Abstract
Phagocytic cells such as neutrophils and macrophages are potential components of the immune defense that protects mammals against Candida albicans infection. We have tested the interaction between the mouse macrophage cell line RAW 264.7 and a variety of mutant strains of C. albicans. We used an end point dilution assay to monitor the killing of C. albicans at low multiplicities of infection (MOIs). Several mutants that show reduced virulence in mouse systemic-infection models show reduced colony formation in the presence of macrophage cells. To permit analysis of the macrophage-Candida interaction at higher MOIs, we introduced a luciferase reporter gene into wild-type and mutant Candida cells and used loss of the luminescence signal to quantify proliferation. This assay gave results similar to those for the end point dilution assay. Activation of the macrophages with mouse gamma interferon did not enhance anti-Candida activity. Continued coculture of the Candida and macrophage cells eventually led to death of the macrophages, but for the RAW 264.7 cell line this was not due to apoptotic pathways involving caspase-8 or -9 activation. In general Candida cells defective in the formation of hyphae were both less virulent in animal models and more sensitive to macrophage engulfment and growth inhibition. However the nonvirulent, hypha-defective cla4 mutant line was considerably more resistant to macrophage-mediated inhibition than the wild-type strain. Thus although mutants sensitive to engulfment are typically less virulent in systemic-infection models, sensitivity to phagocytic macrophage cells is not the unique determinant of C. albicans virulence.
Candida albicans can cause life-threatening infections in immunocompromised patients but is limited to causing primarily superficial mucosal infections in immunocompetent individuals (11, 38). This observation emphasizes that the mammalian immune system is a powerful barrier to Candida infections. Recent evidence has also suggested that the distinct morphological forms exhibited by C. albicans cells are critical for its virulence, as strains trapped in either the yeast or filamentous state in vitro are less virulent in murine infection models than cells capable of undergoing morphological changes (4, 11, 25, 26, 33, 37). It is possible that the hyphal form is required to penetrate the epithelial barrier while the yeast form allows for efficient dissemination through the bloodstream (6). However, the mutants that have been examined in these studies are pleiotropic, and characteristics unrelated to cellular morphology may determine the relative levels of virulence of the strains (7, 18, 22). Ultimately, dissection of the roles of morphology and various virulence factors in the infection process will involve the use of both in vitro and in vivo assays.
A powerful in vitro assay for the analysis of the host-pathogen interaction is the coculture of cells of the mammalian immune system with C. albicans cells (1, 3, 6). The usefulness of this approach is enhanced by the isolation of many mutant versions of C. albicans that are defective in components implicated in the virulence process (3, 33, 41). Initial studies showed that cells capable of forming hyphae were able to escape from phagocytosis through the formation of germ tubes and hyphae, while cells unable to undergo the morphological switch remained engulfed (26, 33). Mutations in several components of signaling pathways have been shown to modify the regulation of this morphological switch and to influence virulence. These components include proteins involved in the production of cyclic AMP (cAMP) such as adenylylcyclase (Cdc35p) (33) and Ras1p (24), as well as proteins required for the function of a mitogen-activated protein (MAP) kinase cascade that includes the p21-activated kinase (PAK) homolog Cst20p (23), the MEK Hst7p (23), and the MAP kinase Cek1p (12). The cAMP regulatory circuit appears to play a much more important role in the control of the hyphal switch than the MAP kinase pathway (33). However, coordinate loss of both pathways blocks the yeast-to-hypha transition under most growth conditions (26). In addition to Cst20p, a second PAK homolog, Cla4p, plays a critical role in the control of cellular morphogenesis (25).
Although the infection process is complex and involves interactions between the pathogen and many host cell types, in vitro studies involving specific cultured immune system cells can permit the analysis of interactions under controlled conditions. These studies can measure the influence of priming factors such as gamma interferon (IFN-γ) and cytokines and can provide the opportunity to monitor and compare host cell behaviors upon challenge with wild-type or mutant strains of the pathogen. A variety of methods for assessing the consequences of the host cell-pathogen interaction have been described, including CFU determination of harvested wells (1, 43). However, the pleiotropic nature of C. albicans, which can take the form of multicellular hyphae or cell aggregates, can bias such CFU measurements. Metabolic assays such as those involving radioisotope incorporation (35) and incorporation of tetrazolium salt indicators such as 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (20) and 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-([phenylamino]carbonyl)-2H-tetrazolium hydroxide (XTT) (40) involve further growth of Candida after the lysis of the host cells and would not be appropriate for low-multiplicity-of-infection (MOI) experiments or for strains with growth defects.
Processes such as the induction of apoptosis within cells of the host immune system can also be monitored during the interaction of the pathogen and the phagocytic cells. Apoptosis occurs via two main pathways, the death ligand receptor pathway mediated through caspase-8 activation and the stress pathway mediated through caspase-9 activation (16, 17). Some pathogens such as Shigella and Salmonella spp. (28, 32) induce apoptosis of phagocytic cells. This can trigger severe inflammation via the production and release of proinflammatory cytokines and can favor the dissemination of the pathogen deeper into host tissues. On the other hand, by undergoing apoptosis, macrophages may be able to expose the invaders to more-potent bactericidal cells such as neutrophils, as well as to the humoral arm of the immune system. Apoptosis can also prevent pathogens from using the phagocytic cells as a vehicle to evade the immune system (28); this apoptotic strategy helps control infections caused by Mycobacterium tuberculosis (14). It has been recently reported that apoptosis could be induced both in mouse macrophages by live C. albicans (36) and in human neutrophils upon challenge with killed C. albicans (34). However, other data suggested that Candida could inhibit apoptosis in human monocytic cells (19).
To be able to compare different Candida strains to each other in their interactions with host cells, we recently developed a rapid in vitro technique which allows for the comparison of Candida survival in the presence or absence of host cells at low MOIs without cell harvesting (33). We also introduced a luciferase reporter gene into five genetically defined C. albicans mutants and monitored their behavior in the presence of RAW 264.7 mouse macrophage cell lines at low and high MOIs. This allowed a characterization of host-pathogen interactions on a wide range of cells regardless of the pathogen phenotype observed at 37°C. We investigated the role of IFN-γ priming in enhancing the ability of macrophages to phagocytose and kill Candida. We also tested whether apoptosis of the RAW 264.7 macrophage cells is induced by wild-type or mutant C. albicans strains by using measurement of caspase-8 and -9 activity on specific substrates.
MATERIALS AND METHODS
Reagents.
