Abstract
We address theoretically aggregation of DNA segments by multivalent polyamines such as spermine and spermidine. In experiments, the aggregation occurs above a certain threshold concentration of multivalent ions. We demonstrate that the dependence of this threshold on the concentration of DNA has a simple form. When the DNA concentration cDNA is smaller than the monovalent salt concentration, the threshold multivalent ion concentration depends linearly on cDNA, having the form αcDNA + β. The coefficients α and β are related to the density profile of multivalent counterions around isolated DNA chains, at the onset of their aggregation. This analysis agrees extremely well with recent detailed measurements on DNA aggregation in the presence of spermine. From the fit to the experimental data, the number of condensed multivalent counterions per DNA chain can be deduced. A few other conclusions can then be reached: 1), the number of condensed spermine ions at the onset of aggregation decreases with the addition of monovalent salt; 2), the Poisson-Boltzmann theory overestimates the number of condensed multivalent ions at high monovalent salt concentrations; and 3), our analysis of the data indicates that the DNA charge is not overcompensated by spermine at the onset of aggregation.
INTRODUCTION
Condensation and aggregation of DNA, induced by multivalent counterions, have been extensively studied in the past two decades (for a review, see Bloomfield et al., 2000, and references therein). The term condensation usually refers to the collapse of a single, long DNA chain. Condensation plays an important role in storage and packing of DNA; for example, in viral capsids (Gelbart et al., 2000). Aggregation of DNA is a closely related phenomenon, where multiple chains attract each other and form a variety of condensed mesophases of complex structure (Pelta et al., 1996a,b). In both phenomena multivalent counterions play a crucial role, screening the electrostatic repulsion between charged strands of DNA and mediating an effective attraction.
A variety of tri- and tetravalent ions can induce aggregation and condensation, among them the polyamines spermidine (3+) and spermine (4+) (Chattoraj et al., 1978; Gosule and Schellman, 1978; Tabor and Tabor, 1984), as well as cobalt-hexamine (Widom and Baldwin, 1980, 1983). In typical experiments on aggregation (Pelta et al., 1996b; Raspaud et al., 1998; Saminathan et al., 1999) multivalent ions are gradually added to a solution with fixed concentration of DNA segments and monovalent salt. Two such examples for spermine and spermidine are reproduced in Fig. 1 (Pelta et al., 1996b). As the multivalent ion concentration is raised above a certain threshold, DNA segments begin to aggregate, and precipitate from the solution. Above the aggregation threshold, the DNA concentration decreases gradually or abruptly, depending on various parameters such as the monovalent salt concentration and total DNA concentration. Further addition of multivalent ions at higher concentrations reverses the aggregation. Above a second, redissolution threshold, all the DNA is redissolved in the solution (Fig. 1). The redissolution threshold (above which all the DNA redissolves) is almost independent on the DNA concentration. Its value can be attributed to screening of electrostatic interactions by multivalent ions (Raspaud et al., 1998).
FIGURE 1.
Percent of solubilized DNA, as function of polyamine concentration. Squares, spermine; circles, spermidine. Solid and dashed lines are guides for the eye. DNA and NaCl concentrations are 3 mM and 25 mM, respectively. Below the aggregation threshold, caggr, and above the redissolution threshold, credissol, all the DNA is dissolved. The data is adapted from Pelta et al. (1996b).
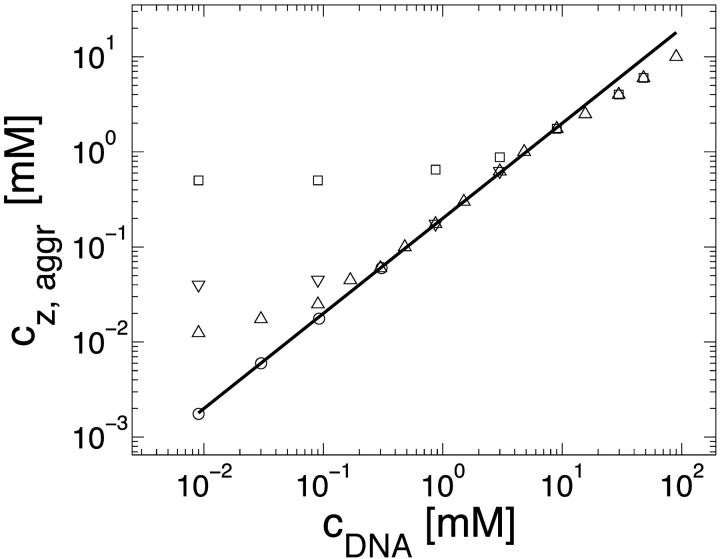
The aggregation threshold, where the onset of aggregation occurs, is the main experimental phenomenon addressed in our theoretical article. The multivalent ion concentration at the onset depends strongly on the monovalent salt and DNA concentrations. This dependence has been recently measured in detail for short (150 basepair) DNA segments in presence of spermine (Raspaud et al., 1998), and is reproduced in Fig. 2. The figure shows measurements of spermine concentrations at the onset of aggregation, for DNA concentrations ranging over four orders of magnitude and for four different monovalent salt concentrations: 2, 13, 23, and 88 mM. At very low DNA concentration, the spermine concentration depends strongly on the monovalent salt concentration. At higher DNA concentration it has only a weak dependence on the monovalent ion concentration but the spermine concentration is proportional to the DNA concentration, indicating that a certain number of spermine counterions are required, per DNA base, to induce aggregation. The solid line in Fig. 2, adapted from Raspaud et al. (1998), corresponds to a ratio: cz,aggr/cDNA = 0.20, where cz,aggr is the spermine concentration at the aggregation onset and cDNA is the DNA concentration. This linear relation fits a large number of the experimental points in the intermediate DNA concentration range. It has been suggested by Raspaud et al. (1998, 1999) that the deviations from this line, at low and high DNA concentrations, represent two distinct physical regimes that need to be analyzed separately from the intermediate regime, where the linear fit works well.
FIGURE 2.
Spermine concentration, cz,aggr, at the onset aggregation, as a function of DNA monomer concentration cDNA. Data is shown for four monovalent salt concentrations: 2 mM (○); 13 mM (Δ); 23 mM (∇); and 88 mM (□). Solid line corresponds to the fixed ratio of cz,aggr/cDNA = 0.20. The data is adapted from Raspaud et al. (1998).
In this work we focus on the onset of aggregation, and specifically on its dependence on the DNA concentration. We show that this dependence is simple for all the range of DNA concentration. Furthermore, for cDNA smaller than the monovalent salt concentration we show that this dependence is linear: cz,aggr = αcDNA + β. The coefficient β is the multivalent counterion concentration far away from the DNA chains, whereas α accounts for the excess of multivalent ions around each chain. These quantities can be extracted, e.g., from the four experimental curves of Fig. 2. Several further conclusions are then drawn on the onset of DNA aggregation and on the counterion distribution around each double-stranded DNA.
THEORETICAL CONSIDERATIONS
Consider an aqueous solution containing monovalent (1:1) salt, multivalent (z:1) salt, and DNA segments below their threshold for aggregation. Throughout this article, the DNA solution is assumed to be dilute enough such that the DNA segments do not overlap. We also assume that these DNA segments can be regarded as rigid rods. The concentrations of added monovalent salt, multivalent salt, and DNA monomers are denoted by cs, cz, and cDNA, respectively. These are the solute concentrations per unit volume as controlled and adjusted in experiments. We will assume that the monovalent and multivalent salts have the same type of co-ion, so that altogether there are three ion species in the solution:
A multivalent counterion contributed from the z:1 multivalent salt, of concentration cz.
A monovalent counterion contributed by monovalent salt of concentration cs, and by counterions dissociated from the DNA, of concentration cDNA: in total cDNA + cs.
Co-ions coming from both z:1 and 1:1 salts, of concentration cs + zcz.
Each DNA segment attracts a layer of oppositely charged counterions referred to as the condensed counterions. As long as the typical distance between segments is large compared to the electrostatic screening length κ−1, the electrostatic potential decays exponentially to zero far away from the DNA segments. In turn, the concentrations of the three ion species decay to well-defined bulk values denoted by 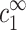 for the monovalent ions and
for the monovalent ions and 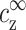 for those that are z-valent. These concentrations should be distinguished from the concentrations cs and cz introduced above, which are the average concentrations of added salts regulated experimentally.
for those that are z-valent. These concentrations should be distinguished from the concentrations cs and cz introduced above, which are the average concentrations of added salts regulated experimentally.
