Abstract
A variety of strains of the genus Lactobacillus was investigated with respect to the structure, softness, and interactions of their outer surface layers in order to construct structure-property relations of the Gram-positive bacterial cell wall. The role of the conformational properties of the constituents of the outer cell-wall layers and their spatial distribution on the cell wall is emphasized. Atomic force microscopy was used to resolve the surface structure, interactions, and softness of the bacterial cell wall at nanometer-length scales and upwards. The pH-dependence of the electrophoretic mobility and a novel interfacial adhesion assay were used to analyze the average physicochemical properties of the bacterial strains. The bacterial surface is smooth when a compact layer of globular proteins constitutes the outer surface, e.g., the S-layer of L. crispatus DSM20584. In contrast, for two other S-layer containing strains (L. helveticus ATCC12046 and L. helveticus ATCC15009), the S-layer is covered by polymeric surface constituents which adopt a much more extended conformation and which confer a certain roughness to the surface. Consequently, the S-layer is important for the overall surface properties of L. crispatus, but not for the surface properties of L. helveticus. Both surface proteins (L. crispatus DSM20584) and (lipo)teichoic acids (L. johnsonii ATCC332) confer hydrophobic properties to the bacterial surface whereas polysaccharides (L. johnsonii DSM20533 and L. johnsonii ATCC 33200) render the bacterial surface hydrophilic. Using the interfacial adhesion assay, it was demonstrated that hydrophobic groups within the cell wall adsorb limited quantities of hydrophobic compounds. The present work demonstrates that the impressive variation in surface properties displayed by even a limited number of genetically-related bacterial strains can be understood in terms of established colloidal concepts, provided that sufficiently detailed structural, chemical, and conformational information on the surface constituents is available.
INTRODUCTION
The surface properties of a microorganism are largely determining its interactions with the environment, including that with other microbes and host organisms, its infectiousness, the exchange of nutrients and waste products, and the resistance to external stresses as caused by mechanical, chemical, thermal, and osmotic factors. Interactions of microorganisms with their environment are of major importance during many of the important stages of the life cycle of a microorganism, like their growth, cell division, protection against a hostile environment, and the infection of a host. Interactions of microorganisms can be specific or nonspecific. Specific interactions involve the recognition of a specific site or ligand by a receptor on the microorganism (Savage and Fletcher, 1985), whereas nonspecific interactions are governed by the overall physicochemical properties of the bacterial cell wall, in particular its outer constituents. Microbial interactions have been studied in particular in relation to their role in bacterial infections (Savage and Fletcher, 1985), bacterial adhesion in environmental systems (Marshall, 1976; Savage and Fletcher, 1985) and in the biomedical field (Ofek and Doyle, 1994).
The vast diversity in microbial surface structure and properties has been investigated using a range of approaches including microscopy, microbiology, immunology, and molecular biology. For a long time, it has been recognized that important aspects of microbial behavior are controlled by the physicochemical properties of the cell wall (Eggerth, 1923; Webster, 1925; Marshall, 1976; Wadström, 1990). Detailed analyses of the relation between cell-wall structure and its physicochemical properties are only gradually emerging, however (Busscher et al., 2000; Boulbitch et al., 2000). Physicochemical analysis of the interactions of the bacterial surface is usually limited to the overall electric properties as represented by the ζ-potential (Eggerth, 1923; Marshall, 1976; Poortinga et al., 2001; Van der Mei and Busscher, 2001) and the hydrophobicity of the surface as determined by classical partitioning analysis (Albertsson, 1986), interfacial adhesion assays (Van Loosdrecht et al., 1987; Reid et al, 1992; Tomeczek et al., 1992; Daffonchio et al., 1995), contact angle methods (Van Loosdrecht et al., 1987; Van Oss, 1994; Reid et al., 1992; Gallardo-Moreno et al., 2002), or hydrophobic interaction chromatography (Makin and Beveridge, 1996). Knowledge of these two surface properties is used to correlate or model the interactions of the bacterial cell wall with external surfaces or hosts. Information on the chemical composition of the outer layers of the microbial cell wall can be obtained from x-ray photoelectron spectroscopy (Mozes and Lortal, 1995; Dufrêne and Rouxhet, 1996; Dufrêne et al., 1997) or from Fourier transform infrared spectroscopy (Curk et al., 1994; Amiel et al., 2000).
The general features of bacterial surface physicochemistry are known and the role of hydrophobic and electrostatic interactions in, for instance, bacterial adhesion, is well established (Marshall, 1976; Van Loosdrecht et al., 1987; Van Oss, 1994). It is clear, however, that, apart from classical colloidal concepts like the electrostatic potential and the hydrophobicity, the conformation of the surface constituents plays a major role, in particular if they have significant degrees of freedom. In addition, the structural organization of the various constituents within the cell wall is reflected in the bacterial surface properties. How the structural organization of the cell wall, the chemical properties of the surface constituents and, in particular, the conformation of the surface macromolecules determine the physicochemical properties of the cell wall are still largely open questions.
Although the conformation of surface polymers is of major importance for the overall physicochemical properties of bacteria, these conformations are only rarely studied, in part because few techniques allow the determination, directly or indirectly, of such conformational properties. Dynamic light scattering is probably the most useful indirect technique (Van der Mei et al., 1994, 2001). Atomic force microscopy (AFM) is the most suitable technique to directly study the conformational properties of surface polymers (Razatos et al., 1998; Razatos, 2001; Boonaert et al., 2000; Dufrêne, 2000, 2001; Van der Mei et al., 2001; Velegol and Logan, 2002; Schär-Zammaretti and Ubbink, 2003), but sample preparation techniques like the use of the crosslinker glutaraldehyde (Razatos et al., 1998; Razatos, 2001) could give rise to artifacts.
Lactic acid bacteria from the genus Lactobacillus could well serve as model systems to study the structure-property relations of the bacterial cell wall, inasmuch as they have the relatively simple cell-wall structure associated with Gram-positive microorganisms (Delcour et al., 1999), are little known for specific interactions potentially interfering with the overall physicochemical properties of the cell wall, and are devoid of long appendages strongly influencing the bacterial surface properties. Moreover, they are nonmotile and a large number of microbiologically and genetically well-characterized strains is available. Lactobacilli are rodlike with a length of between ∼1 and 1.5 μm and a diameter of ∼0.7 to 1 μm.
Lactobacilli are of considerable technological and commercial importance because of their role in the manufacturing and preservation of many fermented food products, but they also play an important role in the control of undesirable microorganisms in the intestinal and urogenital tract (Wood, 1992). Beside indigenous Lactobacilli, which reside in the human gastrointestinal tract, several Lactobacillus strains from fermented food products have shown beneficial effects on gut health (Fuller, 1992). The surface properties of lactic acid bacteria are of major importance in fermentation technology (Mozes and Rouxhet, 1990; Boonaert and Rouxhet, 2000) but they are also thought to play an important role in the adhesion of the bacteria to the gastrointestinal epithelium which is considered to be a prerequisite for, e.g., exclusion of enteropathogenic bacteria (Bernet et al., 1993, 1994; Mack et al., 1999) or immunomodulation of the host (Isolauri et al., 1999; Blum et al., 2002). The adhesive properties of lactic acid bacteria have been extensively tested using many in vitro models, like adhesion tests to Caco-2 or HT-29 cells (Bernet et al., 1993, 1994; Karjavainen et al., 1998; Ouwehand et al., 1999; Tuomola and Salminen, 1998). A deeper understanding of the factors influencing the surface properties of lactic acid bacteria will definitely promote the selection and evaluation of strains having the desired characteristics for food processing and health benefits.
The Gram-positive cell wall of lactic acid bacteria consists mainly of peptidoglycans, (lipo)teichoic acids, proteins and polysaccharides (Delcour et al., 1999). The inner layer of the cell wall consists of a peptidoglycan network, the sacculus, which is made up of linear polysaccharide chains which are themselves made up of alternating n-acetylglucosamine and n-acetyl-muramic acid units extensively crosslinked by two short peptides (Streyer, 1981; Delcour et al., 1999). Because of the high crosslinking density and the limited conformational flexibility allowed by the β 1→4 linkage of the n-acetylglucosamine and n-acetyl-muramic acid units, the sacculus is fairly stiff and rigid and is able to accommodate the significant stretching forces resulting from the bacterial turgor pressure.
The peptidoglycan layer of the cell wall of lactic acid bacteria is covered by a variety of substances. The most important of these substances are (lipo)teichoic acids, neutral and acidic polysaccharides, and (surface) proteins (Delcour et al., 1999). Teichoic acids form a diverse class of substances whose basic structure is a linear polymer of a polyol (such as glycerol or various monosaccharides) linked by phosphodiester bridges (Streyer, 1981; Delcour et al., 1999). Lipoteichoic acids are anchored into the cytoplasmic membrane by their lipidic tail whereas teichoic acids are covalently attached to the sacculus. As its phosphate groups are strong acids, (lipo)teichoic acids display a pronounced polyelectrolyte character.
The polysaccharides associated with the bacterial cell wall and the extracellular polysaccharides of lactic acid bacteria are either neutral or acidic (Delcour et al., 1999; Ricciardi and Clementi, 2000). Because of their abundance and their presence at the outer surface of the cell wall, extracellular and cell-wall associated polysaccharides are expected to determine to a large extent the surface properties of microorganisms.
The most abundant surface proteins in many Lactobacillus species are the S-layer proteins (Mozes and Lortal, 1995; Delcour et al., 1999; Smit et al., 2001). Up to now, S-layers have been found in strains of the species L. brevis, L. acidophilus, L. crispatus, L. helveticus, L. amylovorus, and L. gallinarum (Delcour et al., 1999; Smit et al., 2001; Ventura et al., 2002) but not in species like L. johnsonii and L. gasseri (Ventura et al., 2002). S-layer proteins are usually small proteins of 40–60 kDa with generally highly stable tertiary structures (Engelhardt and Peters, 1998). S-layer proteins are noncovalently bound to the cell wall and assemble into surface layers with high degrees of positional order often completely covering the cell wall (Lortal et al., 1992; Engelhardt and Peters, 1998; Sleytr et al., 2000). In contrast to most bacterial species, the S-layer proteins in lactobacilli are highly basic, with an isoelectric point above pH = 9 (Smit et al., 2001; Ventura et al., 2002; unpublished data). Because it fully covers the cell wall and because of the high isoelectric point of the S-layer protein, the S-layer may be expected to have appreciable effects on the properties of the cell wall of many Lactobacillus strains although its precise functionality is not known (Delcour et al., 1999; Smit et al., 2001).
