Abstract
The hepatocyte nuclear factor (HNF)-3 homologous DNA binding domain is a highly conserved motif that contains a well-folded helix-turn-helix motif and two highly dynamic wings. Although the function and the properties of this motif have been intensively studied, the role of the internal wing (wing 1) is not well understood. In this study, amino acid substitutions were introduced into wing 1 of a conserved HNF-3 homologous protein, Genesis, and heteronuclear NMR, circular dichroism, DNA gel-shift assay, and fluorescent methods were employed to study and compare the properties of both wild-type and variant Genesis proteins. The data indicate that even though the substitutions are located on a dynamic wing outside the hydrophobic core sequences, they still globally influence biophysical properties of DNA-free Genesis and its DNA complex.
INTRODUCTION
Well-controlled transcriptional regulation determines embryonic development and cell differentiation. These processes depend in part on the transcriptional regulation exerted by transcription factors that bind to cis-acting sequences in gene control regions where they recruit specific coactivators. Transcription factors can be grouped into families according to their conserved DNA binding motifs. One of these highly conserved motifs is the winged helix DNA-binding motif which was initially identified in the DNA binding domains of rat hepatocyte nuclear factor (HNF)-3 and Drosophila forkhead homeotic protein (fkh) (Lai et al, 1990; Knochel and Kaufmann, 1997). Since then, many proteins containing this motif were identified in various organisms ranging from yeast to human (Knochel and Kaufmann, 1997). These proteins have been shown to play critical roles in developmental regulation, especially in organogenesis and tissue specific differentiation (Knochel and Kaufmann, 1997; Hebbar and Curtis, 2000).
Previous structural determinations have shown that the winged helix motif is constructed from a tightly packed core, constituted from three helices and a three-strand β-sheet and two dynamic wings (Clark et al., 1993; Jin et al., 1999; Lai et al., 1993; van Dongen et al., 2000; Weigelt et al., 2001). The DNA recognition of winged helix proteins is achieved mainly through the contacts made by helix 3 and wing 2. The structure and dynamics of a conserved HNF-3 homolog, Genesis, in complex with a strong DNA binding site indicate that even though both wings contact DNA, wing 1 still shows large amplitude of motions in the complex (Jin et al, 1998; Jin and Liao, 1999). Thus, it is not clear what is the role of this wing in the DNA recognitions. Furthermore, the amino acid sequence of wing 1 is one of the divergent sequences in the HNF-3 homologous proteins and these divergent sequences may play important roles for the functions of the HNF-3 family members. In a recent study, the DNA binding domain of HNF-3β alone was shown to interact with the cut-homeo DNA binding domains of transcription factor HNF-6 (Rausa et al., 2003). This interaction stimulates the HNF-3β activity on a promoter, while blocking HNF-6 binding to DNA. In another study, the DNA binding domain of Genesis was recognized by the transcription factor Oct4, which represses the activity of Genesis on a promoter (Guo et al., 2002). These interactions are highly specific. For example, even though the sequences of HNF-3β and HNF-3α are almost identical, HNF-6 recognizes HNF-3β as well as Genesis, but not HNF-3α (Rausa et al., 2003). Apparently, the relatively divergent sequences such as wing 1, in the winged helix proteins have to function as recognition markers for the recognition of other cofactors in the transcriptional regulation. To understand potential roles of wing 1 in protein-protein, protein-DNA, and DNA-binding dependent protein-protein interactions, a systematic study of wing 1 is necessary.
As the first step in understanding the roles of these divergent sequences, two wing 1 mutants were constructed, and the biophysical and biochemical properties of the mutants and the wild-type Genesis were compared. In this report, we demonstrate that the amino acid substitutions (70NG73KG and 70NP73KP) in the eight-residue wing 1 sequence influence thermodynamic and internal dynamic properties of the winged helix protein Genesis and its DNA complex. Thus, our data demonstrate important roles for this long, divergent, and flexible sequence in the winged helix DNA binding motif.
