Abstract
Based on the structure of FKBP12 complexed with FK506 or rapamycin, with computer-aided design, two neurotrophic ligands, (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-Leucine ethyl ester and (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-phenylalanine benzyl ester, were designed and synthesized. Fluorescence experiments were used to detect the binding affinity between FKBP12 and these two ligands. Complex structures of FKBP12 with these two ligands were obtained by x-ray crystallography. In comparing FKBP12-rapamycin complex and FKBP12-FK506 complex as well as FKBP12-GPI-1046 solution structure with these new complexes, significant volume and surface area effects and obvious contact changes were detected which are expected to cause their different binding energies—showing these two novel ligands will become more effective neuron regeneration drugs than GPI-1046, which is currently undergoing phase II clinical trail as a neurotrophic drug. Analysis of volume and surface area effects also gives a new clue for structure-based drug design.
INTRODUCTION
The immunophilins are a family of phylogenically conserved binding proteins possessing peptidal prolyl isomerase (PPI) activity (rotamase), including cyclophilin and FK506 binding protein (FKBPs) (Hauske et al., 1992). Immunosuppressant drugs such as cyclosporin A, FK506, and rapamycin can inhibit their PPI activity. Furthermore, associated with immunosuppressant binding, they can interact with calcineurin to block calcineurin-mediated T-cell activation, resulting in the immunosuppressant effects (Clipstone and Crabtree 1992; Griffith et al., 1995; Liu et al., 1991). FKBPs play multiple roles in cell physiology. Especially, besides the PPIase activity and immunosuppressant function, FKBP12 takes the control of signaling modulation via the membrane-bound EGF/TGF-β receptor and also controls intracellular calcium ion flow by binding inositol(1,4,5) trisphosphate(IP3)/ryanodine receptor (Herdegan et al., 2000). Several FKBP12 complex structures have been determined, such as FKBP12-FK506-calcineurin (Griffith et al., 1995; Kissinger et al., 1995), FKBP12-rapamycin-FRAP-binding domain (Choi et al., 1996), and FKBP12-TGFI-β receptor cytoplasmic domain (Huse et al., 1999). Interestingly, recent studies have demonstrated that the level of FKBP12 in brain is 50× higher than those in tissues of the immune system, and at the same time FK506 and rapamycin take neurotrophic effects by binding to FKBP12 (Dawson et al., 1994; Lyons et al., 1994; Steiner et al., 1992). Although there is little understanding of its neuroprotective or neurogenerative mechanism, FKBP12 was still identified as a drug target and several FK506 analogs which have neurotrophic activity without immunosuppressive action have been designed and synthesized (Adalsteinsson and Bruice, 2000; Becker et al., 2000; Gold, 2000; Gold et al., 1997; Guo et al., 2001; Sabatini et al., 1997; Sauer et al.,1999; Sich et al., 2000; Steiner et al., 1997a). For a long time, it has proved difficult to cure stroke sequelae, central and peripheral nerve injury, and neurodegenerative disorders such as Alzheimer's disease and Parkinson's disease. These findings provide a promising future prospect for the treatment of those diseases.
FK506 possesses one binding domain and one effector domain, respectively, and binds to FKBP12 via the binding domain and calcineurin via the effector domain. Its neurotrophic effect was only determined by binding domain, unrelated to the effector domain (Kissinger et al., 1995; Snyder et al., 1998). So several FK506 analogs with neuroregenerative properties but lacking immunosuppressive effects, such as GPI-1046 (Steiner et al., 1997b) and V-10, 367 (Gold et al., 1997) have been synthesized and described.
Research has been done upon the neuroprotective and neurogenerative effects of GPI-1046 (Zhang et al., 2001), which will become a widely used drug for neuron injury therapy. However, there are still some arguments regarding its suitability for therapeutic use. For example, Winter and co-workers found that GPI-1046, when compared with FK506, did not protect against neuronal death and inhibit c-Jun expression in the substantia nigra pars compacta after transection of the rat medial forebrain bundle (Winter et al., 2000). The mechanism of FK506 as a neurotrophic drug is still obscure, but designing drugs based on the hydrophobic pocket conformation of FKBP12 is always an effective method. Some experiments and our calculations reveal that the binding constant of GPI-1046 to FKBP12 is 1000-fold smaller than FK506 (Graziani et al., 1999), which might explain the results of experiments by Winter and co-workers. So, new drugs with small molecular weight, easy synthesis, and higher affinity are still in need of designing and synthesizing.
High-resolution complex structures of FKBP12 provide a solid basis for designing new compounds. After structure analysis and computer-aided drug design (Structure and Activity Relationship Calculation, i.e., SAR), we designed and synthesized two new neurotrophic compounds: (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-Leucine ethyl ester (which we called compound 308); and (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-phenylalanine benzyl ester (which we called compound 107), which are a little larger than GPI-1046. These two compounds show better neurotrophic effect according to neuron growth experiments in vitro than GPI-1046 (data was provided by Professor Song Li). Fluorescence measurements showed these two compounds can bind to FKBP12 effectively, controlled by the rapamycin binding effect. Crystal structures of FKBP12 complexed with these two compounds were obtained to investigate in more detail the interaction between FKBP12 and these two ligands. From structure analysis and comparison, obvious effects of volume and surface area and considerable changes of atom contacting numbers were observed, presumably causing different binding energy for different compounds. As a result, we conclude that these new designed compounds show a better aspect in neuron-protective drug development than GPI-1046. Our work also reveals that volume and surface area changes resulting from ligand binding to protein are a distinguished factor for designing efficient compounds to serve as protein inhibitors.