The Debye screening length, κ−1, characterizing the exponential decay of the electrostatic potential, is determined by the bulk concentrations of all three ionic species:
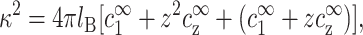 |
(1) |
where the third term is the co-ion concentration. It is equal to 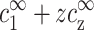 due to charge neutrality far from the DNA where the potential decays to zero. The above equation makes use of the Bjerrum length, lB = e2/(ɛkBT), equal to ∼7 Å in aqueous solution at room temperature. kBT is the thermal energy, e is the electron charge, and ɛ = 80 is the dielectric constant of water. The Debye length as well as
due to charge neutrality far from the DNA where the potential decays to zero. The above equation makes use of the Bjerrum length, lB = e2/(ɛkBT), equal to ∼7 Å in aqueous solution at room temperature. kBT is the thermal energy, e is the electron charge, and ɛ = 80 is the dielectric constant of water. The Debye length as well as 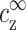 are shown schematically in Fig. 3. Other quantities that will be defined below are also indicated in this figure.
are shown schematically in Fig. 3. Other quantities that will be defined below are also indicated in this figure.
FIGURE 3.
Schematic representation of the multivalent density profile, nz(r), between two neighboring DNA segments, each modeled as a cylinder of radius d. Here r is the distance from the axis of the left DNA strand. The radius r = R corresponds to the interstrand mid-distance and is the unit cell radius. The density decays to its bulk value 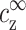 on distances larger than κ−1, where κ−1 is the Debye length defined in Eq. 1. The excess density of multivalent ions, ρz, is indicated by the shaded areas.
on distances larger than κ−1, where κ−1 is the Debye length defined in Eq. 1. The excess density of multivalent ions, ρz, is indicated by the shaded areas.
In dilute solutions different DNA segments do not overlap. Following previous works, we introduce a cell model, also shown schematically in Fig. 3. Note that the model serves to illustrate the subsequent derivations but is not essential for the validity of our main results. In the cell model, each segment, of a cylindrical cross-section, is at the center of a cylindrical cell of radius R and area A = πR2 such that
 |
(2) |
Namely, each DNA monomer occupies a specific volume 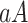 , where a ≃ 1.7 Å is the average charge separation on the chain taken hereafter as the monomer length.
, where a ≃ 1.7 Å is the average charge separation on the chain taken hereafter as the monomer length.
We will assume below that the DNA solution is dilute enough so that R is large compared to the Debye length κ−1. This assumption is essential for our derivation and can be verified for all the experimental data considered in this article. Density profiles of the three ion species are then practically identical to those near an isolated DNA segment with the same bulk concentrations 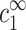 ,
, 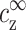 . In other words, the profiles are determined uniquely by
. In other words, the profiles are determined uniquely by  and
and 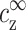 , with practically no dependence (or, more precisely, an exponentially small dependence) on the DNA monomer concentration. A demonstration of this claim is presented in Fig. 4, using the Poisson-Boltzmann theory in a cell model. For two very different values of R corresponding to different cDNA, the counterion profiles match perfectly when the values of
, with practically no dependence (or, more precisely, an exponentially small dependence) on the DNA monomer concentration. A demonstration of this claim is presented in Fig. 4, using the Poisson-Boltzmann theory in a cell model. For two very different values of R corresponding to different cDNA, the counterion profiles match perfectly when the values of  and
and  are the same. Note that the average concentrations of added salts, cs and cz, have different values in the two cells because of the contribution of condensed ions.
are the same. Note that the average concentrations of added salts, cs and cz, have different values in the two cells because of the contribution of condensed ions.
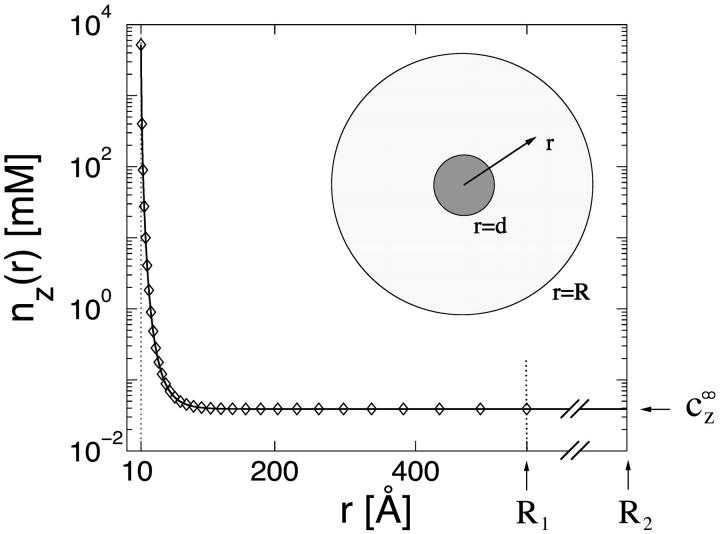
FIGURE 4.
Density profile nz(r) of 4-valent ions as function of r, the distance from the DNA axis, on a semilog plot, calculated using the Poisson-Boltzmann equation in a cell model, where the DNA segment is modeled as a uniformly charged cylinder. The cell model is shown schematically in the inset. Two cell sizes are shown, with outer radii R1 = 560 Å (cDNA = 1 mM) and R2 = 1.8 × 104 Å (cDNA = 10−3 mM), indicated by arrows. In both cases, the radius of closest approach of ions to the charged chain is at r = d, where d = 10 Å, as indicated by a dotted vertical line. The boundary condition at the inner cylinder matches the linear charge density of DNA (1e/1.7 Å). The bulk densities of monovalent and multivalent ions, 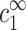 and
and  , are chosen to be the same in the two cells, leading to practically identical density profiles. The solid line represents the larger cell (R2), and diamonds are used for the smaller cell (R1). Density profiles of monovalent counterions and co-ions are not shown but are also practically identical in the two cells. Average salt concentrations are cs = 22 mM and cz = 0.21 mM in the smaller cell, and cs = 23 mM, cz = 0.039 mM in the larger cell. Bulk concentrations are
, are chosen to be the same in the two cells, leading to practically identical density profiles. The solid line represents the larger cell (R2), and diamonds are used for the smaller cell (R1). Density profiles of monovalent counterions and co-ions are not shown but are also practically identical in the two cells. Average salt concentrations are cs = 22 mM and cz = 0.21 mM in the smaller cell, and cs = 23 mM, cz = 0.039 mM in the larger cell. Bulk concentrations are 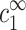 = 23 mM and
= 23 mM and  0.039 mM. Note that these bulk concentrations are practically identical to the salt concentrations in the larger cell. Note also that
0.039 mM. Note that these bulk concentrations are practically identical to the salt concentrations in the larger cell. Note also that  in the smaller cell, reflecting the contribution of the counterions released by the DNA.
in the smaller cell, reflecting the contribution of the counterions released by the DNA.
The total number of z-valent counterions, per cell unit length, is given by:
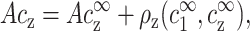 |
(3) |
where ρz is the excess number of z-valent ions per unit length near the DNA. Throughout the article we use the symbol c to denote concentrations per unit volume and ρ for concentrations per DNA unit length. The excess ρz can be evaluated in the limit of infinite cell radius, corresponding to an isolated chain,
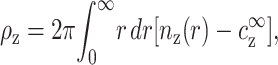 |
(4) |
where nz(r) is the z-valent local counterion concentration at distance r from the axis of symmetry, and nz(∞) = 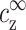 . Following the discussion in the previous paragraph, the excess ρz is determined uniquely by
. Following the discussion in the previous paragraph, the excess ρz is determined uniquely by 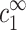 and
and 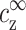 . Its exact functional dependence on these variables is generally not known, although it can be evaluated approximately, e.g., using the Poisson-Boltzmann equation or in computer simulations.
. Its exact functional dependence on these variables is generally not known, although it can be evaluated approximately, e.g., using the Poisson-Boltzmann equation or in computer simulations.