Dulbecco's modified Eagle's medium (DMEM), Leibovitz's L15 medium (L15), and Dulbecco's phosphate-buffered saline (PBS) were purchased from Invitrogen/Gibco (Rockville, Md.). Fetal bovine serum (FBS) was purchased from HyClone (Logan, Utah) and was heat inactivated at 56°C for 30 min.
Strains and cell lines.
The C. albicans strains used in this study are listed in Table 1. Ura+ strains were grown in yeast extract-peptone-dextrose medium (YPD). Ura− strains were grown in YPD supplemented with 25 μg of uridine/ml. The RAW 264.7 mouse macrophage cell line was kindly provided by A. Descoteaux (IAF, Laval, Canada). These cells were grown in DMEM supplemented with 10% FBS (D-10). When required, RAW 264.7 cells were treated with 100 U of mouse IFN-γ (Invitrogen Canada Inc.)/ml for 18 h prior to incubation with Candida cells.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Wild type | 13 |
| CAI4 | ura3::λ imm434/ura3::λ imm434 | 13 |
| CDH107 | CAI4 ras1::hisG-URA3-hisG/ras1::hisG | 24 |
| CDH108 | CAI4 ras1::hisG/ras1::hisG | 24 |
| CaLJ1 | CAI4 cla4::hisG-URA3-hisG/cla4::hisG | 25 |
| CaLJ5 | CAI4 cla4::hisG/cla4::hisG | 25 |
| CDH22 | CAI4 cst20::hisG-URA3-hisG/cst20::hisG | 23 |
| CDH25 | CAI4 cst20::hisG/cst20::hisG | 23 |
| CR216 | CAI4 cdc35::hisG-URA3-hisG/cdc35::hisG | 33 |
| CR276 | CAI4 cdc35::hisG/cdc35::hisG | 33 |
| CAM 1.1 | CAI4 ura3 (R. reniformis luciferase gene-URA3) | This study |
| CAM 3.1 | CDH108 ura3 (R. reniformis luciferase gene-URA3) | This study |
| CAM 5.2 | CaLJ5 ura3 (R. reniformis luciferase gene-URA3) | This study |
| CAM 23.3 | CDH25 ura3 (R. reniformis luciferase gene-URA3) | This study |
| CAM 26.8 | CR276 ura3 (R. reniformis luciferase gene-URA3) | This study |
Plasmid construction and transformation.
An ADH1 promoter-driven Renilla reniformis luciferase expression plasmid was constructed as follows. A 1.5-kb NotI-EcoRV fragment from pYPB1-ADHpL (23) was blunted with Klenow fragment and subcloned into SmaI-digested pCRW3 (39). The resulting plasmid, pAM1.3, was digested with NcoI and SacI and blunted, and the 2.8-kb fragment containing the ADH1 promoter, luciferase gene, and WH11 terminator sequence was subcloned into the SmaI-digested pVEC (27). The resulting plasmid, pAM2.5, also contained the Candida autonomous replicating sequence (CARS) and the URA3 selectable marker. It was transformed in Candida URA3− strains by the rapid lithium acetate method (8). Transformants were streaked on Ura− plates and replica plated on YPD. After two rounds on Ura− plates followed by replica plating to YPD, clones that stably maintained the selectable marker were identified, and plasmid integration of several of these clones was confirmed by Southern analysis (data not shown).
End point dilution survival assay.
The end point dilution survival assay was performed as described previously (33). Briefly, the RAW 264.7 cells were seeded the day before at 1.5 × 105 cell per well in flat-bottom 96-well plates (Costar). When required, macrophages were treated with 100 U of mouse IFN-γ/ml (Invitrogen Canada Inc.). They were washed three times with D-10 medium before incubation with Candida. Overnight cultures of Candida cells were washed in PBS, sonicated for 1 min to disrupt any clumped cells, and resuspended at 107 cells/ml in cold D-10. Fifty microliters of the suspension was added to 150 μl of D-10 in each of the first 8 wells (first column) of 96-well plates containing either medium only or medium with macrophages. Serial fourfold dilutions were done in subsequent columns, and plates were incubated first on ice for 30 min and then at 37°C and 5% CO2 for 48 h. One whole plate per strain with or without macrophages was considered one experiment. Colonies were visualized with a Nikon TMS inverted microscope at ×20 or ×100 magnification. Passive lysis buffer (Promega, Madison, Wis.) at 1× final concentration was added immediately before counting the wells containing macrophages to facilitate colony visualization. Dilutions (without macrophages) where colonies could be distinctively visualized were counted toward the lower limit and compared to the same dilutions with macrophages. Colonies from a total of at least 48 wells per condition were used to provide data. Survival assays using luciferase-expressing strains were done similarly with serial twofold dilutions, starting at a 1:1 ratio for a final volume of 150 μl per well. Wells in triplicate were harvested after 12 h of incubation.
Renilla luciferase detection.
Microplates were centrifuged at 3,000 × g for 5 min, 75 μl of supernatant was removed, and 25 μl of 4× passive lysis buffer was added to each well. After two freeze-thaw cycles at −80°C, luciferase activity was assayed with a Promega Dual-Luciferase reporter kit in accordance with the manufacturer's instructions and detected with a Turner Designs model TD20/20 luminometer with a 20-s measurement period.
Time-lapse microscopy.
Bioptechs petri dishes were segmented into three parts with silicone (Dow Corning Corp., Midland, Mich.), left to dry for 2 to 3 days, washed three times with PBS, and sterilized with 70% ethanol. RAW 264.7 cells were seeded the day before at 3 × 105 cells per section of 1 cm2. Candida cells were added at the desired ratio in the CO2-independent L15 medium containing 10% FBS and 2.5 μg of propidium iodide (Molecular Probes, Eugene, Oreg.)/ml. Phase-contrast and epifluorescence pictures were taken every 30 min with a DMIRE2 inverted microscope (Leica Microsystemes Canada) equipped with a Hamamatsu cooled charge-coupled device camera, a Bioptechs temperature-controlled stage adapter, and a Ludl motorized stage at ×400 magnification. Openlab software (Improvision) was used for image acquisition.
Caspase activity determination.