MATERIALS AND METHODS
Molecular design
Conformations of FK506 and rapamycin complexed to FKBP12, respectively, have been discussed and compared in detail earlier (Denesyuk et al., 1998), and many compounds have been designed according to the structure of FK506 or rapamycin (Becker et al., 1999, 1993; Christner et al., 1999; Hauske et al., 1992). However, there are a few weak arguments upon structure analysis in detail when designing new drugs. Our strategy was to first make molecular design with further analysis based on the complex structures of FKBP12-FK506 and FKBP12-rapamycin. After that, we introduce two potential compounds as new inhibitors of FKBP12. The affinities of these two potential compounds were then compared to the affinities of FK506, rapamycin, and GPI-1046 using Quantitative Structure-Activity Relationships (QSAR) calculations.
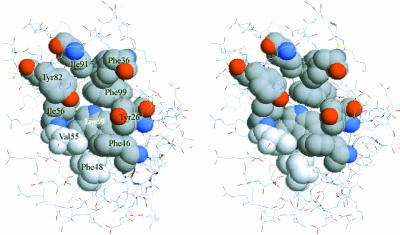
Most FKBPs possess peptidal prolyl cis/trans isomerase (PPIase) activity and might play important roles as chaperones in the restructuring of proteins (Schiene and Fischer, 2000). The conformation of the FKBP binding pocket is specific and facile for Leu-Pro-X peptide segment (where X is a hydrophobic residue such as Leu, Ile, or Val) to bind and change the cis/trans conformation of prolyl, helping the protein to fold well. In FKBP12, the hydrophobic pocket is formed by Tyr82, Ile92, Phe36, Phe99, Tyr26, Phe46, Phe48, Val55, Ile56, and Trp59, and its volume can only accommodate a five-membered or six-membered ring (Fig. 1). Inspecting the complex structures of FKBP12-FK506 and FKBP12-rapamycin, the correspondent part fitting this pocket is obviously the six-membered pipecolinyl ring of FK506 or rapamycin (Denesyuk et al., 1998), so it is necessary to keep this important structure character in new designed ligands.
FIGURE 1.
Stereo view of the binding pocket of FKBP12 formed by the hydrophobic residues Ile91, Phe36, Phe99, Tyr26, Phe46, Phe48, Val55, Ile56, Tyr82, and Trp59.
Further design was based on the detailed investigation of close contacts between FKBP12 and known inhibitors (FK506 and rapamycin). We extracted close atom pairs (Table 1) between protein and ligand according to the cutoff criterion. That is to say, if the distance between two atoms is smaller than the sum of three radii, i.e., the two atoms' radii and the one water molecule radius (2.8 Å), this atom pair will be included. From the listing of close contacts, many contacts are formed among Tyr82, Ile56, Val55, Glu54, and Asp37, and several atoms around the pipecolinyl ring. To design a more efficient compound, these close contacts/interactions should be preserved and more close contacts need to be designed to enhance the binding affinity.
TABLE 1.
Count of contact atom pairs between FKBP12 and inhibitors from five complexes
| 16 Pairs
|
23 Pairs
|
22 Pairs
|
27 Pairs
|
33 Pairs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FKBP12 | FK506 | FKBP12 | Rapamycin | FKBP12 | GPI1046 | FKBP12 | 107 | FKBP12 | 308 | |||||
| Tyr26 | CE2 | O4 | Tyr66 | CE1 | O4 | Tyr26 | CE2 | C5 | Tyr26 | CZ | C7 | Tyr26 | CE1 | O5 |
| Phe36 | CE1 | O4 | Phe36 | CE1 | O4 | CZ | C4 | Phe46 | CE1 | S1 | CE2 | S1 | ||
| Asp37 | CB | O6 | Asp37 | CB | O6 | OH | C5 | Glu54 | C | O5 | CZ | S1 | ||
| CG | O6 | CG | O6 | OH | C4 | O | O5 | Phe36 | CE2 | O4 | ||||
| OD2 | O4 | OD2 | O4 | Phe46 | CE2 | C3 | Val55 | N | O5 | Glu54 | O | C6 | ||
| OD2 | O5 | OD2 | O5 | CZ | C3 | CA | O2 | Val55 | CA | O1 | ||||
| OD2 | O6 | OD2 | O6 | Glu54 | O | O1 | CA | O5 | CB | O1 | ||||
| Glu55 | O | O10 | Phe46 | CZ | O11 | Val55 | CG1 | O1 | CB | O2 | C | O1 | ||
| Val55 | CA | O2 | Glu53 | O | O13 | Tyr82 | CE1 | C15 | C | O2 | C | O10 | ||
| C | O2 | Glu54 | CA | O10 | CZ | C15 | C | O5 | Ile56 | N | O1 | |||
| CB | O2 | C | O10 | OH | O2 | Ile56 | N | O2 | N | C9 | ||||
| Ile56 | N | O2 | O | O10 | OH | C15 | CG2 | C22 | N | C10 | ||||
| CG2 | O2 | O | C30 | OH | C7 | TRP59 | CD2 | S1 | CA | C10 | ||||
| Trp59 | CE2 | C3 | Val55 | CA | O2 | OH | O7 | CE2 | C6 | CB | C10 | |||
| Tyr82 | OH | C42 | C | O2 | His87 | CG | C11 | CE2 | S1 | CG2 | O2 | |||
| OH | O3 | C | O41 | ND1 | C11 | NE1 | C6 | CG2 | C9 | |||||
| CB | O2 | CD2 | C10 | Tyr2 | CE2 | O1 | CG2 | C10 | ||||||
| CG1 | O11 | CD2 | C11 | CE2 | C22 | CG1 | O1 | |||||||
| Ile56 | N | O2 | CE1 | C10 | CZ | O1 | TRP59 | CD2 | C4 | |||||
| Tyr82 | OH | C1 | CE1 | C11 | OH | C1 | CE2 | C4 | ||||||
| OH | O1 | NE2 | C10 | OH | N1 | NE1 | C4 | |||||||
| OH | O3 | NE2 | C11 | OH | O1 | CZ2 | C4 | |||||||
| OH | C49 | OH | C5 | Tyr82 | CE2 | O2 | ||||||||
| OH | C10 | CE2 | C9 | |||||||||||
| His87 | CD2 | C11 | CZ | O2 | ||||||||||
| Phe99 | CE2 | O4 | CZ | C9 | ||||||||||
| CZ | O3 | OH | C3 | |||||||||||
| OH | C5 | |||||||||||||
| OH | N2 | |||||||||||||
| OH | O2 | |||||||||||||
| OH | C9 | |||||||||||||
| OH | C18 | |||||||||||||
| Phe99 | CZ | O4 | ||||||||||||
Each residue/atom label is in accordance with the respective PDB files.