The objective of this article is to investigate the relationship between the organization of the various constituents within the cell wall and the colloidal properties of the bacterium. In particular, we attempt to assess the impact of the conformational degrees of freedom of the surface constituents on the physicochemical behavior. Our approach is to construct structure-property relations of the cell wall by combining biological, microscopic, and physicochemical information at a number of levels. For this purpose, we have selected strains of lactic acid bacteria representing a considerable variation in cell-wall composition. Average information on the effective charge of the bacterium is obtained via electrophoretic mobility measurements. The overall bacterial hydrophobicity is determined using a novel interfacial adhesion assay for which a theoretical foundation is provided. AFM is used to resolve the surface structure, interactions and softness of the bacterial cell wall at nm-length scales and upwards, and, in combination with the physicochemical data, models of the outer layers of the bacterial cell wall are elaborated. The relevance of the main results for bacterial interactions is discussed.
MATERIALS AND METHODS
Growth and preparation of bacterial cultures
The strains used in this study were obtained from the American Type Culture Collection (ATCC) and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM). The following strains were used: L. johnsonii DSM20533; L. johnsonii ATCC332; L. johnsonii ATCC33200; L. crispatus DSM20584; L. helveticus ATCC15009, and L. helveticus ATCC12046. Bacteria were grown overnight under anaerobic conditions in test tubes containing 10 ml MRS broth (Difco, Le Pont de Claix, France) at 40°C and harvested in late-stationary phase (12–14 h). The cells were harvested by centrifugation (∼5000× g, 10 min; 4°C) and washed either 2× with a 0.9% NaCl solution (AFM analysis) or 3× with a 10 mM KH2PO4 buffer (electrophoretic experiments, interfacial adhesion assay). The pellet from a 10 ml fermentation was resuspended in 4 ml 0.9% NaCl solution (pH = 7.0, AFM analysis) or in 1 ml 10 mM KH2PO4 buffer at pH = 5 (ζ-potential and interfacial adhesion experiments) and stored at 4°C until use. Cultures were stored for a maximum of 72 h, as during this period no significant changes were observed in ζ-potential or adhesion behavior. Cultures of L. helveticus ATCC12046 were used within the first day after preparation because of their propensity to autolysis in low-ionic strength buffers (Lortal et al., 1991). For AFM, electrophoresis, and interfacial adhesion analysis, the cells were resuspended in 10 mM KH2PO4 buffer, the pH of which was adjusted by either 1 N HCl or 1 N NaOH to the desired value. The approximate cell count of the final suspensions was 107–108 colony-forming units/ml (CFU/ml).
Determination of electrophoretic mobility and ζ-potential
Electrophoretic mobility was measured by laser Doppler velocimetry with a ZetaSizer 4 (Malvern Instruments, Malvern, UK). A quartz capillary (ZET5104, diameter 4 mm) was used as the electrophoresis cell. Between 5 and 10 ml of the bacterial suspension was injected into the electrophoresis cell using a disposable syringe and the temperature was left to stabilize at 25±0.2°C. Before injection of the bacterial suspension, the measurement cell was flushed with ultrapure water (MilliQ, Millipore, Billerica, MA) or with 10 mM KH2PO4 buffer. Electrophoretic mobilities were converted to the ζ-potential using the Helmholtz-Schmoluchowski equation, which is valid for particles much larger than the Debye screening length (Hiemenz, 1986; Evans and Wennerström, 1994),
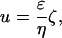 |
(1) |
where u is the electrophoretic mobility, η the viscosity, and ɛ = ɛrɛ0 is the dielectric constant of the medium, with ɛr the relative dielectric constant of water and ɛ0 the permittivity of vacuum. The Helmholtz-Schmoluchowski approximation is valid as the typical size of a bacterium is ∼1 μm and the Debye length κ−1 is of the order of a few nm. The Debye length κ−1 is defined by 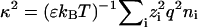 , where kB is Boltzmann's constant, T the absolute temperature, q the elementary charge, and zi and ni the valency and bulk number density of the ith ionic species.
, where kB is Boltzmann's constant, T the absolute temperature, q the elementary charge, and zi and ni the valency and bulk number density of the ith ionic species.
Hydrophobicity through interfacial adhesion
The classical Microbial Adhesion To Hexadecane test (MATH) (Rosenberg, 1984) was carried out largely following the method described by Reid et al. (1992). In brief, to 10 ml of the 10-mM KH2PO4 buffer at pH = 7, a quantity of the bacterial suspension was added such that the resulting optical density (OD) was 0.5 ± 0.05. This usually required the addition of an aliquot of bacterial suspension of 100–200 μl to the 10-ml buffer solution. After homogenization, 3.0 ml of the suspension was pipetted into a 15-ml sealable plastic test tube (Falcon, BD Biosciences, Allschwil, Switzerland). Subsequently, 150 μl hexadecane (purity > 98%; Fluka, Buchs, Switzerland) was added and, after hermetically closing the tube, the mixture was vortexed at maximum speed for 30 s using a Vortex Genie 2 (Scientific Instruments, Bohemia, NY). This was repeated for 30 s after an interval of 1 min. The OD of both the initial and the extracted solution was determined at λ = 600 nm using an Uvikon 810 UV/VIS spectrophotometer (BioTek, Basel, Switzerland) and disposable polystyrene cuvettes with an effective volume of 1 ml. A blank value was determined for the phosphate buffer without added bacteria. A waiting period of between 10 min and 25 min was employed to achieve complete phase separation between the water and hexadecane phases while ensuring that significant sedimentation of the bacteria still in solution did not occur. The interfacial adhesion assay was carried out at room temperature (22 ± 1°C).
The fraction of bacteria adhering to the hexadecane/water interface is calculated as
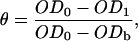 |
(2) |
where OD0, OD1, and ODb are the optical densities of the initial bacterial suspension, the extracted solution, and the blank, respectively. Our procedure deviates from established ones (Reid et al., 1992) in that we subtract a blank value for the buffer solution. We will thus generally report slightly higher values for the degree of bacterial adhesion. However, the range of interfacial adhesion values now spans the full range from 0 to 1, as it should.
The MATH test was modified to study the effects of hexadecane on bacterial interfacial adhesion. Instead of one adhesion value for a fixed aliquot of hexadecane, a series of adhesion values was determined by varying the amount of hexadecane between 0.5 μl and 3000 μl (always on3-ml bacterial suspension with a cell count of 107–108 CFU/ml). The pH of the buffer was kept at pH = 7. The curves obtained by plotting the fraction of bacteria adhering to the hexadecane/water interface as a function of the volume ratio ϕ = Vo / Vw, with Vo the volume of the organic phase and Vw the volume of the aqueous buffer, are called interfacial adhesion curves.
Atomic force microscopy (AFM)
Before AFM analysis, bacteria were adhered to a poly-L-lysine covered glass slide. The bacterial adhesion was carried out at room temperature (22 ± 1°C) by depositing a drop of the bacterial suspension buffered at pH = 7 on a poly-L-lysine covered glass slide and incubating up to 1 h at room temperature. The poly-L-lysine covered surfaces were prepared by adsorption of poly-L-lysine (Mw = 70–100 kDa; Sigma Diagnostics, St. Louis, MO) from a 0.1% w/v solution for a minimum of 12 h and the slides were stored in the same solution. The slides were washed with ultrapure water (MilliQ, Millipore) immediately before use. AFM measurements were performed at 20°C in a 10 mM KH2PO4 buffer adjusted to pH = 7 using a Dimension 3100 atomic force microscope (Digital Instruments, Santa Barbara, CA). Contact mode images were taken in constant force mode with the applied force maintained <1 nN. The scan rate varied between 1 and 2.5 Hz. Si3N4 microfabricated Nanoprobes cantilevers (Digital Instruments) with a nominal spring constant of 0.06 N m−1 were used. The AFM tips were plasma-treated immediately before use.
Analysis of AFM data
For all samples, force volumes were obtained by collecting force-distance curves on a regular two-dimensional grid spanning the sample surface of 32 × 32 force vs. distance curves. Adhesion and elasticity maps were calculated from the force volume. The elasticity map was calculated using the method Force Integration to Equal Limits (FIEL), originally developed by A-Hassan et al. (1998) and implemented in a MatLab (The MathWorks, Natick, MA) worksheet. With this method, the elasticity is calculated as the area w determined by the force-distance curve and the base line from the point of contact of the tip to the sample and a defined force point. The relative elasticity of samples 1 and 2 is then defined as (A-Hassan et al., 1998)
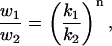 |
(3) |
where k1 and k2 are elastic constants (A-Hassan et al., 1998). The value of n is dependent on the tip geometry, for a parabolic tip n = 2/3. Although the FIEL method provides only a relative measure of the surface elasticity, large inaccuracies in the absolute value of the surface elasticity because of deviations of the tip from the ideal shape and the unknown value of the Poisson ratio of the material are avoided. In addition, tedious tip calibrations are superfluous. In assessing the relative elasticity of the surface of the various bacterial strains, which are present on various sample surfaces and which are scanned with different tips, the elasticity of the poly-L-lysine adsorbed on the substrate serves as a reference. Force-volume matrices also deliver information on the magnitude and spatial distribution of adhesion forces. The magnitude of these forces is calculated from the depth of the adhesion peak. The shape of the adhesion curves provides information on the type of interaction force between sample and AFM tip. In this article, we call the maps obtained with this method force maps.
Transmission electron microscopy (TEM)
The bacteria were suspended in a mixture of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer at pH = 7.0 containing 0.04% Ruthenium Red and incubated at 4°C. After 1 h, the sedimented part of the suspensions were microencapsulated in agar gel tubes. The samples were fixed by incubation in 2.5% glutaraldehyde in sodium cacodylate buffer at pH = 7.0 containing 0.04% Ruthenium Red and incubated for 16 h at 4°C. The samples were washed 3× with sodium cacodylate buffer at pH = 7.0 containing 0.04% Ruthenium Red followed by an incubation in 2% osmium tetroxide in sodium cacodylate buffer at pH = 7.0 containing 0.04% Ruthenium Red for 2 h at room temperature. The samples were next washed as described above before dehydration in a series of solutions with an ethanol concentration increasing from 50% to 100%. The samples were then embedded by three successive incubations for 16 h at 4°C in 50% Spurr resin in ethanol, 75% Spurr resin in ethanol, and finally in 100% Spurr resin. After polymerization of the resin (70°C, 48 h), ultra-thin sections were cut with a Reichert OMU2 ultra-microtome (Reichert-Jung, Austria). Ultra-thin sections (thickness 70 nm), stained with aqueous uranyl acetate and lead citrate, were examined under an transmission electron microscope (Philips CM12 (Philips, Eindhoven, The Netherlands), 80 kV, magnification 128,000×).