RAW 264.7 cells were cultured overnight from an initial inoculum of 2.5 × 106 cells per well in six-well plates to obtain a monolayer at 80% confluence. Medium was then removed and replaced by 2 ml of fresh medium just before the experiment. Overnight culture of C. albicans strain SC5314 and cla4/cla4 and cdc35/cdc35 strains were prepared as described earlier. One milliliter of a Candida suspension containing 5 × 107 or 5 × 106 cells was added to appropriate wells and incubated at 37°C and 5% CO2. Cells were collected with a cell scraper (Sarstedt) and centrifuged at 3,000 × g. The supernatant was removed, and the pellet was frozen at −80°C until the caspase activity determination. Macrophages without Candida cells were used as negative controls. Positive controls consisted of UV treatment of 10 mJ/cm2 (CL-1000 UV cross-linker; UVP). Caspase-8 and caspase-9 activities were assayed with ApoAlert caspase fluorescence assay kits (Clontech) in accordance with the manufacturer's instructions. Caspase-8 specifically cleaves the peptidic substrate IETD-7-amino-4-trifluoromethyl coumarin, and the release of 7-amino-4-trifluoromethyl coumarin was measured. Similarly, caspase-9 activity was detected through the release of 7-amino-4-methyl coumarin from LEHD-7-amino-4-methyl coumarin. Samples were incubated in black microtiter plates (Labsystems) and read with a Spectramax Gemini fluorimeter (Molecular Devices) at the appropriate excitation and emission wavelengths.
Statistical analysis.
For end point dilutions, percentages of survival were compared to those for a wild-type strain (SC5314 or CAM 1.1) according to the Dunnett multiple-comparison test with the PRISM program (GraphPad Software). P values <0.01 were considered significant.
RESULTS
End point dilution analysis of macrophage-C. albicans interaction.
Cell viability assays provide a measure of the consequences of the interaction between phagocytic cells such as macrophages and pathogens such as C. albicans. We used an end point dilution assay to monitor the survival of C. albicans cells in the presence of RAW 264.7 macrophage cells. Candida colonies were counted starting from the MOI where they could be distinctly visualized and at all dilutions below this point that still contained cells. Survival was expressed as the number of colonies in the presence of macrophages divided by the number of colonies in the absence of macrophages. Several mutant lines that compromise hyphal formation in vitro were tested. Their respective phenotypes and levels of virulence in mouse studies are shown in Table 2. This end point survival assay showed that the colony-forming capacity of C. albicans cells defective in components of the cAMP signaling pathway, such as Ras1p and Cdc35p, was reduced in the presence of the macrophage cells (Table 2). Mutant strains with CST20 deleted, which fail to form hyphae under some growth conditions but which form hyphae normally during serum stimulation (23), showed a survival similar to that of the wild type. Surprisingly, mutant cells with CLA4 deleted, which are defective in filament formation and avirulent in a mouse systemic-infection model (25), survived better than even the wild-type cells when subjected to the macrophage challenge.
TABLE 2.
Virulence, phenotype, and end point dilution survival assay
| Parental strain | Lueiferase-expressing strain | Virulencea | Phenotypeb | End point survival (%)c of:
|
|
|---|---|---|---|---|---|
| Parental strain | Luciferase-expressing strain | ||||
| SC5314 | CAM 1.1 | Virulent | Hyphal | 32.2 ± 2.5 (5) | 27.6 ± 5.0 (5) |
| ras1/ras1 | CAM 3.1 | Reduced | Pseudohyphal | 9.9 ± 4.9 (3)∗ | 11.2 ± 1.6 (3)∗ |
| cla4/cla4 | CAM 5.2 | Avirulent | Pseudohyphal, aberrant | 48.5 ± 9.9 (5)∗ | 56.8 ± 11.3 (3)∗ |
| cst20/cst20 | CAM 23.3 | Reduced | Hyphal | 22.0 ± 3.5 (4) | 25.2 ± 4.1 (6) |
| cdc35/cdc35 | CAM 26.8 | Avirulent | Yeast | 7.0 ± 2.1 (4)∗ | 7.0 ± 4.4 (4)∗ |
Hematogenously disseminated murine model for parental strains.
At 37°C, in D-10 medium.
Data represent percentages of survival ± standard deviations of the means and are expressed as the numbers of colonies in the presence of RAW264.7 macrophages divided by the number of colonies in the absence of macrophages. Numbers in parentheses indicate numbers of experiments. ∗, significantly different from the wild-type strain of each group (P < 0.01).
Luciferase assays.
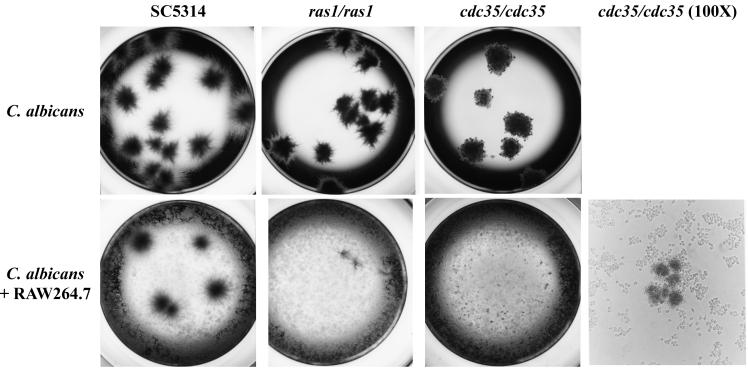
The end point dilution assay provides a measure of the loss of CFU due to macrophage killing of C. albicans cells cocultured at low MOIs. Figure 1 shows the appearance of colonies in the end point dilution assay after 48 h of incubation in D-10 at 37°C. However, as can be seen in Fig. 1, the macrophages can also greatly inhibit the proliferation of cells that are capable of forming colonies since the colony sizes are reduced, and thus the macrophage effect is not limited to reducing colony number due to initial phagocytosis and cell killing. To provide an assay that would allow investigation of the macrophage-C. albicans interaction at higher MOIs and that could measure the influence of the macrophages on Candida cell proliferation, we inserted the Renilla luciferase reporter gene into the C. albicans cells. The strains containing the luciferase reporter have phenotypes similar (cell shape and colony morphology) to those of their parental strains (data not shown) and have similar survival patterns in the end point dilution assay (Table 2).
FIG. 1.
Appearance of C. albicans colonies in the absence (top) or presence (bottom) of RAW 264.7 macrophage cells after a 48-h incubation time in 96-well plates. Photomicrographs were taken at ×20 magnification except where indicated.
We selected the inoculation conditions for the C. albicans cells by determining the initial cell densities that permitted exponential growth of the Candida cells in the absence of macrophages for the length of the experiment. All the cell lines were capable of exponential growth over the 12-h period of the experiments when they were inoculated at an initial cell density of 3.75 × 105 cells/well (150 μl). Shorter periods of incubation did not allow for evaluation of the slow-growing cdc35/cdc35 strain, and longer periods of incubation result in saturation and an eventual decrease in luciferase activity for several of the strains (data not shown).
Macrophages inhibit Candida growth.
