Abstract
Mitochondrial and nuclear DNA are packaged by proteins in a very different manner. Although protein-DNA complexes called “nucleoids” have been identified as the genetic units of mitochondrial inheritance in yeast and man, little is known about their physical structure. The yeast mitochondrial protein Abf2p was shown to be sufficient to compact linear dsDNA, without the benefit of supercoiling, using optical and atomic force microscopy single molecule techniques. The packaging of DNA by Abf2p was observed to be very weak as evidenced by a fast Abf2p off-rate (koff = 0.014 ± 0.001 s−1) and the extremely small forces (<0.6 pN) stabilizing the condensed protein-DNA complex. Atomic force microscopy images of individual complexes showed the 190-nm structures are loosely packaged relative to nuclear chromatin. This organization may leave mtDNA accessible for transcription and replication, while making it more vulnerable to damage.
INTRODUCTION
Mitochondrial (mt) DNA is packaged into discrete units known as “nucleoids” in both yeast and man, and these structures have been shown to be the genetic units of mitochondrial inheritance (Garrido et al., 2003; Jacobs et al., 2000; MacAlpine et al., 2000). However, little is known about nucleoid formation or composition (MacAlpine et al., 2000). The density with which mtDNA is packaged affects both its maintenance and its accessibility during regulatory processes such as replication and transcription. Indeed, mtDNA is thought to be more susceptible to free radical induced damage than nuclear DNA (O'Rourke et al., 2002; Richter et al., 1988) in part because of a lack of protective histone packaging, and this type of damage has been proposed to lead to late-onset neurodegenerative disorders (Menzies et al., 2002; O'Rourke et al., 2002).
Abf2p is a mitochondrial (mt) protein that has been hypothesized to play a major role in packaging mtDNA into nucleoids (Caron et al., 1979; MacAlpine et al., 2000; Megraw and Chae, 1993; Newman et al., 1996) in the yeast Saccharomyces cerevisiae. Abf2p also contributes to mtDNA maintenance (Contamine and Picard, 2000), copy number (Cho et al., 2001; Zelenaya-Troitskaya et al., 1998), transcription (Diffley and Stillman, 1991, 1992), and recombination (MacAlpine et al., 1998). It is closely related in sequence and function to the vertebrate nuclear high-mobility group (HMG) protein HMG1 (Diffley and Stillman, 1992) and is a homolog of human mitochondrial transcription factor h-mtTFA (Fisher et al., 1992; Parisi et al., 1993), a protein implicated in human mitochondrial disease (Wallace, 2002; Wredenberg et al., 2002). Recently, h-mtTFA was identified as the protein responsible for human mtDNA packaging (Alam et al., 2003). Both HMG1 and h-mtTFA bend DNA and introduce supercoils into circular DNA molecules.
We used two different experimental techniques to demonstrate and provide complementary information about the compaction of DNA by Abf2p. Optical trapping of single DNA molecules extended by flow and visualized by fluorescence microscopy has been used to obtain information about the kinetics of binding and force with which the Abf2p-DNA complex is packaged. Atomic force microscopy (AFM) confirmed that DNA is bent by Abf2p, and provided a high resolution view and the dimensions of the compact Abf2p-DNA complex.
MATERIALS AND METHODS
Purification of Abf2p
The Saccharomyces cerevisiae ABF2 gene in a pMalc2X fusion vector (Kao et al., 1993) (New England BioLabs, kindly provided by the Nunnari Lab, University of California Davis) was cloned into pET28b(+) (Novagen, Madison, WI). Inverse polymerase chain reaction was used to fuse an initiator methionine and six histidine codons to the ABF2 reading frame encoding mature Abf2p, residues 21 to 177 with his tag, Mr = 19,484 Daltons. Protein was purified from BL21(DE3)(pET28b-His6ABF2) cultures (5–10 mg/L culture), essentially as described for Cre recombinase (Martin et al., 2002) except 500 mM NaCl was used in the initial lysis buffer, and the solution was dialyzed to low salt buffer before ion exchange chromatography. Concentrated and filtered samples, 20–70 mg/mL (Millipore Centricon-10) (Billerica, MA) in 20 mM Na-HEPES pH 7.5, 1 mM Na-EDTA, 4 mM DTT, 0.1 w/v Na-azide, were diluted with 10 mM Tris-Cl pH 7.8, 4 mM DTT, 1 mM Na-EDTA. Abf2p concentrations were determined using an extinction coefficient at 280 nm of 1.29 mg−1mL, or 24,180 M−1. DNA binding activity was established with a gel-retardation assay (Cho et al., 2001) using either supercoiled pLitmus-38(+) or linear lambda-phage DNA (New England BioLabs, Beverly, MA) as a substrate.