For monovalent counterions we have, in a similar fashion,
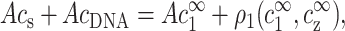 |
(5) |
where ρ1, the excess of monovalent counterions per unit length, is defined as in Eq. 4, and AcDNA = 1/a is the DNA charge density per unit length. The extra term in the left-hand side of Eq. 5 originates from monovalent counterions contributed by the DNA monomers. Using Eq. 2 we can rewrite Eqs. 3 and 5 as
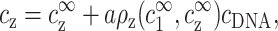 |
(6) |
and
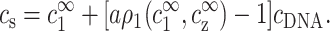 |
(7) |
These two equations relate the experimentally adjustable cs, cz, and cDNA to the bulk densities 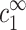 and
and 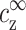 , that, in turn, are important because they determine the ion density profiles.
, that, in turn, are important because they determine the ion density profiles.
In the limit of infinite DNA dilution, cDNA = 0, and therefore  and
and 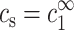 . At any finite DNA concentration, cz and cs are not equal to
. At any finite DNA concentration, cz and cs are not equal to 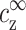 and
and  , respectively, because each segment captures some of the multivalent ions and releases a number of monovalent ones. Equations 6 and 7 express the correction to cs, cz at given
, respectively, because each segment captures some of the multivalent ions and releases a number of monovalent ones. Equations 6 and 7 express the correction to cs, cz at given 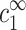 ,
, 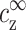 for both mono- and multivalent counterion species. The dimensionless quantities aρ1, aρz are the excess of the mono- and multivalent counterion species, respectively, per DNA monomer.
for both mono- and multivalent counterion species. The dimensionless quantities aρ1, aρz are the excess of the mono- and multivalent counterion species, respectively, per DNA monomer.
We would like to emphasize the generality of Eqs. 6 and 7. They do not depend on the assumption of parallel DNA residing in the middle of oriented cylindrical unit cells, or on any mean-field approximation for the distribution of counterions. The only assumption required to derive Eqs. 6 and 7 is that the average distance between DNA segments is large compared with the Debye length. Although Eqs. 6 and 7 are correct for any cs, cz, and cDNA below the onset of DNA aggregation, we will be interested below specifically in the aggregation onset.
Onset of aggregation
Our aim now is to find how the value of cz at the onset of aggregation, cz,aggr, depends on cDNA. We will assume that this aggregation onset depends on 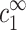 and
and 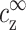 , but not on the average distance between DNA chains. We motivate this assumption by the fact that
, but not on the average distance between DNA chains. We motivate this assumption by the fact that  and
and 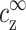 determine the density profile of multivalent counterions around the DNA chains, which, in turn, mediate the attraction necessary for aggregation. Before discussing this assumption in more detail, let us first consider its consequences. We can imagine an experiment where
determine the density profile of multivalent counterions around the DNA chains, which, in turn, mediate the attraction necessary for aggregation. Before discussing this assumption in more detail, let us first consider its consequences. We can imagine an experiment where  is gradually increased while
is gradually increased while 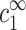 is kept fixed. Aggregation will start, in this experiment, above a certain threshold value of
is kept fixed. Aggregation will start, in this experiment, above a certain threshold value of  . Our assumption is that this threshold does not depend on cDNA. In real experiments, however, cz is adjusted rather than
. Our assumption is that this threshold does not depend on cDNA. In real experiments, however, cz is adjusted rather than 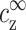 , and cs is kept fixed rather than
, and cs is kept fixed rather than 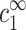 . To find the threshold value in terms of the experimentally available cz, we need to map
. To find the threshold value in terms of the experimentally available cz, we need to map 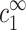 ,
, 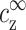 onto cs, cz. This mapping is described by Eqs. 6–7, and involves cDNA. It is only through this mapping that cDNA will affect the threshold of aggregation.
onto cs, cz. This mapping is described by Eqs. 6–7, and involves cDNA. It is only through this mapping that cDNA will affect the threshold of aggregation.
The limit of cDNA ≪ cs
The limit cDNA ≪ cs offers a particularly simple dependence of cz,aggr on cDNA and is considered first. Most models and experiments indicate that monovalent counterions cannot overcharge DNA segments. Hence the monovalent excess, aρ1, in Eq. 7, is a number between zero and one, because the excess monovalent charge is smaller than that of DNA. From Eq. 7, |cs−c1∞| ≪ cs as long as cDNA ≪ cs. It is then possible to replace 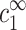 by cs, leading to a simplification of Eq. 6:
by cs, leading to a simplification of Eq. 6:
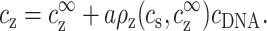 |
(8) |
Note that cDNA is indeed smaller than cz in most of the experimental points in Fig. 2. However a similar simplification cannot be applied for cs because it is typically much smaller than cs, and often smaller than cDNA.
According to our principal assumption, aggregation starts at a threshold value 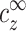 = cz*, which does not depend on cDNA (whereas cz,aggr, the average multivalent salt concentration, does depend on cDNA through Eq. 8). Similarly, the density profile at the threshold does not depend on cDNA, because it is determined by
= cz*, which does not depend on cDNA (whereas cz,aggr, the average multivalent salt concentration, does depend on cDNA through Eq. 8). Similarly, the density profile at the threshold does not depend on cDNA, because it is determined by  = cs and cz*. The excess of z-valent counterions, as determined from this profile, is equal to:
= cs and cz*. The excess of z-valent counterions, as determined from this profile, is equal to:
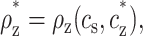 |
(9) |
with no dependence on cDNA. Using the threshold values cz* and ρz* in Eq. 8, we find that the average concentration of z-valent ions at the onset of aggregation is
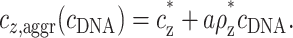 |
(10) |
This is the threshold concentration that was measured experimentally in Raspaud et al. (1998). Note that, in Eq. 10, cz* as well as ρz* depend on the monovalent salt concentration, cs, but the explicit dependence is omitted for clarity.
The simple relationship expressed by Eq. 10 is one of our main results. As a visualization of this result we refer again to Fig. 3. The quantities ρz, 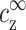 , and the density profile nz(r) are indicated in this figure. At the onset of aggregation,
, and the density profile nz(r) are indicated in this figure. At the onset of aggregation, 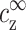 is equal to cz* and does not depend on cDNA (or equivalently, on the spacing between DNA segments, R). As cDNA is increased the distance between DNA strands decreases. The onset values of
is equal to cz* and does not depend on cDNA (or equivalently, on the spacing between DNA segments, R). As cDNA is increased the distance between DNA strands decreases. The onset values of  and ρz do not change, but the contribution of ρz to the average concentration increases, leading to an increase in cz,aggr.
and ρz do not change, but the contribution of ρz to the average concentration increases, leading to an increase in cz,aggr.
The coefficients aρz* and cz* of the linear dependence in Eq. 10 are the coefficients α and β defined in the introduction section. They can be easily found from the experimental data: cz* is the value of cz,aggr in the limit of infinite DNA dilution, cDNA → 0, since in this limit cz =  = cz*. The excess at the onset, ρz*, can be found from the slope of cz,aggr as function of cDNA. Before presenting a detailed comparison with experiments, we generalize the treatment for small cDNA to arbitrary values.
= cz*. The excess at the onset, ρz*, can be found from the slope of cz,aggr as function of cDNA. Before presenting a detailed comparison with experiments, we generalize the treatment for small cDNA to arbitrary values.
The case of cDNA ≥ cs
When cDNA is of the same order as cs or larger, corrections to 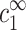 must be taken into account, as expressed by Eq. 7, and the linear relation of Eq. 10 no longer holds. The ion density profiles as well as cs and cz are now determined by the two variables
must be taken into account, as expressed by Eq. 7, and the linear relation of Eq. 10 no longer holds. The ion density profiles as well as cs and cz are now determined by the two variables  and
and 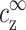 . The relation between
. The relation between 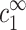 and
and  and the experimentally controlled cs, cz, and cDNA is given by Eqs. 6–7. In terms of
and the experimentally controlled cs, cz, and cDNA is given by Eqs. 6–7. In terms of 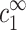 ,
,  the criterion for aggregation remains the same as in the previous case:
the criterion for aggregation remains the same as in the previous case:
 |
(11) |
Equations 6, 7, and 11, with the three unknowns  ,
, 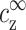 , and cz, lead to a unique solution for cz,aggr. Note that
, and cz, lead to a unique solution for cz,aggr. Note that  is larger than cs because of counterions coming from the DNA as can be seen in Eq. 7, where aρ1−1 is negative. In Eq. 10, cs is replaced by
is larger than cs because of counterions coming from the DNA as can be seen in Eq. 7, where aρ1−1 is negative. In Eq. 10, cs is replaced by 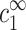 , which is larger than cs for large cDNA. Hence, increasing cDNA has an effect similar to addition of monovalent salt. As noted above, this effect is significant only for cDNA > cs.
