and centrifuged at 10,000 × g for 15 min to remove cellular debris.
Affinity purification of the fusion proteins was carried out on glutathione-Sepharose columns (Promega). Columns were initially washed with 50 ml of PBS containing 3 M NaCl, followed by 50 ml of 6 M guanidine-HCl, and equilibrated with 20 ml of PBS. Prior to use, the columns were prepared by the following series of washes: 50 ml of 70% ethanol, 20 ml of PBS, 20 ml of 0.1 M Tris-HCl containing 0.5 M NaCl (pH 8.5), and 20 ml of 0.1 M sodium acetate containing 0.5 M NaCl (pH 4.5). The 0.1 M Tris-0.5 M NaCl and 0.1 M sodium acetate-0.5 M NaCl washes were repeated twice, and the columns were then equilibrated with 50 ml of loading buffer (50 mM Tris-HCl containing 100 mM KCl, pH 7.5). B-PERII protein extracts were loaded onto the columns, and unbound material was removed with 50 ml of loading buffer. Elution of the fusion proteins was carried out with 20 ml of 50 mM Tris-HCl containing 5 mM glutathione (Sigma Chemical Co.), pH 8.0. The eluate was dialyzed against 5 liters of loading buffer and lyophilized.
Cleavage of fusion proteins.
Prior to cleavage, a small portion of the fusion protein samples was removed for analysis by gel electrophoresis (see below). The remaining lyophilized fusion protein was suspended in 1 ml of distilled H2O, and the solution was adjusted to a final concentration of 1 mM EDTA, 1 mM dithiothreitol, and 0.01% Triton X-100. Cleavage of the fusion proteins was carried out overnight at 4°C after the addition of 40 μl of PreScission protease (2 U per μl; Amersham Pharmacia Biotech). Purification of the AR-A, AR-B, and AR-P peptides from glutathione S-transferase and the PreScission protease was accomplished with glutathione-Sepharose. Columns were prepared and loaded as described above. Since both glutathione S-transferase and PreScission protease bind to glutathione-Sepharose, the flowthrough from loading the column and the subsequent wash with loading buffer contained the desired streptococcal peptides and was collected. The collected material was dialyzed against 10 mM Tris and lyophilized. Fusion proteins and the purified AR peptides were evaluated by electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) gels.
Column chromatography of AR peptides.
Purified peptides were chromatographed by fast protein liquid chromatography (FPLC) on a Superdex 75 column (1.6 by 40 cm) that was equilibrated in 50 mM Tris-HCl containing 100 mM KCl, pH 7.5. All chromatography experiments were carried out with a flow rate of 0.67 ml/min at 37°C in a water-jacketed column. Proteins were detected at OD280. Prior to chromatography of the purified AR peptides, the column was first calibrated with a mixture of blue dextran (10 mg/ml) and p-nitrophenol (5 mM) and subsequently calibrated with commercial protein size standards (Sigma Chemical Co.) bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), cytochrome c (12.4 kDa), and aprotinin (6.4 kDa).
Inhibition of gp340 interaction with streptococci.
The interaction of salivary gp340 with antigen I/II on the surface of intact oral streptococci leads to the formation of bacterial aggregates. To monitor the ability of the AR peptides to interact with gp340, the peptides were used as competitive inhibitors of gp340-mediated streptococcal aggregation. Bacterial aggregation was followed essentially as described by Demuth et al. (12) with the exception that purified gp340 was used in place of parotid saliva. Briefly, S. gordonii M5 cells (for reactions with the A region polypeptides from SspA or SspB) and S. mutans KPSK2 cells (for reactions with the A region peptide from SpaP) were diluted with PBS to an optical density of 1.0 at 675 nm in a disposable cuvette. For the positive-control assays, purified gp340 was added to diluted cells to a final concentration of 30 μg per ml, and the mixture was incubated at 37°C for up to 90 min.
For inhibition assays, various amounts of the AR peptides were added to the assay mixtures prior to the addition of gp340. For some experiments, purified full-length SspA, SspB, or SpaP protein was used as the inhibitor at a concentration of 30 μg/ml. Full-length protein was purified from recombinant E. coli strains as previously described (17). The final volume of all assay mixtures was kept constant at 1 ml. The optical density at 675 nm of the reactions was recorded at 15-min intervals. A blank assay, containing streptococci in buffer alone (i.e., no gp340), was run for each experiment and represented a measure of the incidental settling of bacteria over the time course of the experiment. Incidental settling was ≤0.05 OD675 unit for all experiments. After subtracting the blank from the experimental values, percent inhibition was calculated by the following equation: % inhibition = [(OD at t0 − OD at tx) determined experimentally divided by (OD at t0 − OD at tx) in the control] × 100, where t0 is time zero and tx is the time of determination.
Circular dichroism spectroscopy.
Circular dichroism spectroscopy was carried out on an Aviv 60DS spectrophotometer with operating software version 4.1t. All scans were carried out with a bandwidth of 1 nm and a protein sample concentration of 3 μM in PBS at the desired pH. Wavelength scans at a constant temperature (37°C) and temperature scans at a constant wavelength (222 nm) were carried out for all protein samples. For wavelength scans, samples were analyzed from 320 nm to 195 nm with a scan step of 1 nm and a data acquisition time of 1 s. For temperature scans, the temperature of the sample chamber was varied from 5°C to 80°C in steps of 1°C. An equilibration time of 0.2 min was used at each temperature point, and the θ222 was measured with a data collection time of 1 s for a total of 1 min. Thus, the data at each temperature point are the averages of 60 individual θ222 readings.
Sedimentation equilibrium centrifugation.
