Abstract
Interactions of Na+, K+, Rb+, and Cs+ ions within the selectivity filter of a potassium channel have been investigated via multiple molecular dynamics simulations (total simulation time, 48 ns) based on the high resolution structure of KcsA, embedded in a phospholipid bilayer. As in simulations based on a lower resolution structure of KcsA, concerted motions of ions and water within the filter are seen. Despite the use of a higher resolution structure and the inclusion of four buried water molecules thought to stabilize the filter, this region exhibits a significant degree of flexibility. In particular, pronounced distortion of filter occurs if no ions are present within it. The two most readily permeant ions, K+ and Rb+, are similar in their interactions with the selectivity filter. In contrast, Na+ ions tend to distort the filter by binding to a ring of four carbonyl oxygens. The larger Cs+ ions result in a small degree of expansion of the filter relative to the x-ray structure. Cs+ ions also appear to interact differently with the gate region of the channel, showing some tendency to bind within a predominantly hydrophobic pocket. The four water molecules buried between the back of the selectivity filter and the remainder of the protein show comparable mobility to the surrounding protein and do not exchange with water molecules within the filter or the central cavity. A preliminary comparison of the use of particle mesh Ewald versus cutoff protocols for the treatment of long-range electrostatics suggests some difference in the kinetics of ion translocation within the filter.
INTRODUCTION
Potassium channels are central to many properties of cell membranes. They are membrane proteins that form potassium selective transmembrane pores. K channels help to control the electric potential across cell membranes by enabling rapid and selective diffusion of K+ ions (Hille, 1992) at rates of ∼107 ions s−1channel−1. K channels play an important role in a wide range of physiological processes such as electrical signaling in the nervous system, regulation of cardiac excitability, and regulation of insulin release. K channels are of biomedical importance in that they provide potential targets for novel drugs, and because their malfunctioning has been shown to generate a number of disorders, referred to as channelopathies (Ashcroft, 2000).
All K channels are thought to share the same core structure. They differ in the presence/absence of additional TM helices, and of additional nonmembrane domains and/or subunits that control their gating. The structure of the K channel core domain was revealed by x-ray diffraction studies of a bacterial channel, KcsA (Doyle et al., 1998). The channel-forming domain is composed of two TM helices separated by a re-entrant loop made up of a short pore (P) helix plus a more extended region of polypeptide that forms the selectivity filter (F). K channels have tetrameric symmetry. Thus in KcsA the subunits are arranged around a central fourfold axis that is coincident with the axis of the central pore. The extracellular mouth leads into a narrow selectivity filter lined by backbone carbonyl oxygens that can accommodate two or three K+ ions (Fig. 1 A). Beyond this there is a central water-filled cavity that can accommodate a single K+ ion. There is then a narrow hydrophobic region formed by the closely packed M2 helices, which forms the main channel gate. This structure corresponds to that of the closed form of the channel. On channel opening the M2 helices move apart so as to widen the intracellular mouth of the channel (Perozo et al., 1998, 1999; Liu et al., 2001; Jiang et al., 2002b; Shrivastava and Sansom, 2002; Biggin and Sansom, 2002). This may be accompanied by bending of the M2 helices in the vicinity of a conserved glycine residue, as suggested by comparisons with the structure of MthK (Jiang et al., 2002b), a bacterial Ca-activated K channel that has been crystallized in an open state and whose structure has been determined to 3.5 Å resolution (Jiang et al., 2002a).
FIGURE 1.
(A) Schematic diagram of the simulation systems, illustrating the channel protein (PDB code 1k4c) of which only two subunits are shown, ions within the filter and cavity, and the approximate location of the lipid bilayer (broken lines). (B) Overview of the simulations. Location of the ions within the channel structure. On the left-hand side a diagram of the selectivity filter region (from Thr75 to Asp80) is given along with the ion and water binding sites defined in the x-ray structure. For simplicity only two subunits are shown. Sites S0–S4 of the selectivity filter region and cavity site (Cav) are shown, along with their approximate z coordinates. The right-hand side of the diagram defines the locations of the cations and the water molecules included in the initial configuration of the simulations.
The region of the amino-acid sequence associated with the selectivity filter (i.e., the P and F regions) is highly conserved between different K channels, and contains a sequence motif (TVGYG) characteristic of K channels. The conformation adopted by this motif, in which the main chain parts of successive residues are enantiomeric (Watson and Milner-White, 2002) in the tetrameric channel protein, is essential to the permeation and selectivity mechanism of the channel (Doyle et al., 1998; Morais-Cabral et al., 2001; Zhou et al., 2001). There is also a conserved glycine residue in the M2 helix (G99 in KcsA) that plays an important role in helix flexibility during channel gating (Jiang et al., 2002b; Biggin and Sansom, 2002).
The emergence of the 3.2 Å structure of KcsA enabled a large number of simulation studies (Guidoni et al., 1999, 2000; Guidoni and Carloni, 2002; Bernèche and Roux, 2000, 2001; Allen et al., 1999, 2000; Allen and Chung, 2001; Mashl et al., 2001; Åqvist and Luzhkov, 2000; Luzhkov and Åqvist, 2000, 2001; Jordan, 2000; Garofoli et al., 2002; Shrivastava and Sansom, 2000, 2002; Capener et al., 2000; Ranatunga et al., 2001a,b; Biggin et al., 2001; Sansom et al., 2000, 2002; Tieleman et al., 2001; Capener and Sansom, 2002) of the relationship between K channel structure and function. These studies suggest that ion permeation through the filter may be best described via a concerted mechanism whereby an ion-water-ion group moves through the filter in a single-file fashion. Initial simulation studies of the nature of the selectivity of KcsA between K+ and Na+ have also been conducted (Åqvist and Luzhkov, 2000; Shrivastava et al., 2002). However, the medium resolution of the x-ray structure, and in particular the absence of experimental data for possible water molecules associated with the outer ring of the filter, has impacted upon the accuracy of such simulations.
The recent determination of the structure of KcsA at 2 Å resolution (Zhou et al., 2001) provides an opportunity for considerably more detailed computational and theoretical investigations of aspects of K channel function such as permeation and ion selectivity. For example, although many aspects of the KcsA channel have already been studied at both structural (Cortes et al., 2001; Jiang and MacKinnon, 2000; Perozo et al., 1998, 1999, 2000; Zhou et al., 2001) and functional (Cuello et al., 1998; Meuser et al., 1999; Splitt et al., 2000; Heginbotham et al., 1999; LeMasurier et al., 2001) levels, several questions remain unaddressed or need to be readdressed on the basis of the newer structure. In particular, it is of particular interest to probe some aspects of ion conduction and selectivity by comparing the behavior of the alkali cation series. K+ ions permeate readily through KcsA. Under physiological conditions Na+ ions are impermeable. Selectivity sequences for permanent ions through KcsA have been determined by symmetrical solution conductance and by reversal potentials under bi-ionic or mixed-ion conditions (LeMasurier et al., 2001). These and other experiments (Morais-Cabral et al., 2001) suggest that Rb+ is permeant (albeit with an approximately fourfold lower conductance than K+ at 200 mM) and is also a strong promoter of channel opening. Cs+ behaves primarily as a blocker, having an ionic conductance two orders-of-magnitude lower than that of K+ (Heginbotham et al., 1998, 1999).