Optical trapping measurements
To obtain information about the binding kinetics of Abf2p to DNA, single, linear, lambda-phage DNA molecules attached to beads were held by an optical trap and extended by flowing buffer in a two-channel flow cell (Fig. 1, a and b) as previously described (Brewer et al., 1999). The buffer used in these experiments, 100 mM NaHCO3 (pH 8), contained no sucrose.
FIGURE 1.

(a) Side view of the flow cell showing the trapping and excitation laser beams. (b) Top view of the flow cell. An individual DNA molecule held by an optical trap (orange) via its attached bead, and extended by flowing buffer, is moved into protein solution by translating the stage holding the flow cell perpendicular to the direction of flow. DNA was stained with YOYO-1 dye, allowing the compaction to be observed using fluorescence microscopy. The molecule was then moved back to the DNA side of the flow cell (which was protein-free), and the decompaction of the molecule was observed as protein left it, ultimately returning to its original length. (c) Time-lapse images of a lambda-phage DNA dimer (35 μm contour length) undergoing compaction by Abf2p. The Abf2p concentration is 2 μM. The time interval between successive frames is 0.5 s. The buffer flow speed is 63 μm/s.
Measurement of buffer flow velocity
Flow cell buffer velocities were determined by releasing the bead from the optical trap at the conclusion of each compaction/decompaction measurement and determining the distance the bead moved in a given time.
Measurement of maximal tether force
A value for the tether force (Stigter and Bustamante, 1998; Zimm, 1998) of the compacted DNA-Abf2p complex was determined by first reducing the trapping laser power so that the maximal force, sometimes called the escape force, the optical trap could exert on a 1-μm-diameter bead was 1.4 pN. A maximal value for the “tether force” (the force with which Abf2p-compacted DNA pulls on its attached bead due to hydrodynamic friction) was determined by subtracting the Stokes force on the bead attached to the compacted DNA molecule from the calibrated trap force, as long as the DNA molecule plus bead could be held by the trap while it compacted. The trap force was determined by moving the microscope stage holding a sample cell containing buffer and 1-μm-diameter beads at successively faster velocities until a trapped bead was released from the optical trap. The Stokes force,  , was then used to calculate the force on the bead at the point of release, where
, was then used to calculate the force on the bead at the point of release, where 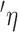 is the buffer viscosity, r is the bead diameter, and v is the stage velocity. The bead was held at a position 10–15 μm beneath the surface of the coverslip, so that no corrections had to be made for the surface. The sample cell consisted of a microscope slide, a drop of buffer containing beads, and a coverslip supported by two 50 μm thick plastic shims. The four sides of the coverslip were attached to the slide by nail polish.
is the buffer viscosity, r is the bead diameter, and v is the stage velocity. The bead was held at a position 10–15 μm beneath the surface of the coverslip, so that no corrections had to be made for the surface. The sample cell consisted of a microscope slide, a drop of buffer containing beads, and a coverslip supported by two 50 μm thick plastic shims. The four sides of the coverslip were attached to the slide by nail polish.
Data analysis
The rate equation describing the binding of Abf2p to DNA for a first order process can be written as follows:
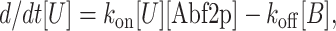 |
(1) |
where [U], [B], and [Abf2p] represent the concentration of unbound DNA sites, bound DNA sites, and Abf2p, respectively, and kon and koff are the rate constants for protein binding and releasing from a single DNA molecule. The length of the DNA molecule itself is proportional to the amount of protein bound to the DNA molecule since protein binding is the rate-limiting step (see Results and Discussion). In Eq. 1, [Abf2p] = 0 when the DNA molecule is on the DNA side of the flow cell. Also, as we shall see, koff is much smaller than kon so we can neglect the term koff[B] when describing binding. The sum of the unbound and bound sites on the DNA molecule is equal to the total number of binding sites:
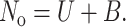 |
(2) |
Using Eqs. 1 and 2 we can solve for the rates and find for the compaction process that
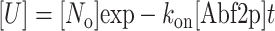 |
(3) |
and similarly for the decompaction process
 |
(4) |
The length versus time plots (Fig. 2) for the compaction and decompaction processes for an individual DNA molecule of length L were then fit to the following equations:
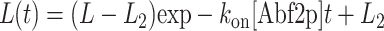 |
(5) |
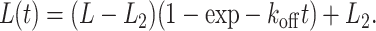 |
(6) |
L2 is the length of the compacted Abf2p-DNA complex, which can be directly measured; L is the initial length of the DNA molecule, which is measured, so the fit is really only a one-parameter fit to koff, or kon[Abf2p] where [Abf2p] is known.