Sedimentation equilibrium experiments were carried out with a Beckman XL-A analytical ultracentrifuge. Samples were loaded into six-channel Epon charcoal-filled centerpieces with quartz windows. Protein samples were analyzed at concentrations of 10 μM, 1.0 μM, and 0.1 μM. Experiments were performed at room temperature with speeds of 20,000, 24,000, 28,000, and 32,000 rpm, with a detection wavelength of 280 nm. Solvent density was assumed to be 1.003 g/ml, and the partial specific volumes for each of the AR peptides were calculated from their amino acid compositions with the SEDNTERP program (http://www.cauma.uthscsa.edu/software). Data were fit by nonlinear least-squares regression with the NONLIN program supplied with the Optima XL-A data analysis software.
RESULTS
Expression of antigen I/II AR peptides.
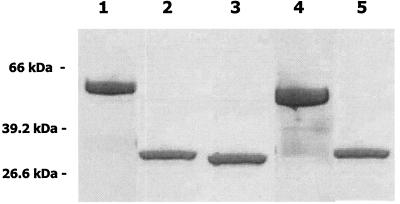
To obtain peptides comprising the alanine-rich repetitive regions of the S. gordonii (SspA and SspB) and S. mutans (SpaP) antigen I/II proteins, our initial strategy was to express DNA fragments encoding both the N-terminal globular region and the alanine-rich repeats of each protein in E. coli (39). With this approach, the purified peptides that were obtained from the recombinant strains routinely migrated as three to four bands in SDS-PAGE (not shown), suggesting that they were cleaved by the host E. coli cells. We therefore amplified DNA fragments encoding only the alanine-rich repeat sequences (A regions) from each of the antigen I/II genes and cloned these fragments into pGEX6-2P (10). The resulting glutathione S-transferase-antigen I/II fusion proteins each contained 248 amino acids from antigen I/II, encompassing the three complete 82-residue repeats that are present in SspA, SspB, and SpaP. The fusion proteins were purified and cleaved from glutathione S-transferase. As shown in Fig. 1, the purified peptides representing the alanine-rich repetitive domain of SspA (AR-A), SspB (AR-B), and SpaP (AR-P) migrated as single bands of approximately 29 kDa in SDS-PAGE, in good agreement with the molecular masses of the polypeptides predicted from their deduced amino acid sequences.
FIG. 1.
Expression and cleavage of glutathione S-transferase-antigen I/II fusion proteins. DNA fragments encoding the N-terminal repetitive domains of SspA, SspB, and SpaP were cloned into pGEX-6P2 and expressed in E. coli. The resulting fusion proteins were purified on glutathione-Sepharose and cleaved with PreScission protease as described in Materials and Methods. The resulting antigen I/II peptides were purified by FPLC on Sephacryl G-75 and evaluated on SDS-10% PAGE gels stained with Coomassie blue. Lanes: 1, SspB/glutathione S-transferase fusion protein; 2, AR-B peptide; 3, AR-P peptide; 4, SspA/glutathione S-transferase fusion protein; 5, AR-A peptide. Positions of molecular size markers are indicated on the left.
Functional analysis of antigen I/II AR peptides.
The A region of SpaP was previously shown to interact with salivary glycoprotein gp340, the salivary isoform of the lung surfactant protein D scavenger receptor. SspA and SspB also interact with gp340 but differ from SpaP in the specificity of their interaction (14). Currently, it is not known what role, if any, the alanine-rich regions of SspA and SspB play in their interactions with gp340.
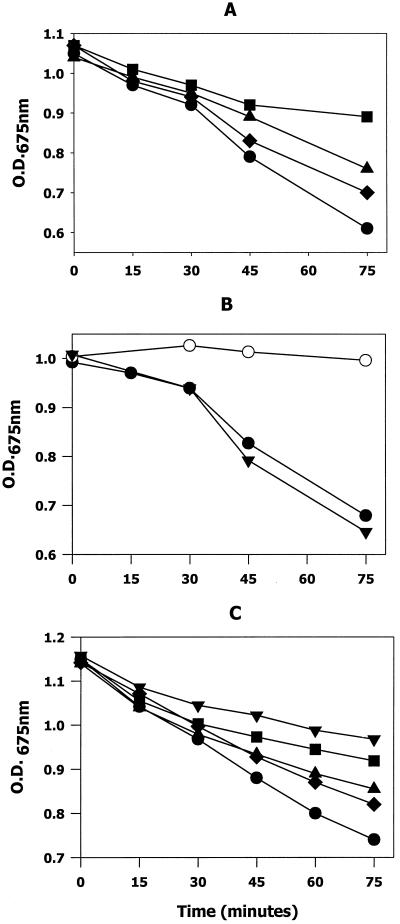
To determine if AR-A and AR-B interact with gp340, we asked if the purified peptides competed for gp340 with intact SspA and SspB contained on the surface of streptococcal cells. This was accomplished by monitoring peptide-dependent inhibition of gp340-mediated streptococcal aggregation. As shown in Fig. 2, coincubation of 10 to 50 μg of the AR-A peptide (Fig. 2A) per ml with bacteria in the presence of 30 μg of gp340 per ml resulted in a dose-dependent inhibition of gp340-mediated aggregation, with 64% inhibition observed with 50 μg of AR-A per ml. The soluble full-length SspA protein at 30 μg/ml was also an effective inhibitor of gp340-mediated streptococcal aggregation (>90% inhibition; not shown).
FIG. 2.