MATERIALS AND METHODS
The mutagenesis and expression of Genesis
The two substitutions designated as 70NG73KG and 70NP73KP (Shiyanova and Liao, 1999) were introduced onto wing 1, which contacts the minor groove DNA in a Genesis-DNA complex (Jin et al., 1999).. Gly and Pro residues are used in the substitutions, since these two types of residue represent the extremes. Gly substitution generates the least restraint and allows the largest freedom in wing 1 conformations, while Pro substitution introduces the most restraint in wing 1 conformation. Therefore, the mutants will be used in this study to examine the interaction between wing 1 and DNA and the changes in the motif that are due to the substitution in this flexible wing. The amino acid substitutions were introduced by the standard PCR-mediated mutagenesis procedure (Shiyanova and Liao, 1999). The mutant proteins used in nuclear Overhauser effect spectroscopy (NOESY)-heteronuclear single quantum coherence (HSQC) experiments (for confirmation of assignments and backbone dynamics) were grown in 15N enriched M9 media and had concentrations ∼1 mM determined with BioRad protein assay (BioRad, Hercules, CA). The concentration of the wild-type Genesis was ∼0.2 mM. The proteins used for circular dichroism (CD) and fluorescent measurements were grown in LB-rich media.
Thermodynamic stability measurements
CD measurements were performed on a Jasco J-710 spectropolarimeter (Jasco, Victoria, BC, Canada) in 1-mm cells using temperature-scanning mode. The denaturation temperatures of these proteins were calculated using the standard analysis program supplied by the manufacturer.
Fluorescence spectra were obtained in a double spectrometer using 1-cm cells at a protein concentration of 1 μM. The excitation wavelength was set to 284 nm and the bandwidth of excitation and emission was 4 nm. Urea-dependent fluorescence spectra were recorded to follow the unfolding progress and to calculate the Gibbs free energy difference of the proteins unfolding at 0 M urea (ΔGu(H2O)). The fluorescence originated mainly from two Trp residues W47 and W77. Their emission maximum shifts from 330 nm to 360 nm during the unfolding process. The ratio of the fluorescence intensity of these two wavelengths (F) is normalized and is a measure of the unfolding progress.
Since F = kn/ku
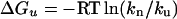 |
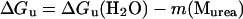 |
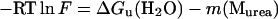 |
Where kn and ku are equilibrium constants of native and unfolding states of protein.
ΔGu(H2O) can be calculated from extrapolation of the unfolding curve to 0 M urea.
NMR experiments
The HSQC and 15N-NOESY-HSQC (mixing time = 180 ms), 15R1, 15N R2, and {1H}-15N NOE spectra were recorded on a Bruker DRX 600 MHz spectrometer. The measurements were performed at 17°C for the DNA-free mutant proteins and at 27°C for the complexes, since the DNA-free proteins are much less stable than the complexes, and at 17°C the signal-to-noise ratios of the spectra of the complexes are not sufficient for accurate determinations of relaxation parameters under the current experimental condition. Standard pulse sequences were used in these measurements (Kay, 1997; Farrow et al., 1995). The intensities of peaks in the two-dimensional spectra were determined by a peak-picking macro in the commercial software SYBYL (Tripos Inc., St. Louis, MO). The relaxation rate constants were determined by fitting the measured peak heights to two-parameter single exponential functions by using the linear least-square method as described previously (Press et al., 1989).
Determination of the stability of the Genesis-DNA complexes by gel-shift assay
Binding reaction mixture contained 20 mM Hepes (pH 7.9), 40 mM KCl, 2 mM MgCl2, 0.05 mM DTT, 0.01% NP-40, 5 μg (∼5 μM) BSA, 1.5 μg (∼0.7 μM) poly(dI-dC), 0.5 ng (∼5 nM) Genesis probe HFH-2#12, and either 5 ng (∼5 nM) of Genesis or 70NG73KG or 70NP73KP mutant. Total volume was set to 19 μl, and reaction mixtures were incubated for 20 min at room temperature before adding 100-fold of the unlabeled probe. After the incubation for an indicated length for a reaction at room temperature, 4 μl of 20% Ficoll was added and 8 μl of the reaction mixture was immediately analyzed on a 9% nondenaturing acrylamide gel, which was constantly running. The radioactivity of each gel shift band was determined with a PhophorImager (Molecular Dynamics, Sunnyvale, CA) for quantitative analysis.