, which is larger than cs for large cDNA. Hence, increasing cDNA has an effect similar to addition of monovalent salt. As noted above, this effect is significant only for cDNA > cs.
COMPARISON WITH EXPERIMENT
Raspaud et al. (1998) measured the spermine (z = 4) concentration cz at the onset of aggregation as a function of cDNA for four values of cs and with cDNA ranging over four orders of magnitude—from 10−2 to 102 mM. We fitted the data (E. Raspaud and J.-L. Sikorav, private communication) for each cs to a straight line according to Eq. 10. The least square fit presented in Fig. 5 takes into account the experimental error bars and data points up to cDNA = 10 mM. In Fig. 5 a the fit is shown using a linear scale which covers the range of cDNA only up to cDNA = 1.5 mM for clarity purposes. Due to the large range of cDNA it is impossible to show all the data on the linear scale of Fig. 5 a. Instead, the same data and linear lines are shown in Fig. 5 b on a log-log scale over the full experimental range of cDNA.
FIGURE 5.
Spermine concentration at the onset of aggregation cz,aggr as a function of cDNA, fitted to the form derived in Eq. 10 (different line types are used for different salt concentrations). Value of cs (in mM) is indicated next to each curve. Experimental data is adapted from Raspaud et al. (1998) and shown in the following symbols: cs = 2 mM (○); 13 mM (Δ); 23 mM (∇); and 88 mM (□). Experimental error bars (E. Raspaud, private communication) are indicated by vertical lines. The fitted lines and experimental points are shown using a linear scale in a, up to cDNA = 1.5 mM, and a log-log scale in b, up to cDNA = 100 mM, allowing all data points to be shown on the same plot. Only the data up to cDNA = 10 mM was used for the linear fit. The crossover values of cDNA, as defined by Eq. 14, are indicated by arrows in b.
The linear fit is very good for all four values of monovalent salt concentration cs. Note that for cs = 88 mM the fit is very good up to the largest value of cDNA = 48 mM reported in the experiment, although our fit takes into account only data points up to cDNA = 10 mM. It was previously suggested (Raspaud et al., 1998) that a separate regime exists for cDNA ≳ 10 mM, characterized by a power law relation between cz and cDNA with an exponent smaller than unity. Our analysis suggests a different conclusion. The fit clearly demonstrates that the relation is linear all the way up to cDNA = 48 mM, as predicted by Eq. 10. Note also that even at cDNA = 48 mM we have cDNA < cs, so the assumptions leading to Eq. 10 are still valid.
The only points in Fig. 5 b that deviate significantly from the fit are the three points where cs = 13 mM (triangles) and cDNA > 20 mM (two of these points coincide with points having cs = 88 mM, shown using square symbols.) This deviation is easily explained by the fact that cDNA ≫ cs so that corrections to 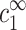 must be taken into account. For example, at cDNA = 90 mM the nominal monovalent counterion concentration is 103 mM, taking into account counterions contributed by the DNA. To find c1∞ we need to subtract the condensed counterions, as determined by ρ1. We can estimate ρ1 at this point by solving the Poisson-Boltzmann equation in a unit cell with the appropriate radius. The chemical potentials of the three ion species are tuned such that their concentrations match the known values of cz and cs. This leads to an estimate:
must be taken into account. For example, at cDNA = 90 mM the nominal monovalent counterion concentration is 103 mM, taking into account counterions contributed by the DNA. To find c1∞ we need to subtract the condensed counterions, as determined by ρ1. We can estimate ρ1 at this point by solving the Poisson-Boltzmann equation in a unit cell with the appropriate radius. The chemical potentials of the three ion species are tuned such that their concentrations match the known values of cz and cs. This leads to an estimate:  mM. Hence, cz at the onset of aggregation should lie a little below the continuation of the cs = 88 mM line which is, indeed, where it is found. The trend for cs = 13 mM can probably be seen already at the point cDNA = 15 mM, although the deviation at this point is still within the range of experimental error. The few other experimental points with cDNA ≈ cs deviate slightly from the straight line as well (still within experimental error bars). In all these cases the deviation is in the direction corresponding to a higher value of cs, as expected.
mM. Hence, cz at the onset of aggregation should lie a little below the continuation of the cs = 88 mM line which is, indeed, where it is found. The trend for cs = 13 mM can probably be seen already at the point cDNA = 15 mM, although the deviation at this point is still within the range of experimental error. The few other experimental points with cDNA ≈ cs deviate slightly from the straight line as well (still within experimental error bars). In all these cases the deviation is in the direction corresponding to a higher value of cs, as expected.
A linear relation of the form cz,aggr = αcDNA + β, was previously suggested on empirical basis for aggregation induced by spermidine (3+), on a smaller range of DNA concentrations (Osland and Kleppe, 1977; Pelta et al., 1996b). Although this result looks similar to our prediction on the onset of aggregation, it is not directly related to our analysis because cz,aggr was taken in those works to be the transition midpoint. This is the point where half of the maximal precipitation of DNA is reached. Our analysis does not apply at the transition midpoint since it requires all the DNA segments to be well separated from each other. Indeed, the coefficient α, related to the transition midpoint, was found in Osland and Kleppe (1977) and Pelta et al. (1996b) to be of order 102, much larger than unity. Such a value of α cannot be interpreted as the excess of spermidine ions per monomer near isolated chains.
The parameters of the linear fit in Fig. 5 are summarized in Table 1 for the four experimentally used values of cs.
TABLE 1.
Fit parameters used in Fig. 5
| cs [mM] | cz* [mM] | aρz* |
|---|---|---|
| 2 | 0 ± 0.0003 | 0.194 ± 0.020 |
| 13 | 0.011 ± 0.002 | 0.191 ± 0.013 |
| 23 | 0.031 ± 0.005 | 0.173 ± 0.025 |
| 88 | 0.52 ± 0.05 | 0.135 ± 0.026 |
Crossover in the log-log plot
For presentation purposes we plot in Fig. 5 b, cz,aggr vs. cDNA on a log-log scale, as appeared in Raspaud et al. (1998). The linear relation that was found between these two quantities is not clearly manifested on the log-log plot, because a linear dependence of the form y = αx + β is not easily recognized in such a plot. Furthermore, such a linear relation appears on a log-log plot to be artificially characterized by two distinct behaviors, at low and high values of the independent variable. These two behaviors were mentioned in Raspaud et al. (1998) and can be seen in Fig. 5 b. However, they do not represent in our opinion two real physical regimes and can be understood by taking the logarithm of Eq. 10. For small cDNA (large R),
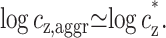 |
(12) |
That is, cz does not depend on cDNA as is seen in Fig. 5 b in the small cDNA limit. In the opposite limit of large cDNA (small R):
 |
(13) |
Here, the linear dependence of cz,aggr on cDNA yields a line of slope 1 in the same figure.
The crossover between these apparent behaviors occurs when the number of bulk and excess ions are the same:
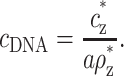 |
(14) |
When cDNA is much smaller than this crossover value, the number of excess multivalent ions near DNA segments is negligible compared to their total number. In the other extreme of cDNA much larger than the crossover value, the number of free multivalent ions is negligible compared to the excess ions, and nearly all multivalent ions are bound to the DNA.
For the experimental data in Fig. 5 the crossover value is equal to 0.06, 0.18, and 3.9 mM for cs = 13, 23, and 88 mM, respectively, and smaller than 1.5 × 10−3 mM for cs = 2 mM. The first three crossover points are indicated by arrows in Fig. 5 b.