FIGURE 2.

(a) Compaction of a single DNA molecule by Abf2p in 1.43 μM Abf2p (kon = 0.45 ± 0.1 μM−1 s−1). (b) Linear variation of Abf2p binding rate (kon[Abf2p]) with Abf2p concentration (kon = 0.36 ± 0.1 μM−1 s−1). (c) Decompaction of the same DNA molecule as in a (koff = 7.0 ± 1.3 × 10−3 s−1). The data in a and c are given by black points, and the exponential fit is represented by a red line.
Effect of YOYO-1 dye on Abf2p binding
The extension of 16 lambda-phage DNA molecules stained with YOYO-1 dye in buffer without protein were measured at different flow velocities. Extensions were calculated from different DNA contour lengths (Stigter and Bustamante, 1998) to compare with the measured values. The contour length which best fit the data was 17.6 μm (mean error in extension fit was 4%) using a persistence length of 55 nm. The contour length of unstained lambda-phage DNA is 16.4 μm, and values of the length of stained DNA have been reported as high as 22 μm (Brewer et al., 1999; Perkins et al., 1995). We infer that the amount of YOYO-1 dye bound to the DNA is much less than the saturating ratio of 1 YOYO-1 dye molecule per 4 bp and do not expect YOYO-1 to affect the binding of Abf2p protein to DNA.
AFM measurements
High resolution atomic force microscopy (AFM) was used to observe the interactions between Abf2p and linear dsDNA (pBR322 was linearized by digesting with BamH1. The DNA was then washed four times in a Centricon-100 concentrator in 10 mM Tris, 1 mM EDTA pH 8 to remove enzyme and twice in 100 mM NaHCO3 to buffer exchange). The microscope, a Nanoscope IIIa (Digital Instruments, Woodbury, NY), was used in tapping mode with Si FESP probes (NanoWorld, Neuchâtel, Switzerland). Abf2p and DNA in buffer (100 mM NaHCO3 pH 8) were mixed together for 5 min before depositing on a substrate. The substrate was prepared by applying 3 μL of 0.1% aqueous solution of poly-l-lysine (Sigma, St. Louis, MO) to a freshly cleaved mica surface for 1 min. The sample was then rinsed with distilled water and dried with nitrogen. The concentration of DNA in the mixed solution remained fixed at 1 μg/mL, and the Abf2p concentration varied from 1.5 μg/mL to 25 μg/mL. Data analysis was performed using IgorPro software (Lake Oswego, OR).
RESULTS AND DISCUSSION
Kinetics of Abf2p binding
The compaction and decompaction of single DNA molecules by Abf2p is shown in Fig. 1 c and Fig. 2. Notable features of the compaction and decompaction plots (Fig. 2, a and c) are their exponential shape, which is indicative of a first order kinetic process, and the finite length of the compacted DNA molecule. The percent compaction varied between 50% and 89% and increased as the length of the DNA molecule (Fig. 3) and flow velocity (data not shown) decreased.
FIGURE 3.
DNA compaction versus initial extension for 67 molecules in 2 μM Abf2p, taken at buffer flow speeds between 50 and 110 μm/s. The compaction is defined as the difference between the initial (no protein) and final (in protein) DNA extensions divided by the initial DNA extension. For the range of DNA contour lengths and buffer flow velocities used in our experiment, the fractional extension of the DNA was fairly constant: 76 ± 5% (Perkins et al., 1995; Stigter and Bustamante, 1998). The incomplete compaction is thought to be due to the extension of the Abf2p-DNA complex by the flowing buffer. The longer the initial DNA contour length, the greater the hydrodynamic drag of the compacted Abf2p-DNA complex, and the less compaction is observed. The linear least squares fit is represented by a green line.