RESULTS
Thermal unfolding and urea-induced denaturation
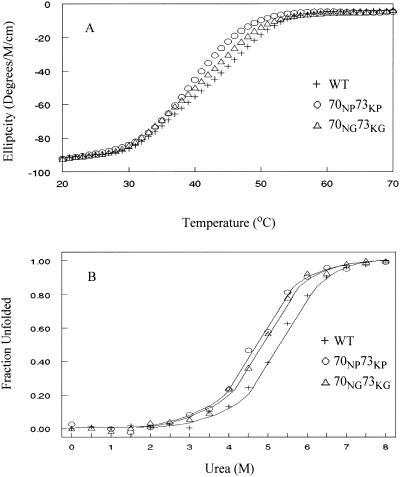
Genesis contains a tightly packed hydrophobic core and two dynamic wings (Fig. 1). Even though the substitutions are introduced into the flexible wing 1 sequence, it is possible that they influence the thermodynamic stability of the Genesis core. Thermal denaturation temperature Tm is an important parameter of the thermodynamic stability of a protein. The thermal denaturing curve of the wild-type Genesis and the amino acid substituted Genesis proteins were recorded using the temperature-scanning mode and following changes in the ellipticity in the far-UV region of the spectrometer (Fig. 2 A). The content of α-helix was monitored at 222 nm. Tm values of wild-type Genesis, and two wing 1 mutants, 70NG73KG and 70NP73KP, were determined at 45.2°C, 42.2°C, and 39.6°C ± 0.2°C, respectively. Even though the reverse curves of the thermal denaturation could not be determined reliably due to the fast precipitation of protein at temperatures above Tm, the result still indicates that the wild-type Genesis is slightly more stable than both the mutants, and that the Gly substituted Genesis is more stable than the Pro substituted Genesis.
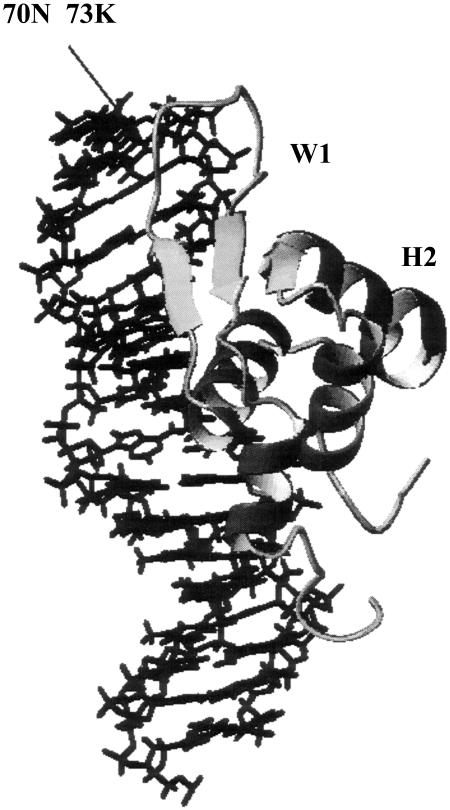
FIGURE 1.
The structure of wild-type HNF-3/fhk protein Genesis. Two mutations were introduced in wing 1 as indicated.
FIGURE 2.
(A) Thermal denaturation curves of 70NG73KG and 70NP73KP mutants and wild-type (WT) Genesis measured by far-UV spectroscopy. (B) Plot of the fraction of unfolded molecules of 70NG73KG and 70NP73KP mutants and wild-type Genesis as a function of urea concentration.
To confirm that the substitutions reduce the thermodynamic stability of the Genesis protein, fluorescence spectra of the three proteins in increasing urea concentrations were recorded. Unfolding progress was followed and the Gibbs free energy differences of the proteins unfolding at 0 M urea, ΔGu(H2O), were extracted (Fig. 2 B) (Pace et al., 1992). The fluorescence from two Trp residues, W47 and W77, changed the emission intensity and the maximum from 330 nm to 360 nm during urea-dependent unfolding as the two residues became more exposed. The ratio of the fluorescence intensity of these two wavelengths follows the unfolding progress as shown in Fig. 2 B. Extrapolation of the unfolding curves to 0 M urea allows the calculation of the Gibbs free energy of unfolding ΔGu(H2O) (18) of 18.5 kJ mol−1, 17.0 kJ mol−1, and 19.5 kJ mol−1 ± 0.5 kJ mol−1 for the 70NG73KG and 70NP73KP mutants and the wild-type Genesis, respectively. This shows that substitution of residues in a surface-exposed and flexible loop sequence influences the thermodynamic stability of Genesis. The data also show that the Pro mutant decreases the Gibbs free energy of unfolding slightly more than the Gly mutant.