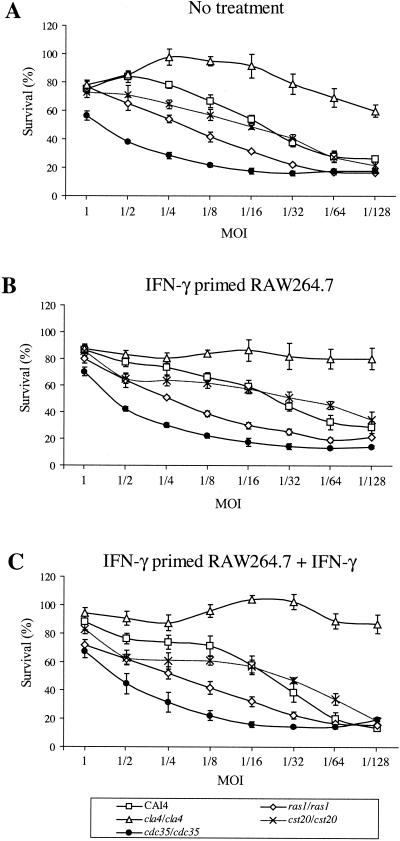
We used the strains with the luciferase reporter gene to investigate growth inhibition in the presence of the mouse macrophage cell line RAW 264.7 at different MOIs. After 12-h incubation periods, wells in triplicate were harvested as described in Materials and Methods. The inhibition of growth of the wild-type strain in the presence of macrophages is illustrated in Fig. 2A and was determined by comparison with luciferase units measured from the same Candida strain grown in the absence of macrophages. The wild-type reference strain CAI4 showed a growth inhibition that is dependent on the MOI used, with an 80% survival rate for MOIs of 1 and 1/2, which is reduced to 30% for MOIs of 1/64 and 1/128. At MOIs higher than 1/16, the cst20/cst20 strain is more sensitive than the wild-type CAI4. However, below this MOI, the behavior of that strain is similar to that of the control. Growth of the ras1/ras1 strain and that of the cdc35/cdc35 strain are more inhibited than that of CAI4 at MOIs lower than 1. The cdc35/cdc35 strain is the most sensitive strain, with a growth inhibition of 55% at an MOI of 1. The cla4/cla4 strain is up to two times more resistant than the wild-type CAI4 strain; the cla4/cla4 strain forms aberrant pseudohyphae compared to the hyphae observed in CAI4.
FIG. 2.
Survival rate of C. albicans strains in the presence of RAW 264.7 mouse macrophage cells. Macrophages untreated (A), IFN-γ primed (B), or IFN-γ primed and left in media (C) were incubated with twofold dilutions of C. albicans strains for 12 h and harvested as indicated in Materials and Methods. The percentage of survival was determined as residual Renilla luciferase activity compared to that for the same C. albicans strains grown in the absence of macrophages. Values obtained are the averages for three wells assayed. Bars, standard deviations. Experiments were done at least three times with similar results.
IFN-γ treatment of macrophages does not enhance Candida growth inhibition.
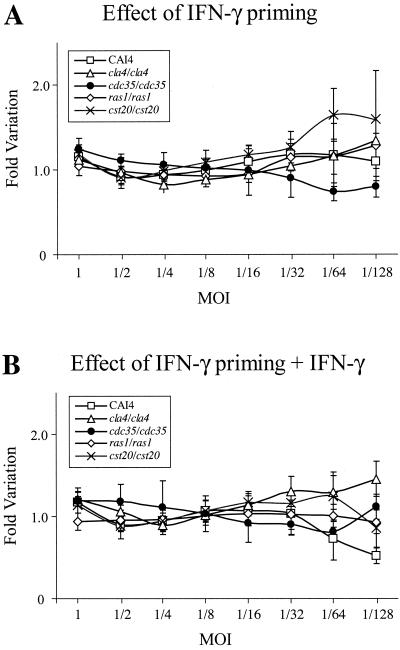
Candida phagocytosis and killing activity have been shown to increase upon IFN-γ priming of macrophages, alone or in combination with bacterial lipopolysaccharides (LPS) (5, 15, 30, 42). Figure 2B shows Candida survival when RAW 264.7 mouse macrophages were primed the day before with IFN-γ. The efficiency of IFN-γ priming could be observed by changes in RAW 264.7 cell morphology (data not shown). Surprisingly, this treatment seemed less efficient in causing growth inhibition of Candida cells than that of not priming RAW 264.7 cells, especially for the cla4/cla4 strain at MOIs lower than 1/16, where macrophages seemed inefficient in growth inhibition (80 to 100% survival). As shown in Fig. 2C, leaving IFN-γ in the medium during Candida challenge did not improve growth inhibition for MOIs higher than 1/32 for all the strains tested. However, the CAI4 strain was more inhibited at lower MOIs (1/64 and 1/128). IFN-γ had no effect on Candida cell proliferation when added directly to the medium (data not shown). Figure 3 shows the effect of those treatments on survival of C. albicans strains at different MOIs, compared to survival in untreated macrophages. A value of 1 indicates identical survival rates. No substantial variation could be visualized for MOIs at or above 1/32. Below this MOI, results are more variable due to lower luciferase activities detected in wells containing macrophages (near detection limit) than in wells without macrophages.
FIG. 3.
Effect of IFN-γ on the survival of C. albicans strains when incubated with macrophages that were IFN-γ primed (A) or IFN-γ primed and left in media (B) at different MOIs, compared to their survival when incubated with unprimed RAW 264.7 macrophages. Bars, standard deviations.
Macrophages are killed by virulent Candida strains.
