The chromosomal DNA in the bacterium Escherichia coli is thought to be organized and compacted at least in part as a consequence of the interaction with so-called histone-like or nucleoid-associated proteins. The groups of Stavans and Oppenheim have recently embarked on an ambitious project which aims to quantify the compactive effects of the various members of this group of proteins using magnetic tweezers (Ali et al., 2001). Eventually this could lead to a better understanding of how these proteins work together in the formation of a compact nucleoid.
In their most recent study (Amit et al., 2003), they describe the structural effects of H-NS on lambda DNA at the single-molecule level. Interestingly, their data seem to indicate that H-NS does not induce DNA compaction. Rather, the DNA molecule attains an extended structure upon interaction with H-NS and becomes less flexible. In fact, the effective persistence length is about three times higher than that of naked DNA.
The data of Amit et al. (2003) are in striking contrast with recent models about the interaction of H-NS with DNA, which are based both on insights into the structure of the H-NS dimer and microscopic (electron microscopy (EM) and scanning force microscopy (SFM)) observations. H-NS exists as a dimer, which has the ability to self-associate and form large oligomers (Smyth et al., 2000). The formation of dimers is a result of a leucine zipper-kind of interaction among the N-terminal regions of the two identical monomeric subunits of the protein (Esposito et al., 2002). DNA binding takes place through the C-terminal region (Shindo et al., 1995, 1999). Obviously, within the context of the dimer, two separate DNA binding domains are exposed. It is not exactly clear how H-NS interacts with DNA, but the presence of two DNA binding domains could allow the protein to bind to two DNA strands simultaneously. Large oligomers are thought to be formed by association of dimers in a head-to-tail fashion (Esposito et al., 2002). The DNA binding domains are probably exposed in opposite directions (Esposito et al., 2002), both at the level of a single dimer and at the level of these oligomeric forms of H-NS. Therefore, it is likely that upon initial binding of H-NS oligomers to DNA, only half the number of these domains is used, whereas the others protrude from the opposite side of the H-NS oligomer. A large interaction “surface” is thus still available for binding to another stretch of DNA (within the same or on another DNA molecule—see Fig. 1). Early electron microscopy images suggested coating of DNA by H-NS, but also showed the formation of DNA loops in which distant tracts are apparently brought together by the action of H-NS (Tupper et al., 1994). Subsequently, a number of SFM studies provided further evidence for H-NS as a “DNA bridge” (Dame et al., 2000, 2001, 2002) and showed the functional significance of such bridging (Dame et al., 2001, 2002). A more recent EM study also confirmed these data (Schneider et al., 2001). What does this mean? Should the microscopic data be considered as artifacts, or could there be something particular happening in the magnetic tweezers studies?
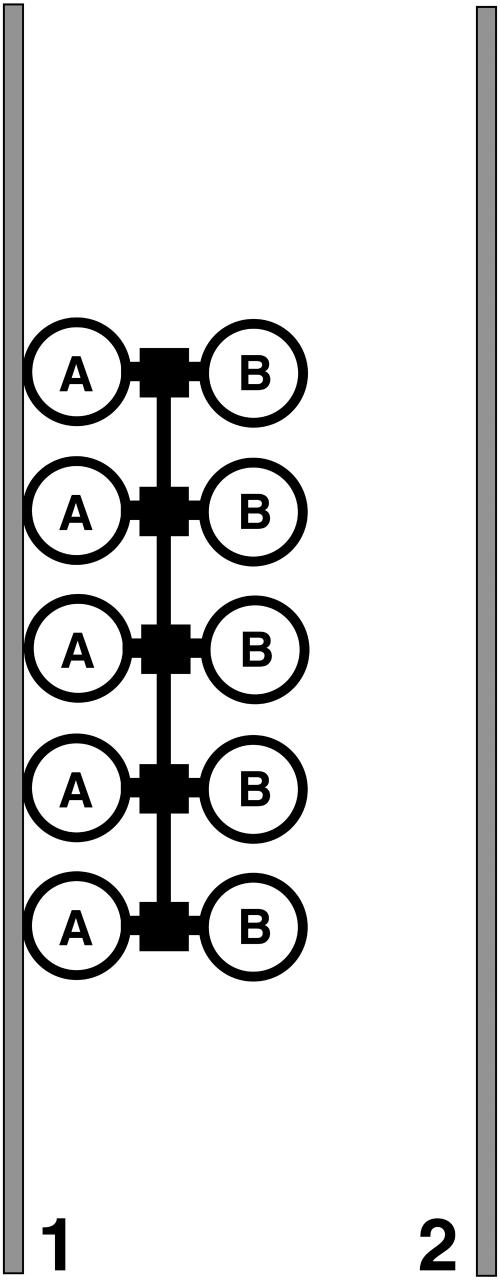
FIGURE 1.