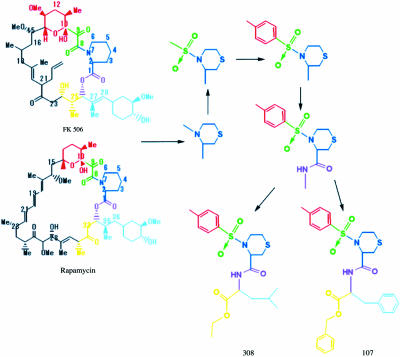
Based on the principle discussed above, we tried to design two compounds as shown (Fig. 2). One is (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-Leucine ethyl ester and the other one is (3R)-4-(p-Toluenesulfonyl)-1,4-thiazane-3-carboxylic acid-L-phenylalanine benzyl ester. For brevity, we called these two compounds 107 and 308, respectively. Following molecular design, these two compounds were synthesized as described. (Data was provided by Professor Song Li and preserved in Patent No. 01142744-2, China.)
FIGURE 2.
The schematic procedure of designing new inhibitors only with neurotrophic effect from the structures of FK506 and rapamycin. The part marked by blue refers to the core structure fitting to the binding pocket; the parts marked by green and red have many interactions with protein, which were polished for stronger binding; and the left parts, marked by other colors, have great changes after designing to increase the hydrophobic interactions between ligands and protein.
Docking
The crystal structures of FKBP12 in complex with FK506 (Wilson et al., 1995), rapamycin (Wilson et al., 1995), and GPI-1046 (Sich et al., 2000) were taken from the Brookhaven Protein Data Bank (entries 1FKJ, 1FKL, and 1F40, respectively). The polar hydrogen model was chosen for FKBP12. The charges of proteins and ligands were assigned with Kollman-united and Gasteiger-Hückel charges, respectively. The hydrogens and charges were added using molecular modeling software SYBYL, rel. 6.7 (SYBYL 2000, Tripos, St. Louis, MO).
The advanced docking program AutoDock 3.01 (Morris et al., 1998) was used to evaluate the binding free energy of these five inhibitors (FK506, GPI-1046, rapamycin, 107, and 308) with FKBP12 (Table 2). AutoDock 3.01 includes a new genetic algorithm search engine and an empirical free energy function for estimating binding free energies and inhibition constants (Ki). Four binding energy terms were included in the score function: electrostatic, Van der Waal, hydrogen bonding, and desolvation effect. The binding free energy was empirically calculated based on these energy terms and a set of coefficient factors (Morris et al., 1998).
TABLE 2.
Calculated binding energy, molecular weight, and dissociation constant for five inhibitor compounds
| Compound | Calculated binding energy (kcal/mol) | Molecular weight (Da) | Dissociation constant from literature (μM)* |
|---|---|---|---|
| GPI-1046 | −4.94 | 359.5 | 1.6 |
| FK506 | −9.23 | 820 | 0.0016 |
| Rapamycin | −12.52 | 1040.5 | 0.0035† |
| 107 | −9.39 | 538.7 | N/A |
| 308 | −8.3 | 442.6 | N/A |
All dissociation constants refer to the data from Graziani et al. (1999).
This is the IC50 of rapamycin with the order of its dissociation constant.
The whole docking process could be summarized as follows. Firstly, the macromolecules were checked for polar hydrogens and assigned for partial atomic charges, and the atomic solvation parameters were also assigned for the macromolecules. Meanwhile, the torsion angles of the inhibitors during molecular docking were defined. Secondly, the three-dimensional grid was created by AutoGrid algorithm (Morris et al., 1998) to evaluate the binding energies based on macromolecular coordinates. At this stage, the FKBP12 was embedded in the three-dimensional grid and a probe atom was placed at each grid point. The affinity and electrostatic potential grid were calculated for each atom type present in the inhibitors. The energetics of a given inhibitor configuration was found by tri-linear interpolation of affinity values and electrostatic interaction of the eight grid points surrounding each of the atoms in an inhibitor. Finally, AutoDock 3.01 (Morris et al., 1998, 1999) was used to calculate the binding energy of a given ligand conformation in the crystal/NMR structure while the probable structure inaccuracies were ignored in the calculations. All computer simulations were performed on Silicon Graphics R10000 workstations.