In this article, we take advantage of the higher resolution structure of KcsA to explore filter-ion interactions for alkali metal cations ranging from Na+ to Cs+. In particular we focus on the short timescale motion of the ions in the filter and on the filter flexibility and the behavior of the water molecules that form a band around the outside of the filter.
METHODS
Simulation system
The simulation system consisted of the 2 Å resolution high [K+] structure (1k4c; Zhou et al., 2001) of KcsA, embedded in a palmitoyloleoyl phosphatidylcholine (POPC) lipid bilayer. Each of the four subunits consists of 102 residues, from residue 22 to 123—the atomic coordinates of residues 1–21 and 124–160 not being present in the x-ray structure. An acetyl group was attached to residue 22 to mimic the preceding peptide bond and the C-terminal carboxylate was protonated. The side chain of Glu71 was constructed in a protonated state to form a diacid hydrogen bond with the carboxylate group of Asp80, following the results presented in Ranatunga et al. (2001a) and Bernèche and Roux (2002). The rest of the ionizable residues were in their default protonation state. A fully equilibrated POPC lipid bilayer was used as the starting point for generating the lipid bilayer into which KcsA was embedded. The lipid bilayer consists of 115 molecules of POPC in the extracellular side and 127 in the intracellular side. The protocol to place the protein within the bilayer was previously described in detail in Shrivastava and Sansom (2000). The ions and crystallographic waters were then added (see below).
The system was solvated with SPC water molecules (Berendsen et al., 1981). The central cavity was solvated 1), by retaining all the pore water molecules present in the x-ray structure; and 2), by overlaying with a pre-equilibrated box of SPC water, rejecting any waters within van der Waals overlap distance of a protein atom. This yielded ∼34 water molecules in the final system. To preserve the electric neutrality of the system sufficient Cl− ions (∼15) were added to the bulk solvent on either side of the bilayer. These ions replaced water molecules at the lowest Coulomb potential positions. The total number of atoms in the system was ∼41,000.
Simulation details
Once the system was set up, an energy minimization was carried out followed by a 0.2-ns equilibration period where the protein and the cation positions were restrained. After this, unrestrained molecular dynamics simulations were performed in the NPT ensemble. A cutoff was used for longer-range interactions: 10 Å for van der Waals interactions and 18 Å for electrostatic interactions. Note that the distances between consecutive cations at the start of the simulation do not exceed this cutoff. We have also performed a simulation using particle mesh Ewald (PME) (Darden et al., 1993) to treat the longer-range interactions. The time step was 2 fs with the LINCS algorithm (Hess et al., 1997) to constrain bond lengths. A constant pressure of 1 bar independently in all three directions was used with a coupling constant of τp = 1.0 ps. Water, protein, and lipid were coupled separately to a temperature bath at 300K using a coupling constant τT = 1.0 ps. Coordinate sets were saved every 0.1 ps for analysis. For comparison purposes, a simulation where no cations were included has been performed.
Lipid parameters were based on those by Berger et al. (1997) and Marrink et al. (1998). The lipid-protein interactions used GROMOS parameters, and parameters derived from those of Åqvist (1990) for use in GROMACS were used for the ions. All simulations used GROMACS v3 (Berendsen et al., 1995; Lindahl et al., 2001) (www.gromacs.org). All the simulations were performed on a 64 CPU. Analysis used programs in the GROMACS suite plus locally written code. The HOLE program (Smart et al., 1993) was used to calculate the pore radius profiles. Structural diagrams were prepared using VMD (Humphrey et al., 1996).
Initial system configurations
Three different configurations of cations in the selectivity filter and the cavity were used (Fig. 1 B). The definition of each of the sites in KcsA is as follows: sites S1–S4 form the selectivity region per se; an additional site, S0, at the external mouth was recently reported (Zhou et al., 2001), and was also seen in simulation studies (Bernèche and Roux, 2001; Sansom and Shrivastava, 2002; Sansom et al., 2002). Each of these sites is defined as the center of two rings of four oxygen atoms. Site S1 is formed by the carbonyl oxygens of residues Y78 and G77 of each of the monomers: S2 by the carbonyl oxygens of G77 and V76, and S3 by the carbonyl oxygens of residues V76 and Y75. Site S4, next to the central cavity, is formed by one ring of carbonyl oxygen (from T75) and another ring of hydroxyl oxygens from the side chains of the same residue T75. Finally, S0 is defined by the carbonyls of G79, which provide four oxygen atoms with the remaining four oxygens being donated by water molecules at the extracellular mouth.
By exploring different configurations of ions and water molecules within the filter we initiated simulations from different intermediates along the proposed pathway of ion conduction through the selectivity filter (Doyle et al., 1998; Morais-Cabral et al., 2001; Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001; Tieleman et al., 2001). Each of the three configurations was repeated for the four cations (Na+, K+, Rb+, and Cs+). A further simulation was run in which all of the filter sites were occupied by water molecules. A summary of all the simulations performed is provided in Table 1. Thus, the total simulation time was 48 ns.
TABLE 1.
Summary of simulations
| Ion | Initial configuration* | Cutoff versus PME | Duration (ns) |
|---|---|---|---|
| K+ | Configuration 1 | cutoff | 9 |
| Configuration 1 | PME | 9 | |
| Configuration 2 | cutoff | 2 | |
| Configuration 3 | cutoff | 2 | |
| Na+ | Configuration 1 | cutoff | 4 |
| Configuration 2 | cutoff | 2 | |
| Configuration 3 | cutoff | 2 | |
| Rb+ | Configuration 1 | cutoff | 4 |
| Configuration 2 | cutoff | 2 | |
| Configuration 3 | cutoff | 2 | |
| Cs+ | Configuration 1 | cutoff | 4 |
| Configuration 2 | cutoff | 2 | |
| Configuration 3 | cutoff | 2 | |
| None | − | cutoff | 2 |
For definitions of the configurations, see Fig. 1 B.
RESULTS AND DISCUSSION
Structural drift
A simple measure of the drift in structure of the protein from its initial, crystallographic conformation is provided by the root-mean-square deviation (RMSD) between the channel structure at a given time and the initial structure. Analysis of all atom RMSD and alpha carbon atom RSMD for the entire tetramer versus time revealed a similar pattern for each of the simulations over the first 1–2 ns (Fig. 2). There is a jump of ∼1 Å at the start of each simulation followed by a small drift to a plateau value of ∼2 Å which is maintained for the initial period (∼1–2 ns) of the simulation. This is a little lower than the plateau value of ∼2.5 Å after 1 ns seen in comparable simulations based on the lower resolution structure of KcsA (Shrivastava and Sansom, 2000). The initial structural drift is thought to reflect the fact that the restraints imposed during the equilibration period have been switched off and that the protein is free to relax within the lipid bilayer and the solvent. The overall RMSD over the first 2 ns is the same for all of the simulations, regardless of the ionic species or configuration. This suggests that nature and configuration of the ions does not influence the overall conformational stability of the protein at this timescale.