The binding of Abf2p to DNA was shown to be the rate-limiting step in the compaction of DNA. By moving the translation stage holding the flow cell in the same direction as the flowing buffer we were able to increase the effective velocity of buffer containing Abf2p and observe the length of a partially compacted DNA molecule (100% compaction was never achieved; see Fig. 3) increase significantly. Upon stopping the stage, the protein-DNA complex rapidly returned to its initial length in ∼1 s, which was much faster than the timescale observed for compaction. This result shows that the rate-limiting step in the compaction of the Abf2p-DNA complex is the rate of Abf2p binding to DNA. From this experiment we also deduced that at least part of the incomplete compaction of the Abf2p-DNA complex was due to the elongation of the protein-DNA complex by the flowing buffer.
The data and fit (using Eq. 5 and Eq. 6) for the compaction and decompaction of a single DNA molecule by Abf2p are shown in Fig. 2, a and c. We measured the on-rate at three different Abf2p concentrations (Fig. 2 b) and the binding rate determined from the exponential fit, kon[b] varied linearly with concentration, as expected. The on-rate constant, kon = 0.36 ± 0.1 μM−1 s−1, did not depend on either the initial DNA extension (at constant flow speed) or flow velocity. Decompaction measurements (10 molecules) allowed us to determine koff = .014 ± 0.001 s−1. The ratio of these rates, kon/koff, provided the binding constant Kb = 2.57 ± 0.74 × 107 M−1.
Limit on binding from the McGhee-von Hippel theory
The maximal fractional coverage of DNA by Abf2p, which can be calculated from the equilibrium constant, Kb, Abf2p footprint (∼27 bp (Diffley and Stillman, 1991, 1992), and Abf2p concentration (2 μM) using the McGhee-von Hippel theory (McGhee and von Hippel, 1974) equals 87%. The greatest observed compaction of DNA by Abf2p was 89% (Fig. 3). Measurements obtained from AFM images (Fig. 4) of individual compact Abf2p-DNA complexes showed a reduction in length of DNA from 1.5 μm to 190 nm, a compaction of 87%. These results indicate that 13% of each DNA molecule was not bound with Abf2p.
FIGURE 4.

AFM images (image scale is 1 μm along each axis, height scale (f) is in nm) of Abf2p protein bound to single linear dsDNA molecules (pBR322, 1.5 μm contour length, 4361 bp). The DNA is bent and compacted as the concentration of Abf2p increases, ultimately forming a compact, 190 ± 90 nm diameter, round object. The ratio of Abf2p molecules to DNA bp is as follows with Abf2p concentrations indicated in parenthesis: (a) 0.0 Abf2p/bp (0 μM), (b) 0.04 Abf2p/bp (0.075 μM), (c) 0.1 Abf2p/bp (0.175 μM), (d) 0.2 Abf2p/bp (0.35 μM), (e) 0.45 Abf2p/bp (0.75 μM), and (f) 0.75 Abf2p/bp (1.25 μM). For an Abf2p footprint of 27 bp, the DNA coverage is complete for an Abf2p-to-bp ratio of 0.037.
Abf2p compaction force
The most direct approach to determine the strength with which Abf2p packages DNA is to pull on both ends of the compacted DNA molecule and measure the force versus extension curve (Baumann et al., 2000; Wang et al., 1997). Another approach, which is suitable for measuring very small compaction forces, is to use flowing buffer to extend the compacted Abf2p-DNA complex (held by an optical trap) and measure its tether force (Stigter and Bustamante, 1998; Zimm, 1998). The tether force is the hydrodynamic drag the flowing buffer exerts on the protein-DNA complex, and it provides an estimate of the forces needed to elongate DNA bound by Abf2p. Fig. 3 shows how the percent compaction DNA by Abf2p decreases as the length of the DNA molecule increases. This is what one would expect if the tether, or drag, force of the Abf2p-DNA complex is sufficient to overcome the forces that maintain the complex in a compacted state. Using this approach, we have measured the maximal tether force for 15 different DNA molecules bound with Abf2p. Initial DNA extensions ranged from 5 to 15 μm, and compactions range from 72% to 89%. We find that the mean tether force ≤0.60 ± 0.12 pN. This suggests it takes very little force to elongate DNA compacted by Abf2p and the forces that stabilize the compacted protein-DNA complex are quite weak.