RESULTS AND DISCUSSION
Electrophoretic mobility
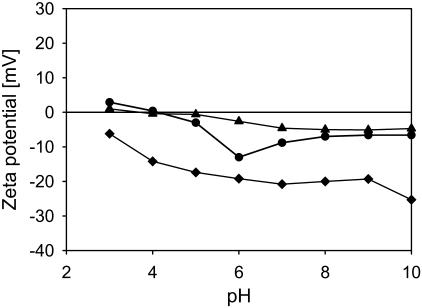
The values for the isoelectric point (pI) of the strains determined by linear interpolation of the ζ-potential data as a function of pH (Figs. 1 and 2) are given in Table 2. The isoelectric point of most strains is very close, ∼3.5–4, with the exception of L. johnsonii ATCC332, which has an isoelectric point <pH = 3 and L. crispatus DSM20584, with an isoelectric point of 4.9. The values of the isoelectric point of the S-layer-containing strains are surprisingly low, given the abundance of S-layer proteins and their high isoelectric point (Smit et al., 2001; Ventura et al., 2002; unpublished data). In particular for the two L. helveticus strains, this would mean that the S-layer is not exposed on the outer surface of the bacterium. The ζ-potential profile of L. crispatus DSM20584 is dominated toward low pH by the basic groups of the surface proteins. The steep decrease in ζ-potential between pH = 3 and 7 is likely to be caused by the increase in the dissociation of weak acidic groups, of both the polysaccharide constituents of the cell wall and the surface proteins.
FIGURE 1.
The ζ-potential of the L. johnsonii strains as a function of pH in a 10-mM potassium phosphate buffer. All data points are the average of two measurements with independently fermented cultures. L. johnsonii DSM20533 (triangles); L. johnsonii ATCC332 (diamonds); and L. johnsonii ATCC33200 (circles). Error bars are not shown, as they are generally smaller than the symbols.
FIGURE 2.
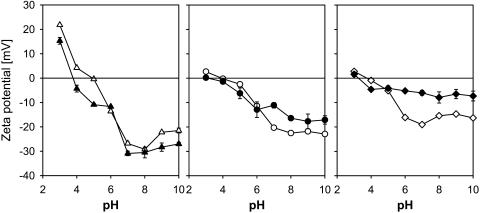
The ζ-potential of the S-layer containing strains harvested in the late-stationary and exponential growth phases. The bacteria are suspended in a 10-mM KH2PO4 buffer. Late-stationary phase (unfilled symbols); logarithmic phase (solid symbols). L. crispatus DSM20584 (triangles); L. helveticus ATCC12046 (circles); and L. helveticus ATCC15009 (diamonds). Error bars are shown, but are generally smaller than the symbols for bacteria harvested in late-stationary phase.
TABLE 2.
Physicochemical characteristics of the bacterial strains and dominant constituents of their outer surfaces
| ζ-potential (pH = 3) | ζ-potential (min) | Outer surface
|
||||
|---|---|---|---|---|---|---|
| Strain | pI* | [mV]* | [mV]* | θ0* | Polymer properties | Dominant surface constituents |
| L. johnsonii DSM20533 | 3.7 | 1.0 | −5.1 | 0.07 | Heterogenous, crosslinked | Neutral polysaccharides |
| L. johnsonii ATCC332 | <3 | −6.2 | −25.3 | 0.53 | Heterogeneous | (Lipo) teichoic acids |
| L. johnsonii ATCC33200 | 4.1 | 2.9 | −13.0 | 0.08 | Short, single polymers | Anionic polysaccharides |
| L. crispatus DSM20584 | 4.9 | 21.8 | −29.2 | 0.75 | Compact conformation | S-layer proteins |
| L. helveticus ATCC12046 | 3.9 | 2.7 | −22.9 | 0.10 | Heterogeneous, long, extended polymers | Anionic polysaccharides |
| L. helveticus ATCC15009 | 3.7 | 2.8 | −16.3 | 0.01 | Compact conformation, extended surface polymers | Anionic polysaccharides |
In 10 mM KH2PO4-buffer.
Interestingly, the dependence of the ζ-potential profile on the growth phase differs substantially for the L. crispatus strain on the one hand and both L. helveticus strains on the other (Fig. 2, unfilled vs. solid symbols). Whereas the ζ-potential as a function of pH does not significantly change from the logarithmic growth phase to the late stationary growth phase for L. crispatus DSM20584, the differences become pronounced for L. helveticus ATCC12046 and even more so for L. helveticus ATCC15009, in particular at pH > 6. Therefore, we infer that, in the later growth phases, L. helveticus strains express non-proteinaceous constituents at the outer layers of the cell wall, covering the S-layer.
The low value of the isoelectric point of L. johnsonii ATCC332 implies that the outer surface of the cell wall has a different composition than the other two L. johnsonii strains. The ζ-potential of L. johnsonii ATCC332 is negative for the whole pH range, changing rather steeply from ∼−6 mV at pH = 3 to a plateau at ∼−20 mV for pH values of 6 and above. This behavior can be understood in terms of a cell wall of which the majority of the ionic groups is anionic. The saturation of the ζ-potential at pH = 6–7 is likely to be caused by weakly acidic groups arriving at full dissociation (for an effective pKa of 4.5, which is a typical value for, e.g., carboxylic acid groups, the degree of dissociation would be ∼80% at pH = 6). In addition, we expect that a substantial amount of phosphate-based acidic groups are present at the outer layers of the cell wall, likely in the form of (lipo) teichoic acids, which have a low pKa (Table 1). The phosphate groups in the (lipo)teichoic acids constitute the only strong acids occurring in significant quantities in the cell wall of Gram-positive bacteria.
TABLE 1.
Estimated electrostatic parameters of important cell-wall constituents
| Constituent | pKa | pI |
|---|---|---|
| Anionic polysaccharides | ∼4.5* | – |
| (Lipo) teichoic acids | ∼2.1† | – |
| S-layer proteins | – | ∼9–11‡ |
Smit et al. (2001); unpublished results using data from Lortal et al. (1992) and Ventura et al. (2002).
The quantitative interpretation of the ζ-potential in terms of surface charge densities is difficult even for simple colloidal particles (Hunter, 1981; Evans and Wennerström, 1994; Van Oss, 1994). For microorganisms, the situation is considerably more complicated as the charges in the system are not present at a well-defined surface, but are distributed throughout the cell wall and on the plasma membrane. However, in the electrophoretic experiments, only the outer constituents of the cell wall heavily contribute to the ζ-potential, whereas the effect of the inner cell-wall layers will be very limited because of electroneutrality and electrostatic screening (Van der Wal et al., 1997; Wasserman and Felmy, 1998; Poortinga et al., 2001). Therefore, conclusions as to which surface constituent is present on the outer surface can be drawn, but solely on a qualitative level.
Interfacial adhesion assay
In Fig. 3, the interfacial adhesion curves at pH = 7 are shown for all six strains. Although the interfacial adhesion assay using hexadecane as hydrophobic phase is widely used (Reid et al., 1992), the advantages of a systematic variation in the volume of organic phase do not appear to have been exploited except for a few initial experiments (Olsson and Westergen, 1982; Bohach and Snyder, 1983; Hogt et al., 1983). In particular, a description of the bacterial adhesion in terms of an adsorption or binding process, as outlined in the Appendix, was never put forward.
FIGURE 3.
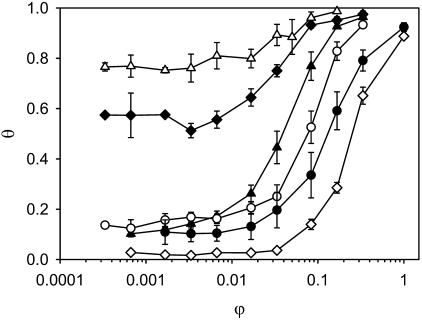
Interfacial adhesion curves of the six strains in 10 mM potassium phosphate buffer at pH = 7. Plotted are the ratio of the volume of hexadecane to aqueous buffer on the horizontal axis, and the fraction of microorganisms adhered at the water-hexadecane interface on the vertical axis. L. johnsonii DSM20533 (solid triangles); L. johnsonii ATCC332 (solid diamonds); L. johnsonii ATCC33200 (solid circles); L. crispatus DSM20584 (unfilled triangles); L. helveticus ATCC12046 (unfilled circles); and L. helveticus ATCC15009 (unfilled diamonds). Initial cell count of the buffers is such that the OD is between 0.45 and 0.55. All data points are the average of three measurements with three independently fermented cultures. Error bars denote ±1 SD.
The interfacial adhesion curves are plotted on a logarithmic x-axis to illustrate the behavior at low volume ratios of hexadecane to aqueous buffer. As usual for affine adsorption curves plotted in this way, the interfacial adhesion curves display a pronounced sigmoidal shape. However, as the initial part of the curves is virtually flat for all strains (with the exception of L. johnsonii DSM20533; see Fig. 3), the overall adhesion and adsorption process is more complicated than a simple affine adhesion to the interface between the hexadecane and aqueous phases.
The degree of interfacial adhesion θ of the two L. helveticus strains and two of the three L. johnsonii strains (DSM20533 and ATCC33200) starts at a low value of 0–0.1, increases rapidly between ∼50 and 500 μl of added hexadecane, and plateaus at values close to 1 (complete interfacial adhesion) at the largest volumes tested. Differences between the two L. johnsonii strains and the L. helveticus strains are observed, with L. johnsonii ATCC332000 and L. helveticus ATCC15009 being more hydrophilic than L. johnsonii DSM20533 and L. helveticus ATCC12046 strain, but the general characteristics of the adhesion curves are very similar. The interfacial adhesion behavior of L. crispatus DSM20584 and L. johnsonii ATCC332 is completely different, however. Already at the smallest hexadecane volumes tested (0.5 and 1 μl), the degree of interfacial adhesion of L. crispatus DSM20584 and L. johnsonii ATCC332 is very high, ∼0.8 and 0.6, respectively. The degree of interfacial adhesion remains virtually constant for a relatively large increase in the amount of added hexadecane, and then slowly approaches the 100%-adhesion plateau in a weak-sigmoidal curve.
Our interpretation of the interfacial adhesion behavior of L. crispatus DSM20584 and L. johnsonii ATCC332 is that, at low quantities of added hexadecane, the bacteria do not adhere to the water-hexadecane interface, but that, conversely, the hexadecane adsorbs at sites on the bacterial cell wall. These adsorption sites are most likely the hydrophobic moieties of surface proteins and (lipo)teichoic acids close to the outer layers of the cell wall. Therefore, even minute quantities of hexadecane completely change the surface characteristics of L. crispatus DSM20584 and L. johnsonii ATCC332, rendering it very hydrophobic. Consequently, this leads to extensive aggregation of the bacteria and subsequently to a rapid precipitation of large bacterial clusters.