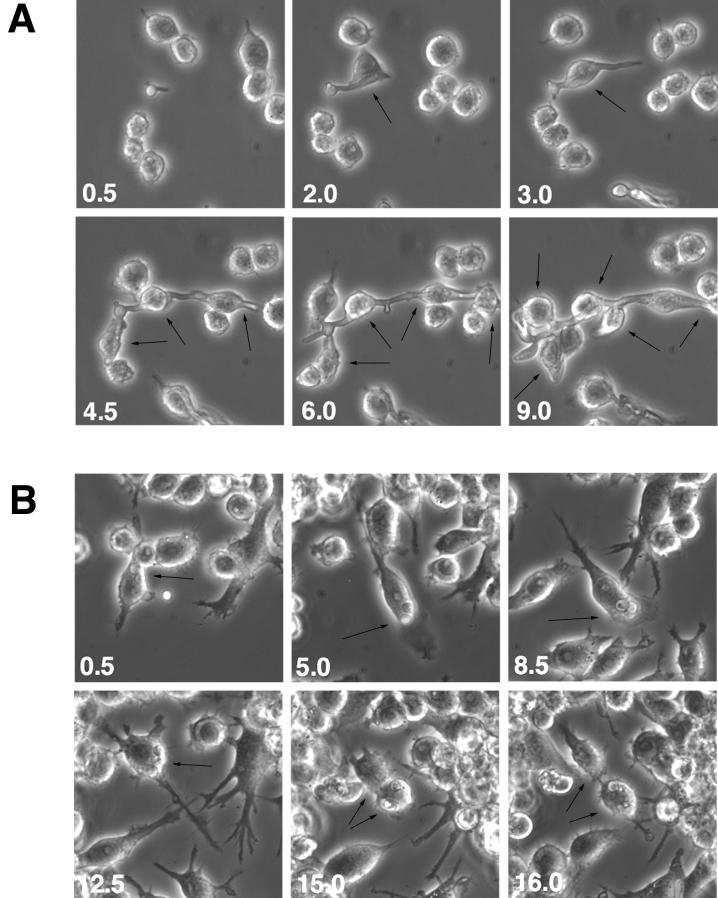
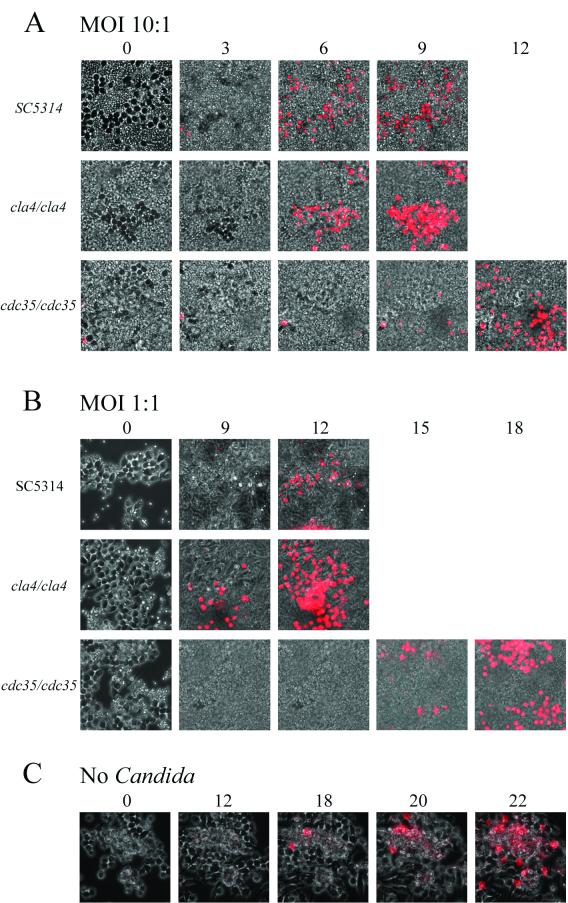
Time-lapse microscopy experiments were performed to monitor the macrophage-Candida interactions as described in Materials and Methods. The ability of Candida to form hyphae or pseudohyphae allows its escape from the macrophage phagolysosome (21, 26). As shown in Fig. 4A, at low MOIs (1/20) cla4/cla4 strain pseudohyphae could impale many macrophage cells without any apparent damage to either cell type for over a 9-h period or longer (not shown). The cdc35/cdc35 mutant is unable to form pseudohyphae or hyphae and therefore could not escape from the macrophage upon phagocytosis. In some observations (Fig. 4B), both macrophage (at 15 h) and ingested cdc35/cdc35 cells (at 8.5 h) were able to undergo cellular division. Propidium iodide staining was used to monitor macrophage cell death. As shown in Fig. 5, Candida at high MOIs (1 or 10) rapidly overgrew the macrophage monolayer. When RAW macrophages were challenged with SC5314 or cla4/cla4 Candida strains at an MOI of 10 (Fig. 5A), propidium iodide staining indicates that they died after 6 h; they died after 9 to 12 h when challenged by strains at an MOI of 1 (Fig. 5B). When macrophages were challenged with the avirulent, nonhyphal cdc35/cdc35 strain, macrophage death still occurred at MOIs of 10 and 1 but at later time points (10 to 12 h and 15 h, respectively) (Fig. 5A and B). At an MOI of 1/20, macrophage death occurred after 15 to 18 h of incubation (data not shown). The control RAW 264.7 macrophage cell line cultured without Candida died after 18 to 22 h under these culture conditions (Fig. 5C).
FIG. 4.
Time-lapse microscopy of RAW 264.7 mouse macrophage cells in the presence of cla4/cla4 (A) or cdc35/cdc35 (B) C. albicans strains at low MOI (1/20). Times (in hours) are indicated at lower left corners. Arrows, Candida-macrophage interactions.
FIG. 5.
Killing of RAW 264.7 mouse macrophage cells by C. albicans strains. Macrophages were incubated with C. albicans SC5314, cla4/cla4, or cdc35/cdc35 strain at MOIs of 10 (A) and 1 (B) or without Candida (C) and monitored by time-lapse microscopy at ×400 magnification for the indicated times (top, in hours) in the presence of propidium iodide. Killing of macrophages is indicated by appearance of propidium iodide-stained cells.