FIGURE 2.
All atom RMSDs from the starting structure as a function of time. (A) Simulations with K+ (black line) or Na+ (gray line) ions within the filter (both initially in Configuration 1—see Fig. 1 B). (B) Simulations with Rb+ (black line) or Cs+ (gray line) ions within the filter (again, both initially in Configuration 1; see Fig. 1 B).
When the Configuration 1 simulations were extended to 4 ns, a more complex picture emerged. For all four ionic species a further rise in RMSD was observed. These increases were due to the exit of one or more ions from the selectivity filter (see below). For example, for the Cs+ Configuration 1 simulation, a substantial rise in RMSD was seen at ∼2.8 ns; at that time there is just one ion at site S4 and the filter started to distort. For the K+ and Na+ Configuration 1 simulations, their RMSDs remained lower than for Rb+ and Cs+ because ions remained within the filter. In Fig. 3 we show snapshots of the selectivity filter at 4 ns in simulations with K+, Rb+, and Cs+. In the case of K+, ions remain at sites S2 and S4, with water molecules at S1 and S3. For Rb+ and Cs+, only one ion remains close to the filter, in the vicinity of S4. In both cases the filter has become occupied by a number of water molecules and has distorted substantially.
FIGURE 3.
Snapshots of the filter at 4 ns for the simulations with the following ions initially in Configuration 1: (A) K+; (B) Rb+; and (C) Cs+.
We also ran a short (2-ns) simulation in the absence of any ions within the channel. This resulted in a small but significant increase in the overall RMSD (to ∼2.5 Å) toward the end of the 2-ns period. Detailed examination of the filter (data not shown) reveals substantial distortions of the filter backbone, movements of the crystallographic waters “behind” the filter (see below), and entry of several (∼8) water molecules into the volume created by the distorted filter. Together with the distortion of the filter late in the Rb+ and Cs+ simulations, this suggests that occupancy of the selectivity filter by cations is an essential component of its conformational stability, even on a nanosecond timescale.
In the remainder of this article we will focus on analysis of the 2-ns simulations during which the filters remained at least partly occupied by ions. This was the main reason why many simulations of 2 ns were performed rather than longer simulations; the average timescale before these ions vacate the filter is ∼2 ns. It is possible if we had a high concentration of Rb+ or Cs+ in the bulk solution that further ions might have entered the filter. Alternatively, one may place ions at the extracellular mouth of the filter after the filter empties to run longer simulations. However, the latter was judged to be a somewhat artificial approach and so we have focused on the behavior of the filter while it is occupied by the different ionic species, rather than on entry/exit of ions.
Given these considerations, it is perhaps more informative to examine the RMSD values over the first 2 ns, i.e., while the filters remain occupied. In particular, we have examined the RMSDs for the different regions of the KcsA molecule (Table 2). Similar trends are seen for the different configurations of a given ionic species. In particular, the magnitude of structural drift is generally M1 > M2, P > F (filter). Thus, as might be anticipated, the structural drift is largest for the surface elements (M1) of the molecule and smallest for the core (filter). If one compares the structural drift of the filter between different simulations, then the general pattern that emerges is [no ions] > Na+, Cs+ > Rb+, K+. Thus we would expect to see some effect of the presence or absence of ions and the ionic species on the conformational dynamics of the channel. In particular, we note that, in the absence of ions, the filter RMSD tends to be the highest of that of any of the (nonloop) regions of the protein, i.e., the opposite of what is seen in the presence of ions.
TABLE 2.
RMSDs for simulations
| No ion
|
Na+
|
K+
|
Rb+
|
Cs+
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Structural element | Cα | All | Cα | All | Cα | All | Cα | All | Cα | All |
| Configuration 1 | ||||||||||
| Tetramer | 1.9 | 2.1 | 1.7 | 2.1 | 1.5 | 1.9 | 1.7 | 2.1 | 1.7 | 2.1 |
| M1 | 1.1 | 1.5 | 1.4 | 1.9 | 1.0 | 1.5 | 1.4 | 1.9 | 1.0 | 1.4 |
| M2 | 0.9 | 1.5 | 1.0 | 1.2 | 0.9 | 1.2 | 0.8 | 1.1 | 0.9 | 1.3 |
| P-helix | 0.9 | 1.6 | 0.9 | 1.3 | 0.9 | 1.2 | 0.8 | 1.1 | 0.8 | 1.3 |
| Filter | 1.5 | 1.7 | 0.9 | 1.4 | 0.6 | 1.0 | 0.6 | 1.0 | 1.2 | 1.3 |
| Configuration 2 | ||||||||||
| Tetramer | 1.9 | 2.1 | 1.8 | 2.1 | 1.7 | 2.2 | 1.7 | 2.0 | 1.7 | 2.0 |
| M1 | 1.1 | 1.5 | 1.5 | 1.9 | 0.9 | 1.4 | 1.3 | 1.5 | 1.3 | 1.5 |
| M2 | 0.9 | 1.5 | 1.1 | 1.6 | 0.8 | 1.3 | 0.9 | 1.3 | 0.9 | 1.3 |
| P-helix | 0.9 | 1.6 | 0.9 | 1.4 | 0.8 | 1.3 | 1.0 | 1.5 | 1.0 | 1.5 |
| Filter | 1.5 | 1.7 | 0.9 | 1.2 | 0.5 | 1.0 | 0.8 | 1.1 | 0.8 | 1.0 |
| Configuration 3 | ||||||||||
| Tetramer | 1.9 | 2.1 | 1.5 | 1.9 | 1.6 | 1.9 | 1.6 | 2.1 | 1.6 | 2.1 |
| M1 | 1.1 | 1.5 | 1.1 | 1.5 | 0.9 | 1.5 | 1.3 | 1.6 | 1.2 | 1.6 |
| M2 | 0.9 | 1.5 | 0.8 | 1.2 | 0.9 | 1.4 | 0.9 | 1.2 | 1.0 | 1.2 |
| P-helix | 0.9 | 1.6 | 0.8 | 1.2 | 0.8 | 1.4 | 0.9 | 1.2 | 1.0 | 1.2 |
| Filter | 1.5 | 1.7 | 0.9 | 1.3 | 0.6 | 1.0 | 0.8 | 1.1 | 0.9 | 1.2 |
The RMSD at 2 ns (i.e., while ions remain in the filter; see text) of all atoms and the Cα atoms (Å) of the full tetramer, the outer helix (M1), the inner helix (M2), the pore helix (P), and the selectivity filter (Filter) from their starting structure from the initial model with the ions arranged in each of the three configurations.
A slightly more complex pattern is seen if one examines the structural dynamics of the selectivity filter by calculating the RMSD versus time for each of the simulations over the first 2 ns (Fig. 4). Again, the presence of K+ ions in the selectivity filter stabilizes its conformation. If Na+, Rb+, or Cs+ are present in place of K+, the filter conformation tends to be somewhat distorted. This effect is much greater if no ions are present within the filter—within 1 ns, the filter RMSD jumps from ∼0.8 to ∼1.5 Å. Thus it seems that, even in simulations starting with the high resolution structure of the channel, the filter conformation is not maintained in the absence of cations. This may also be correlated with the changes in filter conformation observed if crystals are grown in the presence of a much lower concentration of K+ ions (Zhou et al., 2001).