For the other L. johnsonii strains and the L. helveticus strains, adsorption of hexadecane on the bacterial cell wall is also likely to occur, as an initial plateau was observed as for L. crispatus DSM20584 and L. johnsonii ATCC332 (Fig. 3). An exception is possibly L. johnsonii DSM20533, of whose degree of interfacial adhesion increases continuously with increasing amount of hexadecane. As the degree of clustering for these four strains is fairly low at the initial plateau, it is likely that the adsorption sites for hexadecane reside in the inner parts of the cell wall. In this case, the adsorption of limited quantities of hexadecane does not significantly change the surface characteristics of the bacteria. If now the quantity of hexadecane increases beyond the saturation limit of the cell wall, macroscopic hexadecane droplets will appear and the hydrophobic parts of the cell wall will start to adhere to the hexadecane/aqueous buffer interface. Further increasing the amount of hexadecane increases the level of bacterial adsorption, which continues until all bacteria are effectively extracted from the solution at the highest hexadecane volumes. Our observation of the adsorption of small quantities of hexadecane by the bacterial cell wall, and its impact on the aggregation and interfacial adhesion as depending on the location of the hydrophobic moieties within the cell wall, explain several growth-phase-dependent phenomena observed long ago (Neufeld et al., 1980).
Apart from the immediate relevance of adhesion curves as shown in Fig. 3, there is also an important principal argument to using a varying amount of organic phase instead of just one fixed aliquot. As in all adsorption and binding phenomena, the imperative quantity describing the adsorption process is not so much the amount adsorbed at a given solution concentration or partial pressure, but the values of the parameters describing the adsorption isotherm or binding curve (Tanford, 1980). This is particularly true close to the surface or site saturation limit, where large differences in the adsorption or binding constant lead to small and often experimentally insignificant changes in the degree of adsorption or binding. In addition, in the interfacial adhesion assays, the experimentally accessible parameter is the optical density in the aqueous phase. Therefore, hydrophilic microbial strains cannot reliably be distinguished if the volume of organic phase is too small to induce appreciable levels of adhesion (Reid et al., 1992). We expect that this recognized disadvantage of the MATH test is considerably reduced using our interfacial adhesion assay.
The precise mechanism of interaction between hexadecane and the bacterial surface is complex and presumably dependent on the microbial strain. To have a fitting relation which is both simple and sufficiently broad in its application and which, in addition, is based on a theoretical foundation, we have developed a simple model. This model assumes a two-stage process: an initial plateau determined by microbial clustering caused by the adsorption on the cell wall of a very small amount of hexadecane and an interfacial adhesion of the microorganisms at higher volumes of hexadecane. For a detailed discussion we refer to the Appendix, but the simplest relation describing such a two-stage process is
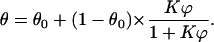 |
(4) |
The fitting equation is characterized by two constants: θ0 representing the initial plateau (for the formation of which various mechanisms are discussed in the Appendix), and K the interfacial adhesion constant. Values of both parameters for the six bacterial strains are reported in Table 3.
TABLE 3.
Fitting parameters of the interfacial adhesion curves
| Strain | ϕ50* | θ0 | K | ϕ50, model*† |
|---|---|---|---|---|
| L. johnsonii DSM20533 | 4.8 × 10−2 | 7.0 × 10−2 | 29 | 3.0 × 10−2 |
| L. johnsonii ATCC332 | <1.7 × 10−4 | 0.53 | 33 | – |
| L. johnsonii ATCC33200 | 0.14 | 0.08 | 7.0 | 0.12 |
| L. crispatus DSM20584 | <1.7 × 10−4 | 0.75 | 38 | – |
| L. helveticus ATCC12046 | 7.8 × 10−2 | 0.10 | 13 | 6.2 × 10−2 |
| L. helveticus ATCC15009 | 0.27 | 1.0 × 10−2 | 3.0 | 0.33 |
The bacterial suspensions are in 10 mM KH2PO4 at pH = 7. Cell count of the buffers is 107–108 CFU/ml.
Volume ratio of hexadecane at which 50% adhesion occurs.
Calculated from θ0 and K.
Combined colloidal properties
From the combination of ζ-potential and interfacial adhesion properties, we can infer several important aspects of the composition of the bacterial surface (Table 2). L. johnsonii DSM20533 is rather hydrophilic and possesses only a very weak surface charge. Therefore, we surmise that this strain is covered by a layer of essentially neutral polysaccharides, which can be either cell-wall associated or extracellular, the distinction often being difficult to make (Delcour et al., 1999). L. johnsonii ATCC332 is likely to be covered by (lipo)teichoic acids, as the strain is both strongly negatively charged and rather hydrophobic. L. johnsonii ATCC33200 could be covered by anionic polysaccharides, given its low ζ-potential at low pH and its hydrophilic character. The nature of its surface polymers is quite different from L. johnsonii DSM20533, because the bacterium is more highly negatively charged at high pH.
The combination of a strongly positive ζ-potential at low pH and a highly negative surface charge at high pH combined with a high hydrophobicity of L. crispatus DSM20584 hints at a surface covered by proteins, potentially the S-layer. In the case of the two other S-layer containing strains, L. helveticus ATCC12046 and L. helveticus ATCC15009, the surface properties are clearly not determined by a surface protein, as the surface charge at low pH is only weakly positive and both strains are strongly hydrophilic. It is most likely that the two strains are covered by a polysaccharide layer. For L. helveticus ATCC12046, this was indeed concluded from a direct determination of the chemical composition of the outer layers of the cell wall using x-ray photoelectron spectroscopy (Boonaert and Rouxhet, 2000).
AFM contact imaging
Contact-mode images were taken with minimal force in retraction mode. From the error-signal deflection mode images (Fig. 4), we obtained information about the dimensions of the microorganisms and qualitative data on the structure of their surfaces. Whereas L. johnsonii ATCC33200 (Fig. 4 c) and L. crispatus DSM20584 (Fig. 4 d) display a smooth, homogenous surface, the surfaces of the L. johnsonii strains DSM20533 (Fig. 4 a) and ATCC332 (Fig. 4 b) and the surface of both L. helveticus strains (Fig. 4, e and f) are more heterogeneous and rough.
FIGURE 4.
AFM deflection images showing the surface morphology of the Lactobacillus strains. The microorganisms are adhered to a poly-L-lysine-covered substrate. Imaging is performed in a 10 mM KH2PO4 buffer at pH 7. (a) L. johnsonii DSM20533; (b) L. johnsonii ATCC332; (c) L. johnsonii ATCC33200; (d) L. crispatus DSM20584; (e) L. helveticus ATCC12046; and (f) L. helveticus ATCC15009. The surfaces of the L. johnsonii strains exhibit a fuzzy character, whereas the definition of the surfaces of the S-layer-containing strains is higher, and the surfaces appear smoother.
The surface of L. crispatus DSM20584 does not have any fuzzy or heterogeneous character at the length scales probed in the AFM experiments, which hints at the absence of extended, loosely crosslinked polymer networks on the outside of the cell wall or isolated polymer chains protruding out of the cell wall. In line with the physicochemical data, we conclude that the outer layer of this bacterium is fully covered by a compact protein layer. Conversely, the two other S-layer containing strains, both of the L. helveticus species, have a surface structure showing locally some bumps on an otherwise fairly smooth surface. Consistent with the physicochemical analysis, we conclude that the S-layer is covered by polymeric substances adopting extended conformations.
The surface of L. johnsonii ATCC33200 is also smooth, but on the left edge of the bacterium in Fig. 4 c, deformable material is observed. Because of the direction of scanning of the bacterial surface, the deformable material can only be seen at the edge of the bacterium where the AFM tip is leaving the bacterial surface (the specimens were horizontally scanned in two directions: from left to right and backward; only the backward or “retrace” scan is used in the images shown in Fig. 4). Because of the relative high definition of imaging of the surface of L. johnsonii ATCC33200, we suspect that the polysaccharides extending from the bacterial surface are not or only very slightly crosslinked so that we can penetrate through them to reach the underlying surface which is more robust. The surface has stretchable molecules, and many surface molecules could be laterally moved during scanning, without affecting the attachment of the bacterium to the substrate. Therefore, we conclude that the outer surface of L. johnsonii ATCC33200 is formed by a layer of fairly low surface density consisting of flexible polymers extending into the solution.
The other two L. johnsonii strains have more heterogeneous surfaces. L. johnsonii DSM20533 (Fig. 4 a) is fairly rough and patchy and its long surface polymers could be laterally moved during the AFM analysis. By combining the deflection image with the physicochemical analysis (summarized in Table 2), we infer that the surface consists of a heterogeneous polymeric network, most likely made up of polysaccharides. The surface of L. johnsonii ATCC332 is very rough (Fig. 4 b) and could well be chemically highly heterogeneous. The surface composition cannot be determined from the AFM analysis, but, in combination from the physicochemical analysis, we infer that (lipo)teichoic acids are expressed on the surface.
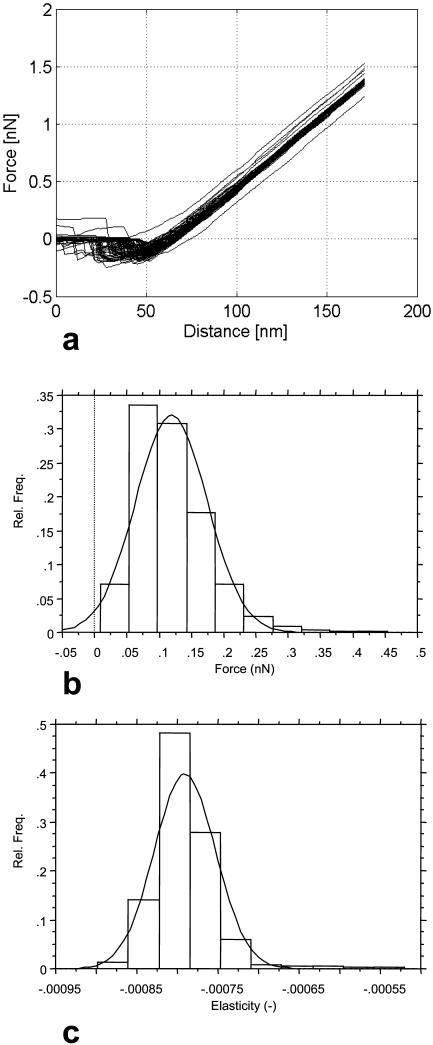
A confirmation for the surface structure emerging from the deflection images is given by the force-distance curves obtained on the same bacterial surfaces (Fig. 5). Again, we see major qualitative differences between the L. crispatus strain, the L. helveticus strains and the L. johnsonii strains. Whereas the L. johnsonii strains show clear adhesion peaks upon retraction of the AFM tip from the bacterial surface (Fig. 5, a and b), no such peaks are seen for L. crispatus DSM20584 (Fig. 5 c). For the other two S-layer containing strains, L. helveticus ATCC12046 and L. helveticus ATCC15009, adhesion peaks were not or only infrequently observed (Fig. 5, e and f).
FIGURE 5.