Competitive inhibition of salivary gp340-mediated streptococcal aggregation by antigen I/II peptides AR-A (A), AR-B (B), and AR-P (C). The AR-A and AR-B peptides were incubated at 37°C with S. gordonii M5 cells in the presence of 30 μg of gp340 per ml in a 1-ml volume. The AR-P peptide was similarly incubated with S. mutans KPSK2 (Fig. 2C). gp340-mediated bacterial aggregation was followed by monitoring the decrease in OD675 as cell aggregates formed and settled. Incubations were carried out in the presence of 10 (♦), 25 (▴), 50 (▪), or 90 μg (▾) of each peptide per ml or in the presence of 30 μg of full-length SspB per ml (•). Positive control reactions (○) consisted of bacteria and gp340 in the absence of AR peptide. Nonspecific settling of cells was monitored in blank reactions containing only bacteria and was ≤0.05 OD675 units for all reactions. All reactions were carried out in triplicate.
In contrast, AR-B (Fig. 2B) did not function as a competitive inhibitor of gp340-mediated aggregation even at the highest peptide concentration that was tested (90 μg/ml). This concentration of AR-B represented an 18-fold molar excess over the positive control reaction, which contained 30 μg of soluble full-length SspB per ml. The positive control inhibited the gp340 interaction with bacteria by >95% (see Fig. 2B). Finally, the AR-P peptide (10 to 90 μg/ml) inhibited gp340-mediated aggregation of S. mutans cells in a dose-dependent manner, consistent with the previous results of Crowley et al. (10). These results suggest that the A regions of SspA and SpaP interact with salivary gp340, whereas the corresponding sequences of SspB do not.
Structure and stability of antigen I/II AR peptides.
The A region polypeptides from SspA, SspB, and SpaP exhibit greater than 60% primary sequence identity and exhibit a seven-residue periodicity of hydrophobic amino acids. This suggests that the overall structure of this region is well conserved in all three proteins. However, our previous studies suggested that local structural differences may exist in highly conserved regions of antigen I/II proteins and that these differences may influence antigen I/II function (15). Therefore, to determine if structural differences exist in the conserved alanine-rich repetitive regions of SspA, SspB, and SpaP, a series of studies were carried out to examine the structure and stability of the three AR peptides.
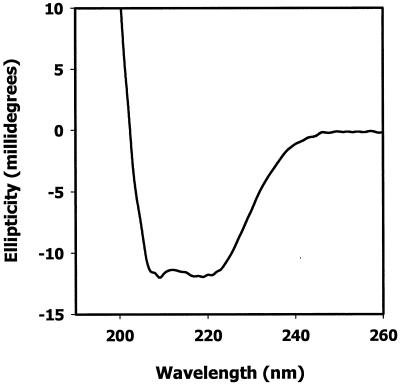
AR-A, AR-B, and AR-P were initially analyzed by circular dichroism spectroscopy. A representative plot of circular dichroism data collected for AR-B at 37°C at pH 7.5 is shown in Fig. 3. Similar profiles were obtained for AR-A and AR-P (not shown). The spectra for all three of the peptides exhibited prominent ellipticity minima at 208 nm and 222 nm and a positive transition at ≤200 nm, indicating that the peptides exist in solution with a structure comprising significant α-helical content. In addition, the ellipticity minima at 208 nm and 222 nm were of similar magnitude, suggesting that the peptides formed a coiled coil (20).
FIG. 3.
Circular dichroism spectrograph of the antigen I/II peptide AR-B. Circular dichroism spectra of AR-B (100 μg/ml) were recorded from 300 nm to 200 nm on an Aviv 60DS circular dichroism spectrophotometer at 37°C with a scan rate of 1 nm/s. The contents of α-helix, β-sheet, and random coil were deduced from the spectrograph data with the K2d program (1).
The distribution of α-helix, β-sheet, and random coil as determined from the circular dichroism data with the K2d program (1, 38) is shown in Table 2. At pH 7.5, the deduced secondary structures of the three peptides were very similar; each of the peptides comprised approximately 60% α-helix, 7% β-sheet, and 33% random structure. Relatively little change in the overall content of α-helix and β-sheet was observed in the SspB and SpaP A region peptides when circular dichroism spectra were recorded at pH 5.5. The α-helical content of the SspA peptide increased to 65% at pH 5.5 (Table 2 and Fig. 4). However, under basic conditions (pH 8.5), the AR-B peptide exhibited a loss of α-helical content and a concomitant increase in β-sheet structure (from 9 to 19%). In contrast, the α-helix content dropped only slightly for AR-A and AR-P at pH 8.5 but remained at levels that were similar to those observed for AR-B at pH 7.5. These results suggest that the structures of AR-A and AR-P are relatively stable from pH 5.5 to pH 8.5, whereas AR-B appears to be less stable at pH 8.5 and undergoes a conformational shift resulting in loss of α-helix.
TABLE 2.
Deduced secondary structures of AR peptidesa
| Peptide | Structure | % of molecule at:
|
||
|---|---|---|---|---|
| pH 5.5 | pH 7.5 | pH 8.5 | ||
| AR-A | α-Helix | 65 | 61 | 58 |
| β-Sheet | 5 | 7 | 8 | |
| Random | 30 | 32 | 33 | |
| AR-B | α-Helix | 58 | 57 | 46 |
| β-Sheet | 8 | 9 | 19 | |
| Random | 33 | 34 | 35 | |
| AR-P | α-Helix | 61 | 60 | 57 |
| β-Sheet | 7 | 7 | 9 | |
| Random | 32 | 33 | 34 | |
Data were deduced with the K2d program (1) and are expressed as the percentage of the molecule that assumed the indicated structure. Values have been rounded off to the nearest integer.
FIG. 4.