FIGURE 4.
Filter region (defined as residues T75 to G79) Cα atom RMSDs from the starting structure as a function of time. (A) Simulations with K+ (solid black line) or no ions were present in the filter or cavity (water, solid gray line). (B) Simulations with Rb+ (solid black line) or Cs+ (solid gray) ions within the filter. In all cases the ions were initially in Configuration 2.
Transitions of ions along the pore axis: K+, Rb+, and Cs+
Movements of the cations and crystallographic water molecules were examined in terms of their z coordinates (i.e., their projection onto the pore axis) as a function of time. Note that z = 0 corresponds to center of the selectivity filter, i.e., site S2. A sample of such ion trajectories is provided in Fig. 5. Visual inspection of trajectories was used to identify transitions between the ion binding sites in the filter. The results of this analysis are presented in Table 3.
FIGURE 5.
Trajectories along the pore (z) axis of K+, Rb+, Cs+, and Na+ ions (solid black lines). Trajectories of selected water molecules within the filter are shown (solid gray lines). The thin horizontal lines correspond to the binding sites of the selectivity filter, S0–S4. The center of the filter (site S2) is at z = 0 Å and the intracellular gate of the channel at z ∼ −20 Å.
TABLE 3.
Transitions between ion binding sites
| Configuration 1 | Na+
|
K+ | Rb+ | Cs+ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| t event (ns) | Initial | 0.002 | 0.3 | 0.3 | 0.3 | 0.02 | ||||
| S0 | ||||||||||
| S1 | + | |||||||||
| S2 | + | + | + | + | + | |||||
| S3 | + | |||||||||
| S4 | + | + | + | + | ||||||
| Cavity | + | + | 2+ | + | + | + | ||||
| Configuration 2 | Na+
|
K+ | Rb+
|
Cs+ | ||||||
| t event (ns) | Initial | 0.002 | 1.0 | 0.23 | 0.2 | 0.5 | 0.125 | |||
| S0 | + | |||||||||
| S1 | + | + | ||||||||
| S2 | + | + | + | + | + | |||||
| S3 | + | + | ||||||||
| S4 | + | + | + | + | + | |||||
| Cavity | + | + | + | + | + | + | ||||
| Configuration 3 | Na+
|
K+
|
Rb+ | Cs+
|
||||||
| t event (ns) | Initial | 0.1 | 1.25 | 0.05 | 1.7 | 0.2 | 0.125 | 1.1 | 1.35 | |
| EC | + | + | ||||||||
| S0 | ||||||||||
| S1 | + | |||||||||
| S2 | + | + | + | + | + | + | ||||
| S3 | + | + | + | |||||||
| S4 | + | + | + | + | + | + | ||||
| Cavity | + | 2+ | + | + | + | 2+ | + | |||
This table records the location of the M+ ions (for each of the three initial configurations) at preferred sites during each simulation. Approximate times at which concerted motion of the ions and water molecules occurred are given. S0–S4 represent the binding sites in the filter (see Fig. 1), and EC is the extracellular mouth of the channel, adjacent to the filter.
Configurations 1 and 2 were selected as representing the two low energy intermediates in the permeation mechanism for ions discussed by Åqvist and Luzhkov (2000), Bernèche and Roux (2001), and Morais-Cabral et al. (2001). At first sight, it appears that K+, Rb+, and Cs+ have some preference to remain in sites S2 and S4. For both the starting configurations where the ions initially occupied sites S1 and S3 (Configuration 1) and where they initially occupied sites S0, S2, and S4 (Configuration 2), there is in general either a single movement or sometimes a double translocation (in the case of Rb+ in Configuration 2) that leads to a state where ions occupy S2 and S4. In the case of the K+ ion, these transitions take place within the first ∼0.3 ns. The K+ ions remain then in these positions for the next ∼8 ns (see below) of the simulation. A similar pattern was observed for Cs+, but the transition to S2 and S4 occurred somewhat sooner, within the first ∼0.1 ns.
When Rb+ initially occupies sites S1 and S3, there is a single translocation of the ions and water molecules toward sites S2 and S4 that takes place at ∼0.3 ns. This is similar to what happens with K+ in Configuration 2. However, when Rb+ ions are initially distributed according to Configuration 2, the first displacement of ions and water molecules takes place at ∼0.2 ns such that Rb+ ions in the filter go to sites S1 and S3 and the third ion, initially in site S4, makes its way toward the central cavity. After a further 0.3 ns, another concerted motion takes place and the filter ions go to sites S2 and S4. The third rubidium ion already in the cavity travels ∼5 Å down it at the same time. This later configuration remains the same for the remaining 1.5 ns of the simulation (Fig. 6).
FIGURE 6.
Snapshots of the Rb+ (Configuration 2) and Na+ (Configuration 1) simulations. In both cases the filter regions (backbone atoms only) of two subunits plus the ions and water molecules within the filter are shown. For the Na+ simulation, the flip of the carbonyl of V76 is indicated with an arrow.
The third configuration was selected as a putative “high energy” arrangement of the ions in the filter (Doyle et al., 1998), as it is difficult to envisage two M+ ions occupying consecutive positions (i.e., sites S3 and S4). Thus we wished to examine whether, starting from this configuration, the cavity could be populated by more than one ion—thus possibly driving exit of an ion through the closed gate.
Simulations starting from Configuration 3 are the most “dynamical” in the sense that only Rb+ ions perform a single motion during the entire simulation moving from sites S1, S3, and S4, to sites S2, S4, and the central cavity, respectively. In simulations where K+ and Cs+ ions are involved, some “rare” events occur—i.e., an ion initially in the central cavity exits through the intracellular mouth. Analysis of the pore radius profile of this region as a function of time and any possible correlation with the transitions of the ions will be reported later.
There are also differences between the K+ and Cs+ ions. After the first displacement of K+ and Cs+ takes place at ∼0.1 ns the ions occupy sites S2 and S4 and the central cavity. Subsequently, the K+ ions in the cavity exit through the intracellular “gate.” In contrast, in the Cs+ Configuration 3 simulation, a subsequent concerted motion leads toward a situation where two ions are found in the cavity separated by ∼15 Å. Interestingly, subsequently the Cs+ situated lower down in the cavity interacts with Cβ of A108 and A111 for ∼0.5 ns before exiting the cavity at 1.35 ns.
Transitions of ions along the pore axis: Na+
The behavior of Na+ ions is somewhat different. Overall, it seems that the Na+ ion has a special preference for site S2, albeit in a distorted conformation (see Fig. 6 and below). Indeed, there are some general differences between this ion and the others. In the three different simulations carried out there is a double concerted motion in contrast with the single motion described before. When Na+ is initially in sites S1 and S3, or in S0, S2, and S4, there is a fast displacement toward the next set of sites (S2 and S4 or S1 and S, respectively), which occurs within ∼2 ps from the removal of restraints. Afterwards, in the first simulation considered (Configuration 1), the one Na+ ion remains in site S2 for the rest of the simulation while the ion in site S4 moves on toward the cavity. Thus, after 0.3 ns, two Na+ ions are coexisting in the central cavity. From the analysis of the trajectories along the pore axis versus time, it can be observed that these two cavity Na+ ions can be as far as 9 Å from each other or as close as 4 Å.