DNA AGGREGATION AND COUNTERION CONDENSATION
We separate the discussion following our results in three parts. The first addresses the conditions required for DNA aggregation. The coefficients of the linear relation in Eq. 10, cz* and ρz*, have a definite physical meaning. Their values, as extracted from the experimental data, provide insight on these conditions. The second part deals with condensation of counterions on DNA (to be distinguished from condensation of DNA chains). The general relation ρz = ρz ( ,
, 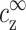 ) that was introduced in Eqs. 3–4 is a property of counterion condensation on isolated chains. By extracting the values of ρz,
) that was introduced in Eqs. 3–4 is a property of counterion condensation on isolated chains. By extracting the values of ρz,  , and
, and  at the onset of DNA aggregation, we can learn about exact density profiles of spermine around DNA, and compare our findings with approximations such as Poisson-Boltzmann theory. Finally, we comment on our main assumption, which was used in the theoretical considerations section.
at the onset of DNA aggregation, we can learn about exact density profiles of spermine around DNA, and compare our findings with approximations such as Poisson-Boltzmann theory. Finally, we comment on our main assumption, which was used in the theoretical considerations section.
Conditions at the onset of aggregation
Most of the proposed theoretical models for interchain attraction and aggregation (see, for example, Arenzon et al., 1999; Borukhov et al., 2001, 2002; Ha and Liu, 1997; Nguyen et al., 2000; Olvera de la Cruz et al., 1995; Raspaud et al., 1998; Wittmer et al., 1995) regard the charged chain as surrounded by a layer of condensed ions which is usually modeled as a one-dimensional gas. This layer mediates an interchain attraction, and the models predict the number of condensed ions required to initiate aggregation of the chains. In the current work we do not address this theoretical problem, but rather concentrate on what can be inferred from the experimental results using the analysis presented in the previous section. This analysis provides insight on the conditions prevailing at the onset of aggregation. In particular, the excess ρz* characterizes the number of condensed multivalent counterions that are present near each chain at the onset. Although, in general, the notion of condensed counterions is somewhat ill-defined as it depends on which ions are regarded as bound to the DNA, we show in the Appendix that in our case it does have a reasonably well-defined meaning. Furthermore, the number of condensed multivalent ions per monomer is practically the same as aρz*.
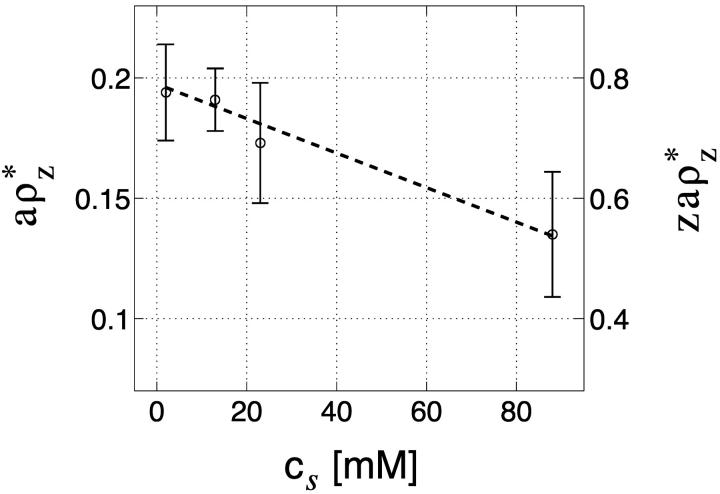
The excess of multivalent counterions per monomer, aρz*, is shown in Fig. 6 as a function of cs. All values are taken from Table 1, as extracted from the experimental data. The dashed line is a linear fit. Two different axis scales are used on the left and right of the plot. The left axis shows the value of aρz*. The right one shows the part of DNA charge that is compensated by condensed multivalent ions, zaρz*, where z = 4 for spermine. From the plot we deduce the following two conclusions:
The number of condensed multivalent ions (per DNA monomer) aρz* at the onset decreases as the monovalent salt concentration increases, with variation between 0.19 and 0.14. A possible reason for this trend may be that the bare electrostatic repulsion between chains is decreased due to increased screening. Hence a smaller number of multivalent ions is required to overcome this repulsion. The change in ρz* may also be related to the competition between monovalent and multivalent ions in the aggregated DNA state.
The data indicates that there is no over-charging of the DNA by spermine at the onset (see also Nguyen et al., 2000) since zaρz* < 1. At higher concentration of spermine, beyond the threshold, we do not rule out the possibility of DNA overcharging, as was suggested by Nguyen et al. (2000).
FIGURE 6.
Excess of multivalent counterions per monomer at the onset of aggregation, aρz*, as function of cs. All values are taken from Table 1, as extracted from the experimental data of Raspaud et al. (1998). Error bars are indicated by vertical bars and the dashed line is a linear fit to be used as a guide to the eye. On the right axis, zaρz* is shown, where z = 4 for spermine. This value is equal to the fraction of DNA charge compensated by the condensed multivalent ions. Note that, according to the Manning condensation theory, the same quantity is equal to 0.94, for tetravalent ions and no added salt.
Although ρz* decreases with increase of cs, it is of the same order of magnitude for all the cs values in Table 1. In contrast, cz* varies in Table 1 over more than three orders of magnitude. As was previously suggested (Olvera de la Cruz et al., 1995; Raspaud et al., 1998), this large variation in cz* is a result of competition between monovalent and multivalent counterions. We discuss the relation between ρz* and cz* to some extent in the following subsection. A more detailed analysis of this relation, emphasizing the role of competition between the two counterion species, will be presented in a separate publication (see also Belloni et al., 1984; Wilson and Bloomfield, 1979; Wilson et al., 1980).
Counterion condensation
We now turn to analyze the condensation of monovalent and multivalent ions around DNA. Each line in Table 1 provides a measurement of the excess ρz at certain values of  and
and 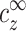 . The general relation ρz(
. The general relation ρz(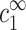 ,
,  ) is a property of counterion density profiles around isolated DNA segments. Hence, the data in Table 1 can be used to test any particular theory used to calculate such ion distributions.
) is a property of counterion density profiles around isolated DNA segments. Hence, the data in Table 1 can be used to test any particular theory used to calculate such ion distributions.
The most simple model to consider is the Poisson-Boltzmann (PB) theory (see Andelman, 1994; Guéron and Weisbuch, 1980; Le Bret and Zimm, 1984; Oosawa, 1971). In Table 2 we compare the excess predicted by PB theory with the experimental result, by solving the PB equation such that  and
and  match the experimental values of cs and cz* from Table 1. The excess is then calculated from the PB density profile, and compared with the experimental value of aρz (equal to aρz* of Table 1). The DNA is modeled as a uniformly charged cylinder of radius d = 10 Å.
match the experimental values of cs and cz* from Table 1. The excess is then calculated from the PB density profile, and compared with the experimental value of aρz (equal to aρz* of Table 1). The DNA is modeled as a uniformly charged cylinder of radius d = 10 Å.
TABLE 2.
Excess of 4-valent ions near DNA compared with PB theory
| c1∞ [mM] | cz∞ [mM] | aρz (exp) | aρz (PB) |
|---|---|---|---|
| 2 | 0 ± 0.0003 | 0.194 ± 0.020 | 0.186 ± 0.005 |
| 13 | 0.011 ± 0.002 | 0.191 ± 0.013 | 0.178 ± 0.002 |
| 23 | 0.031 ± 0.005 | 0.173 ± 0.025 | 0.172 ± 0.002 |
| 88 | 0.52 ± 0.05 | 0.135 ± 0.026 | 0.164 ± 0.002 |
Inspection of the results in Table 2 shows that there is a reasonable agreement with experiment (within the error bars) for the three smaller values of cs = 2, 13, and 23 mM. However, for cs = 88 mM there is a 30% deviation. The two data points with cDNA > 10 mM that were not taken into account in the linear fit of Fig. 5 suggest that ρz is closer to the lower bound of the experimental error range, whereas the PB value is larger than the upper bound.
Overall, the agreement with PB theory (Table 2) is surprisingly good, considering that PB theory does not work so well for bulky multivalent ions. Deviations from PB theory have several sources. One of these sources is specific molecular details such as the geometrical shape of ions, DNA structure, and short-range interactions. Another source for deviations are ion-ion correlations between spermine molecules, computed in theories which go beyond the mean-field approximation. However, these correlations tend to increase the number of bound multivalent counterions (Lyubartsev and Nordenskiöld, 1997), whereas for cs = 88 mM, the number of bound multivalent counterions is decreased. We conclude that correlation effects by themselves are not the main source of the deviations seen in Table 2. In addition the data analysis does not indicate overcharging of the DNA. Such an effect may be expected if correlation effects are strong (Nguyen et al., 2000).