The amino acid substitutions modify the internal dynamics of Genesis
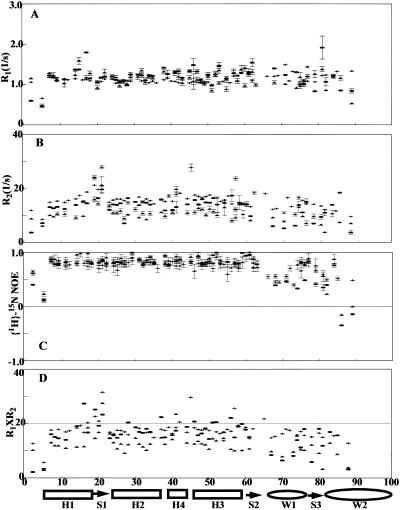
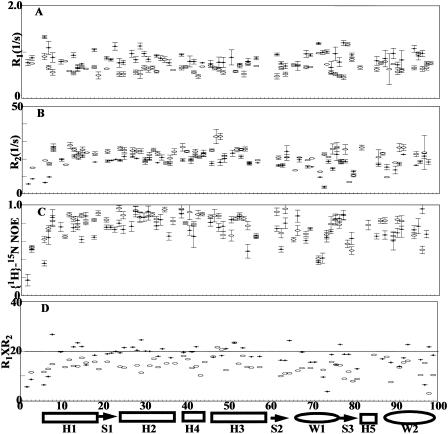
The amino acid substitutions introduced into a long flexible loop reduce the thermodynamic stability of the protein. Whether the substitutions are influencing the stability of the protein by having local or global effects is not clear. To address this question, the relaxation data were acquired on the two DNA-free wing 1 mutants and the wild-type Genesis protein. Due to the resonance overlap, relaxation data were collected only for 54, 56, and 58 residues in the 70NG73KG, the 70NP73KP and the wild-type DNA-free Genesis proteins respectively (Fig. 3). Judged on the data, the wild-type protein and its loop mutants show similar patterns of relaxation parameters. The residues in wing 1 and wing 2 show weaker than average NOE, and the residues between H1 and H2 show longer than average R2 values. In general the relaxation parameters obtained outside of these three regions (wing 1, wing 2, and between H1 and H2) are close to the average value. Therefore, the relaxation data indicate that individual secondary structural elements are not severely perturbed by the loop mutations.
FIGURE 3.
Relaxation parameters of the DNA-free wild-type Genesis (crosses), 70NG73KG (open circles), and 70NP73KP (open triangles) at 17°C. (A) 15N R1, (B) 15N R2, (C) heteronuclear NOE, and (D) R1 × R2. The secondary structure of the proteins is indicated at the bottom of the figure.
It is worth pointing out that a low concentration NMR sample (∼0.25 mM) of DNA-free Genesis was used in the data collection, since the τm value of the DNA-free Genesis is protein concentration-dependent. A possible cause for this is protein-protein interaction at high concentrations used in the NMR studies. Although, the relaxation parameters for each individual residue are influenced by the protein concentration, the trends of the parameters obtained from various protein concentrations are the same. Therefore, the conclusion drawn from the previous study of Genesis is still valid (Jin et al., 1999). Interestingly, the wing 1 mutant does not show obvious aggregations even at higher concentrations. It is thus possible that the wild-type wing 1 is involved in the forming of possible Genesis dimers.