AFM images
AFM images of Abf2p-DNA complexes (Fig. 4, a–f) show sharp bending of DNA by Abf2p. As the concentration of Abf2p increases, the DNA becomes increasingly compact, finally forming a round object with diameter 190 ± 90 nm. The ratio of Abf2p molecules to DNA bp (Abf2p/bp) for the complexes imaged in Fig. 4 b is 0.04, allowing Abf2p to fill all available binding sites on the DNA molecule for a 27-bp footprint. However, complete DNA compaction was not observed until the Abf2p/bp ratio reached 0.45. This is consistent with our calculations of the equilibrium fractional coverage of DNA by Abf2p, given by kon[Abf2p]/koff, where [Abf2p] denotes the concentration of Abf2p, kon = 0.36 ± 0.1 μM−1 s−1, and koff = 0.014 ± 0.001 s−1. For [Abf2p] = 0.04 μM (Fig. 4 b), the fractional coverage equals 50% and some bending is seen, whereas when [Abf2p] = 0.35 μM (Fig. 4 d), the fraction coverage equals 90%, and almost complete compaction of the individual DNA molecules is observed. However, the McGhee-von Hippel theory limits the fractional coverage to 85% for [Abf2p] = 1 μM, and this may explain why the degree of compaction in images Fig. 4, e and f look similar.
We have shown that Abf2p obeys first order binding kinetics to linear DNA. The measured on-rate constant, (kon = 0.36 ± 0.1 μM−1 s−1), together with an estimated concentration of Abf2p in the mitochondria, can be used to determine the compaction time for the mitochondrial genome. For a mitochondria with ellipsoidal dimensions of 0.5 μm (radius) by 1.0 μm, and 50 genome equivalents of mtDNA (MacAlpine et al., 2000), the concentration of Abf2p is 540 μM (assuming complete coverage of the DNA by Abf2p). This concentration multiplied by the measured on-rate constant gives an exponential time constant of 194 s−1. Clearly, mtDNA will be rapidly packaged by Abf2p. The off-rate of Abf2p from DNA was measured to be koff = 0.014 ± 0.001 s−1. This implies that in the absence of free Abf2p, the time needed for all the bound protein to dissociate from mtDNA would be several minutes. Therefore, Abf2p is not strongly bound to DNA compared to the histones (Brower-Toland et al., 2002) that organize nuclear DNA or protamine (Brewer et al., 1999), which packages DNA in the sperm cell. This implies that a significant concentration of free Abf2p must be maintained in the mitochondria to keep the mtDNA in a compact state, which is consistent with the calculation above of the Abf2p concentration in the mitochondria.
The tether force measured for DNA molecules compacted by Abf2p was extremely small, ≤ 0.60 ± 0.12 pN, showing that the forces holding the Abf2p-DNA complex together are weak. AFM measurements of the spatial extent of single DNA molecules bound with Abf2p showed that the DNA is loosely packaged. Both the high off-rate of the Abf2p protein and the limited compaction of the complex should aid in allowing enzymes and proteins access to regulatory sites where Diffley and Stillman have showed that “phased” binding of Abf2p occurs (Diffley and Stillman, 1991, 1992). At the same time, this would leave the mtDNA more vulnerable to damage by shear forces, free radicals (Menzies et al., 2002), and nucleases. It is also important to note that the mitochondrial genome in S. cerevisiae is circular, rather than linear, and the binding of Abf2p to circular DNA has been shown to form negative supercoils (Diffley and Stillman, 1992) which may impact the compaction of mtDNA through the formation of plectonemes (Strick et al., 2000). Further studies comparing the compaction of DNA by Abf2p and its homolog, h-mtTFA, using both linear and circular DNA, should provide insight into how mtDNA packaging impacts human mitochondrial genome-related diseases.
Acknowledgments
We would like to thank Dr. Stigter for his calculation of DNA contour lengths, Professor Jodi Nunnari for her comments on the manuscript, and Dr. Michael Colvin for helpful discussions.
Work was performed at Lawrence Livermore National Laboratory (LLNL) under the auspices of the U.S. Department of Energy under contract W-7405-ENG-48 and partially supported by the National Science Foundation Biophotonics Center at University of California at Davis under Agreement No. PHY 0120999. (R.J.B.).