In Fig. 7 we compare the DNA aggregation data with PB predictions at finite DNA concentrations. For each DNA concentration the PB equation is solved in a cylindrical cell of the appropriate radius. The multivalent counterion concentration cz is gradually increased until the onset is reached, and its onset value, cz,aggr, is plotted as function of cDNA. Two different criteria are used to determine the onset cz,aggr. In Fig. 7 a it is chosen as the point where  is equal to the experimental value cz* of Table 1, whereas in Fig. 7 b, the onset is chosen as the point where ρz = ρz*. To span all the data range we use, for convenience, a log-log plot, as in Fig. 5 b.
is equal to the experimental value cz* of Table 1, whereas in Fig. 7 b, the onset is chosen as the point where ρz = ρz*. To span all the data range we use, for convenience, a log-log plot, as in Fig. 5 b.
FIGURE 7.
Spermine concentration (in mM) as a function of DNA monomer concentration (mM) at the onset of aggregation, calculated using the PB equation. Two different criteria are used in a and b to determine the onset: in a,  , as calculated using the PB equation, is equal to the experimental value of cz* from Table 1. In b, ρz of PB theory is equal to ρz* from Table 1. The radius of DNA is taken as d = 10 Å. Log-log plot is used to show the five decades of DNA concentrations. For each cs the plot covers experimental data up to cDNA = cs. For larger cDNA, corrections due to changes in
, as calculated using the PB equation, is equal to the experimental value of cz* from Table 1. In b, ρz of PB theory is equal to ρz* from Table 1. The radius of DNA is taken as d = 10 Å. Log-log plot is used to show the five decades of DNA concentrations. For each cs the plot covers experimental data up to cDNA = cs. For larger cDNA, corrections due to changes in  should be taken into account, as was discussed in the preceding section. All notations are the same as in Fig. 5.
should be taken into account, as was discussed in the preceding section. All notations are the same as in Fig. 5.
On a linear scale, all the lines in Fig. 7, a and b, are straight lines. This fact serves as additional confirmation of our general analysis in the Theoretical Considerations section. In accordance with our analysis, both cz* and ρz are constant along each line, and the slope of each line is equal to aρz. Note that the relation between cz* and ρz is determined in Fig. 7 within the PB approximation, while in Fig. 5 both of these coefficients are related to the actual counterion density profiles in the experimental system. The use of the PB equation is the source of deviations from experimental data in Fig. 7.
On first inspection the match with experiment in Fig. 7 a is very good, whereas the match in Fig. 7 b is not as good. On closer inspection it is seen that the fit in Fig. 7 b is not good for small values of cDNA, while it is actually better than in Fig. 7 a for large cDNA. With the PB equation it is not possible to obtain a perfect fit for both small and large cDNA because the values of 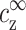 and ρz are not independent. Fixing
and ρz are not independent. Fixing 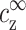 = cz* (as in Fig. 7 a) sets a value of ρz that is different from ρz*, and the opposite happens in Fig. 7 b. The fit in Fig. 7 a is quite good even for large cDNA because the values of ρz* are of similar order of magnitude for all four lines.
= cz* (as in Fig. 7 a) sets a value of ρz that is different from ρz*, and the opposite happens in Fig. 7 b. The fit in Fig. 7 a is quite good even for large cDNA because the values of ρz* are of similar order of magnitude for all four lines.
Deviations as in Fig. 7 are inevitable if any approximations are used to model the distribution of counterions around DNA. Note, however, that within such approximate models our general theoretical considerations should apply, as long as the total number of ions in the system is counted properly. Such a model that goes beyond PB was proposed in Nguyen and Shklovskii (2001). Indeed, within this model a linear relationship similar to Eq. 10 was found.
The experimental results analyzed in this section may be influenced, to a certain degree, by the fact that there was more than one type of monovalent counterion in the system. For the three higher salt concentrations, except for cs = 2 mM, the solution contained 10 mM of Tris–H+ ions in addition to Na+ (Raspaud et al., 1998). For the largest salt concentration, 88 mM, where significant deviations from PB theory are found, this effect is probably negligible. Another detail regarding the TE buffer is that the Tris ions may be only partly ionized. If only 80% of Tris is ionized, as suggested in Tang et al. (1997), the concentrations cs = 13 mM, 23 mM, and 88 mM should be reduced by 2 mM. Although this will have only a small effect on our results, it will improve both the comparison with PB and the fit with the dashed line in Fig. 6, for the point cs = 13 mM. For the two other concentrations of 23 mM and 88 mM the effect will be negligible.
Further comments on underlying model assumption
Our underlying assumption, that the onset of aggregation depends uniquely on 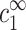 and
and  (but not on cDNA), is an approximation that can be justified on several different levels but deserves further and more thorough investigation. The most simple motivation for this assumption is that multivalent ions, in the vicinity of the chains, mediate the attraction necessary for aggregation. In turn, the number of condensed multivalent ions near each chain is determined by
(but not on cDNA), is an approximation that can be justified on several different levels but deserves further and more thorough investigation. The most simple motivation for this assumption is that multivalent ions, in the vicinity of the chains, mediate the attraction necessary for aggregation. In turn, the number of condensed multivalent ions near each chain is determined by 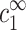 and
and 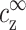 .
.
Let us first suppose that aggregation starts when a net attraction appears between two chains. This assumption may be justified if chains are sufficiently long and their translational entropy can be neglected. To find the onset of two-chain attraction the free energy of a two-chain complex should be calculated as a function of the distance between the two chains. This free energy represents the effective interaction between the two chains, mediated by the ionic solution. The counterion distribution near each chain will not be the same for close-by and for isolated chains. However, in both cases, the concentrations must decay to their bulk values throughout the solution, 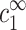 and
and  . This requirement serves as a boundary condition, imposed at a large distance from the two chains. It will determine uniquely the counterion distribution between the chains, as well as the free energy associated with the two-chain complex. Hence
. This requirement serves as a boundary condition, imposed at a large distance from the two chains. It will determine uniquely the counterion distribution between the chains, as well as the free energy associated with the two-chain complex. Hence  and
and 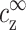 determine the effective interaction between chains, and in particular whether an attraction occurs at a certain range of interchain separations; in terms of these variables the onset of two-chain attraction does not depend on cDNA.
determine the effective interaction between chains, and in particular whether an attraction occurs at a certain range of interchain separations; in terms of these variables the onset of two-chain attraction does not depend on cDNA.
Strictly speaking, the onset of aggregation and the onset of two-chain attraction are not the same. The aggregate phase involves interactions between multiple chains, whereas chains in the dilute phase interact very weakly with each other. Aggregation starts when the free energy per chain is equal in the dilute and aggregate phases. Note that the chemical potential of each ion species must be the same in the two phases, and that in the dilute phase these chemical potentials are directly related to 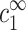 and
and  . Hence
. Hence 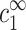 and
and  determine the free energy per chain in the two phases. The approximation of independence on cDNA neglects the translational entropy of DNA segments, which can be justified for long enough and rigid segments. It also neglects contributions from interactions between chains in the dilute phase, which are assumed to be small compared to the free energy of the single DNA-counterion complexes.
determine the free energy per chain in the two phases. The approximation of independence on cDNA neglects the translational entropy of DNA segments, which can be justified for long enough and rigid segments. It also neglects contributions from interactions between chains in the dilute phase, which are assumed to be small compared to the free energy of the single DNA-counterion complexes.
SUMMARY
We have shown that the onset of aggregation at finite (nonzero) DNA concentration, cz,aggr, is determined by the onset in the limit of infinite DNA dilution. For DNA monomer concentration smaller than that of monovalent salt, cDNA ≲ cs, the multivalent counterion concentration at the onset, cz,aggr, depends linearly on cDNA. The coefficients of this linear dependence are the bulk concentration of multivalent counterions and their excess relative to the bulk near each DNA segment. Both of these coefficients are of theoretical interest and can be extracted from the available experimental data.
Our main assumption is that the onset of aggregation can be related to the ion density profiles around each chain. Hence, it is uniquely determined by  and
and  , the bulk concentrations of the two counterion species, respectively. Our results and fit to experiment strongly support this assumption. Nevertheless, we believe that more detailed theoretical and experimental investigations are needed to fully understand its range of validity. For example, it will be of interest to test experimentally the equilibration of a DNA solution through a dialysis membrane, with a cell containing only counterions (Braunlin et al., 1982; Plum and Bloomfield, 1988; Subirana and Vives, 1981). This procedure allows a direct control of the ionic bulk concentrations.