To determine whether the destabilizing effects of the wing 1 amino acid substitution are passed onto the entire sequences, the multiple R1R2 values are used to analyze the internal motions of the proteins and the complexes (Kneller et al., 2001), which are likely to have complicated and severe anisotropic motions. This criterion requires ωnτm ≫ 1 (ωn is the Lamor frequency of 15N, τm is the rotational correlation time of protein DNA complex). In this study the data were acquired at the magnetic field strength of 14.0 T. At this strength, the upper limit of R1R2 (represented as R1R2max) for a residue is 20 as illustrated in the previous study, when a residue is restricted in motions characterized by S2 = 1 and does not have chemical exchange (Kneller et al., 2001). (0 ≤ S2 ≤ 1 is the order parameter of a residue. S2 describes the degree of spatial restriction of the internal motions of an intranuclear 15N-1H vector). Due to the relatively large τm values (Table 1) estimated from R1/R2 ratios, this limit is approached for the DNA-free proteins. As suggested (Kneller et al., 2001), a smaller S2 value reduces the R1R2 value of a residue, while a nonzero Rex value increases the R1R2 value of a residue. Therefore, in this study, any residue with a R1R2 value higher than R1R2max contains chemical exchange, while without the effect of anisotropy a residue with a small R1R2 value should have a small S2 value (Fig. 3 D). Due to the large τm values, the average 〈S2〉 can also be estimated from 〈R1R2〉(〈S2〉 = (〈R1R2〉/R1R2max)½), where R1R2max is the calculated maximum value) (Kneller et al., 2001). In this study, the 〈S2〉 of 70NP73KP is 0.72 and is obviously lower than the 〈S2〉 values of 0.85 for the wild-type Genesis and 0.80 for 70NG73KG. Thus, the data indicate that the modification of the wing 1 sequence of Genesis increases the overall motional freedom of the folded sequences, probably by destabilizing the hydrophobic core packing, as suggested by the thermodynamic measurement. The data also indicate that the destabilizing effects are passed on to the entire sequence of Genesis.
TABLE 1.
Average S2 values for Genesis and its mutants
| *τm (ns) | 〈R1 R2〉 | 〈S2〉 | |
|---|---|---|---|
| Wild-type Genesis at 17°C | 10.7 | 17.49 | 0.85 |
| 70NG73NG at 17°C | 11.2 | 15.86 | 0.80 |
| 70NP73NP at 17°C | 10.3 | 12.29 | 0.72 |
| Genesis complex at 27°C | 15.2 | 17.61 | 0.85 |
| 70NP73NP complex at 27°C | 16.7 | 13.86 | 0.77 |
The τm values are estimated from 〈R1/R2〉 from well-structured sequences.
The DNA binding property of the wing 1 mutants
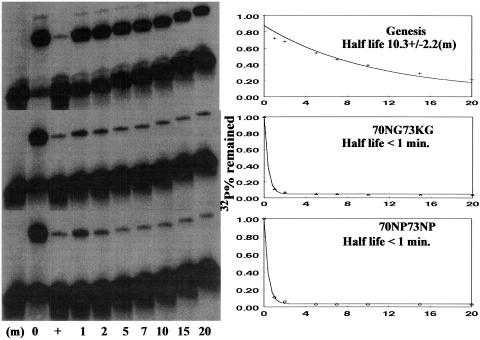
The Genesis-DNA interaction also modifies the internal motions of the protein (Jin et al., 1999). The question then is whether the protein-DNA interaction can counter the destabilizing effect caused by the wing 1 mutations. Wing 1 has strong internal motions even when it binds to a high-affinity binding site, HFH-2#12. Apparently, wing 1 is not a major DNA contact element of Genesis. First the influences of the amino acid substitutions on the DNA binding property of Genesis are studied. The kinetic stability of the DNA complexes of the two mutants and the wild-type Genesis were determined using the gel shift method (Shiyanova and Liao, 1999). Our data show that the half-lives are reduced from ∼10 min for the wild-type DNA complex to <1 min for the mutant DNA complexes (Fig. 4). Therefore, even though wing 1 is highly dynamic in the wild-type protein Genesis DNA complex, it still contributes to the kinetic stability of the complex. Even though the complex is destabilized due to the wing 1 substitution, the forming of the complex band in the gel shift assay indicates that Genesis still binds to the DNA site. The question that remains unanswered is whether the protein-DNA interaction will increase the stability of the hydrophobic core of the mutants.
FIGURE 4.
The determination of half lives of Genesis and its wing 1 mutants. The determination of the stability of the Genesis-DNA complexes: The 0 time value was the initial value without the unlabeled probe added. The equilibrium value (+) was obtained by premixing the labeled and unlabeled probe before adding the corresponding protein in a reaction.