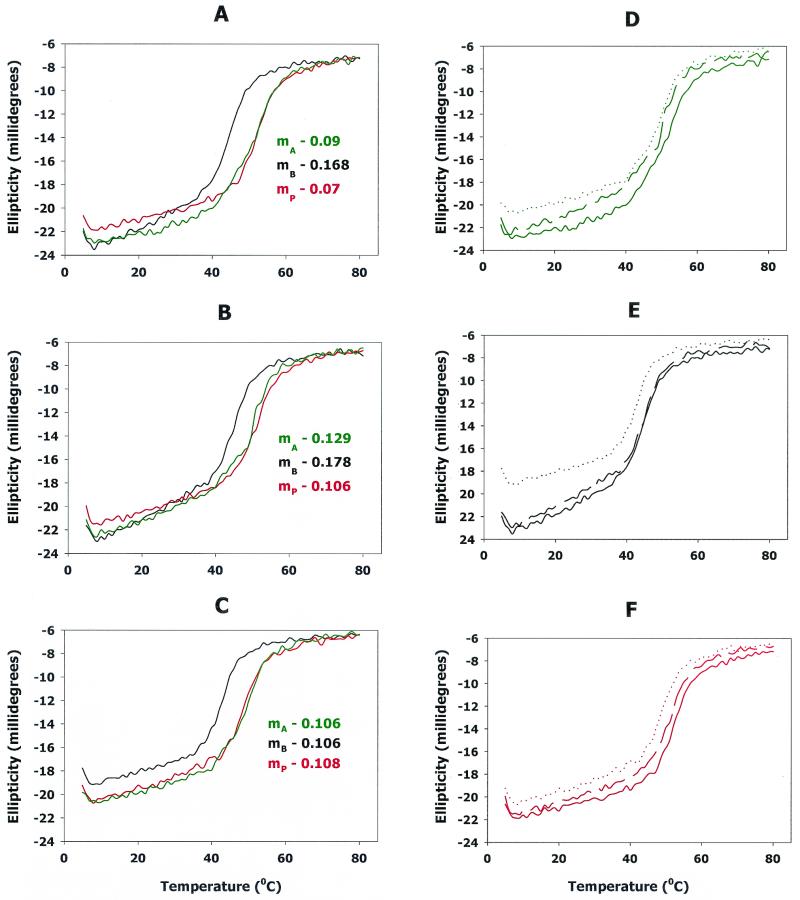
Effects of temperature (A to C) and pH (D to F) on the stability of AR peptides. To compare the stability of the AR-A (green), AR-B (black), and AR-P (red) peptides as a function of temperature, the ellipticity at 222 nm was determined from 5°C to 80°C in buffers of pH 5.5 (A), pH 7.5 (B), and pH 8.5 (C). Data were acquired with a temperature interval of 1°C. The slope for each plot was determined by linear regression of a data set encompassing the temperature points from 15°C to 37°C for each peptide. The calculated slopes are shown in panels A to C. A positive slope represents an increase in ellipticity at 222 nm, indicating a decrease in the α-helix content. To more clearly represent the influence of pH on the stability of the peptides, the melt curves for AR-A (D), AR-B (E), and AR-P (F) in each buffer were plotted separately. In each panel, the solid lines represent pH 5.5, the dashed lines represent pH 7.5, and the dotted lines represent pH 8.5.
To further compare the stability of the three A region peptides, a series of circular dichroism spectra were collected at 222 nm to follow α-helical content as a function of temperature. These experiments were carried out in the three buffers used above, at pH 5.5, pH 7.5, and pH 8.5. Interestingly, under all of the conditions that were tested, the AR-B peptide denatured (as indicated by the rapid linear increase in ellipticity at 222 nm) at a temperature that was 7 to 9°C lower than that required to denature AR-A and AR-P, as shown in Fig. 4A to C. Furthermore, between 15°C and 37°C at pH 5.5 and pH 7.5, AR-B exhibited a more rapid loss of α-helical structure than did AR-A or AR-P (compare the slopes of the plots within this range of temperatures for each of the peptides in Fig. 4A and 4B).
However, although AR-B possessed less α-helical structure than AR-A and AR-P at pH 8.5 (see Fig. 4C and Table 2), the rate of loss of α-helix was similar for all three of the peptides as the temperature increased from 15°C to 37°C (compare the slopes in Fig. 4C). The effect of pH on the individual A region peptides is more clearly shown in Fig. 4D, 4E, and 4F. Each of the peptides exhibited the highest α-helix content under acidic conditions and the least α-helical structure at pH 8.5, the loss of α-helix being greatest for AR-B (Fig. 4E). Together, these data suggest that the structure and stability of the A region of SspB differ from those of the corresponding A regions of SspA and SpaP.
Higher-order structure of AR peptides.
Little is known about the three-dimensional structure of streptococcal antigen I/II proteins. The A region of antigen I/II possesses long segments of sequence that exhibit a seven-residue periodicity of hydrophobic and hydrophilic amino acids. As a result, predictive programs (e.g., PAIRCOIL) rank the A region and the three AR peptides as having a high probability of forming an intermolecular dimeric α-helical coiled coil. To determine if the AR peptides exist as coiled-coil multimers in solution, we first examined the elution of the peptides from Superdex 75 and compared their migration to that of known protein standards.