Force-distance curves showing the interactions of the AFM tip with the bacterial surfaces. (a) L. johnsonii DSM20533; (b) L. johnsonii ATCC332; (c) L. johnsonii ATCC33200; (d) L. crispatus DSM20584; (e) L. helveticus ATCC12046; and (f) L. helveticus ATCC15009. The microorganisms are adhered to a poly-L-lysine covered substrate. The force-distance curves are obtained in a 10 mM KH2PO4 buffer at pH 7. The L. johnsonii strains (a–c) show clear adhesion peaks upon retraction of the AFM tip from the bacterial surface. The adhesion curves of L. johnsonii DSM20533 and L. johnsonii ATCC332 show multiple unbinding events upon retraction, whereas for L. johnsonii ATCC33200 the unbinding appears to proceed via a single event. For the L. crispatus DSM20584, no significant adhesion between the bacterial surface and the AFM tip is observed upon retraction; adhesion events are also rarely recorded for the two L. helveticus strains. From the force-distance curves, elasticity data are calculated (Table 4).
In the case of L. crispatus DSM20584, we are inclined to believe that we are directly probing the S-layer of the bacterium on the outside of the bacterial cell wall. Our reasoning is that for compact layers of essentially unextensionable molecules (like the globular S-layer proteins arranged in a two-dimensional para-crystalline lattice) significant adhesion forces are only found over the range of molecular interactions, which is several nm at most (i.e., below the range which can meaningfully be analyzed using the current AFM setup). In contrast, for flexible polymers, like most polysaccharides, adhesion peaks range over length scales of tens of nanometers because of the interplay of entropy, elasticity, and interactions. Moreover, the absence of surface features (i.e., smoothness) could also be seen as a confirmation of the highly regular, and thus probably the para-crystalline character of outer layer of the cell wall, hinting at the presence of the bacterial S-layer at this outer layer (Lortal et al., 1991). This is in line also with the results of our physicochemical analyses.
An interesting feature of the two L. helveticus strains is that a low-density, highly extended, soft outer layer is detected. This polymeric layer repels the AFM tip upon approach, most likely by entropic repulsion. For L. helveticus ATCC12046, only a small fraction of the force-distance curves shows this behavior, and we therefore tentatively conclude that this soft layer consists of long, flexible molecules grafted with a low surface density on an underlying surface which is more robust. The typical behavior of the force-distance curves is in agreement with the physicochemical analysis, from which we concluded that the S-layer is covered by an outer polymer layer. Soft polymer layers have also been observed for a fibrillated Streptococcus (Van der Mei et al., 2000). For L. helveticus ATCC15009, a similar soft layer is observed, but for this strain, the spatial extension of this layer is lower and the surface density of the polymers is presumably somewhat higher.
A reviewer has incited us to think about a measure of the smoothness and fuzziness of a surface as probed by AFM (see e.g., Colton et al., 1998), in particular as we use the concepts smoothness and fuzziness to loosely distinguish between the characteristics of the surfaces of the various bacterial strains. We do not attempt here to provide quantitative measures, but we merely note that, in the images obtained in error-signal deflection mode (which is particularly sensitive to spatial variations in surface structure), heterogeneities are observed for a number of strains (see Fig. 4, a, b, e, and f), whereas for others they are not (see Fig. 4, c and d). The characteristic heterogeneities observed show up at a length scale which is typically much smaller than the characteristic size of a bacterium, but which is, at the same time, larger than typical molecular dimensions (a surface protein, a single surface polymer). In effect, in the current AFM experiments, we do not probe very small scale heterogeneities like the structure of the S-layer (which, given the soft nature of a bacterium, is best done on S-layers reconstituted in vitro; see Scheuring et al., 2002), or variations in contour length of surface polymers, but only those associated with larger, multimolecular assemblies.
Concerning variations in surface structure, a second, interesting aspect shows up in the AFM analysis of soft matter in the native state (but not in microscopic techniques in which a fixed specimen is analyzed, like e.g., in electron microscopy) and that is the effect of thermal fluctuations on the observed surface structure. Whereas the bacteria in both Fig. 4, c and d, appear smooth in the sense that no heterogeneities appear on a length scale between molecular dimensions and the characteristic size of the bacteria, it is clear that there is a difference in surface structure between the two. This difference is essentially related to the degrees of freedom of the surface constituents. Whereas the configuration of the surface constituents in Fig. 4 d remains unaltered during the time frame of the experiment (the packing of the proteins in the S-layer lattice is preserved), thermal fluctuations perturb the conformation of the surface polymers of the bacterium shown in Fig. 4 c. Even at low forces, the AFM tip will influence the conformations of the bacterial surface polymers and vice versa. This is what we denote as the “fuzziness” of the surface.
Force-distance curves
It is also worthwhile to compare the force-distance curves of the L. johnsonii strains (Fig. 5, a–c). A first observation is that the adhesion forces of L. johnsonii DSM20533 and L. johnsonii ATCC33200 are much higher than for L. johnsonii ATCC332. Whereas the highest adhesion forces registered for L. johnsonii DSM20533 are ∼0.3 nN (Fig. 5 a), the maximum values are ∼0.4 nN for L. johnsonii ATCC33200 (Fig. 5 b) and only ∼0.07 nN for L. johnsonii ATCC332. The magnitude of the adhesion forces between the AFM tip and the surfaces of L. johnsonii DSM20533 and L. johnsonii ATCC33200 are typical of the magnitude of forces observed for polysaccharide molecules (Rief et al., 1997). It should be borne in mind, however, that the magnitude of the forces registered is dependent not only on the polymer (Magonov and Reneker, 1997; Rief et al., 1997), but also on the buffer solution, its pH, ionic strength, and temperature. Also, variations in molecular weight, charge density, and hydrophobicity of the polymer will be reflected in the magnitude of the adhesion peaks.
The shape of the force-distance curves is different for the L. johnsonii strains. The force-distance curves for L. johnsonii DSM20533 and L. johnsonii ATCC332 show broad minima, indicative of the release of the AFM tip from the surface proceeding via multiple unbinding events, whereas the force-distance curve of L. johnsonii ATCC33200 shows a sharp minimum, which strongly suggests that the separation of the bacterial surface and the AFM tip proceeds via a single unbinding event.
Both the difference in magnitude of the adhesion forces and the shape of the unbinding curves point at the following interpretation. It is likely that the surface of L. johnsonii DSM20533 is covered by a rather dense, crosslinked network of flexible polymers, probably polysaccharides. The same is the case for L. johnsonii ATCC332, but the nature of the polymers is different given the distinct physicochemical properties of the bacterial surface (Table 2) and the lower adhesion forces. Although the surface polymers are probably also polysaccharides, the structure of the surface of L. johnsonii ATCC33200 is different from L. johnsonii DSM20533 and consists probably largely of single polymers, protruding into the solution. However, as the unbinding curves of L. johnsonii ATCC33200 do not show the typical single-polymer stretching shape (Rief et al., 1997), we are probably not probing single polymers with the AFM tip, but more likely a combined effect of the stretching of a number of non-crosslinked polymers of almost equal contour length and, at the same time, a local deformation of the bacterial surface (Velegol and Logan, 2002). This is also what we observe in the contact mode imaging of L. johnsonii ATCC33200 (Fig. 4 c).
TEM analysis
Several of the important observations from AFM are confirmed by the micrographs shown in Fig. 6. In particular, the presence of the S-layer on the outer surface of L. crispatus DSM20584 is clearly demonstrated (Fig. 6 c). In addition to a thick, dark, proteinaceous band hidden about halfway up the cell wall (indicated by the white arrow), a very thin, dark layer can be observed at the outer edge of the cell wall (indicated by the black arrow; see also the inserted enlargement). We conclude that this thin layer is the S-layer, as the thick band is observed also for L. johnsonii DSM20533 (Fig. 6 a) and L. johnsonii ATCC33200 (Fig. 6 b, white arrows), which do not possess an S-layer.
FIGURE 6.
Transmission electron micrographs of the Lactobacillus strains harvested in late-stationary phase. (a) L. johnsonii DSM20533; (b) L. johnsonii ATCC33200; and (c) L. crispatus DSM20584. In all three images, a dark band can be observed ∼20–40 nm below the surface (white arrows). The dark staining hints at a high protein content. Only in the case of L. crispatus DSM20584 (c), a thin dark band can be seen at the outer surface (black arrow and insert). It is inferred that this protein-rich layer largely determines the surface properties of the strain. Bar = 250 nm. Magnification of the insert is 2.2× the magnification of the main image.
In addition, several findings from the AFM analysis on the structure of the outer polymer layer of the cell wall are confirmed. For L. johnsonii ATCC33200, the outer cell-wall layer appears to be very loose but rather homogeneous (Fig. 6 b), whereas the outer layer of L. johnsonii DSM20533 is very thick and heterogeneous (Fig. 6 a).
Cell-wall elasticity
Recently, there has been an increase in interest in the determination of the elastic properties of the bacterial cell wall (Xu et al., 1996; Yao et al., 1999; Boulbitch et al., 2000). It is generally argued that the resistance to mechanical stress of a microorganism is determined by the bacterial turgor pressure and the stretching elasticity of the peptidoglycan network, the bending of the cell wall under influence of external forces making only a small contribution (Yao et al., 1999). These conclusions are in line with our force-distance curves, which show Hookian behavior upon indentation for all bacterial strains (Fig. 5). An exception is possibly formed by L. crispatus DSM20584, but even in this case, the deviations from linearity are fairly modest. At constant volume of the cytoplasm, the indentation of the microorganism by the probe tip will lead to stretching of the cell wall as a whole, in addition to the large local deformation in the immediate vicinity of the AFM tip. We do not attempt to calculate the elastic effects of bacterial deformation, as it is of little relevance for understanding the role of molecular forces in bacterial interactions. In any case, such an analysis is significantly more straightforward and less prone to artifacts if the tip of the probe is flat and larger in size than the microorganism, like in classical cytotensiometry (Petersen et al., 1982). However, if the indentation by the AFM tip is sufficiently local (i.e., the radius of the region of deformation is much smaller than the typical size of the bacterium), and if we assume that the bacterium is essentially spherical, the principal effect of the indentation is to displace a volume ΔV from the region of deformation to the bulk of the cytoplasm. The volume increase will lead to an increase of the average size Δr and an increase in surface area ΔS via ΔV = 4/3π ((r + Δr)3 − r3) ≈ 4π r2 Δr and ΔS = 4π ((r + Δr)2 − r 2) ≈ 8 π r Δr. This qualitative argumentation leads to the expected Hookian relation between the indentation distance and the force experienced by the AFM tip because for longitudinal deformations of plates the stress is proportional to the strain (Landau and Lifshitz, 1970).