In contrast, in Configuration 2, after the first concerted displacement, there is a second displacement at ∼1.0 ns whereby Na+ ions subsequently occupy sites S2 and S4 and the ion in the cavity travels ∼5 Å toward the gate region. When the Na+ ions are initially arranged in Configuration 3, there is a first movement at 0.1 ns whereby each of the ions moves down one site (thus locating one Na+ at S2). At 1.25 ns the ion in site S4 enters the cavity. As soon as this ion enters the cavity, the ion already in the cavity is displaced by ∼5 Å down toward the intracellular region in a concerted motion.
Overall, it therefore seems that K+, Rb+ and Cs+ ions prefer sites S2 and S4 whereas Na+ seems to have some preferences only for site S2 (in a somewhat distorted conformation—Fig. 6). K+ and Rb+ ions seem to share the strongest similarities regarding their residence times at the filter sites. Configuration 3 seems to be relatively unstable, provoking multiple occupancy of the central cavity and followed by exit of ions through the intracellular gate.
Coordination numbers of the ions
There are many definitions for the term “coordination number,” although there is no one simple and unambiguous definition that works in all cases. In the current context, it will be defined as the number of oxygen atoms either from a water molecule or from the protein surrounding each of the ions. Therefore, the coordination number is to be considered to be a dynamic parameter as it changes with time. In general terms, the bigger the size of the ion, the larger the number of neighbors that will be able to accommodate around itself and therefore, it should be expected that the coordination number will increase from Na+ toward Cs+. In the present study, an O atom is considered to be coordinating an ion if the distance of this oxygen atom to the ion is <2.7 Å for Na+, <3.2 Å for K+, <3.9 Å for Rb+, and <4.0 Å for Cs+. These distances are based on data for ion/ligand interactions (Tieleman et al., 2001).
The coordination of the ions within the filter at each of the four sites S1–S4 is made up of eight oxygen atoms, at most, from the protein arranged at the corners of a twisted cube with an ion in the center and the oxygen of the water in the nearest sites. Once in the cavity, the coordination number will be dominated by the number of oxygens provided by the waters surrounding the ion. Between four and eight oxygen atoms from the protein coordinate the K+ ion while in the selectivity filter, and up to nine oxygen atoms coordinate the K+ if the oxygen atoms from the water molecules are taken into account. Once in the cavity, the K+ ion is 5–7 coordinated by the water molecules. Na+ ions are in general coordinated by a smaller number of atoms as expected due to geometrical considerations. For example, six oxygen atoms coordinate the Na+ ion once in the selectivity filter, 4–6 of which belong to the protein and the rest, up to six, belonging to the water molecules. As can be seen from Fig. 6, this involves some local deformation of the filter such that a Na+ ion close to S2 sits in the center of a ring of four oxygens from G77 carbonyls. The same coordination number (from water oxygens) is observed once a Na+ ion has moved to the cavity. On the other hand, as Rb+ is larger, it can accommodate more oxygen atoms around it. For most of the time, up to 12 water oxygen atoms surround this ion when it is in the cavity. However, we have observed a rare event where 10 of oxygen atoms coordinating Rb+ belonged to the protein. During the Configuration 3 simulation while a Rb+ ion was in transition between S1 and S2, the carbonyl oxygens of Y78, G77, and V76 all contributed to the coordination of Rb+. This event did not coincide with any particular deformation of the selectivity filter. In the other cases, the oxygen atoms belonged both to the protein and the water molecules.
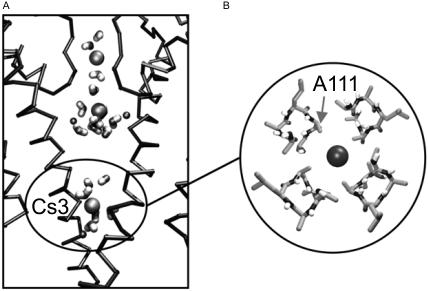
Finally, Cs+ is most of the time coordinated by 10 oxygen atoms; sometimes it can be seen surrounded by up to 14 water molecules when in the cavity. When in the filter region, eight of these oxygen atoms belong to the protein and the rest belong to the water molecules. However, as in the case of Rb+, there are particular situations in which nine or 10 oxygen contributions are from the protein. For example, a Cs+ ion at S2 was coordinated by eight carbonyl oxygens from G77 and V76 plus extra carbonyl oxygens from some of the Y78 residues. Within the cavity, the coordination number of the Cs+ varies from six to 14. It is important to mention here that in two of the simulations, Cs+ spends an important amount of time in the hydrophobic region close to the intracellular mouth. This is not surprising, as Cs+ is considered the most hydrophobic ion in the alkali series. While in this region, Cs+ also interacts with the Cβ atoms of A108 and A111 (Fig. 7).
FIGURE 7.
Cs+ ions in the cavity. (A) Filter and cavity region (two subunits only) of simulation Cs+ Configuration 3, showing ions and water molecules within the channel. Time, 0.95 ns, The lower Cs+ ion in the cavity (labeled Cs3) is located in a hydrophobic region created by a ring of A111 side chains. This is shown in more detail in B.
Thus it would seem that within the simulations the various ions have, on average, a full first coordination shell. The exception to this is within the filter. Here the coordination number is generally eight (except for Na+). A higher coordination number may be transiently achieved during transition of, for example, Rb+ between sites, associated with only minor local deformations of the peptide backbone. Within the cavity full coordination by water molecules occurs, although Cs+ shows some propensity for interacting with more hydrophobic regions of the cavity.
Water molecules forming part of the filter
Recent experimental work extending the resolution of the KcsA structure from 3.2 Å to 2.0 Å revealed four oxygen atoms from four water molecules sitting “behind” the filter, i.e., between the filter backbone and the remainder of the protein. These water molecules H-bond to the amide nitrogen of G79 and participate in a H-bond network between residues G79 and D80 (Zhou et al., 2001). As these four waters (Fig. 8) thus seem to form an essential component of the filter we were interested in their behavior over the course of the simulations with the different ionic species.
FIGURE 8.
Diagram of KcsA (viewed from the extracellular mouth looking down the pore axis), highlighting the four “filter” water molecules located “behind” the filter. The axes define the plane perpendicular to the pore (see Fig. 9).
The movement of these crystallographic waters (which are fully buried within the protein) has been followed during the simulation and no appreciable correlation of their rather limited motions with the concerted translocations of the ions and the water in the selectivity filter has been observed. There is no significant change in the coordinates of these water molecules in the simulations with Na+ ions. In the simulations with K+, the water molecules remain in their initial position and the small oscillations (RMSF ≤ 0.1 Å) around these initial positions are somewhat smaller than the oscillations observed in the carbonyl oxygen atoms of the selectivity filter (RMSF ∼ 0.5 Å). However, there is a single event worth mentioning during the course of the simulation Configuration 1 (Fig. 9 A). The x coordinate of one of the buried water molecules changes by ∼5 Å for ∼0.15 ns before returning to its initial value. This water movement is correlated with the changes in the backbone torsion angles of residues Y78 and G77 which occur in one of the monomers in the selectivity filter region at this time. This transient peptide backbone “flip” does not appear to correlate with an ion-water translocation event within the filter.