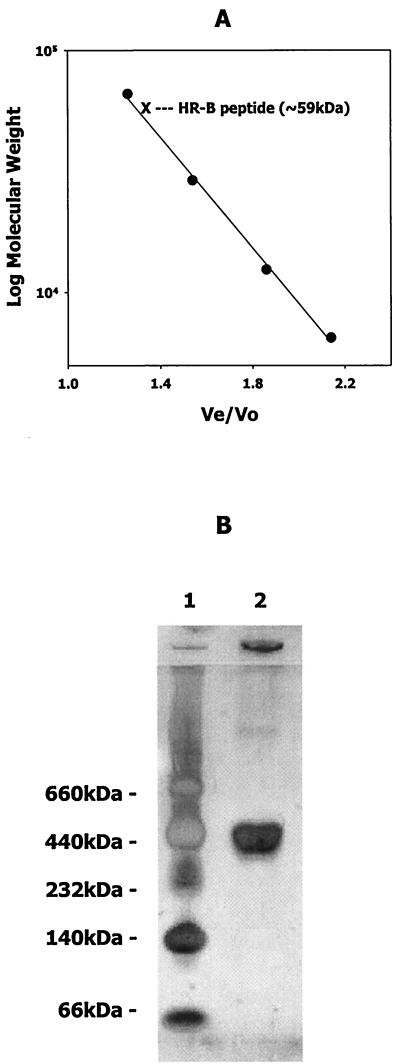
As shown in Fig. 5A, AR-B consistently eluted near the bovine serum albumin standard and exhibited an apparent molecular mass in solution of approximately 59 kDa. Similar elution profiles were also observed for AR-A and AR-P (data not shown). The molecular masses of the AR peptides were approximately 29 kDa, as calculated from the deduced amino acid sequence encoded by the cloned DNA fragments in pGEX-6P2. Furthermore, the full-length SspB, SspA, and SpaP proteins (predicted sizes of 165 to 170 kDa) eluted from Sepharose 6B columns prior to α-amylase (200 kDa) and exhibited apparent native molecular masses of between 300 kDa and 500 kDa (not shown). Consistent with this, purified full-length SspB protein migrated with an apparent molecular mass of approximately 370 kDa in native polyacrylamide gel electrophoresis, as shown in Fig. 5B. Thus, the apparent molecular masses of the A region polypeptides and the full-length antigen I/II proteins are 2- to 2.5-fold higher than the molecular mass predicted from their deduced amino acid sequences. This suggests that they exist as dimers in solution or, alternatively, assume a nonglobular monomeric conformation that results in the anomalous migration observed during electrophoresis and chromatography.
FIG. 5.
(A) Elution profile of standard proteins and AR-B peptide from Superdex 75. Purified AR-B was chromatographed on Superdex 75 (1.6 by 40 cm) as described in Materials and Methods and compared to the elution of the standard proteins bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), cytochrome c (12.4 kDa), and aprotinin (6.4 kDa). (B) Native gel electrophoresis of S. gordonii SspB protein. Purified SspB protein (15 μg) was electrophoresed in a 4 to 15% gradient PhastGel with native gel buffer strips. Native size standards were thyroglobulin (660 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (66 kDa).
To distinguish between these possibilities, the peptides were analyzed by sedimentation equilibrium ultracentrifugation. The results from these analyses are summarized in Table 3. In all experiments, the average molecular mass calculated for the A region polypeptides was approximately 31 kDa. Since the predicted molecular mass of the A region polypeptides is 29 kDa, the equilibrium centrifugation results best fit the sedimentation of a monomeric peptide species exhibiting a molecular mass of approximately 31 kDa. This suggests that under the experimental conditions used, the A region peptides may form monomeric intramolecular coiled-coil structures which may be nonglobular.
TABLE 3.
Sedimentation equilibrium ultracentrifugation of AR peptides
| Peptide | Mass, kDa (spin speed, krpm) | Avg mass (kDa) |
|---|---|---|
| AR-A | 30 (20) | 34 |
| 32 (24) | ||
| 36 (28) | ||
| 36.5 (32) | ||
| AR-B | 34.5 (20) | 32.5 |
| 35.5 (24) | ||
| 30.5 (28) | ||
| 29.5 (32) | ||
| AR-P | 34 (20) | 31 |
| 32 (24) | ||
| 30 (28) | ||
| 27 (32) |
DISCUSSION
The antigen I/II family of streptococcal cell surface polypeptides comprises highly conserved multifunctional adhesins that mediate interactions of oral streptococci with salivary glycoproteins, e.g., gp340 (10, 12, 39, 40), collagen, and other matrix proteins (35, 48), and various oral bacteria (7, 15). However, despite the high degree of sequence and structural conservation that exists among these proteins, the individual members of the antigen I/II family of polypeptides are functionally distinct (14, 45). Furthermore, while many of the functional domains that have been mapped in antigen I/II polypeptides reside in highly conserved regions of the proteins, several recent studies have suggested that the variable residues within these conserved regions define antigen I/II function (15, 28).
Our studies illustrate another example of differential function in the antigen I/II family of proteins and show that the alanine-rich region (A region) of the two antigen I/II proteins expressed by S. gordonii (SspA and SspB) differ in their interaction with salivary gp340 and exhibit different structural properties. A recombinant peptide encompassing the A region of SspB did not interact with purified gp340, nor did it competitively inhibit the interaction of gp340 with intact S. gordonii cells. In contrast, the corresponding A region peptides from SspA and from SpaP of S. mutans were effective inhibitors of gp340-mediated streptococcal aggregation. These results suggest that the interaction of SspB with gp340 does not require the A region repeats and may be fundamentally different from that of SspA/SpaP. Thus, SspB may possess only the C-terminal gp340-interactive sequences that are analogous to the Ad1/Ad2 region identified in SpaP by Munro et al. (40).
One possible outcome of this is that SspB may not interact with gp340 as effectively as SspA and SpaP, which contain both the A region and C-terminal binding domains. Consistent with this, the inactivation of sspA in S. gordonii DL1 results in a disproportionately large loss of gp340 binding activity in mutant cells (13). Furthermore, Holmes et al. (22) have shown that recombinant Lactococcus lactis cells expressing SspA bind more efficiently to immobilized gp340 than do recombinant cells expressing equivalent levels of SspB. Studies to directly compare the kinetics of the SspA and SspB interaction with gp340 are currently under way.