The resistance of bacterial surfaces to external forces varies to some extent, as is clear from the slopes of the force-distance curves (Fig. 5; Table 4), and the FIEL maps (Fig. 7). The slopes and the elasticity constants reported in Table 4 for the six Lactobacillus strains are in fact very close to values recently reported for (Gram-negative) E. coli bacteria (Velegol and Logan, 2002). The variations in slope are presumably largely caused by variations in turgor pressure. In fact, variations in turgor pressure span at least one order of magnitude for Gram-negative bacteria and are thought to be even higher for Gram-positive bacteria (Poolman et al., 2002), but are dependent on the composition of the medium. Structural features of the cell wall could also play a role in the observed deformation behavior. It would be tempting to conclude that the bacterial S-layers play a role in the overall elasticity of the cell wall, in particular because the two stiffest strains contain S-layers. The stiffening effect of a protein sheath on the cell wall was already established before for an archaebacterium (Xu et al., 1996). However, the stiffening effect of an S-layer would be small for lactobacilli given the small variation between the surface elasticity of the six strains (Table 4).
TABLE 4.
Evaluation of the bacterial surface elasticity
| Strain | Indentation [nm] | Max. applied force [nN] | Elastic constant [Nm−1]* | Relative elasticity† |
|---|---|---|---|---|
| L. johnsonii DSM20533 | 84.4 | 1.39 | 1.6 × 10−2 | 1.4 |
| L. johnsonii ATCC332 | 77.7 | 1.59 | 2.0 × 10−2 | 1.8 |
| L. johnsonii ATCC33200 | 85.5 | 1.47 | 1.7 × 10−2 | 1.5 |
| L. crispatus DSM20584 | 40.1 | 2.11 | 5.3 × 10−2 | 4.6 |
| L. helveticus ATCC12046 | 80.3 | 1.57 | 2.0 × 10−2 | 1.7 |
| L. helveticus ATCC15009 | 58.5 | 1.80 | 3.1 × 10−2 | 2.7 |
Slope of the force-indentation curves.
The poly-L-lysine surface is taken as reference (slope of the force-distance curves = 1.1 × 10−2 Nm−1).
FIGURE 7.
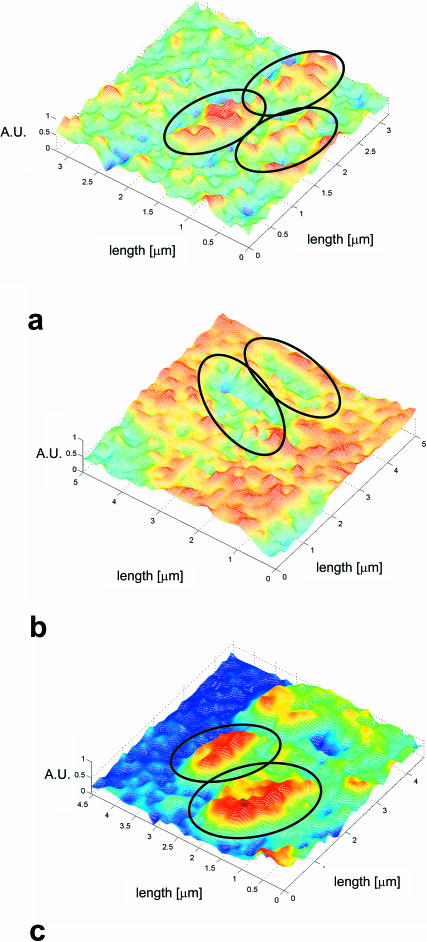
Elasticity maps of Lactobacillus strains harvested in late stationary phase. (a) L. johnsonii DSM20533; (b) L. johnsonii ATCC33200; and (c) L. crispatus DSM 20584. The microorganisms are adhered to a poly-L-lysine covered substrate. The elasticity data are obtained in a 10 mM KH2PO4 buffer at pH 7. The elasticity is plotted on a relative scale from 0 to 1 (AU, arbitrary units). Comparison of the surface stiffness of the various bacteria is possible as the stiffness of the poly-L-lysine adsorbed on the substrate slides serves as a reference. The elasticity of the surface of L. crispatus DSM 20584 is high and spatially fairly homogeneous (c). The surfaces of the two L. johnsonii strains are much softer and the surface elasticity is heterogeneous (a and b).
Microelastic mapping of the spatial variation in elasticity is of more interest for understanding the way a bacterium mediates interactions with external surfaces, as such variations in stiffness are caused by surface constituents decorating the peptidoglycan network. In Fig. 7, FIEL maps are shown for L. johnsonii DSM20533 (Fig. 7 a), L. johnsonii ATCC33200 (Fig. 7 b) and L. crispatus DSM20584 (Fig. 7 c). These plots are calculated from the force-distance curves following the procedure outlined by A-Hassan et al. (1998). Interestingly, the surface of L. crispatus DSM20584 appears to be fairly highly regular in its elasticity, whereas the distribution of elasticity or softness over the two L. johnsonii strains is much more heterogeneous. In particular, the surface of L. johnsonii DSM20533 is very irregular and heterogeneous, which is probably caused by tufts of extracellular polysaccharides.
We use the properties of the adsorbed poly-L-lysine layer as a reference to evaluate the bacterial surface elasticity and interactions. In Fig. 8, the relevant characteristics are shown. The AFM measurements on the poly-L-lysine layers are highly reproducible in the analysis of both adhesion forces (Fig. 8 b) and repulsive, elastic forces (Fig. 8 c) if sufficient repetitions are carried out. Therefore, poly-L-lysine substrates are well-suited as reference material and allow the comparison of the (relative) elasticity of biological and colloidal samples (see the last column of Table 4 for relative elasticity values for the six strains).
FIGURE 8.
AFM-characterization of the poly-L-lysine substrates. (a) Force-distance curves. (b) Distribution of adhesion forces as determined from the minima of the force-distance curves. (c) Relative elasticity according to the FIEL method. In the analysis for b and c, 1024 force-distance curves were used.
Bacterial surface constituents and conformations
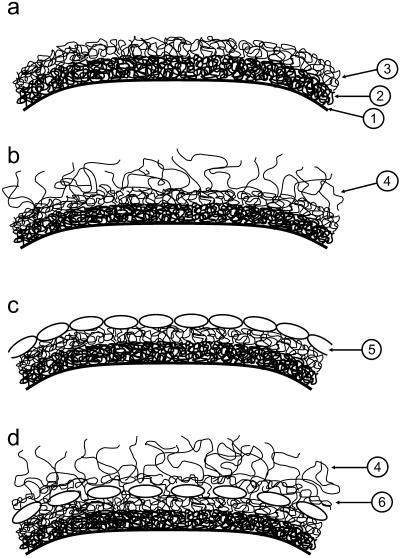
Our findings on the properties of the cell wall of the investigated Lactobacillus strains can be interpreted within the context of simple models of the bacterial cell wall (Fig. 9). Fig. 9 a shows a cell wall consisting of peptidoglycan (inner layer) and crosslinked polysaccharides (outer layer). This model would apply to L. johnsonii DSM20533. The model depicted in Fig. 9 b is similar to the cell-wall model in Fig. 9 a, but contains single polymers on the outer surface. This model would explain our observations made on L. johnsonii ATCC33200. Bacterial S-layers can be envisaged both on the outside of the cell wall (Fig. 9 c) as in the case of L. crispatus DSM20584 or covered by polymers extending into the solution (4) (like for the L. helveticus strains) or a polymer network (6) (Fig. 9 d). L. johnsonii ATCC332 is somewhat ambiguous with respect to the models shown in Fig. 9, but its surface would probably be reasonably well-represented by a schematic diagram as in Fig. 9 a, but with the outer layer (largely) consisting of (lipo)teichoic acids.
FIGURE 9.
Structural models of the bacterial cell wall of the Lactobacillus strains, utilized in the interpretation of the various experiments. (1) Cell membrane. (2) Inner, protein-rich layer of the cell wall. (3) Outer layer of the cell wall, rich in various polymers like polysaccharides and lipoteichoic acids. (4) Extracellular polysaccharides and other polymeric compounds attached to the cell wall protruding into the buffer. (5) Surface proteins (S-layer); even if the surface proteins form a close packing, a significant fraction of the surface is open to the outside. (6) Crosslinked polymer layer outside of the layer containing the surface proteins.
CONCLUDING REMARKS
Bacterial surfaces are soft systems, which display an impressive variation in physicochemical properties. Apart from the chemical nature of the surface constituents and the organization of these constituents within the cell wall, these properties are determined by the conformational degrees of freedom of the polymeric surface constituents. We have probed the surface properties of a number of strains of lactic acid bacteria using a variety of microscopic and physicochemical techniques with emphasis on the elucidation of the global physicochemical nature of the outer layer of the cell wall, the conformation of the surface macromolecules, and the susceptibility of the surface toward external perturbations (interfacial behavior, micromechanical forces). These three factors essentially determine the propensity of a bacterium to adhere to surfaces and to bind polymeric constituents of the growth medium; and they are also implied in bacterial auto- and co-aggregation and clustering.
Cell-wall heterogeneities can strongly influence the colloidal properties of the bacteria. Such heterogeneities are difficult to detect using classical physicochemical techniques, but AFM is particularly suitable to analyze their nature. The relevant aspects of bacterial surface roughness show up at a length scale which is typically much smaller than the characteristic size of a bacterium, but which is, at the same time, much larger than the typical dimensions of a surface protein or a single surface polymer. In the case where the outer surface is made up of a regular lattice of globular proteins, like an S-layer, the surface is smooth on length scales larger than the typical size of the surface protein (a few nm). When the outer surface is made up of single polymers of fairly equal contour length, the surface is also smooth at these length scales, but may appear fuzzy because of thermal fluctuations of the surface polymers. Spatially varying distributions of surface polymers, which are also possibly crosslinked, result in heterogeneous and rough surfaces. This is the case if the outer surface contains polysaccharides and (lipo)teichoic acids.
The presence of a dominant surface constituent can be inferred by combining the various physicochemical and microscopic analyses. The presence of surface proteins in lactobacilli can be deducted from the elevated isoelectric point and the high hydrophobicity of the surface. (Lipo)teichoic acids render the surface strongly negatively charged and hydrophobic at the same time. Surfaces rich in polysaccharides are generally weakly charged and are hydrophilic. Hydrophobic compounds like hexadecane can adsorb on sites on or within the cell wall. If the absorbing moieties are at the outer surface, this will render the bacterial surface very hydrophobic.
In summary, we have found that the diversity in surface properties of lactobacilli strains can be fruitfully analyzed using a combination of classical physicochemical techniques and advanced microscopic techniques. In particular, AFM is a tool, which is highly suitable to study bacterial surface properties because spatial heterogeneities in surface structure, softness, and interaction forces can be detected at the same time. We expect that our findings will be helpful in increasing the understanding of the structure-property relations of the bacterial cell wall—in particular, with respect to bacterial interactions.