Fluorescence quenching
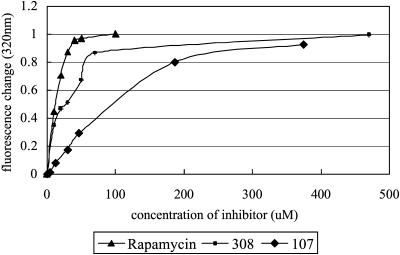
Recombinant human FKBP12 was expressed in Escherichia coli and purified as described (Pei et al., 2000). The solution of FKBP12 can fluoresce at 310–340 nm when irradiated with ultraviolet light at 295 nm as described (Park et al., 1992). The fluorescence of FKBP12 solution is caused by its buried tryptophan residue Trp59, which is located at the bottom of its binding pocket. Another three tyrosine residues—Tyr82 and Tyr26 located around the pocket, and Tyr80 near the pocket—also make a little fluorescence contribution to this excitation and emission wavelength. After the inhibitor was added and bound to the pocket, the polar environment around Trp59 would change, resulting in fluorescence quenching. 30-μM protein buffered with 5 mM NaH2PO4 of pH 6.5 and 100 mM NaCl was prepared to measure the initial fluorescence at 25°C. Inhibitors (rapamycin, 308, and 107) were added respectively to make the titration for fluorescence quenching. The fluorescence changes at an emission wavelength of 320 nm were calculated and normalized according to each saturated quenching. These changes were then plotted against inhibitor concentration (Fig. 3). All the fluorescence measurements were carried out by using AMINCO-Bowman Series 2 Luminescence Spectrometer (Thermo Electron) and data was processed by Excel 2000 (Microsoft).
FIGURE 3.
The fluorescence quenching at 320 nm against different inhibitor concentrations. All the fluorescence changes were normalized for comparison among rapamycin, 308, and 107.
Crystallization and crystal preparation
The purified protein in 10 mM cacodylic acid sodium (pH 6.5) was incubated with compounds 308 and 107, respectively, for 12 h at 4°C, in 1:1.2 molar ratio. Then the complex protein was concentrated to 20 mg/ml. Crystallization of FKBP12 complexed with 308/107 was carried out using the hanging-drop vapor-diffusion method at 18°C. 2-μl drops of complexed protein solution were mixed with 2-μl reservoir solution containing 15–27% PEG 10,000, 0.1 M HEPES Na (pH 7.5). Rod-like crystals appeared after six days and the microseeding method was used to obtain larger single crystals (Li et al., 2002, 2003).
Data collection and processing
Crystals were flash-frozen with liquid nitrogen, using the mother liquor as cryoprotectant. Data were collected using a MarResearch image plate on a Rigaku 18-kW rotating anode generator, operating at 48 kV and 90 mA with monochromated radiation (λ = 1.5418 Å).
Data processing was performed using the program DENZO and data sets were scaled and merged using SCALEPACK (Otwinowski and Minor 1997). The crystal of FKBP12-308 complex belongs to space group P21, a = 41.2 Å, b = 29.6 Å, c = 41.5 Å, and β = 114°, containing one molecule per asymmetric unit and 36% solvent. And the crystal of FKBP12-107 complex also belongs to space group P21, a = 42.0 Å, b = 30.4 Å, c = 42.4 Å, and β = 110.7°, containing one molecule per asymmetric unit and 42% solvent (Li et al., 2002, 2003). All backbone conformation angles are in the fully allowed region of the Ramachandran Plot. An analysis of side-chain conformation angles for the refined structures shows very good statistics according to the program PROCHECK (Laskowski et al., 1993).
RESULTS AND DISCUSSION
QSAR analysis
To compare the potency of compounds 107 and 308 with FK506, rapamycin, and GPI-1046, the binding free energy for each of these five inhibitors binding with FKBP12 was evaluated (Table 2) with SAR calculation, using advanced docking program AutoDock 3.01 (Morris et al., 1998). Rapamycin has the lowest binding energy and GPI-1046 is the weakest one. QSAR analysis clearly revealed that both compounds 107 and 308 possess a significant higher binding affinity than GPI-1046 when binding to FKBP12. Furthermore, compound 107 has the same binding affinity as FK506, but has a much smaller molecular weight than FK506. Taking binding energy and molecular weight into account, compound 308 would be the best drug candidate with a high affinity and low weight. Above all, the calculation showed a good aspect for these two small molecules and it is necessary to synthesize them and make a more detailed investigation of their function by experiments.
Binding affinity estimation by fluorescence quenching experiment
According to the systematic error of concentration measurements, we opted not to calculate the accurate dissociation constants of these inhibitors using Scatchard analysis. Instead, we normalized these fluorescence-quenching changes to put them on the same scale. On this normalized scale, the same change will refer to the same concentration ratio between bound protein and total protein ([EI]/[E]0). In other words, for [E]0 = [EI] + [E], the same ratio between bound protein and free protein ([EI]/[E]) will occur. According to the equilibrium equation for protein-inhibitor binding (Kd=[E][I]/[EI]), the ratio of dissociation constants for different inhibitors could be estimated by the ratio of inhibitor concentrations. Following this principle, we calculated these ratios. The result indicates that the dissociation constant of 308 is ∼40-fold larger than that of rapamycin and 107 is ∼200-fold larger than rapamycin. According to Graziani's data (Table 2, this article; see also Graziani et al., 1999), the binding affinity of rapamycin to FKBP12 is more than 1000-fold larger than GPI-1046. So, consideration of these data together reveals that the binding affinity of 308 to FKBP12 is at least 25-fold larger than GPI-1046 and 107 is at least fivefold larger.