We cannot exclude the possibility that the inability of the SspB peptide to interact with gp340 arises from improper folding of the recombinant A region peptide. However, we believe that this is unlikely, since each of the three peptides was expressed and isolated under identical conditions. The SspA and SpaP peptides obtained by these methods were active. Furthermore, the activity of our SpaP peptide was consistent with the gp340 binding activity of A region peptides derived from other S. mutans strains, as reported by Crowley et al. (10) and Hajishengallis et al. (21). Therefore, we conclude that the A region of SspB does not contribute to the interaction of intact SspB protein with salivary gp340.
Little is known about the molecular mechanism of the A region interaction with gp340. Previous studies suggested that two discrete amino acid sequences in the A region of SpaP function as binding determinants for the salivary glycoprotein (39, 51). One sequence, TELARVQKANADAKAAY, competitively inhibited the adherence of S. mutans cells to immobilized gp340 (39) and is present in two of the three A region repeats of SpaP (24, 39). The second putative gp340 binding sequence identified in SpaP, TYEAALKQYEADL, is present only in single copy (51). Monoclonal antibodies that react with this sequence (designated KH5 and SH2) inhibit the adherence of streptococci to immobilized salivary proteins and block S. mutans colonization of rat teeth (50).
However, sequences that are identical and/or highly related to the putative gp340 binding determinants above also exist in the corresponding A region repeats of SspA and SspB. Where differences in the primary sequences of SspA and SspB exist, they are usually conservative amino acid substitutions (24). Thus, while we cannot rule out the possibility that variation in the primary sequence of the gp340 binding region may render the A region of SspB inactive, the structural basis by which this might occur is not clear. An alternative explanation is that the interaction of the A region with gp340 is more complex and may involve higher-order structures in the A region of antigen I/II. Indeed, each of the putative gp340 binding sequences above exhibits the heptad periodicity of hydrophobic residues that is characteristic of an α-helical coiled-coil structure. Furthermore, the core B-cell epitopes of the neutralizing antibodies KH5 and SH2 have been reported to be xYxxxLxxYxxxx and xYxxxxxxYxxxx, respectively (50). These epitopes exhibit the same seven-residue periodicity, suggesting that the monoclonal antibodies react with an α-helical or coiled-coil determinant of antigen I/II.
In silico analysis of A region sequences from streptococcal antigen I/II predicts that this region is highly α-helical (14, 26, 31). To our knowledge, the circular dichroism data in this report provide some of the first experimental evidence that supports these predictions. Circular dichroism spectra obtained for each of the A region peptides exhibited large molar ellipticity minima at 222 nm and 208 nm and a positive transition at <200 nm. These traits are characteristic of α-helical structures. Furthermore, the close equivalence of the minima at 208 nm and 222 nm is consistent with circular dichroism spectra obtained for coiled coils (9, 20, 54) and differs from the smaller negative ellipticity at 222 nm that is observed for single-stranded α-helices in peptides and proteins (9). Together, these results suggest that the A region of antigen I/II has significant α-helical coiled-coil structure.
At physiological pH, our data suggest that the A region peptides from SspA, SspB, and SpaP are approximately 60% α-helix, 30 to 35% random (or undetermined) structure, and 5 to 10% β-sheet. Relatively little change in these values was observed when the SpaP and SspA peptides were analyzed under acidic or slightly basic conditions. In contrast, the SspB peptide lost significant α-helical structure at pH 8.5 (Table 2 and Fig. 4C and 4E). This change does not correlate with the isoelectric points of SspA and SspB, since the predicted isoelectric points for AR-A and AR-B are similar, 5.7 and 6.02, respectively. Furthermore, the loss of α-helical structure in SspB was also evident when the peptides were analyzed as a function of increasing temperature. The A region peptide of SspB exhibited a more rapid loss of α-helical structure from 10°C to 40°C and underwent gross denaturation at a temperature 7 to 9°C below that for the SspA and SpaP peptides. These results suggest that the A region α-helical coiled coil of SspB is less stable than the corresponding regions of SspA and SpaP and may more readily undergo structural changes in response to environmental changes.
Very little is known about the structure of antigen I/II proteins in vivo. However, our analysis of the sequences of the A regions with PAIRCOIL (3) predicts that they may exist as a coiled-coil dimer. Consistent with this, the intact SspB protein and all three of the A region polypeptides migrated under nondenaturing conditions with apparent molecular masses that were two- to threefold greater than predicted from their amino acid sequences. In contrast, analytical ultracentrifugation data indicated that the A region peptides sedimented as monomers, suggesting that they exist as intracellular coiled coils under the experimental conditions used.
Their anomalous behavior in gel electrophoresis and column chromatography can be explained if the peptides form nonglobular structures. Sedimentation velocity centrifugation experiments are currently under way to examine the shape of the A region peptides. However, these results do not exclude the possibility that antigen I/II multimers exist in vivo. Indeed, a nonheptad region of the M57 protein has been shown to be essential for the formation of coiled-coil dimers in Streptococcus pyogenes (29), and in the absence of this nonheptad region, the heptad repeat domain of M57 existed as a monomer. It is possible that the A region repeats of antigen I/II exhibit a similar requirement. Alternatively, other domains of antigen I/II may mediate subunit association independently of the A region. The proline-rich repeats (P region) have been suggested to mediate aggregation of antigen I/II (41), and deletion of this domain prevents secretion of the protein (4), suggesting that the P region may be important for the proper folding and assembly of antigen I/II. Obtaining the crystal structure of an antigen I/II polypeptide will begin to address these structural issues and will be invaluable for correlating antigen I/II structure and function.