C. albicans does not induce apoptosis of macrophages.
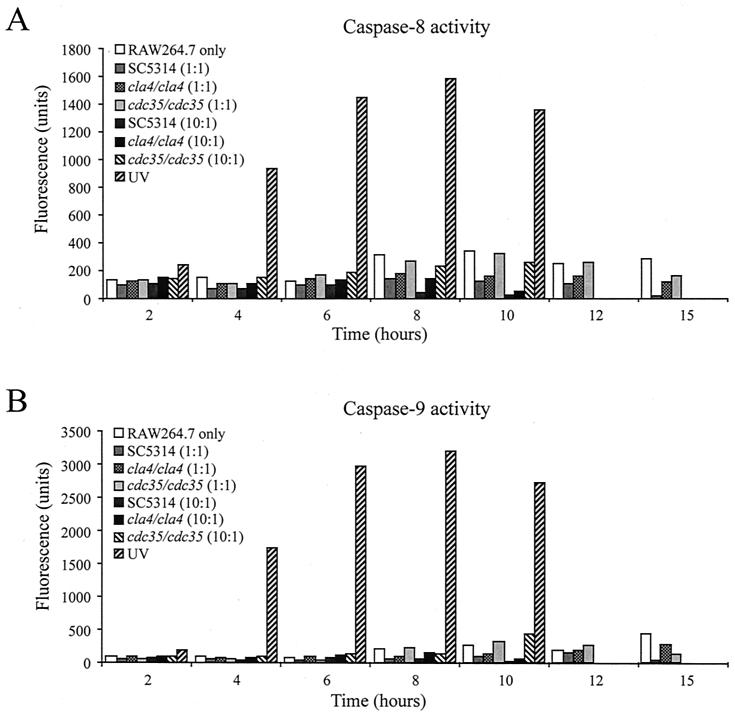
We analyzed whether the macrophage cell death observed (Fig. 5A and B) was mediated through an apoptotic or a necrotic process. Caspase-8 and caspase-9 identify the two main pathways through which apoptosis could be induced, and both proteases are activated early in the apoptotic process. The death receptor pathway, which is mediated by tumor necrosis factor-α or CD95, involves cleavage of procaspase-8 to caspase-8 through a death-inducing signaling complex (17). The stress pathway, induced by nutrient deprivation, oxidant stress, UV, ischemia, toxins, or heat, involves release of cytochrome c from mitochondria to the apoptosome and mediates cleavage of procaspase-9 to caspase-9 (16, 17). The specific activities of these proteases could be evaluated by measurement of the fluorescent compound released through cleavage of their specific peptidic substrates, IETD-AFC for caspase-8 and LEHD-AMC for caspase-9. RAW 264.7 mouse macrophages were incubated with the C. albicans SC5314, cla4/cla4, and cdc35/cdc35 strains at MOIs of 10 or 1 and harvested at the times indicated in Fig. 6. Caspase-8 and caspase-9 activities were measured for each sample. Negative controls consisting of macrophages only and positive controls consisting of macrophages induced with the known caspase inducer UV (44) were also included. Figure 6A and B show caspase-8 and caspase-9 activities, respectively, in RAW 264.7 cells cocultured with Candida strain SC5314 or the mutant cla4/cla4 or cdc35/cdc35 strain at MOIs of 1 or 10. As expected (44), exposure of RAW 264.7 cells to UV generated increased levels of both caspase-8 and -9 activities. For all Candida strains tested, caspase-8 activity was not increased in the presence of Candida compared to that for the control. In fact, caspase-8 activity was lower after macrophage challenge with SC5314 or the cla4/cla4 strain at MOIs of 10 and 1. Small amounts of caspase-9 activity were detected in RAW 264.7 cells challenged with the cdc35/cdc35 strain at an MOI of 1 in the 10- and 12-h samples. Because the RAW 264.7 macrophage cells had extensive mortality within the assessed period (Fig. 5), it appears that macrophage cell death detected does not occur through an apoptotic process.
FIG. 6.
Caspase-8 (A) and caspase-9 (B) activities of RAW 264.7 macrophage cells in the presence of C. albicans strains. Macrophage cells incubated without Candida and those treated with UV correspond to negative and positive controls, respectively. Cell samples were harvested at the indicated times, and caspase-8 and -9 activities were determined as described in Materials and Methods.
DISCUSSION
C. albicans is an important human fungal pathogen that can cause superficial or potentially lethal systemic infections of mammalian hosts. A powerful approach in the dissection of aspects of the infection process has been to monitor the interaction between C. albicans cells and cultured or purified mammalian cells (1, 5). Many of these studies have investigated the consequences of coculture of C. albicans cells and cells comprising elements of the mammalian immune system (15, 19, 36, 41). This approach permits the study of the roles of different immune system cells in killing the pathogen and also allows the analysis of the effects of signaling molecules such as cytokines on specific components of the host-pathogen interaction. These studies require the measurement of the killing of C. albicans as well as the mammalian cells during the interaction; measurements of CFU (3, 43) and metabolic activity by dyes (20, 40) are typical strategies to determine pathogen killing.
The potential of this overall approach for the dissection of the host-pathogen interaction has been enhanced by the construction of C. albicans strains defective in specific molecular functions (3, 4, 23-26, 33). While these mutant strains provide important tools for the analysis of host-pathogen interactions, properties of the strains, such as changes in the ratio of yeast to hyphal forms or changes in cell wall characteristics leading to cell fragility or clumping of cells can bias assays such as CFU determinations. We have recently developed an end point dilution assay that provides a convenient measure of the effect of immune cells on Candida viability at low MOIs without the need for cell harvesting (33). In the present study we have applied this assay to a series of C. albicans mutants (cst20, cdc35, ras1, and cla4 mutants). CDC35 and RAS1 are primarily implicated in control of cAMP levels, while CST20 and CLA4 encode PAK kinase homologs that are required for proper morphogenesis. We selected these mutants because they provide a spectrum of effects on the morphological switching process. We determined that strains defective in ras1 and adenylylcyclase are more sensitive to killing by RAW 264.7 macrophages than are the wild-type C. albicans cells. This sensitivity to macrophage killing correlates well with the reduced virulence exhibited by the mutant strains in systemic-infection assays (24, 33). In contrast, strains with mutated CST20 appear near the wild type in their resistance to macrophage killing in the end point dilution assay and exhibit virulence in the systemic-infection assays similar to that of the wild-type cells. The observation that the wild-type strains are somewhat more resistant than the mutants could reflect minor influences of the Cst20p function. Alternatively, this could be due to the fact that the SC5314 (wild-type) strain contains an intact URA3 locus, while the CAI4-derived strains such as the cst20/cst20 mutant contain the replaced URA3 gene but are still missing the other genes deleted during the construction of CAI4 (13). Intriguingly, strains defective in the CLA4-encoded PAK kinase are avirulent in the systemic-infection model (25) but exhibit a resistance to macrophage killing in the end point dilution assay that was greater than that of the wild-type strain from which the mutant was derived. Thus although sensitivity to macrophage killing is well correlated with reduced virulence in vivo, resistance to macrophages does not ensure virulence in an in vivo infection model.
To confirm the results of the end point dilution assays, as well as to provide an assay that can monitor the macrophage-Candida interaction at higher MOIs, we introduced a Renilla luciferase reporter construct into the wild-type and mutant C. albicans strains. Analysis of the effect of the RAW 264.7 macrophage cells on the various C. albicans strains shows that the inhibition of Candida proliferation caused by macrophages is density dependent. At high MOIs, where the number of C. albicans cells is similar to that of the macrophages, the wild-type C. albicans cells proliferate to almost the same extent as cells growing in the absence of macrophages. However, at levels where the macrophages outnumber the fungal cells by 100 to 1 there is substantial reduction in Candida proliferation relative to that in the cultures without macrophages. At intermediate MOIs the proliferation inhibition is also intermediate; there does not appear to be a threshold of influence but rather a constant dose dependence on the ratio of Candida cells to macrophages. This pattern is not dramatically influenced by treating the macrophages with IFN-γ, suggesting that this cytokine does not play a significant role in activating these mouse macrophage cells against Candida. Previous evidence on the role of IFN-γ in candidacidal activity by macrophages has been controversial. The priming of macrophages with IFN-γ was found to enhance their candidacidal potential (5, 15, 30, 42). However, systemic infection of IFN-γ knockout mice showed that Candida clearance was not impaired (31). Baltch et al. (1) recently demonstrated that, in vitro, this priming was efficient against Candida only when used in combination with fluconazole.