Structure determination and refinement
The two complex structures were solved by molecular replacement by using the program CNS (Brünger et al., 1998), with a starting model from the x-ray structure of FKBP12-DMSO complex (PDB:1D7H). The DMSO molecule and all water molecules were discarded and the temperature factors of all remaining atoms were fixed at 20.0 Å2. The rotation and translation function calculation was carried out using the data within the resolution range 15–3.5 Å.
The model was rebuilt with O and refined with CNS. After a rigid body refinement, σ-A-weighted 2|FO|−|FC| and |FO|−|FC| difference maps were used to locate the ligands and solvent molecules. Using the 40–1.8 Å data, 2|FO|−|FC| and |FO|−|FC| electron density maps were calculated and examined with O. The refinement was completed by alternating between manual building and minimization using data in the resolution range 40–1.8 Å. After refinement, the final R-value and R-free value was 0.19 and 0.22, respectively, for FKBP12-308 complex structure, and 0.21 and 0.26, respectively, for FKBP12-107 complex structure (Table 3).
TABLE 3.
Crystallographic data and refinement statistics
| Data and statistics | FKBP12-308 | FKBP12-107 |
|---|---|---|
| Resolution (Å) | 50–1.8 | 50–1.8 |
| Space group | P21 | P21 |
| Unit cell constants | a = 41.2 Å | a = 42.0 Å |
| b = 29.6 Å | b = 30.4 Å | |
| c = 41.5 Å | c = 42.4 Å | |
| β = 114° | β = 110.7° | |
| Total number of reflections | 83830 | 32811 |
| Number of unique reflections | 9385 (769*) | 9478 (773*) |
| I/σI | 18.6 (6.9*) | 17.2 (4.9*) |
| Redundancy | 8.9 (8.9*) | 3.5 (3.3*) |
| Rmerge† (%) | 7.3 (24.8*) | 5.2 (29.1*) |
| Completeness (%) | 99.9 (99.7*) | 99.6 (98.1*) |
| Rwork‡/Rfree§ | 0.19/0.22 | 0.21/0.26 |
| RMS bond length deviation (Å) | 0.019 | 0.019 |
| RMS bond angle deviation (°) | 2.10 | 1.98 |
| Solvent molecules | 119 | 133 |
| Average B-factor (Å2) | 15 | 17 |
Numbers in parentheses correspond to the highest resolution shell (2.07–2.0 Å).
Rmerge = Σhkl|Ii−Im|/Σhkl Im, where Ii and Im are the observed intensity and the mean intensity of related reflections, respectively.
Rworking = S(‖Fp(obs)|−|Fp(calc)‖)/Σ|Fp(obs)|; Rfree = R-factor for a selected subset (10%) of the reflections that was not included in prior refinement calculations.
10% of the total reflections were used for Rfree calculation.
Crystal structure of FKBP12 complexed with ligands
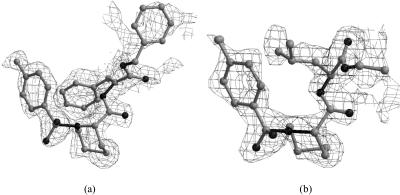
Mass spectrometry was used to observe the specific binding between each compound and FKBP12 as described (data was provided by Professor Song Li). To study the detailed interaction between FKBP12 and its inhibitor, we co-crystallized FKBP12 with each compound and obtained good crystals (Li et al., 2002, 2003). The complexes' structures were solved to a high resolution (1.8 Å) and small ligands showed good occupancy according to the calculated omit map (Fig. 4, a and b).
FIGURE 4.
The 2|Fo| − |Fc| stimulated electron density maps with ligand omitted. (a) Omit map for 107; (b) omit map for 308.
Detailed interactions of the complex structures of FKBP12 with 107, 308, FK506, and rapamycin have been compared. The interactions among them are approximately the same in five/six-membered ring part, which takes accord with the initial design. Three hydrogen bonds are involved in 308 binding to FKBP12 (Table 4): from Ile56 amide (N) to C5 carbonyl (O1), from Tyr82 hydroxyl (O) to C5 acetyl amide (N2), and from Tyr82 hydroxyl (O) to C8 ester (O2). There are also three observed hydrogen bonds between 107 and FKBP12 (Table 4): from Ile56 amide (N) to C1 carbonyl (O2), from Tyr82 hydroxyl (O) to C3 ester (O1), and from Tyr82 hydroxyl (O) to C1 acetyl amide (N1). The above atom labels take accord with the structure of each inhibitor compound in PDB. Six or seven hydrophobic contacts between the protein and 107/308 were observed (Table 4). The contacts between FKBP12 and ligands were counted by the same way described above and listed in Table 1. Compared with FK506 and rapamycin, there are a lot of additional close contacts for 107 and 308, coming from Val55, Ile56, Trp59, and Tyr82, which suggest that the binding is stronger for these two new compounds. Obviously, the additional interactions are mainly due to the additional parts of the designed compounds, which are colored with red, yellow, and cyan, respectively (Fig. 2).
TABLE 4.