, the bulk concentrations of the two counterion species, respectively. Our results and fit to experiment strongly support this assumption. Nevertheless, we believe that more detailed theoretical and experimental investigations are needed to fully understand its range of validity. For example, it will be of interest to test experimentally the equilibration of a DNA solution through a dialysis membrane, with a cell containing only counterions (Braunlin et al., 1982; Plum and Bloomfield, 1988; Subirana and Vives, 1981). This procedure allows a direct control of the ionic bulk concentrations.
To predict precisely the onset of aggregation, the structure of the aggregated phase must be considered. Nevertheless, it is instructive to focus only on single chains at the onset, as is often done. At the aggregation onset the electrostatic repulsion between isolated chains in solution must be overcome by a sufficiently strong attraction mediated by multivalent counterions. This number of counterions is expected to depend only weakly on physical parameters such as the monovalent salt concentration. Our analysis does not address directly the question of the onset origin, but merely supports the fact that the number of condensed multivalent ions at the onset, aρz*, is of the same order of magnitude, regardless of the cs value. A more refined result of our analysis is that aρz* is not constant but decreases with increase of cs. On the other hand, cz*, the value of  at the onset, depends strongly on cs. This is mainly a result of the competition between monovalent and multivalent ions, as will be addressed in a separate publication.
at the onset, depends strongly on cs. This is mainly a result of the competition between monovalent and multivalent ions, as will be addressed in a separate publication.
Our analysis also sheds light on counterion condensation on DNA, which is independent on the criterion for DNA aggregation. The experimental data indicates that for high cs the number of spermine ions in the vicinity of DNA is smaller than the prediction of Poisson-Boltzmann theory. A similar trend was observed in computer simulations (Lyubartsev and Nordenskiöld, 1997) of spermidine (3+) and NaCl in contact with DNA. Spermidine binding was affected by addition of monovalent salt more strongly than the Poisson-Boltzmann prediction. For high salt concentrations spermidine binding was considerably smaller. In the computer simulations both molecular-specific interactions, the geometrical shape of the constituents and interion correlations were taken into account. All these effects, and in particular the geometry of the spermidine molecule, which is similar to that of spermine, were found to play an important role.
The above analysis demonstrates that specific interactions play an important role in determining the threshold of aggregation. In the dilute phase these interactions strongly influence the competition between monovalent and multivalent ions and the free energy of DNA-counterion complexes. Similarly, specific interactions play a prominent role in the dense phase (Strey et al., 1998). Force measurements under osmotic stress (Rau et al., 1984; Rau and Parsegian, 1992a,b) provide a wealth of information on these interactions.
In conclusion, the physical parameters extracted here from experiment on the onset of DNA aggregation provide insight on the conditions required for aggregation, and on condensation of ions around DNA. These parameters may turn out to be of great value in assessment of various theoretical models. Additional detailed experiments may further deepen our understanding of these complex phenomena.
Acknowledgments
We are grateful to E. Raspaud and J.-L. Sikorav for numerous discussions and for providing us with their unpublished experimental data. We also wish to thank I. Borukhov, H. Diamant, M. Kozlov, A. Lyubartsev, G. Manning, T. Nguyen, M. Olvera de la Cruz, A. Parsegian, R. Podgornik, D. Rau, and T. Thomas for discussions and correspondence.
Support from the Israel Science Foundation under grant No. 210/02, to B.S.F., is gratefully acknowledged. D.A. thanks the Alexander von Humboldt Foundation for a research award.
APPENDIX
In this Appendix we discuss the relation between the excess and the number of condensed ions. The latter quantity is not as well-defined as the former, but relates more naturally to the aggregation mechanism. The notion of condensed ions suggests that some ions are bound to the charged chain whereas others are free. In reality there is a density profile that extends all the way from r = d to r = R with no definite separation between condensed and free ions. In the following we define condensed ions rather loosely as the number of ions up to a certain characteristic distance from the chain (Belloni et al., 1984; Wilson et al., 1980). We show that for multivalent ions this number does not depend strongly on the choice of this characteristic distance. Hence, the number of condensed ions is reasonably well defined. Moreover, the excess number of multivalent counterions, which can be directly calculated from the experimental data, is nearly identical to this quantity. This point will be further explained below.
Fig. 8 shows the excess of 4-valent counterions δρz(r) up to a distance r from the DNA axis, as a function of r:
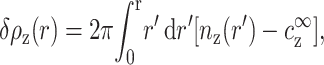 |
(A1) |
with the limit δρz (∞) = ρz of Eq. 4. The density profile was calculated using the Poisson-Boltzmann equation, with the radius of DNA taken as d = 10 Å and with bulk densities of ions as in the last line of Table 1: cs =  = 88 mM, and
= 88 mM, and  = 0.52 mM.
= 0.52 mM.
FIGURE 8.
Excess of 4-valent ions per DNA monomer, up to a distance r from the axis of a charged cylinder of radius d = 10 Å (modeling the DNA) as obtained using the Poisson-Boltzmann equation (solid line). The excess δρz (r) is defined in Eq. A1. The number of charges per unit length on the cylinder is 1/a where a = 1.7 Å to fit DNA values. The bulk densities of monovalent and multivalent ions are 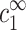 = 88 mM,
= 88 mM,  = 0.52 mM, yielding κ−1 = 10.0 Å. The quantity δρz (solid line) can be compared with the total number of 4-valent ions (dashed line) up to a distance r from the cylinder. The distance d + κ−1 from the DNA axis is indicated by a vertical arrow, and characterizes the decay of the density profile far away from the DNA.
= 0.52 mM, yielding κ−1 = 10.0 Å. The quantity δρz (solid line) can be compared with the total number of 4-valent ions (dashed line) up to a distance r from the cylinder. The distance d + κ−1 from the DNA axis is indicated by a vertical arrow, and characterizes the decay of the density profile far away from the DNA.
Three observations can be made. First, most, but not all, of the excess z-valent ions are localized very close to the DNA, at a distance of order λ/z, where λ is the Gouy-Chapman length (see Andelman, 1994):
 |
(A2) |
where σ is the average charge per unit area on the cylinder surface, σ = ρDNA/2πd, and ρDNA = 1/a is the DNA charge per unit length. At room temperature the Bjerrum length lB ≃ 7 Å, and for DNA with 4-valent counterions λ/z ≃ 0.6 Å. Second, the counterions within a layer of a few times the Debye length (κ−1 = 10.0 Å in Fig. 8) neutralize the DNA charge. Nearly all the excess distribution is in this layer. Third, to estimate the total amount of counterions in the condensed layer of thickness ακ−1, where α is a number of order unity, we need to add δρz to the bulk contribution, πα2κ−2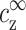 . Using κ from Eq. 1, the latter is equal to:
. Using κ from Eq. 1, the latter is equal to:
 |
(A3) |
In experiment,  is much smaller than
is much smaller than 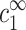 at the onset, and the bulk contribution of Eq. A3 can be neglected relative to ρz, for α of order unity. This can be seen specifically in Fig. 8 by comparing the solid and dashed lines.
at the onset, and the bulk contribution of Eq. A3 can be neglected relative to ρz, for α of order unity. This can be seen specifically in Fig. 8 by comparing the solid and dashed lines.
The outcome of the above discussion is that ρz, defined in Eq. 4 as the excess of counterions throughout the cell, can be regarded, to a good approximation, as the total number of counterions within a condensation layer whose thickness is approximately the Debye length. For typical concentration ranges as considered here we do not expect that this outcome will change, even for models going beyond Poisson-Boltzmann theory.
As a further demonstration, the number of multivalent counterions up to several different distances from the DNA is shown in Table 3, as calculated in a unit cell using the Poisson-Boltzmann equation. For each cs in Table 1 we find the Poisson-Boltzmann density profile such that  = cs and ρz = ρz*, and then calculate the number of multivalent ions (per DNA monomer) up to the following distances from the DNA radius: 10 Å, 20 Å, κ−1, and 2κ−1. The values of κ−1, as obtained from Eq. 1 are equal to 68, 26, 20, and 10 Å for cs = 2, 13, 23, and 88 mM, respectively. These numbers are compared with aρz*. All the different measures in Table 3 yield results that are very close to each other.