Acknowledgments
For the AFM experiments, the support of Alcon Laboratories (Fort Worth, Texas), is acknowledged. In particular, we thank Gerald Cagle for enabling us to work at Alcon, and Andrew Griggs for carrying out a number of AFM experiments. For microbiological support, we are indebted to Marco Ventura and Nicola D'Amico. We are grateful for experimental support received from Marie-Lise Dillmann (TEM analysis) and Christophe Schmitt (electrophoresis). The force-volume data were analyzed using a MatLab macro originally developed by Urs Ziegler (University of Zürich) and adapted with help from Christoph Schär (ETH Zürich). We thank Conrad Woldringh (University of Amsterdam) for discussions on the bacterial cell cycle and Yves Dufrêne (Université Catholique de Louvain) for discussions on the analysis of biological specimens by AFM. Elisabeth Prior is thanked for a critical reading of the manuscript. The management of Nestec Ltd. is acknowledged for the permission to publish the work.
APPENDIX: CONSIDERATIONS ON THE ADSORPTION OF HYDROPHOBIC COMPOUNDS BY THE MICROBIAL CELL WALL AND THE INTERFACIAL ADHESION OF MICROORGANISMS
In a simplified model of the interfacial adhesion process, the initial state is a suspension of volume Vw containing a monodisperse population of microorganisms of number density ρb. The microorganisms all have a maximum capacity to adsorb hydrocarbons qmax and a surface area for adhesion A. The total adsorption capacity of the microorganisms in the buffer Qmax = qmaxρbVw. To the microbial suspension, a volume Vo of hydrocarbons is added. The volume fraction of organic phase in the system is then φ = Vo/(Vo + Vw) and the ratio between the volumes of the organic and aqueous phases is ϕ = Vo/Vw. The effective volume fraction φeff and volume ratio ϕeff of hydrocarbons available for interfacial adhesion is lower because a volume Vq of the hydrocarbons adsorbs on sites on or within the cell wall: Veff = Vo − Vq, where Vq = QMw/ρ with Mw the molar weight of the hydrocarbon and ρ its density. The quantity of hydrocarbons which adsorbs on sites in the cell wall is dependent on the strain and possibly on its growing conditions and is a function of the amount of available organic phase. As the solubility of hydrocarbons in aqueous buffers is very low (<10−6% w/w; Lide, 2000), the system of relevance is essentially a three-phase system (aqueous buffer, organic phase, and microbial surface sites) in which the aqueous phase is fully saturated with the hydrocarbon which transfers to the bacterial surface sites by partitioning from the organic phase into the aqueous phase.
A variety of relations between the quantity of cell-wall-adsorbed hydrocarbons and the quantity of hydrocarbons in the system is possible. Plausible relations are, for instance, a linear isotherm (Eq. A1a), and a Langmuir-type isotherm (Eq. A1b), as
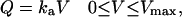 |
(A1a) |
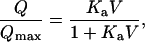 |
(A1b) |
where ka and Ka are constants and Vmax is the volume of added organic phase at which the cell-wall sites become fully saturated. In particular, when ka = ρ/Mw, Vq = V and the adsorption at the cell-wall sites following Eq. A1a proceeds in a stepwise manner. We use this in deriving Eq. 4.
A soon as a macroscopic organic phase appears, microorganisms will start to adhere to the interface with the aqueous buffer. The degree of adhesion will depend on a number of factors, like the available interfacial area, the strength of the interactions between the microbial surface and the interface, the effective surface area taking part in the adsorption of the microorganisms, and the kinetics of microbial transfer to the interface. Again, various relations are feasible and one could also expect a significant dependence of the mechanism of interfacial adhesion on the strain. Because of a lack of detailed information on the interactions between microorganisms and the interface between aqueous and organic phases, and as we are striving for a relation which is sufficiently general that it reasonably well describes the interfacial adhesion as a function of the volume fraction of organic phase without introducing spurious parameters, we simply assume that 1), the amount of interface created during vortexing is proportional to the ratio of the volume of organic phase to aqueous buffer; and 2), the interfacial coverage by the microorganisms is proportional to their number density in the aqueous phase.
The number of microorganisms is conserved and the microorganisms can distribute only over the aqueous phase and the water-hydrocarbon interface,
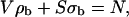 |
(A2) |
where N is the number of microorganisms in the system, S is the interfacial surface area, and σb is the surface density of microorganisms. The change in the distribution of the microorganisms over the two phases upon a change in Vo depends on the assumed relations for S (Vo) and σb(ρo), which in the simplest form are linear: S = k1 Vo and σb = k2 ρo,
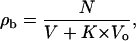 |
(A3) |
where K = k1k2. Due to interfacial adhesion, the number density of microorganisms in the water phase changes by
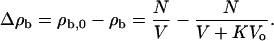 |
(A4) |
The fraction of the microorganisms adhering to the water-hydrocarbon interface is then
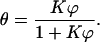 |
(A5) |
If, in Eq. A5, ϕ is replaced by ϕeff, a two-state model is obtained which allows for both adsorption of hydrocarbons by the cell wall and adhesion of the microorganisms to the water-hydrocarbon interface, while taking into account the delayed onset of the appearance of the organic phase.
Finally, a relation between the change in optical density of the microbial suspension and the change in the state of the microorganisms in suspension and the adhesion of the microorganisms to the water-hydrocarbon interface needs to be established. Implicit in relations like Eqs. 3 and A5 is that the optical density is proportional to the number density of microorganisms. This is unlikely to hold true, even if only because many microbial strains cluster at very low volume fractions of added organic phase. However, at the relatively low optical densities of interest, it is plausible that there is a one-to-one correspondence between the optical density of the suspension and the state and number density of the microorganisms. In the experiments, we observe that the clustering and the interfacial adhesion of the microorganisms are well-separated in terms of the required amounts of organic phase. In a fair approximation, we may then split θ into the range from 0 to θ0 (bacterial clustering) and from 1−θ0 to 1 (interfacial adhesion). Therefore, Eqs. 3 and 4 are expected to be useful in the quantification of microbial behavior when exposed to hydrocarbons.
Two additional factors need to be taken into consideration when carrying out the interfacial adhesion assay using an organic solvent: the solubility of the solvent in water and its vapor pressure. Hexadecane is essentially insoluble in water and is also of low volatility, but as we use very small quantities on relatively large volumes of water and air, it could be that a significant fraction of the hexadecane either evaporates or dissolves in the water phase and is thus not available to interact with the bacterial surface. The latter of the two factors may immediately be disregarded, as the aqueous solubility of high-molecular weight alkanes is generally <10−6% w/w (Lide, 2000). In 3 ml of water <∼0.04 μl of hexadecane would therefore dissolve, which is at least one order-of-magnitude lower than the smallest amount of hexadecane used in the adhesion experiments. The risk of a significant loss of hexadecane by volatilization is higher, but still acceptable at hexadecane volumes of 1 μl and larger. By extrapolating the partial pressure data of hexadecane at 41.1°C and 67.4°C (Lide, 2000) using the Claudius-Clapeyron equation (Atkins, 1982), we arrive at a hexadecane vapor pressure of ∼0.2 Pa at 25°C. As the headspace volume in the test tubes is ∼15 − 3 = 12 ml, the amount of hexadecane in the saturated headspace would be ∼1.1 × 10−9 mole, which equals 0.3 μl. Thus, to avoid potential issues of significant dissolution or volatilization of the hexadecane, any hexadecane volume >∼1 μl is acceptable.
Prisca Schär-Zammaretti's current affiliation is the Institute of Biomedical Engineering, ETH, and University of Zürich, Moussonstrasse 18, CH-8044 Zürich, Switzerland.
References
- A-Hassan, E., W. F. Heinz, M. D. Antinik, N. P. D'Costa, S. Nageswaran, C. A. Schoenenberger, and J. H. Hoh. 1998. Relative microelastic mapping of living cells by atomic force microscopy. Biophys. J. 74:1564–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsson, P. A. 1986. Partitioning of Cell Particles and Macromolecules, 3rd Ed. Wiley, New York.
- Amiel, C., L. Mariey, M.-C. Curk-Daubié, P. Pichon, and J. Travert. 2000. Potentiality of Fourier transform infrared spectroscopy (FTIR) for discrimination and identification of dairy lactic acid bacteria. Lait. 80:445–459. [Google Scholar]
- Atkins, P. W. 1982. Physical Chemistry, 2nd Ed. Oxford University Press, Oxford.
- Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1993. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 59:4121–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 35:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, S., D. Haller, A. Pfeifer, and E. J. Schiffrin. 2002. Probiotics and immune response. Clin. Rev. Allergy Immunol. 22:287–309. [DOI] [PubMed] [Google Scholar]
- Bohach, G. A., and I. S. Snyder. 1983. Characterization of surfaces involved in adherence of Legionella pneumophila to Fischerella species. Infect. Immun. 42:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonaert, C. J. P., and P. G. Rouxhet. 2000. Surface of lactic acid bacteria: relationships between chemical composition and physico-chemical properties. Appl. Environ. Microbiol. 66:2548–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonaert, C. J. P., P. G. Rouxhet, and Y. F. Dufrêne. 2000. Surface properties of microbial cells probed at the nanometer scale with atomic force microscopy. Surf. Interf. Anal. 30:32–35. [Google Scholar]
- Boulbitch, A., B. Quinn, and D. Pink. 2000. Elasticity of the rod-shaped Gram-negative eubacteria. Phys. Rev. Lett. 85:5246–5249. [DOI] [PubMed] [Google Scholar]
- Busscher, H. J., R. Bos, H. C. van der Mei, and P. S. Handley. 2000. Physicochemistry of microbial adhesion from an overall approach to the limits. In Physical Chemistry of Biological Interfaces. A. Baszkin, and W. Norde, editors. Marcel Dekker, New York. 431–458.
- Colton, R. J., A. Engel, J. E. Frommer, H. E. Gaub, A. A. Gewirth, R. Guckenberger, J. Rabe, W. M. Heckel, and B. Parkinson. 1998. Procedures in Scanning Probe Microscopies. John Wiley, Chichester.
- Curk, M. C., F. Peladan, and J. C. Hubert. 1994. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillus species. FEMS Microbiol. Lett. 123:241–248. [Google Scholar]
- Daffonchio, D., J. Thaveersi, and W. Verstraete. 1995. Contact angle measurement and cell hydrophobicity of granular sludge from upflow anaerobic sludge bed reactors. Appl. Environ. Microbiol. 61:3676–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek. 76:159–184. [PubMed] [Google Scholar]
- Dufrêne, Y. F., and P. G. Rouxhet. 1996. X-ray photoelectron spectroscopy analysis of the surface composition of Azospirillum brasilense in relation to growth conditions. Coll. Surf. B. 7:271–279. [Google Scholar]
- Dufrêne, Y. F., A. Van der Wal, W. Norde, and P. G. Rouxhet. 1997. X-ray photoelectron spectroscopy analysis of whole cells and isolated cell walls of Gram-positive bacteria: comparison with biochemical analysis. J. Bacteriol. 179:1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne, Y. F. 2000. Direct Characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys. J. 78:3286–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne, Y. F. 2001. Atomic force microscopy of microbial cells. Microscopy Anal. 10:27–29. [Google Scholar]
- Eggerth, A. H. 1923. Changes in the stability and potential of cell suspensions. J. Gen. Physiol. 6:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276–302. [DOI] [PubMed] [Google Scholar]
- Evans, D. F., and H. Wennerström. 1994. The Colloidal Domain: Where Physics, Chemistry, Biology and Technology Meet. VCH Publishers, New York.