The luciferase assay confirms the relative sensitivities of the Candida mutants determined by the end point dilution assay. The cdc35/cdc35 mutant strain was the most sensitive to macrophage-mediated inhibition at essentially all MOIs tested, while the ras1/ras1 mutant was more sensitive than the wild-type cells but less sensitive than the adenylylcyclase-defective strain. The strain defective in CST20 behaved similarly to the wild-type strain, while the cla4/cla4 strain was very resistant to the influence of the macrophages. The resistance of the cla4/cla4 strain was most evident at the lower MOIs, where the wild-type and the other mutant strains were substantially inhibited in proliferation by the macrophages. The activation of the macrophages by IFN-γ did not enhance the ability of the macrophages to inhibit proliferation of the mutant C. albicans cells. In fact the cla4/cla4 strain was essentially unaffected by the IFN-γ-stimulated cells at any MOI, while the untreated macrophages showed a moderate ability to reduce proliferation of these mutant cells at low MOIs. This resistance to macrophages exhibited by the cla4/cla4 mutant strain may reflect the altered morphology exhibited by these cells (25).
The effect of the interaction between the pathogen and the immune system cells is not limited to the growth inhibition of the pathogen cells. Coculture of the C. albicans and RAW 264.7 cells also enhances the death of the RAW macrophage cells. This killing is significant: high levels of wild-type Candida cells (MOI of 10:1) cause macrophage death within 6 h, whereas macrophages cultured without Candida cells die after 18 to 22 h (Fig. 5). This killing is density dependent, in that lower MOIs increase the time before macrophage death, and is also dependent on the virulence of the Candida cells, as the cdc35/cdc35 mutant strains cause macrophage death at 10 h at an MOI of 10:1 and after 15 to 18 h at an MOI of 1:1.
This Candida-induced macrophage death does not appear to be due to apoptosis of the mouse cells. We measured both the caspase-8 and -9 activities that define the two major apoptotic pathways and showed that neither enzyme was significantly induced before the macrophage cells were killed. This observation contrasts with previous work showing that Candida could trigger apoptosis of mammalian cells (34, 36). Schroppel et al. (36) examined decreased mitochondrial potential and chromatin degradation of IFN-γ- and LPS-stimulated mouse peritoneal macrophages challenged with wild-type Candida at a low MOI (1/32). Rotstein et al. (34) examined microscopic manifestations of apoptosis such as nuclear condensation and formation of apoptotic bodies, as well as caspase-8, -9, and -3 activities of human neutrophils upon challenge with killed Candida. There are several possible explanations for the differences among the studies. First, mammalian cells of different origins could respond differentially to a Candida challenge and the response might also be MOI dependent. Second, the presence or absence of LPS priming could have a major influence on the activation of apoptotic pathways in response to Candida. In this work we challenged macrophages with live Candida without prior IFN-γ and LPS priming. IFN-γ and LPS are strong inducers of inducible nitric oxide synthase (2), and a high level of production of NO could potentially induce apoptosis in macrophages through downregulation of antiapoptotic proteins (10). As well, macrophages challenged with Candida have been shown to reduce the level of inducible nitric oxide synthase (9, 36); this reduction could help prevent macrophage apoptosis. Because LPS can bind to Toll-like receptors and because these receptors can activate proapoptotic pathways through activation of caspase-8 (28), LPS priming of macrophages could stimulate apoptotic processes. A third difference among the studies is the use of heat-killed Candida; this treatment modifies Candida-macrophage interactions both biochemically and physiologically (9), and thus comparisons between live and heat-killed cells may not be valid. Finally, apoptosis could be induced by nutrient limitation. In the present study we selected conditions to minimize the effect of nutrient limitation, but this could have influenced other studies.
It is interesting that cultured monolayered RAW 264.7 macrophages are not particularly efficient in killing Candida: up to 30% of the wild-type pathogen cells still survived the interaction even at low MOIs (Table 2 and Fig. 2). In addition, RAW 264.7 cells can both phagocytose Candida and be impaled by Candida filaments without any immediately apparent cell damage (Fig. 4A), and macrophages can continue to divide in the presence of Candida (Fig. 4B). Overall then, the Candida-macrophage interaction is not dramatically detrimental to either cell type; this may reflect the fact that Candida is usually a common but innocuous component of mammalian flora (38). It is also clear from the observation that CLA4 mutants are avirulent but also insensitive to macrophage killing that the whole organism's response to Candida involves much more than macrophages. Other cell types such as neutrophils or dendritic cells, as well as the humoral immune response, presumably play a significant role in host defense against Candida systemic infections (6, 29). Because in vitro experiments allow for the dissection and measurement of individual processes involved in the host-pathogen interaction, further studies should provide insight into the importance of each individual component in the process. Ultimately, a combination of in vitro and in vivo studies will be necessary to provide the framework for understanding this complex interaction.
Acknowledgments
We thank D. R. Soll, B. Magee, and A. Brown for plasmids, W. A. Fonzi, C. Rocha, and E. Leberer for providing C. albicans strains, and Antoine W. Caron for help with time-lapse microscopy. We are especially grateful to Albert Descoteaux for providing the RAW 264.7 macrophage cell line and for helpful discussion and André Nantel for critical reading of the manuscript. Finally, we thank members of the laboratory for fruitful discussions.
Editor: T. R. Kozel
Footnotes
National Research Council of Canada publication 44836.