FIGURE 9.
Trajectories in the plane perpendicular to the pore axis (see Fig. 8) of the “filter” water molecules (shown as distance along x versus time). (A) Simulation K+, Configuration 1; (B) Rb+, Configuration 2; and (C) Cs+, Configuration 1. The black lines are the ion trajectories and the blue lines correspond to centers of the binding sites of the selectivity filter. In B, the red lines show the trajectories of the O atoms of V76 .
A similar picture is observed for Rb+ where there is no significant deviation from the initial positions of the buried waters except in one case (Fig. 9 B). During the Configuration 2 simulation one of the buried molecules is displaced up to 7 Å from its initial position. Again, this movement does not correlate with the concerted translocation of ions and water along the selectivity filter. However, it is correlated with the movement of the carbonyl oxygen of the V76 of one of the monomers in the selectivity filter, again due to a change in backbone torsion angles. This local distortion last for ∼1 ns after which the peptide backbone flips back to its crystal conformation, pulling the bound water back to its initial location.
In relation to the Cs+ simulations, in Configuration 1 two significant displacements in the x-y plane are observed. The magnitude of these displacements is ∼1–3 Å. One of the displacements takes place at ∼0.2 ns and lasts for ∼0.5 ns whereas the second one takes place at ∼1.2 ns. These changes are correlated with the changes in the backbone conformation of the G77 residue of one monomer. The second of the displacements is correlated with the changes in the backbone conformation of residue Y78 of the filter in the same monomer.
Thus, it can be seen that the buried water molecules behave as a component of the filter in that 1), they do not exchange (at least on a 2-ns timescale) with bulk water or water molecules within the filter; and 2), their motions are strongly coupled to those of the surrounding protein. Their motions do not appear to be coupled to the pattern of occupancy of the sites in the filter with cations and water molecules. This behavior is approximately the same regardless of whether PME or cutoff is used to treat the longer-range electrostatics (see below).
Influence of simulation protocol
To facilitate comparison with our previous studies (Shrivastava and Sansom, 2000, 2002; Shrivastava et al., 2002; Sansom et al., 2002) we have used cutoffs to truncate the long-range nonbonded electrostatics interactions (>18 Å) rather than Ewald summation. However, we are aware that such truncation of electrostatic interactions, even at large cutoff distances, may neglect important ion-ion interactions that could result in simulation artifacts (i.e., one ion might not “see” other ions). An alternative is to use the Ewald summation approach. This avoids the possible artifacts due to cutoff. However, an alternative source of simulation artifact is possible when Ewald summation is employed (Hunenberger and McCammon, 1999), namely long-range electrostatic “coupling” between a simulation box and its periodic images, and so a detailed comparison of the effects of different simulation protocol is required (Domene and Sansom, unpublished data). A preliminary comparison using a closely related K channel model did not suggest any profound differences between the simulations run using cutoffs and those run using PME (Capener and Sansom, 2002). For the current (high resolution) KcsA structure we extended the comparison by running K+ Configuration 1 simulations with cutoff versus PME for 9 ns (see Table 1). The major difference seems to be one of the timescales of the concerted transitions between sites (Fig. 10). In the cutoff simulation there is a transition from ions at S1 and S3 to S2 and S4 within the first nanosecond, whereas in the 9-ns simulation we do not see a transition until toward the end of the simulation. Of course, these are only single realizations of a inherently stochastic process and so a more detailed analysis will be required in the future. There are also some differences in the behavior of the K+ ion within the cavity that may merit further examination.
FIGURE 10.
Ion trajectories in the two 9-ns simulations with K+ ions initially in Configuration 1, with longer-range electrostatics treated using an 18 Å cutoff (A) versus particle mesh Ewald (B).
Events at the gate
Based on a variety of theoretical and experimental considerations it is thought that the x-ray structure of KcsA corresponds to the closed form of the channel. Spectroscopic (Perozo et al., 1998, 1999; Liu et al., 2001) and simulation (Shrivastava and Sansom, 2002; Biggin and Sansom, 2002) studies suggest that relatively small displacements of the M2 helices at the intracellular mouth of the channel could switch it to a functionally open conformation. Comparison with the x-ray structure of a Ca-gated K channel, MthK (Jiang et al., 2002b), trapped in an open state also suggest (rather larger) motions of M2 produce channel opening. In the earlier simulations, based on the 3.2 Å resolution structure, rare ion exit events seemed to be associated with opening of the intracellular mouth of the channel. The higher resolution structure has revealed an additional three residues at the C-terminus of each M2 helix. Thus, spontaneous opening of the pore in simulations based on the 2 Å structure might be anticipated to be less likely. We were therefore interested to examine possible changes in the IC mouth of the pore associated with rare ion exit events.
In the crystal structure, the radius of the intracellular mouth (i.e., the gate region) of the pore in the vicinity of the intracellular gate reaches a minimum of ∼1 Å (Fig. 11 A). This means that its dimension is narrower than the radius of most the ions considered in these simulations, namely 0.95 Å for Na+, 1.33 Å for K+, 1.48 Å for Rb+, and 1.69 Å for Cs+. This implies that, for a rare exit event to occur, the radius of the intracellular mouth of the channel should increase. Furthermore, the side chains lining the pore in this region are mainly hydrophobic, thus presenting an energetic barrier to ion exit/entry from/to this region. The fact that this region is hydrophobic can also explain the observation made in these simulations related to the time the Cs+ ions spend there; Cs+ is thought to be a relatively hydrophobic ion (Jungwirth and Tobias, 2001).
FIGURE 11.
Pore radius profiles for simulation Cs+ Configuration 3. In A, the pore radius profiles are compared for the x-ray structure (gray line) and the simulation structure (black line) at t = 1.35 ns, i.e., just as a Cs+ ion is exiting through the intracellular mouth of the pore. In B, the average pore radius profile (black line) over the duration of the simulation is shown, along with error bars (± SD, in gray). The horizontal broken line is the radius of an unhydrated Cs+ ion.
However, dynamic fluctuations in the pore radius occur in the gate region (as they also do in the filter region). For example, for simulation Cs+ Configuration 3, at about the time (t = 1.35 ns) that a Cs+ ion exits the channel through the intracellular mouth, the pore radius in this region is nearly 2 Å (Fig. 11 A)—i.e., just greater than the radius of a (dehydrated) Cs+ ion. If, for the same simulation, one calculates the average pore radius profile over the whole simulation (Fig. 11 B) it is evident that there are considerable fluctuations in the radius in both the gate and filter regions, such that in both cases the radius must transiently exceed that of a Cs+ ion.
In the light of recent structural studies (Jiang et al., 2002b) we have examined those trajectories, wherein an ion exits through the intracellular mouth, for any evidence of M2 helix bending near the conserved glycine residue (G99) which is suggested to play a role as a molecular hinge in channel gating. Although ion exit is associated with small concerted motions of the helices (as reflected in the pore radius profiles), there is no pronounced hinge-bending around the G99 residues. It will be of interest to explore in future studies whether or not different degrees of stable channel opening can occur.