- Fuller, R. 1992. Probiotics: The Scientific Basis. Chapman & Hall, London, UK.
- Gallardo-Moreno, A. M., H. C. Van der Mei, H. J. Busscher, and C. Perez-Giraldo. 2002. The influence of subinhibitory concentrations of ampicillin and vancomycin on physicochemical surface characteristics of Enterococcus faecalis 1131. Coll. Surf. B. 24:285–295. [Google Scholar]
- Hiemenz, P. C. 1986. Principles of Colloid and Surface Chemistry, 2nd Ed. Marcel Dekker, New York.
- Hogt, A. H., J. Dankert, and J. Feijen. 1983. Encapsulation, slime production and surface hydrophobicity of coagulase-negative staphylococci. FEMS Microbiol. Lett. 18:211–215. [Google Scholar]
- Hunter, R. J. 1981. Zeta Potential in Colloid Science. Academic Press, London, UK.
- Isolauri, E., S. Salminen, and T. Mattila-Sandholm. 1999. New functional foods in the treatment of food allergy. Ann. Med. 31:299–302. [DOI] [PubMed] [Google Scholar]
- Karjavainen, P. V., A. C. Ouwehand, E. Isolauri, and S. J. Salminen. 1998. The ability of probiotic bacteria to bind to human intestinal mucus. FEMS Microbiol. Lett. 167:185–189. [DOI] [PubMed] [Google Scholar]
- Lambert, P. A., I. C. Hancock, and J. Baddiley. 1975. The interaction of magnesium ions with teichoic acid. Biochem. J. 149:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau, L. D., and E. M. Lifshitz. 1970. Theory of Elasticity, 2nd Ed. Pergamon, Oxford.
- Lide, D. R. (Editor.) 2000. Handbook of Chemistry and Physics, 81st Ed. CRC Press, Boca Raton, FL.
- Lortal, S., M. Rousseau, P. Boyaval, and J. Van Heijenoort. 1991. Cell wall and autolytic system of Lactobacillus helveticus ATCC12046. J. Gen. Microbiol. 137:549–559. [Google Scholar]
- Lortal, S., J. Van Heijenoort, K. Gruber, and S. Sleytr. 1992. S-layer of Lactobacillus helveticus ATCC12046: isolation, chemical characterization and re-formation after extraction with lithium chloride. J. Gen. Microbiol. 138:611–618. [Google Scholar]
- Mack, D. R., S. Michail, S. Wei, L. McDougall, and M. A. Hollingsworth. 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276:G941–G950. [DOI] [PubMed] [Google Scholar]
- Magonov, S. N., and D. Reneker. 1997. Characterization of polymer surfaces with atomic force microscopy. Annu. Rev. Mater. Sci. 27:175–222. [Google Scholar]
- Makin, S. A., and T. J. Beveridge. 1996. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology. 142:299–307. [DOI] [PubMed] [Google Scholar]
- Marshall, K. C. 1976. Interfaces in Microbial Ecology. Harvard University Press, Cambridge, MA.
- Mozes, N., and S. Lortal. 1995. X-ray photoelectron spectroscopy and biochemical analysis of the surface of Lactobacillus helveticus ATCC12046. Microbiology. 141:11–19. [Google Scholar]
- Mozes, N., and P. G. Rouxhet. 1990. Microbial hydrophobicity and fermentation technology. In Microbial Cell Surface Hydrophobicity. R. J. Doyle, and M. Rosenberg, editors. American Society for Microbiology, Washington, DC.
- Neufeld, R. J., J. E. Zajic, and D. F. Gerson. 1980. Cell surface measurements in hydrocarbon and carbohydrate fermentations. Appl. Environ. Microbiol. 39:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek, I., and R. Doyle. 1994. Bacterial Adhesion to Cells and Tissues. Chapman & Hall, New York.
- Olsson, J., and G. Westergen. 1982. Hydrophobic surface properties of oral streptococci. FEMS Microbiol. Lett. 15:319–323. [Google Scholar]
- Ouwehand, A. C., E. Isolauri, P. V. Karjavainen, and S. J. Salminen. 1999. Adhesion of four Bifidobacterium strains to human intestinal mucus from subjects in different age groups. FEMS Microbiol. Lett. 172:61–64. [DOI] [PubMed] [Google Scholar]
- Petersen, N. O., W. M. McConnaughy, and E. L. Elson. 1982. Dependence of locally measured cellular deformability on position on the cell, temperature, and cytochalasin B. Proc. Natl. Acad. Sci. USA. 79:5327–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman, B., P. Blount, J. H. A. Folgering, R. H. E. Friessen, P. C. Moe, and T. van der Heide. 2002. How do membrane proteins sense water stress? Mol. Microbiol. 44:889–902. [DOI] [PubMed] [Google Scholar]
- Poortinga, A. T., R. Bos, and H. J. Busscher. 2001. Electrostatic interactions in the adhesion of an ion-penetrable and ion-impenetrable bacterial strain to glass. Coll. Surf. B. 20:105–117. [DOI] [PubMed] [Google Scholar]
- Razatos, A., Y. L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 95:11059–11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razatos, A. 2001. Application of atomic force microscopy to study initial events of bacterial adhesion. Methods Enzymol. 337:276–285. [DOI] [PubMed] [Google Scholar]
- Reid, G., P. L. Cuperus, A. W. Bruce, H. C. Van der Mei, L. Tomeczek, A. H. Khoury, and H. J. Busscher. 1992. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poultry Lactobacilli: effect of serial culture passages. Appl. Environ. Microbiol. 58:1549–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi, A., and F. Clementi. 2000. Exopolysaccharides from lactic acid bacteria: structure, production and technological applications. Ital. J. Food Sci. 12:23–45. [Google Scholar]
- Rief, M., F. Oesterhelt, B. Heymann, and H. E. Gaub. 1997. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science. 275:1295–1297. [DOI] [PubMed] [Google Scholar]
- Rosenberg, M. 1984. Bacterial adherence to hydrocarbons: a useful technique for studying cell surface hydrophobicity. FEMS Microbiol. Lett. 22:289–295. [Google Scholar]
- Savage, D. C., and M. Fletcher. 1985. Bacterial adhesion: mechanisms and physiological significance. Plenum Press, New York.
- Schär-Zammaretti, P., and J. Ubbink. 2003. Imaging of lactic acid bacteria with AFM—elasticity and adhesion maps and their relationship to biological and structural data. Ultramicroscopy. 97:199–208. [DOI] [PubMed] [Google Scholar]
- Scheuring, S., H. Stahlberg, M. Chami, C. Houssin, J.-L. Rigaud, and A. Engel. 2002. Charting and unzipping the surface layer of Corynebacterium glutamicum with the atomic force microscope. Mol. Microbiol. 44:675–684. [DOI] [PubMed] [Google Scholar]
- Sleytr, U. B., M. Sara, and D. Pum. 2000. Crystalline bacterial cell surface layers (S-layers): a versatile self-assembly system. In Supramolecular Polymers. A. Ciferri, editor. New York. 177–213.
- Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of Lactobacillus acidophilus ATCC4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245–257. [DOI] [PubMed] [Google Scholar]
- Streyer, L. 1981. Biochemistry, 2nd Ed. Freeman, San Francisco.
- Tanford, C. 1961. Physical Chemistry of Macromolecules. Wiley, New York.
- Tanford, C. 1980. The Hydrophobic Effect. Wiley, New York.
- Tomeczek, L., G. Ried, P. L. Cuperus, J. A. McGroarty, H. C. Van der Mei, A. W. Bruce, A. E. Khoury, and H. J. Busscher. 1992. Correlation between hydrophobicity and resistance to nonoxynol-9 and vancomycin for urogenital isolates of lactobacilli. FEMS Microbiol. Lett. 94:101–104. [DOI] [PubMed] [Google Scholar]
- Tuomola, E. M., and S. J. Salminen. 1998. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 41:45–51. [DOI] [PubMed] [Google Scholar]
- Van der Mei, H. C., J. M. Meinders, and H. J. Busscher. 1994. The influence of ionic strength and pH on diffusion of microorganisms with different structural surface features. Microbiology. 140:3414–3419. [Google Scholar]
- Van der Mei, H., H. Busscher, R. Bos, J. de Vries, C. J. Boonaert, and Y. F. Dufrêne. 2000. Direct probing by atomic force microscopy of the cell surface softness of a fibrillated and a nonfibrillated oral streptococcal strain. Biophys. J. 78:2668–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Mei, H. C., and H. J. Busscher. 2001. Electrophoretic mobility distributions of single-strain microbial populations. Appl. Environ. Microbiol. 67:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Mei, H. C., P. Kiers, J. De Vries, and H. J. Busscher. 2001. Measurements of softness of microbial cell surfaces. Methods Enzymol. 337:270–276. [DOI] [PubMed] [Google Scholar]
- Van der Wal, A., M. Minor, W. Norde, A. J. B. Zehnder, and J. Lyklema. 1997. Electrokinetic potential of bacterial cells. Langmuir. 13:165–177. [Google Scholar]
- Van Loosdrecht, M. C. M., J. Lyklema, W. Norde, G. Schraa, and A. J. B. Zehnder. 1987. The role of bacterial cell wall hydrophobicity in adhesion. Appl. Environ. Microbiol. 53:1893–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss, C. J. 1994. Interfacial Forces in Aqueous Media. Marcel Dekker, New York.
- Velegol, S. B., and B. E. Logan. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir. 18:5256–5262. [Google Scholar]
- Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lacobacillus gasseri. Appl. Environ. Microbiol. 68:6172–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadström, T. 1990. Hydrophobic characteristics of Staphylococci: role of surface structures and role in adhesion and host colonization. In Microbial Cell Surface Hydrophobicity. R. J. Doyle, and M. Rosenberg, editors. American Society for Microbiology, Washington, D.C.
- Wasserman, E., and A. R. Felmy. 1998. Computation of the electrical double layer properties of semipermeable membranes in multicomponent electrolytes. Appl. Environ. Microbiol. 64:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, L. T. 1925. The acid agglutination of mixtures of oppositely charged bacterial cells. J. Gen. Physiol. 7:513–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, B. J. B. 1992. The lactic acid bacteria in health and disease. In The Lactic Acid Bacteria, Vol. 1. Elsevier Applied Science, London, UK.
- Xu, W., P. J. Mulhern, B. L. Blackford, M. H. Jericho, M. Firtel, and T. J. Beveridge. 1996. Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication for methane production. J. Bacteriol. 178:3106–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., M. Jericho, D. Pink, and T. Beveridge. 1999. Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 181:6865–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]