In summary, we have shown that the functional properties of the A region of SspB differ from those of the related antigen I/II proteins SspA and SpaP. Structural analyses of recombinant A region peptides provided experimental evidence to suggest that these polypeptides are highly α-helical and may form a coiled coil. Furthermore, the A region of SspB appears to be structurally less stable than the A region polypeptides of SspA and SpaP, which may explain the functional differences observed.
Acknowledgments
This work was supported by Public Health Service grant DE-12750 from the National Institute of Dental and Craniofacial Research.
Editor: B. B. Finlay
REFERENCES
- 1.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism with an unsupervised neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Bikker, F. J., A. J. Ligtenberg, J. E. van der Wal, P. A. van den Keijbus, U. Holmskov, E. C. Veerman, and A. V. Nieuw Amerongen. 2002. Immunohistochemical detection of salivary agglutinin/gp340 in human parotid, submandibular, and labial salivary glands. J. Dent. Res. 81:134-139. [PubMed] [Google Scholar]
- 3.Bonnie, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. N. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun.. 66:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady, L. J., D. A. Piacentini, P. J. Crowley, and A. S. Bleiweis. 1991. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutans and cross-reactivity with related surface antigens of oral streptococci. Infect. Immun. 59:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady, L. J., D. A. Piacentini, P. J. Crowley, P. C. Oyston, and A. S. Bleiweis. 1992. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 60:1008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, W. O., D. R. Demuth, and R. J. Lamont. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68:6758-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, T. M., and R. W. Woody. 1990. The effect of conformation on the circular dichroism of interacting helices: a theoretical study of tropomyosin. Biopolymers 30:657-676. [DOI] [PubMed] [Google Scholar]
- 10.Crowley, P. J., L. J. Brady, D. A. Piacentini, and A. S. Bleiweis. 1993. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect. Immun. 61:1547-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley, P. J., W. Fischlschweiger, S. E. Coleman, and A. S. Bleiweis. 1987. Intergeneric bacterial coaggregations involving mutans streptococci and oral actinomyces. Infect. Immun. 55:2695-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demuth, D. R., C. A. Davis, A. M. Corner, R. J. Lamont, P. S. Leboy, and D. Malamud. 1988. Cloning and expression of a Streptococcus sanguis surface antigen that interacts with a human salivary agglutinin. Infect. Immun. 56:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 14.Demuth, D. R., E. E. Golub, and D. Malamud. 1990. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J. Biol. Chem. 265:7120-7126. [PubMed] [Google Scholar]
- 15.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demuth, D. R., M. S. Lammey, M. Huck, E. T. Lally, and D. Malamud. 1990. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb. Pathog. 9:199-211. [DOI] [PubMed] [Google Scholar]
- 17.Duan, Y., E. Fisher, D. Malamud, E. Golub, and D. R. Demuth. 1994. Calcium-binding properties of SSP-5, the Streptococcus gordonii M5 receptor for salivary agglutinin. Infect. Immun. 62:5220-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egland, P. G., L. D. Du, and P. E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Sabaeny, A., D. R. Demuth, and R. J. Lamont. 2001. Regulation of Streptococcus gordonii sspB by the sspA gene product. Infect. Immun. 69:6520-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graddis, T. J., D. G. Myszka, and I. M. Chaiken. 1993. Controlled formation of model homo- and heterodimer coiled coil polypeptides. Biochemistry 32:12664-12671. [DOI] [PubMed] [Google Scholar]
- 21.Hajishengallis, G., T. Koga, and M. W. Russell. 1994. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J. Dent. Res. 73:1493-1502. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, A. R., C. Gilbert, J. M. Wells, and H. F. Jenkinson. 1998. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, A. R., R. McNab, and H. F. Jenkinson. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 25.Jenkinson, H. F., S. D. Terry, R. McNab, and G. W. Tannock. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly, C., P. Evans, J. K. Ma, L. A. Bergmeier, W. Taylor, L. J. Brady, S. F. Lee, A. S. Bleiweis, and T. Lehner. 1990. Sequencing and characterization of the 185 kDa cell surface antigen of Streptococcus mutans. Arch. Oral Biol. 35(Suppl.):33S-38S. [DOI] [PubMed] [Google Scholar]
- 27.Kelly, C. G., S. Todryk, H. L. Kendal, G. H. Munro, and T. Lehner. 1995. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect. Immun. 63:3649-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly, C. G., J. S. Younson, B. Y. Hikmat, S. M. Todryk, M. Czisch, P. I. Haris, I. R. Flindall, C. Newby, A. I. Mallet, J. K. Ma, and T. Lehner. 1999. A synthetic peptide adhesion epitope as a novel antimicrobial agent. Nat. Biotechnol. 17:42-47. [DOI] [PubMed] [Google Scholar]
- 29.Khandke, K. M., T. Fairwell, E. H. Braswell, and B. N. Manjula. 1991. The amino terminal region of group A streptococcal M protein determines its molecular state of assembly and function. J. Protein Chem. 10:49-59. [DOI] [PubMed] [Google Scholar]
- 30.Lamont, R. J., D. R. Demuth, C. A. Davis, D. Malamud, and B. Rosan. 1991. Salivary-agglutinin-mediated adherence of Streptococcus mutans to early plaque bacteria. Infect. Immun. 59:3446-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaPolla, R. J., J. A. Haron, C. G. Kelly, W. R. Taylor, C. Bohart, M. Hendricks, J. P. Pyati, R. T. Graff, J. K. Ma, and T. Lehner. 1991. Sequence and structural analysis of surface protein antigen I/II (SpaA) of Streptococcus sobrinus. Infect. Immun. 59:2677-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, S. F., A. Progulske-Fox, and A. S. Bleiweis. 1988. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect. Immun. 56:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ligtenberg, A. J., E. C. Veerman, and A. V. Nieuw Amerongen. 2000. A role for Lewis a antigens on salivary agglutinin in binding to Streptococcus mutans. Antonie van Leeuwenhoek 77:21-30. [DOI] [PubMed] [Google Scholar]
- 34.Ligtenberg, T. J., F. J. Bikker, J. Groenink, I. Tornoe, R. Leth-Larsen, E. C. Veerman, A. V. Nieuw Amerongen, and U. Holmskov. 2001. Human salivary agglutinin binds to lung surfactant protein-D and is identical with scavenger receptor protein gp340. Biochem. J. 359:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love, R. M., M. D. McMillan, and H. F. Jenkinson. 1997. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect. Immun. 65:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love, R. M., M. D. McMillan, Y. Park, and H. F. Jenkinson. 2000. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon binding specificity of streptococcal antigen I/II adhesin. Infect. Immun. 68:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, J. K., C. G. Kelly, G. Munro, R. A. Whiley, and T. Lehner. 1991. Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect. Immun. 59:2686-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merelo, J. J., M. A. Andrade, A. Prieto, and F. Moran. 1994. Proteinotopic feature maps. Neurocomputing 6:443-454.