CONCLUSIONS
In this study we have performed a substantial (48-ns total simulation time) new set of KcsA simulations. These simulations are a substantial advance on a number of previous studies in that: 1), they are based on the higher resolution structure; 2), they incorporate the buried waters that help to stabilize the filter; and 3), they take into account E71-D80 proton-sharing side-chain pairs (we note that the latter feature was included in previous studies by Bernèche and Roux (2002). Encouragingly, the newer simulations support a number of conclusions derived from simulations based on the lower resolution structure, both by our research group and by other investigators. In particular, we see concerted motions of ions and water within the filter, alongside a degree of flexibility of the filter and much more pronounced distortion of filter if no ions are present within it.
An important aspect of the current studies is that it might be expected that by starting with a structure in which interactions around the filter were more clearly resolved, a lesser degree of flexibility would have been observed in this region in simulations. However, this does not seem to be the case, supporting the suggestion that such conformational fluctuations are an important aspect of the functional dynamics of K channels at physiological temperatures. Thus the flexibility of the KcsA channel (and by extension of other K channels) seems to be an inherent property, and not simply a consequence of the low resolution structure and absence of “crystallographic” water molecules from previous studies. This is of evident functional importance in terms of trying to assess the importance of the balance between rigidity and flexibility in the design of a selective yet high throughput channel.
A detailed comparison of the behavior of four different alkali metal cations has been performed. This also extends previous studies, in particular in attempting to address the extent to which ions larger than K+ might be accommodated within the filter. The two most permeant ions, K+ and Rb+, are similar in their interactions with the selectivity filter. This is consistent with their comparable permeation properties. Na+ ions tend to distort the filter “inwards” by binding to one ring of our carbonyl oxygens rather than eight. The larger Cs+ ions result in a small degree of expansion of the filter (see Fig. 9) relative to the x-ray structure. They also seem to interact differently with the gate region of the channel. Interestingly, in our simulations we observe Rb+ ions at site S2 in the filter. This has not been seen in the x-ray studies (Morais-Cabral et al., 2001). This merits further investigation to see whether it reflects shortcomings of the simulation parameters (see below) or a possible difference between ion/filter interactions at 300 K (in the simulations) and at 100 K (in the crystal).
It is of particular interest that, even given the filter flexibility seen in the simulations, the buried water molecules “behind” the filter do not exchange with bulk water (on a nanosecond timescale) but rather appear to function as an integral part of the filter, only moving substantially when rare peptide bond flips occur. This suggests that it will be important to consider equivalent buried water molecules when using KcsA to generate homology models of other K channels (Capener et al., 2003). Of course, this does present a further layer of difficulty when using homology modeling to extrapolate from the x-ray structure of bacterial K channels to models of, for example, human channels.
A limitation of the current study that must be addressed is that of the parameters used for ion-filter and ion-water interactions. These do not allow for a consideration of polarization effects (Roux and Bernèche, 2002), a possible limitation shared with a number of other studies of ion selectivity (Allen et al., 2000; Åqvist and Luzhkov, 2000) and permeation (Bernèche and Roux, 2001) of KcsA. Some results have been presented in other articles concerning the robustness of simulation studies to ion parameters (Shrivastava et al., 2002; Tieleman et al., 2001). Although some sensitivity was reported, a consensus of 2.2–2.3 Å for the Na/O distance versus 2.7–2.8 Å for the K/O distance emerges which is similar to the distances seen for interactions within the selectivity filter in these simulations. However, it is remains likely that electronic polarization could play an effect during movement of the M+ ions between sites (Guidoni and Carloni, 2002) and this will merit further study. In the absence of a fully developed polarizable force field for ions, water, protein and lipids the current study represents an attempt to delineate some of the main features of KcsA permeability and selectivity in terms of filter flexibility vs. rigidity when starting with a high resolution x-ray structure. A more detailed energetic analysis (Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001) will doubtless benefit from an improved force field.
Acknowledgments
Our thanks to all of our colleagues for valuable discussions, especially Charlotte Capener and José Faraldo-Gómez.
This work was funded by the Wellcome Trust, and supported by the Oxford Supercomputer Centre.
References
- Allen, T. W., A. Bliznyuk, A. P. Rendell, S. Kuyucak, and S. H. Chung. 2000. The potassium channel: structure, selectivity and diffusion. J. Chem. Phys. 112:8191–8204. [Google Scholar]
- Allen, T. W., and S. H. Chung. 2001. Brownian dynamics study of an open-state KcsA potassium channel. Biochim. Biophys. Acta. 1515:83–91. [DOI] [PubMed] [Google Scholar]
- Allen, T. W., S. Kuyucak, and S. H. Chung. 1999. Molecular dynamics study of the KcsA potassium channel. Biophys. J. 77:2502–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist, J. 1990. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phys. Chem. 94:8021–8024. [Google Scholar]
- Åqvist, J., and V. Luzhkov. 2000. Ion permeation mechanism of the potassium channel. Nature. 404:881–884. [DOI] [PubMed] [Google Scholar]
- Ashcroft, F. M. 2000. Ion Channels and Disease. Academic Press, San Diego, CA.
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Intermolecular Forces. Reidel, Dordrecht, The Netherlands.
- Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 95:43–56. [Google Scholar]
- Berger, O., O. Edholm, and F. Jahnig. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidycholine at full hydration, constant pressure and constant temperature. Biophys. J. 72:2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche, S., and B. Roux. 2000. Molecular dynamics of the KcsA K+ channel in a bilayer membrane. Biophys. J. 78:2900–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche, S., and B. Roux. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77. [DOI] [PubMed] [Google Scholar]
- Bernèche, S., and B. Roux. 2002. The ionization state and the conformation of Glu-71 in the KcsA K+ channel. Biophys. J. 82:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin, P. C., and M. S. P. Sansom. 2002. Open-state models of a potassium channel. Biophys. J. 83:1867–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin, P. C., G. R. Smith, I. H. Shrivastava, S. Choe, and M. S. P. Sansom. 2001. Potassium and sodium ions in a potassium channel studied by molecular dynamics simulations. Biochim. Biophys. Acta. 1510:1–9. [DOI] [PubMed] [Google Scholar]
- Capener, C. E., and M. S. P. Sansom. 2002. MD Simulations of a K channel model—sensitivity to changes in ions, waters and membrane environment. J. Phys. Chem. B. 106:4543–4551. [Google Scholar]
- Capener, C. E., I. H. Shrivastava, K. M. Ranatunga, L. R. Forrest, G. R. Smith, and M. S. P. Sansom. 2000. Homology modeling and molecular dynamics simulation studies of an inward rectifier potassium channel. Biophys. J. 78:2929–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capener, C. E., P. Proks, F. M. Ashcroft, and M. S. P. Sansom. 2003. Filter flexibility in a mammalian K channel: models and simulations of Kir6.2 mutants. Biophys. J. 84:2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes, D. M., L. G. Cuello, and E. Perozo. 2001. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117:165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello, L. G., J. G. Romero, D. M. Cortes, and E. Perozo. 1998. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 37:3229–3236. [DOI] [PubMed] [Google Scholar]
- Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Cahit, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Garofoli, S., G. Miloshevsky, V. L. Dorman, and P. C. Jordan. 2002. Permeation energetics in a model potassium channel. In Ion Channels: from Atomic Resolution Physiology to Functional Genomics. Novartis Foundation Symposium. 109–126. [PubMed]
- Guidoni, L., and P. Carloni. 2002. Potassium permeation through the KcsA channel: a density functional study. Biochim. Biophys. Acta. 1563:1–6. [DOI] [PubMed] [Google Scholar]
- Guidoni, L., V. Torre, and P. Carloni. 1999. Potassium and sodium binding in the outer mouth of the K+ channel. Biochemistry. 38:8599–8604. [DOI] [PubMed] [Google Scholar]
- Guidoni, L., V. Torre, and P. Carloni. 2000. Water and potassium dynamics in the KcsA K+ channel. FEBS Lett. 477:37–42. [DOI] [PubMed] [Google Scholar]
- Heginbotham, L., L. Kolmakova-Partensky, and C. Miller. 1998. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heginbotham, L., M. LeMasurier, L. Kolmakova-Partensky, and C. Miller. 1999. Single Streptomyces lividans K+ channels: functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- Hille, B. 1992. Ionic Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA.