Hydrogen bonds and hydrophobic interactions between FKBP12 and inhibitors from five complexes
| Compound | Tyr26 | Phe36 | Phe46 | Glu54 | Val55 | Ile56 | Trp59 | Tyr82 | His87 | Ile90 | Ile91 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GPI_1046 | + | + | + | + | hb | + | + | ||||
| FK506 | + | + | + | hb | + | hb | + | + | |||
| Rapamycin | + | + | + | + | + | hb | + | hb | + | + | |
| 107 | + | + | + | hb | + | hb | + | + | |||
| 308 | + | + | + | + | hb | + | hb | + | + |
hb = hydrogen bond; + = hydrophobic interaction.
Compared with the structure of FKBP12-GPI–1046 complex, both 107 and 308 show a similar binding way. They all form a hydrogen bond with Tyr82 hydroxyl. However, for GPI-1046, the lack of conservative hydrogen bonds with Ile56 and fewer hydrophobic contacts (Table 4) make this small molecule a weaker inhibitor of FKBP12, which is highly consistent with our AutoDock score and fluorescence quenching analysis.
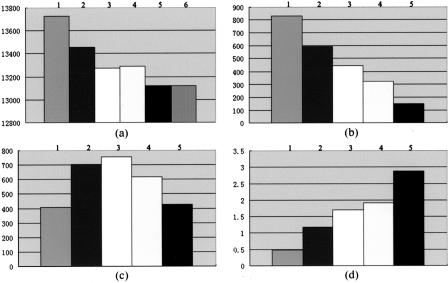
The lower B-factor of the protein when complexed with inhibitors suggests the binding could stabilize the conformation of FKBP12, resulting in good affinity. The average B-factor of the free protein was 31 Å2 and reduced to 17 Å2 after binding with compound 107, 15 Å2 after binding to compound 308, 13 Å2 for FK506, and 21 Å2 for rapamycin. The flexibility of an 80-s loop composed of residues from 82 to 95 in free FKBP12 allows the ligand to enter the binding cavity easily. This would be accompanied with small conformation changes, which might cause obvious surface area, interacting area, and molecular volume effects. To study this area and volume effect, the accessible surface areas of FKBP12 (in free state or complex state), the ligands (FK506, rapamycin, GPI-1046, 107, and 308), and the complexes were calculated, respectively, by the program GRASP (Nicholls et al., 1991). The molecular volume for free protein, ligand, and complex was also calculated in the same way. Comparing these parameters, we can see the molecular volume of FKBP12 and the whole system volume increased (Fig. 5, a and b) on binding to a small ligand, which is obviously disadvantageous for ligand binding since this effect will increase the system energy. However, the accessible area of the system decreased (Fig. 5 c) after ligand binding, which is favorable to reduce the system energy and form a good interaction. To combine these two opposite effects, we introduce a parameter r defined as follows: r = Δarea/Δvolume, where delta-area and delta-volume represent differences in accessible area and molecular volume, respectively. According to this r-value, which could approximate the binding affinity, it seems that 308 owns highest affinity to FKBP12 among GPI-1046, 107, and 308 (Fig. 5 d). From this surface and volume analysis, GPI-1046 was still the worst one when binding to FKBP12, which is also consistent with AutoDock calculation, fluorescence quenching experiments, and contacts analysis.
FIGURE 5.
The volume and area effects of (1) FKBP12-GPI-1046 complex; (2) FKBP12-FK506 complex; (3) FKBP12-rapamycin complex; (4) FKBP12-107 complex; (5) FKBP12-308 complex; and (6) free FKBP12. (a) The volumes (Å3) of FKBP12 in unliganded state or complex state. (b) The delta-volumes (Å3) of FKBP12-ligand systems, before and after ligand binding. (c) The delta-areas (Å2) of FKBP12-ligand systems, before and after ligand binding. (d) The r-values for different ligands used to determine the binding affinity approximately (r = Δarea/Δvolume).
CONCLUSION
Both compounds 107 and 308 were designed based on the complex structures of FKBP12-FK506 and FKBP12-rapamycin by using the following guiding principles: the core structure formed by a six-member ring is essential when binding to protein; more polar atoms, e.g., N and O, were added to try to increase electrical interaction; and more acyclic or alicyclic groups were designed to try to increase hydrophobic interactions.
In principle, volume and surface effect should be taken into consideration when investigating the interactions between protein and ligands, which might be responsible for the binding energy. Assuming the structure determination errors are not large enough to dominate this probably important effect, the analysis above indicates that compounds 308 and 107 had larger volume/surface effects than GPI-1046 in principle. Structure comparisons show that the binding affinity of 308 is >107, which is in accord with fluorescence analysis, but different from AutoDock score. This discrepancy might be due to the limitations of the energy calculation items, which only included electrostatic, Van der Waal, hydrogen bonding energies and desolvation effect, but did not include volume/surface effects resulting from conformational changes. However, when the results of AutoDock calculation, fluorescence quenching, and crystallographic analysis were combined, we conclude that these two compounds are new potent inhibitors of FKBP12 with higher affinity than GPI-1046, and might become alternative drug candidates in place of GPI-1046.
Acknowledgments
We are grateful to Yujia Zhai, Beili Wu, and Zhuhao Wu (of the Z.R. group) for technical assistance. The coordinates of the FKBP12-107 and FKBP12-308 complexes have been deposited in the Protein Data Bank (PDB accession codes 1J4H and 1J4I, respectively). The design and synthesis of compounds 107 and 308, including their corresponding derivatives, have been patent-applied-for in China; the application number is 01142744-2.