= cs and ρz = ρz*, and then calculate the number of multivalent ions (per DNA monomer) up to the following distances from the DNA radius: 10 Å, 20 Å, κ−1, and 2κ−1. The values of κ−1, as obtained from Eq. 1 are equal to 68, 26, 20, and 10 Å for cs = 2, 13, 23, and 88 mM, respectively. These numbers are compared with aρz*. All the different measures in Table 3 yield results that are very close to each other.
TABLE 3.
Number of z-valent counterions, per DNA monomer, up to several different distances from the DNA axis, compared with aρz
| cs [mM] | d + 10 Å | d + 20 Å | d + κ−1 | d + 2κ−1 | aρz |
|---|---|---|---|---|---|
| 2 | 0.191 | 0.193 | 0.194 | 0.194 | 0.194 |
| 13 | 0.187 | 0.190 | 0.190 | 0.191 | 0.191 |
| 23 | 0.171 | 0.172 | 0.172 | 0.173 | 0.173 |
| 88 | 0.134 | 0.135 | 0.134 | 0.135 | 0.135 |
References
- Andelman, D. 1994. Electrostatic properties of membranes: the Poisson-Boltzmann theory. In Handbook of Physics of Biological Systems, Vol. I. R. Lipowsky, and E. Sackmann, editors. Elsevier Science, Amsterdam, pp.603–642.
- Arenzon, J. J., J. F. Stilck, and Y. Levin. 1999. Simple model for attraction between like-charged polyions. Eur. Phys. J. B. 12:79–82. [Google Scholar]
- Belloni, L., M. Drifford, and P. Turq. 1984. Counterion diffusion in polyelectrolyte solutions. Chem. Phys. 83:147–154. [Google Scholar]
- Bloomfield, V. A., D. M. Crothers, and I. Tinoco. 2000. Nucleic Acids: Structures, Properties, and Functions. University Science Books, Sausalito, CA.
- Borukhov, I., R. F. Bruinsma, W. M. Gelbart, and A. J. Liu. 2001. Elastically driven linker aggregation between two semiflexible polyelectrolytes. Phys. Rev. Lett. 86:2182–2185. [DOI] [PubMed] [Google Scholar]
- Borukhov, I., K.-C. Lee, R. F. Bruinsma, W. M. Gelbart, A. J. Liu, and M. Stevens. 2002. Association of two semiflexible polyelectrolytes by interchain linkers: theory and simulations. J. Chem. Phys. 117:462–480. [Google Scholar]
- Braunlin, W. H., T. J. Strick, and M. T. Record, Jr. 1982. Equilibrium dialysis studies of polyamine binding to DNA. Biopolymers. 21:1301–1314. [DOI] [PubMed] [Google Scholar]
- Chattoraj, D. K., L. C. Gosule, and J. A. Schellman. 1978. DNA condensation with polyamines. II. Electron microscopic studies. J. Mol. Biol. 121:327–337. [DOI] [PubMed] [Google Scholar]
- Gelbart, W. M., R. F. Bruinsma, P. A. Pincus, and V. A. Parsegian. 2000. DNA-inspired electrostatics. Phys. Today. 53:38–44. [Google Scholar]
- Gosule, L. C., and J. A. Schellman. 1978. DNA condensation with polyamines. I. Spectroscopic studies. J. Mol. Biol. 121:311–326. [DOI] [PubMed] [Google Scholar]
- Guéron, M., and G. Weisbuch. 1980. Polyelectrolyte theory. I. Counterion accumulation, site-binding, and the insensitivity to polyelectrolyte shape in solutions containing finite salt concentrations. Biopolymers. 19:353–382. [Google Scholar]
- Ha, B. Y., and A. J. Liu. 1997. Counterion-mediated attraction between two like-charged rods. Phys. Rev. Lett. 79:1289–1292. [Google Scholar]
- Le Bret, M., and H. Zimm. 1984. Distribution of counterions around a cylindrical polyelectrolyte and Manning's condensation theory. Biopolymers. 23:287–312. [DOI] [PubMed] [Google Scholar]
- Lyubartsev, A. P., and L. Nordenskiöld. 1997. Monte Carlo simulation study of DNA polyelectrolyte properties in the presence of multivalent polyamine ions. J. Phys. Chem. B. 101:4335–4342. [Google Scholar]
- Nguyen, T. T., I. Rouzina, and B. I. Shklovskii. 2000. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 112:2562–2568. [Google Scholar]
- Nguyen, T. T., and B. I. Shklovskii. 2001. Complexation of DNA with positive spheres: phase diagram of charge inversion and reentrant condensation. J. Chem. Phys. 115:7298–7308. [Google Scholar]
- Olvera de la Cruz, M., L. Belloni, M. Delsanti, J. P. Dalbiez, O. Spalla, and M. Drifford. 1995. Precipitation of highly charged polyelectrolyte solutions in the presence of multivalent salts. J. Chem. Phys. 103:5781–5791. [Google Scholar]
- Oosawa, F. 1971. Polyelectrolytes. Marcel Dekker, New York.
- Osland, A., and K. Kleppe. 1977. Polyamine induced aggregation of DNA. Nucleic Acids Res. 4:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelta, J., D. Durand, J. Doucet, and F. Livolant. 1996a. DNA mesophases induced by spermidine: structural properties and biological implications. Biophys. J. 71:48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelta, J., F. Livolant, and J.-L. Sikorav. 1996b. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 271:5656–5662. [DOI] [PubMed] [Google Scholar]
- Plum, G. E., and V. A. Bloomfield. 1988. Equilibrium dialysis study of binding of hexamine cobalt to DNA. Biopolymers. 27:1045–1051. [DOI] [PubMed] [Google Scholar]
- Raspaud, E., I. Chaperon, A. Leforestier, and F. Livolant. 1999. Spermine-induced aggregation of DNA, nucleosome and chromatin. Biophys. J. 77:1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspaud, E., M. Olvera de la Cruz, J.-L. Sikorav, and F. Livolant. 1998. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 74:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, D. C., B. Lee, and V. A. Parsegian. 1984. Measurement of the repulsive force between poly-electrolyte molecules in ionic solution-hydration forces between parallel DNA double helices. Proc. Natl. Acad. Sci. USA. 81:2621–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, D. C., and V. A. Parsegian. 1992a. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Biophys. J. 61:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, D. C., and V. A. Parsegian. 1992b. Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J. 61:260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan, M., T. Antony, A. Shirahata, L. H. Sigal, T. Thomas, and T. J. Thomas. 1999. Ionic and structural specificity effects of natural and synthetic polyamines on the aggregation and resolubilization of single-, double-, and triple-stranded DNA. Biochemistry. 38:3821–3830. [DOI] [PubMed] [Google Scholar]
- Strey, H. H., R. Podgornik, D. C. Rau, and V. A. Parsegian. 1998. DNA-DNA interactions. Curr. Opin. Struct. Biol. 8:309–313. [DOI] [PubMed] [Google Scholar]
- Subirana, J. A., and J. L. Vives. 1981. The precipitation of DNA by spermine. Biopolymers. 20:2281–2283. [Google Scholar]
- Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53:749–790. [DOI] [PubMed] [Google Scholar]
- Tang, J. X., T. Ito, T. Tau, P. Traub, and P. A. Janmey. 1997. Opposite effects of electrostatics and steric exclusion on bundle formation by F-actin and other filamentous polyelectrolytes. Biochemistry. 36:12600–12607. [DOI] [PubMed] [Google Scholar]
- Widom, J., and R. L. Baldwin. 1980. Cation-induced toroidal condensation of DNA. J. Mol. Biol. 144:431–453. [DOI] [PubMed] [Google Scholar]
- Widom, J., and R. L. Baldwin. 1983. Monomolecular condensation of λ-DNA induced by cobalt hexamine. Biopolymers. 22:1595–1620. [DOI] [PubMed] [Google Scholar]
- Wilson, R. W., and V. A. Bloomfield. 1979. Counterion-induced condensation of deoxyribonucleic acid. A light scattering study. Biochemistry. 79:2192–2196. [DOI] [PubMed] [Google Scholar]
- Wilson, R. W., D. C. Rau, and V. A. Bloomfield. 1980. Comparison of polyelectrolyte theories of the binding of cations to DNA. Biophys. J. 30:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmer, J., A. Johner, and J. F. Joanny. 1995. Precipitation of polyelectrolytes in the presence of multivalent salts. J. Phys. II France. 5:635–654. [Google Scholar]