References
- Alam, T. I., T. Kanki, T. Muta, K. Ukaji, Y. Abe, H. Nakayama, K. Takio, N. Hamasaki, and D. Kang. 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31:1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, C. G., V. A. Bloomfield, S. B. Smith, C. Bustamante, M. D. Wang, and S. M. Block. 2000. Stretching of single collapsed DNA molecules. Biophys. J. 78:1965–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, L. R., M. Corzett, and R. Balhorn. 1999. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 286:120–123. [DOI] [PubMed] [Google Scholar]
- Brower-Toland, B. D., C. L. Smith, R. C. Yeh, J. T. Lis, C. L. Peterson, and M. D. Wang. 2002. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 99:1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, F., C. Jacq, and J. Rouviere-Yaniv. 1979. Characterization of a histone-like protein extracted from yeast mitochondria. Proc. Natl. Acad. Sci. USA. 76:4265–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J. H., Y. K. Lee, and C. B. Chae. 2001. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim. Biophys. Acta. 1522:175–186. [DOI] [PubMed] [Google Scholar]
- Contamine, V., and M. Picard. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64:281–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F., and B. Stillman. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA. 88:7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F., and B. Stillman. 1992. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 267:3368–3374. [PubMed] [Google Scholar]
- Fisher, R. P., T. Lisowsky, M. A. Parisi, and D. A. Clayton. 1992. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267:3358–3367. [PubMed] [Google Scholar]
- Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A. M. Van Der Bliek, and J. N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 14:1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H. T., S. K. Lehtinen, and J. N. Spelbrink. 2000. No sex please, we're mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays. 22:564–572. [DOI] [PubMed] [Google Scholar]
- Kao, L. R., T. L. Megraw, and C. B. Chae. 1993. Essential role of the HMG domain in the function of yeast mitochondrial histone HM: functional complementation of HM by the nuclear nonhistone protein NHP6A. Proc. Natl. Acad. Sci. USA. 90:5598–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. USA. 95:6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are coregulated by the general amino acid control pathway. EMBO J. 19:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. S., E. Pulido, V. C. Chu, T. S. Lechner, and E. P. Baldwin. 2002. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol. 319:107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee, J. D., and P. H. von Hippel. 1974. Theoretical aspects of DNA-protein interactions: cooperative and noncooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86:469–489. [DOI] [PubMed] [Google Scholar]
- Megraw, T. L., and C. B. Chae. 1993. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 268:12758–12763. [PubMed] [Google Scholar]
- Menzies, F. M., P. G. Ince, and P. J. Shaw. 2002. Mitochondrial involvement in amyotrophic lateral sclerosis. Neurochem. Int. 40:543–551. [DOI] [PubMed] [Google Scholar]
- Newman, S. M., O. Zelenaya-Troitskaya, P. S. Perlman, and R. A. Butow. 1996. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 24:386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, T. W., N. A. Doudican, M. D. Mackereth, P. W. Doetsch, and G. S. Shadel. 2002. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 22:4086–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M. A., B. Xu, and D. A. Clayton. 1993. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 13:1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, T. T., D. E. Smith, R. G. Larson, and S. Chu. 1995. Stretching of a single tethered polymer in a uniform flow. Science. 268:83–87. [DOI] [PubMed] [Google Scholar]
- Richter, C., J. W. Park, and B. N. Ames. 1988. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA. 85:6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigter, D., and C. Bustamante. 1998. Theory for the hydrodynamic and electrophoretic stretch of tethered B-DNA. Biophys. J. 75:1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick, T. R., V. Croquette, and D. Bensimon. 2000. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 404:901–904. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C. 2002. Animal models for mitochondrial disease. Methods Mol. Biol. 197:3–54. [DOI] [PubMed] [Google Scholar]
- Wang, M. D., H. Yin, R. Landick, J. Gelles, and S. M. Block. 1997. Stretching DNA with optical tweezers. Biophys. J. 72:1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wredenberg, A., R. Wibom, H. Wilhelmsson, C. Graff, H. H. Wiener, S. J. Burden, A. Oldfors, H. Westerblad, and N. G. Larsson. 2002. Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl. Acad. Sci. USA. 99:15066–15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman, and R. A. Butow. 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 148:1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimm, B. H. 1998. Extension in flow of a DNA molecule tethered at one end. Macromolecules. 31:6089–6098. [Google Scholar]