We also thank Yuming Li (the Instruction Center of the Biology Department, Tsinghua University) for supplying us with the fluorescence spectrometer instrument, which was supported by the Laboratory Fund of Tsinghua University. This work was supported by “863” project grant 2001AA233011 and “973” project grant G1999075602.
References
- Adalsteinsson, H., and T. C. Bruice. 2000. Generation and evaluation of putative neuroregenerative drugs. Part 2: screening virtual libraries of novel polyketides which possess the binding domain of rapamycin. Bioorg. Med. Chem. 8:625–635. [DOI] [PubMed] [Google Scholar]
- Becker, D. B., J. N. Jensen, T. M. Myckatyn, V. B. Doolabh, D. A. Hunter, and S. E. Mackinnon. 2000. Effects of FKBP-12 ligands following tibial nerve injury in rats. J. Reconstr. Microsurg. 16:613–620. [DOI] [PubMed] [Google Scholar]
- Becker, J. W., J. Rotonda, J. G. Cryan, M. Martin, W. H. Parsons, P. J. Sinclair, G. Wiederrecht, and F. Wong. 1999. 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein. J. Med. Chem. 42:2798–2804. [DOI] [PubMed] [Google Scholar]
- Becker, J. W., J. Rotonda, B. M. McKeever, H. K. Chan, A. I. Marcy, G. Wiederrecht, J. D. Hermes, and J. P. Springer. 1993. FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818. J. Biol. Chem. 268:11335–11339. [PubMed] [Google Scholar]
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 54:905–921. [DOI] [PubMed] [Google Scholar]
- Choi, J., J. Chen, S. L. Schreiber, and J. Clardy. 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 273:239–242. [DOI] [PubMed] [Google Scholar]
- Christner, C., R. Wyrwa, S. Marsch, G. Kullertz, R. Thiericke, S. Grabley, D. Schumann, and G. Fischer. 1999. Synthesis and cytotoxic evaluation of cycloheximide derivatives as potential inhibitors of FKBP12 with neuroregenerative properties. J. Med. Chem. 42:3615–3622. [DOI] [PubMed] [Google Scholar]
- Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T- lymphocyte activation. Nature. 357:695–697. [DOI] [PubMed] [Google Scholar]
- Dawson, T. M., J. P. Steiner, W. E. Lyons, M. Fotuhi, M. Blue, and S. H. Snyder. 1994. The immunophilins, FK506 binding protein and cyclophilin, are discretely localized in the brain: relationship to calcineurin. Neuroscience. 62:569–580. [DOI] [PubMed] [Google Scholar]
- Denesyuk, A. I., K. A. Denessiouk, V. P. Zav'yalov, J. Lundell, and T. Korpela. 1998. Analogous conformations of both binding and effector regions in cyclosporin A, FK506 and rapamycin. Comput. Chem. 22:339–344. [DOI] [PubMed] [Google Scholar]
- Gold, B. G. 2000. Neuroimmunophilin ligands: evaluation of their therapeutic potential for the treatment of neurological disorders. Expert Opin. Invest. Drugs. 9:2331–2342. [DOI] [PubMed] [Google Scholar]
- Gold, B. G., M. Zeleny-Pooley, M. S. Wang, P. Chaturvedi, and D. M. Armistead. 1997. A nonimmunosuppressant FKBP-12 ligand increases nerve regeneration. Exp. Neurol. 147:269–278. [DOI] [PubMed] [Google Scholar]
- Graziani, F., L. Aldegheri, and G. C. Terstappen. 1999. High throughput scintillation proximity assay for the identification of FKBP-12 ligands. J. Biomol. Screen. 4:3–7. [DOI] [PubMed] [Google Scholar]
- Griffith, J. P., J. L. Kim, E. E. Kim, M. D. Sintchak, J. A. Thomson, M. J. Fitzgibbon, M. A. Fleming, P. R. Caron, K. Hsiao, and M. A. Navia. 1995. X-ray structure of calcineurin inhibited by the immunophilin- immunosuppressant FKBP12–FK506 complex. Cell. 82:507–522. [DOI] [PubMed] [Google Scholar]
- Guo, X., V. L. Dawson, and T. M. Dawson. 2001. Neuroimmunophilin ligands exert neuroregeneration and neuroprotection in midbrain dopaminergic neurons. Eur. J. Neurosci. 13:1683–1693. [DOI] [PubMed] [Google Scholar]
- Hauske, J. R., P. Dorff, S. Julin, J. DiBrino, R. Spencer, and R. Williams. 1992. Design and synthesis of novel FKBP inhibitors. J. Med. Chem. 35:4284–4296. [DOI] [PubMed] [Google Scholar]
- Herdegan, T., G. Fischer, and B. G. Bold. 2000. Immunophilin ligands as a novel treatment of neurological disorders. Trends Pharmacol. Sci. 21:3–5. [DOI] [PubMed] [Google Scholar]
- Huse, M., Y. G. Chen, J. Massague, and J. Kuriyan. 1999. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell. 96:425–436. [DOI] [PubMed] [Google Scholar]
- Kissinger, C. R., H. E. Parge, D. R. Knighton, C. T. Lewis, L. A. Pelletier, A. Tempczyk, V. J. Kalish, K. D. Tucker, R. E. Showalter, E. W. Moomaw, L. N. Gastinel, N, Habuka, X. Chen, F. Maldonado, J. E. Barker, R. Bacquet, and J. E. Villafranca. 1995. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 378:641–644. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- Li, P., L. Wang, B. Wu, C. Shu, A. Nie, B. Shen, S. Li, and Z. Rao. 2003. Crystallization and preliminary x-ray diffraction analysis of FKBP12 complexed with a new neurotrophic ligand. Progr. Natur. Sci. 13:765–767. [DOI] [PubMed] [Google Scholar]
- Li, P., L. Wang, Y. Ding, B. Wu, C. Shu, A. Nie, S. Li, B. Shen, and Z. Rao. 2002. Crystallization and preliminary x-ray diffraction analysis of FKBP12 complexed with a novel neurotrophic ligand. Prot. Peptide Lett. 9:459–463. [DOI] [PubMed] [Google Scholar]
- Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP- FK506 complexes. Cell. 66:807–815. [DOI] [PubMed] [Google Scholar]
- Lyons, W. E., E. B. George, T. M. Dawson, J. P. Steiner, and S. H. Snyder. 1994. Immunosuppressant FK506 promotes neurite outgrowth in cultures of PC12 cells and sensory ganglia. Proc. Natl. Acad. Sci. USA. 91:3191–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, R, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19:1639–1662. [Google Scholar]
- Morris, G. M., D. S. Goodsell, R. Huey, W. E. Hart, S. Halliday, R. Belew, and A. J. Olson. 1999. AUTODOCK, V. 3.0.3. The Scripps Research Institute, Molecular Graphics Laboratory, Department of Molecular Biology.
- Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. C. W. Carter, Jr., and R. M. Sweet, editors. Academic Press. 307–326. [DOI] [PubMed]
- Park, S. T., R. A. Aldape, O. Futer, M. T. DeCenzo, and D. J. Livingston. 1992. PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J. Biol. Chem. 267:3316–3324. [PubMed] [Google Scholar]
- Pei, W-H, Y.-H. He, X. Chen, S. Li, and B.-F. Shen, 2000. Solube expression and activity research of human FKBP12. J. Cell. Mol. Immunol. (in Chinese). 16:204–206. [Google Scholar]
- Sabatini, D. M., M. M. Lai, and S. H. Snyder. 1997. Neural roles of immunophilins and their ligands. Mol. Neurobiol. 15:223–239. [DOI] [PubMed] [Google Scholar]
- Sauer, H., J. M. Francis, H. Jiang, G. S. Hamilton, and J. P. Steiner. 1999. Systemic treatment with GPI 1046 improves spatial memory and reverses cholinergic neuron atrophy in the medial septal nucleus of aged mice. Brain Res. 842:109–118. [DOI] [PubMed] [Google Scholar]
- Schiene, C., and G. Fischer. 2000. Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 10:40–45. [DOI] [PubMed] [Google Scholar]
- Sich, C., S. Improta, D. J. Cowley, C. Guenet, J. P. Merly, M. Teufel, and V. Saudek. 2000. Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics. Eur. J. Biochem. 267:5342–5355. [DOI] [PubMed] [Google Scholar]
- Snyder, S. H., D. M. Sabatini, M. M. Lai, J. P. Steiner, G. S. Hamilton, and P. D. Suzdak. 1998. Neural actions of immunophilin ligands. Trends Pharmacol. Sci. 19:21–26. [DOI] [PubMed] [Google Scholar]
- Steiner, J. P., T. M. Dawson, M. Fotuhi, C. E. Glatt, A. M. Snowman, N. Cohen, and S. H. Snyder. 1992. High brain densities of the immunophilin FKBP colocalized with calcineurin. Nature. 358:584–587. [DOI] [PubMed] [Google Scholar]
- Steiner, J. P., M. A. Connolly, H. L. Valentine, G. S. Hamilton, T. M. Dawson, L. Hester, and S. H. Snyder. 1997a. Neurotrophic actions of nonimmunosuppressive analogues of immunosuppressive drugs FK506, rapamycin and cyclosporin A. Nat. Med. 4:412–428. [DOI] [PubMed] [Google Scholar]
- Steiner, J. P., G. S. Hamilton, D. T. Ross, H. L. Valentine, H. Guo, M. A. Connolly, S. Liang, C. Ramsey, J. H. Li, W. Huang, et al. 1997b. Neurotrophic immunophilin ligands stimulate structural and functional recovery in neurodegenerative animal models. Proc. Natl. Acad. Sci. USA. 94:2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K. P., M. M. Yamashita, M. D. Sintchak, S. H. Rotstein, M. A. Murcko, J. Boger, J. A. Thomson, M. J. Fitzgibbon, J. R. Black, and M. A. Navia. 1995. Comparative x-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin. Acta Crystallogr. D. 51:511–521. [DOI] [PubMed] [Google Scholar]
- Winter, C., J. Schenkel, E. Burger, C. Eickmeier, M. Zimmermann, and T. Herdegen. 2000. The immunophilin ligand FK506, but not GPI-1046, protects against neuronal death and inhibits c-Jun expression in the substantia nigra pars compacta following transection of the rat medial forebrain bundle. Neuroscience. 95:753–762. [DOI] [PubMed] [Google Scholar]
- Zhang, C., J. P. Steiner, G. S. Hamilton, T. P. Hicks, and M. O. Poulter. 2001. Regeneration of dopaminergic function in 6-hydroxydopamine-lesioned rats by neuroimmunophilin ligand treatment. J. Neurosci. 21:RC156. [DOI] [PMC free article] [PubMed] [Google Scholar]