- 39.Moisset, A., N. Schatz, Y. Lepoivre, S. Amadio, D. Wachsmann, M. Scholler, and J. P. Klein. 1994. Conservation of salivary glycoprotein-interacting and human immunoglobulin G-cross-reactive domains of antigen I/II in oral streptococci. Infect. Immun. 62:184-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munro, G. H., P. Evans, S. Todryk, P. Buckett, C. G. Kelly, and T. Lehner. 1993. A protein fragment of streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect. Immun. 61:4590-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai, M., N. Okahashi, H. Ohta, and T. Koga. 1993. Saliva-binding region of Streptococcus mutans surface protein antigen. Infect. Immun. 61:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogier, J. A., M. Scholler, Y. Leproivre, A. Pini, P. Sommer, and J. P. Klein. 1990. Complete nucleotide sequence of the sr gene from Streptococcus mutans OMZ 175. FEMS Microbiol. Lett. 56:223-227. [DOI] [PubMed] [Google Scholar]
- 43.Ogier, J. A., D. Wachsmann, M. Scholler, Y. Lepoivre, and J. P. Klein. 1990. Molecular characterization of the gene sr of the saliva interacting protein from Streptococcus mutans OMZ175. Arch. Oral Biol. 35(Suppl.):25S-31S. [DOI] [PubMed] [Google Scholar]
- 44.Okahashi, N., C. Sasakawa, M. Yoshikawa, S. Hamada, and T. Koga. 1989. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol. Microbiol. 3:673-678. [DOI] [PubMed] [Google Scholar]
- 45.Petersen, F. C., S. Assev, H. C. van der Mei, H. J. Busscher, and A. A. Scheie. 2002. Functional variation of the antigen I/II surface protein in Streptococcus mutans and Streptococcus intermedius. Infect. Immun. 70:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen, F. C., S. Pasco, J. Ogier, J. P. Klein, S. Assev, and A. A. Scheie. 2001. Expression and functional properties of the Streptococcus intermedius surface protein antigen I/II. Infect. Immun. 69:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakobphol, A., F. Xu, V. M. Hoang, T. Larsson, J. Bergstrom, I. Johansson, L. Frangsmyr, U. Holmskov, H. Leffler, C. Nilsson, T. Boren, J. R. Wright, N. Stromberg, and S. J. Fisher. 2000. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp340. J. Biol. Chem. 275:39860-39866. [DOI] [PubMed] [Google Scholar]
- 48.Sciotti, M. A., I. Yamodo, J. P. Klein, and J. A. Ogier. 1997. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol. Lett. 153:439-445. [DOI] [PubMed] [Google Scholar]
- 49.Senpuku, H., T. Iizima, T. Koga, and T. Nisizawa. 1996. Identification of human antigenic epitopes in an alanine-rich repeating region with sera from hu-PBL-SCID mice immunized with a surface protein antigen of Streptococcus mutans. Oral Microbiol. Immunol. 11:343-349. [DOI] [PubMed] [Google Scholar]
- 50.Senpuku, H., K. Matin, S. M. Abdus, I. Kurauchi, S. Sakurai, M. Kawashima, T. Murata, H. Miyazaki, and N. Hanada. 2001. Inhibitory effects of MoAbs against a surface protein antigen in real-time adherence in vitro and recolonization in vivo of Streptococcus mutans. Scand J. Immunol. 54:109-116. [DOI] [PubMed] [Google Scholar]
- 51.Senpuku, H., T. Miyauchi, N. Hanada, and T. Nisizawa. 1995. An antigenic peptide inducing cross-reacting antibodies inhibiting the interaction of Streptococcus mutans PAc with human salivary components. Infect. Immun. 63:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todryk, S. M., C. G. Kelly, G. H. Munro, and T. Lehner. 1996. Induction of immune responses to functional determinants of a cell surface streptococcal antigen. Immunology 87:55-63. [PMC free article] [PubMed] [Google Scholar]
- 53.Tokuda, M., N. Okahashi, I. Takahashi, M. Nakai, S. Nagaoka, M. Kawagoe, and T. Koga. 1991. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infect. Immun. 59:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, N. E., C. M. Kay, and R. S. Hodges. 1992. Synthetic model proteins: the relative contribution of leucine residues at the nonequivalent positions of the 3-4 hydrophobic repeat to the stability of the two-stranded alpha-helical coiled-coil. Biochemistry 31:5739-5746. [DOI] [PubMed] [Google Scholar]