The effect of the protein-DNA interaction on the dynamic properties of the 70NP73KP-DNA complex
The relaxation data of the 70NP73KP-DNA complex and the wild-type Genesis DNA complex measured at 27°C are compared (Fig. 5). The protein-DNA interaction reduces the motions of highly flexible wing 2 and the chemical exchanges of the sequence between helix 1 and helix 2, as observed previously with the Genesis-DNA complex at different temperatures (Jin and Liao, 1999). These two sequences are directly engaged in the DNA recognition, and the interactions therefore reduce the motional amplitudes of these two sequences. However, the increased motional freedom in DNA-free 70NP73KP can still be detected in the 70NP73KP-DNA complex calculated from the R1R2 products (Fig. 5). The mutant shows an 〈S2〉 of 0.77 at 27°C, while the wild-type Genesis-DNA complex shows 〈S2〉 values of 0.85. The result indicates that the protein-DNA interactions greatly reduce the motional freedoms of the DNA contact residues, which undergo structural transitions in the complex, but have reduced influences on the motional properties of non-DNA contact sequences in the protein.
FIGURE 5.
Relaxation parameters of the wild-type Genesis-DNA complex (crosses) and the 70NP73KP-complex (open circles) at 27°C. (A) 15N R1, (B) 15N R2, (C) heteronuclear NOE, and (D) the products of R1 × R2. The secondary structure of the proteins is indicated at the bottom of the figure.
DISCUSSION
The winged helix DNA binding motif is a highly conserved motif, which contains a well-packed helix-turn-helix motif and two dynamic wings. The motif is engaged in both specific protein-DNA and protein-protein interactions (Clark et al., 1993; Rausa et al., 2003; Guo et al., 2002). It is likely that the multifunctions of this highly conserved motif need the relatively divergent sequences, such as wing 2 and wing 1, to play critical roles in those functions. Wing 2 of the HNF-3 homologous winged helix proteins is indispensable for DNA binding, while the role of wing 1 is not clear. Even though wing 1 contacts DNA in the minor groove, it is still highly dynamic in the complex (Jin et al., 1999) and its interaction with DNA is dispensable. It is also possible that wing 1 participates in more than one function of the winged helix proteins. To fully understand the functions of wing 1, it is important to investigate the contributions of wing 1 to DNA recognition and motif folding.
In this study, two wing 1 mutants were constructed and their properties were studied. Our data show that the substitutions in this 8-residue wing sequence influence the DNA binding properties, the thermodynamics, and the internal dynamics of Genesis and one of its DNA complexes. Even though the amino acid substitutions do not disrupt the helix-turn-helix motif, they reduce the stability and increase the internal motions of the protein. Furthermore, the double Pro substitution has a slightly more destabilizing effect on Genesis than the double Gly substitution. This is reasonable since the proline substitution is expected to introduce restraints in wing 1, which likely influence conformational freedom of strand 2 and strand 3 in the hydrophobic core packing and lead to the destabilization of the protein. The resulting destabilization is small, since the hydrophobic core of Genesis is still intact and the mutants only show a slight drop in the Gibbs free energy of unfolding (ΔΔGu(H2O)) and a small reduction in thermodynamic stability (ΔTm). The winged helix DNA binding motif is a small motif, and is involved in the packing of three helixes and three β-strands. Due to the small hydrophobic core, the winged helix proteins only contain several highly conserved hydrophobic core residues. Thus the ΔGu(H2O) value of the wild-type Genesis is relatively small compared to that of large and well-folded proteins (Ladbury et al., 1993). Also due to this small hydrophobic potential, the structure is prone to perturbation. Thus, even the amino acid substitutions in wing 1 can destabilize the protein.
However, this destabilization has a profound effect on the internal dynamics of Genesis. The 〈S2〉 values for both mutants are lower than that of the wild-type, and the Pro-substituted Genesis shows a considerably reduced 〈S2〉 value. The data indicate that the dynamics of a protein should also be viewed as an entity and can be influenced by a flexible linker sequence outside of the hydrophobic core. Our data also show that the increased internal motions of 70NP73KP cannot be completely reversed by the strong protein-DNA interaction. Our data indicate that amino acid substitutions in this flexible internal wing can influence the dynamic properties of the winged helix domain globally, while protein-DNA interactions may only modify dynamic properties of the domain locally. Furthermore, although, the wing 1 mutations do not disrupt the Genesis-DNA interaction, the altered dynamic properties may influence the protein-protein interactions between Genesis and other transcription factors. This possibility deserves a careful study, especially since our preliminary result implicates the interaction between wing 1 and HNF-6 (Yan and Liao, unpublished).