REFERENCES
- 1.Baltch, A. L., R. P. Smith, M. A. Franke, W. J. Ritz, P. B. Michelsen, and L. H. Bopp. 2001. Effects of cytokines and fluconazole on the activity of human monocytes against Candida albicans. Antimicrob. Agents Chemother. 45:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, K. F., W. Eberhardt, S. Frank, A. Huwiler, U. K. Messmer, H. Muhl, and J. Pfeilschifter. 1999. Inducible NO synthase: role in cellular signalling. J. Exp. Biol. 202:645-653. [DOI] [PubMed] [Google Scholar]
- 3.Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 5.Brummer, E., and D. A. Stevens. 1989. Candidacidal mechanisms of peritoneal macrophages activated with lymphokines or gamma-interferon. J. Med. Microbiol. 28:173-181. [DOI] [PubMed] [Google Scholar]
- 6.Calderone, R., and N. A. Gow. 2002. Host recognition by Candida species, p. 67-86. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 7.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 9.Chinen, T., M. H. Qureshi, Y. Koguchi, and K. Kawakami. 1999. Candida albicans suppresses nitric oxide (NO) production by interferon-gamma (IFN-gamma) and lipopolysaccharide (LPS)-stimulated murine peritoneal macrophages. Clin. Exp. Immunol. 115:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, H. T., H. O. Pae, B. M. Choi, T. R. Billiar, and Y. M. Kim. 2001. Nitric oxide as a bioregulator of apoptosis. Biochem. Biophys. Res. Commun. 282:1075-1079. [DOI] [PubMed] [Google Scholar]
- 11.Corner, B. E., and P. T. Magee. 1997. Candida pathogenesis: unravelling the threads of infection. Curr. Biol. 7:R691-R694. [DOI] [PubMed] [Google Scholar]
- 12.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fratazzi, C., R. D. Arbeit, C. Carini, M. K. Balcewicz-Sablinska, J. Keane, H. Kornfeld, and H. G. Remold. 1999. Macrophage apoptosis in mycobacterial infections. J. Leukoc. Biol. 66:763-764. [DOI] [PubMed] [Google Scholar]
- 15.Gaviria, J. M., J. A. van Burik, D. C. Dale, R. K. Root, and W. C. Liles. 1999. Comparison of interferon-gamma, granulocyte colony-stimulating factor, and granulocyte-macrophage colony-stimulating factor for priming leukocyte-mediated hyphal damage of opportunistic fungal pathogens. J. Infect. Dis. 179:1038-1041. [DOI] [PubMed] [Google Scholar]
- 16.Goyal, L. 2001. Cell death inhibition: keeping caspases in check. Cell 104:805-808. [DOI] [PubMed] [Google Scholar]
- 17.Green, D. R., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 18.Haynes, K. 2001. Virulence in Candida species. Trends Microbiol. 9:591-596. [DOI] [PubMed] [Google Scholar]
- 19.Heidenreich, S., B. Otte, D. Lang, and M. Schmidt. 1996. Infection by Candida albicans inhibits apoptosis of human monocytes and monocytic U937 cells. J. Leukoc. Biol. 60:737-743. [PubMed] [Google Scholar]
- 20.Jahn, B., E. Martin, A. Stueben, and S. Bhakdi. 1995. Susceptibility testing of Candida albicans and Aspergillus species by a simple microtiter menadione-augmented 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay. J. Clin. Microbiol. 33:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaposzta, R., L. Marodi, M. Hollinshead, S. Gordon, and R. P. da Silva. 1999. Rapid recruitment of late endosomes and lysosomes in mouse macrophages ingesting Candida albicans. J. Cell Sci. 112:3237-3248. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, S. D., and J. E. Cutler. 1998. Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol. 6:92-94. [DOI] [PubMed] [Google Scholar]
- 23.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. Gow, A. J. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 25.Leberer, E., K. Ziegelbauer, A. Schmidt, D. Harcus, D. Dignard, J. Ash, L. Johnson, and D. Y. Thomas. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7:539-546. [DOI] [PubMed] [Google Scholar]
- 26.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 27.Magee, B. B., and P. T. Magee. 1997. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143:289-295. [DOI] [PubMed] [Google Scholar]
- 28.Navarre, W. W., and A. Zychlinsky. 2000. Pathogen-induced apoptosis of macrophages: a common end for different pathogenic strategies. Cell. Microbiol. 2:265-273. [DOI] [PubMed] [Google Scholar]
- 29.Newman, S. L., and A. Holly. 2001. Candida albicans is phagocytosed, killed, and processed for antigen presentation by human dendritic cells. Infect. Immun. 69:6813-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puliti, M., D. Radzioch, R. Mazzolla, R. Barluzzi, F. Bistoni, and E. Blasi. 1995. Influence of the Bcg locus on macrophage response to the dimorphic fungus Candida albicans. Infect. Immun. 63:4170-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian, Q., and J. E. Cutler. 1997. Gamma interferon is not essential in host defense against disseminated candidiasis in mice. Infect. Immun. 65:1748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raupach, B., and S. H. Kaufmann. 2001. Immune responses to intracellular bacteria. Curr. Opin. Immunol. 13:417-428. [DOI] [PubMed] [Google Scholar]
- 33.Rocha, C. R., K. Schroppel, D. Harcus, A. Marcil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotstein, D., J. Parodo, R. Taneja, and J. C. Marshall. 2000. Phagocytosis of Candida albicans induces apoptosis of human neutrophils. Shock 14:278-283. [DOI] [PubMed] [Google Scholar]
- 35.Scaringi, L., E. Blasi, P. Cornacchione, C. Bietta, and F. Bistoni. 1991. A rapid Candida albicans hyphal-form growth inhibition assay: determination of myelomonocytic-mediated antifungal activity. Mycoses 34:119-123. [DOI] [PubMed] [Google Scholar]
- 36.Schroppel, K., M. Kryk, M. Herrmann, E. Leberer, M. Rollinghoff, and C. Bogdan. 2001. Suppression of type 2 NO-synthase activity in macrophages by Candida albicans. Int. J. Med. Microbiol. 290:659-668. [DOI] [PubMed] [Google Scholar]
- 37.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 38.Soll, D. R. 2002. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 81:101-110. [DOI] [PubMed] [Google Scholar]
- 39.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tellier, R., M. Krajden, G. A. Grigoriew, and I. Campbell. 1992. Innovative end point determination system for antifungal susceptibility testing of yeasts. Antimicrob. Agents Chemother. 36:1619-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torosantucci, A., P. Chiani, F. De Bernardis, A. Cassone, J. A. Calera, and R. Calderone. 2002. Deletion of the two-component histidine kinase gene (CHK1) of Candida albicans contributes to enhanced growth inhibition and killing by human neutrophils in vitro. Infect. Immun. 70:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vazquez-Torres, A., and E. Balish. 1997. Macrophages in resistance to candidiasis. Microbiol. Mol. Biol. Rev. 61:170-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vonk, A. G., C. W. Wieland, M. G. Netea, and B. J. Kullberg. 2002. Phagocytosis and intracellular killing of Candida albicans blastoconidia by neutrophils and macrophages: a comparison of different microbiological test systems. J. Microbiol. Methods 49:55-62. [DOI] [PubMed] [Google Scholar]
- 44.Vu, C. C., C. D. Bortner, and J. A. Cidlowski. 2001. Differential involvement of initiator caspases in apoptotic volume decrease and potassium efflux during Fas- and UV-induced cell death. J. Biol. Chem. 276:37602-37611. [DOI] [PubMed] [Google Scholar]