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Hunenberger, P. H., and J. A. McCammon. 1999. Ewald artifacts in computer simulations of ionic solvation and ion-ion interaction: a continuum electrostatics study. J. Chem. Phys. 110:1856–1872. [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002a. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002b. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. X., and R. MacKinnon. 2000. The barium site in a potassium channel by x-ray crystallography. J. Gen. Physiol. 115:269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, P. 2000. Ionic energetics in narrow channels. In Proceedings of the IMA Workshop on Membrane Transport and Renal Physiology. H. Layton, editor. Springer-Verlag.
- Jungwirth, P., and D. J. Tobias. 2001. Molecular structure of salt solutions: a new view of the interface with implications for heterogeneous atmospheric chemistry. J. Phys. Chem. B. 105:10468–10472. [Google Scholar]
- LeMasurier, M., L. Heginbotham, and C. Miller. 2001. KcsA: It's a potassium channel. J. Gen. Physiol. 118:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- Liu, Y., P. Sompornpisut, and E. Perozo. 2001. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8:883–887. [DOI] [PubMed] [Google Scholar]
- Luzhkov, V. B., and J. Åqvist. 2000. A computational study of ion binding and protonation states in the KcsA potassium channel. Biochim. Biophys. Acta. 1481:360–370. [DOI] [PubMed] [Google Scholar]
- Luzhkov, V. B., and J. Åqvist. 2001. Mechanisms of tetraethylammonium ion block in the KcsA potassium channel. FEBS Lett. 495:191–196. [DOI] [PubMed] [Google Scholar]
- Marrink, S. J., O. Berger, D. P. Tieleman, and F. Jahnig. 1998. Adhesion forces of lipids in a phospholipid membrane studied by molecular dynamics simulations. Biophys. J. 74:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashl, R. J., Y. Z. Tang, J. Schnitzer, and E. Jakobsson. 2001. Hierarchical approach to predicting permeation in ion channels. Biophys. J. 81:2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuser, D., H. Splitt, R. Wagner, and H. Schrempf. 1999. Exploring the open pore of the potassium channel from Steptomyces lividans. FEBS Lett. 462:447–452. [DOI] [PubMed] [Google Scholar]
- Morais-Cabral, J. H., Y. Zhou, and R. MacKinnon. 2001. Energetic optimization of ion conduction by the K+ selectivity filter. Nature. 414:37–42. [DOI] [PubMed] [Google Scholar]
- Perozo, E., D. M. Cortes, and L. G. Cuello. 1998. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 5:459–469. [DOI] [PubMed] [Google Scholar]
- Perozo, E., D. M. Cortes, and L. G. Cuello. 1999. Structural rearrangements underlying K+-channel activation gating. Science. 285:73–78. [DOI] [PubMed] [Google Scholar]
- Perozo, E., Y. S. Liu, P. Smopornpisut, D. M. Cortes, and L. G. Cuello. 2000. A structural perspective of activation gating in K+ channels. J. Gen. Physiol. 116:5a. [Google Scholar]
- Ranatunga, K. M., I. H. Shrivastava, G. R. Smith, and M. S. P. Sansom. 2001a. Side-chain ionization states in a potassium channel. Biophys. J. 80:1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranatunga, K. M., G. R. Smith, R. J. Law, and M. S. P. Sansom. 2001b. Electrostatics and molecular dynamics of a homology model of the Shaker K+ channel pore. Eur. Biophys. J. 30:295–303. [DOI] [PubMed] [Google Scholar]
- Roux, B., and S. Bernèche. 2002. On the potential functions used in molecular dynamics simulations of ion channels. Biophys. J. 82:1681–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom, M. S. P., and I. H. Shrivastava. 2002. Ion channels: frozen motion. Curr. Biol. 12:R65–R67. [DOI] [PubMed] [Google Scholar]
- Sansom, M. S. P., I. H. Shrivastava, J. N. Bright, J. Tate, C. E. Capener, and P. C. Biggin. 2002. Potassium channels: structures, models, simulations. Biochim. Biophys. Acta. 1565:294–307. [DOI] [PubMed] [Google Scholar]
- Sansom, M. S. P., I. H. Shrivastava, K. M. Ranatunga, and G. R. Smith. 2000. Simulations of ion channels—watching ions and water move. Trends Biochem. Sci. 25:368–374. [DOI] [PubMed] [Google Scholar]
- Shrivastava, I. H., and M. S. P. Sansom. 2000. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 78:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava, I. H., and M. S. P. Sansom. 2002. Molecular dynamics simulations and KcsA channel gating. Eur. Biophys. J. 31:207–216. [DOI] [PubMed] [Google Scholar]
- Shrivastava, I. H., D. P. Tieleman, P. C. Biggin, and M. S. P. Sansom. 2002. K+ vs. Na+ ions in a K channel selectivity filter: a simulation study. Biophys. J. 83:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, O. S., J. M. Goodfellow, and B. A. Wallace. 1993. The pore dimensions of gramicidin A. Biophys. J. 65:2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splitt, H., D. Meuser, I. Borovok, M. Betzler, and H. Schrempf. 2000. Pore mutations affecting tetrameric assembly and functioning of the potassium channel from Steptomyces lividans. FEBS Lett. 472:83–87. [DOI] [PubMed] [Google Scholar]
- Tieleman, D. P., P. C. Biggin, G. R. Smith, and M. S. P. Sansom. 2001. Simulation approaches to ion channel structure-function relationships. Quart. Rev. Biophys. 34:473–561. [DOI] [PubMed] [Google Scholar]
- Watson, J. D., and E. J. Milner-White. 2002. The conformations of polypeptide chains where the main-chain parts of successive residues are enantiomeric. Their occurrence in cation and anion-binding regions of proteins. J. Mol. Biol. 315:183–191. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]