Acknowledgments
We thank Dr. Jin for his works on DNA-free Genesis. Thanks to Dr. Lee for his help on CD spectroscopy. Thanks to Marija Backovic for reviewing the manuscript. We thank Dr. Sheng and Dr. Marco for their help on NMR experiments.
This research was supported by a National Institutes of Health grant to X.L. The Bruker DRX600 was purchased with funds from the University of Illinois at Chicago and grants from the National Science Foundation Academic Research Infrastructure Program.
References
- Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 364:412–420. [DOI] [PubMed] [Google Scholar]
- Farrow, N. A., O. Zhang, A. Szabo, D. A. Torchia, and L. E. Kay. 1995. Spectral density function mapping using 15N relaxation data exclusively. J. Biomol. NMR. 6:153–162. [DOI] [PubMed] [Google Scholar]
- Guo, Y., R. Costa, H. Ramsey, T. Starnes, G. Vance, K. Robertson, M. Kelley, R. Reinbold, H. Scholer, and R. Hromas. 2002. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc. Natl. Acad. Sci. USA. 99:3663–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar, P. B., and S. E. Curtis. 2000. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J. Bacteriol. 182:3572–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C., and X. Liao. 1999. Backbone dynamics of a winged helix protein and its DNA complex at different temperatures: changes of internal motions in genesis upon binding to DNA. J. Mol. Biol. 292:641–651. [DOI] [PubMed] [Google Scholar]
- Jin, C., I. Marsden, X. Chen, and X. Liao. 1999. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J. Mol. Biol. 289:683–690. [DOI] [PubMed] [Google Scholar]
- Jin, C., I. Marsden, X. Chen, and X. Liao. 1998. Sequence specific collective motions in a winged helix DNA binding domain detected by 15N relaxation NMR. Biochemistry. 37:6179–6187. [DOI] [PubMed] [Google Scholar]
- Kay, L. E. 1997. NMR methods for the study of protein structure and dynamics. Biochem. Cell Biol. 75:1–15. [DOI] [PubMed] [Google Scholar]
- Kneller, J. M., M. Lu, and C. Bracken. 2001. An effective method for the discrimination of motional anisotropy and chemical exchange. J. Am. Chem. Soc. 124:1852–1853. [DOI] [PubMed] [Google Scholar]
- Knochel, W., and E. Kaufmann. 1997. Transcription factors and induction in Xenopus laevis embryos. Cell. Mol. Life Sci. 53:362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E., K. L. Clark, S. K. Burley, and J. E. J. Darnell. 1993. Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA. 90:10421–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury, J. E., R. Wynn, H. W. Hellinga, and J. M. Sturtevant. 1993. Stability of oxidized Escherichia coli thioredoxin and its dependence on protonation of the aspartic acid residue in the 26 position. Biochemistry. 32:7526–7530. [DOI] [PubMed] [Google Scholar]
- Lai, E., V. R. Prezioso, E. Smith, O. Litvin, R. H. Costa, and J. E. J. Darnell. 1990. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 4:1427–1436. [DOI] [PubMed] [Google Scholar]
- Pace, C. N., D. V. Laurents, and R. E. Erickson. 1992. Urea denaturation of barnase: pH dependence and characterization of the unfolded state. Biochemistry. 31:2728–2734. [DOI] [PubMed] [Google Scholar]
- Press, W. H., B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling. 1989. Numerical Recipes. Cambridge University Press, Cambridge, U.K.
- Rausa, F. M., Y. Tan, and R. H. Costa. 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol. 23:437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiyanova, T., and X. Liao. 1999. The dissociation rate of a winged helix protein-DNA complex is influenced by non-DNA contact residues. Arch. Biochem. Biophys. 362:356–362. [DOI] [PubMed] [Google Scholar]
- van Dongen, M. J., A. Cederberg, P. Carlsson, S. Enerback, and M. Wikstrom. 2000. Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11. J. Mol. Biol. 296:351–359. [DOI] [PubMed] [Google Scholar]
- Weigelt, J., I. Climent, K. Dahlman-Wright, and M. Wikstrom. 2001. Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4). Biochemistry. 40:5861–5869. [DOI] [PubMed] [Google Scholar]