Model for binding of H-NS to DNA. The two identical DNA binding domains (A and B) of each H-NS dimer (large circles) are directed in opposite directions and have the ability to interact independently with a stretch of double-stranded DNA (within the same or on another DNA molecule). Two stretches of dsDNA (1 and 2) are indicated in gray. H-NS dimers are held together through the oligomerization domain (small squares). In addition (a different region on) the same domain is responsible for oligomerization of adjacent H-NS dimers.
The most obvious difference between these studies is that the microscopy studies were carried out in bulk, whereas the magnetic tweezers experiments are carried out with one single DNA molecule. As a consequence, the DNA concentration in the single-molecule experiment is extremely low, whereas the H-NS concentrations used for both types of experiments are in the same range (∼10−7/10−6 M). The fraction of independent binding sites on the DNA molecules occupied by H-NS is determined by the concentrations of protein and DNA and the affinity of the protein for DNA, and follows directly from Le Chatelier's principle of mass action (Le Chatelier, 1888). The difference between single-molecule and bulk experiments can be analyzed quantitatively following an approach based on this principle as described in Rippe (1997) or McGhee and von Hippel (1974), depending on the type of binding in the given system (single site, multiple adjacent sites, and cooperative binding).
Following Linus Pauling's adage, “the student (or the scientist) would be wise to refrain from using the mathematical equation unless he understands the theory that it represents, and can make a statement about the theory that does not consist just in reading the equation. It is fortunate that there is a general qualitative principle, called Le Chatelier's principle, that relates to all the applications of the principles of chemical equilibrium. When you have obtained a grasp of Le Chatelier's principle, you will be able to think about any problem of chemical equilibrium that arises, and, by use of a simple argument, to make a qualitative statement about it….” (L. Pauling 1964, College Chemistry, 3rd ed., Freeman, San Francisco, CA, 437–438).
In following that adage, we limit ourselves to a general and qualitative evaluation of these differences. It follows directly from Le Chatelier's principle that a different ratio between protein and DNA results in a different degree of saturation of the binding sites on the DNA. In single-molecule studies such as described here, there is an enormous excess of protein present when compared to bulk studies assuming that the protein concentrations are similar in both cases. As a consequence, a single DNA molecule held between tweezers will reach the same degree of saturation at much lower protein concentrations than each of the single molecules in a bulk experiment (see also Fig. 2 for an illustration of this effect). It is possible that the apparent difference between both types of data thus stems from a very different degree of saturation of the DNA with H-NS molecules.
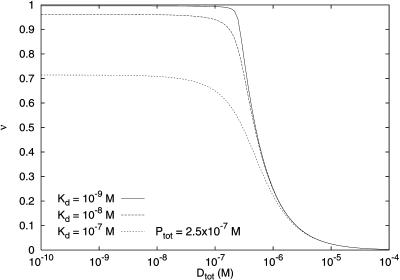
FIGURE 2.
Theoretical binding curves for the degree of saturation with ligands as dependent on the concentration of available binding sites on the DNA. ν denotes the fractional occupation of DNA binding sites, Dtot the total concentration of DNA binding sites (in M), Ptot is the total concentration of ligand (in M), and Kd is the equilibrium dissocation constant for the binding reaction (in M). In this example, one ligand occupies 15–20 bp and binds to equivalent sites without cooperativity (Rippe, 1997). Clearly, at a fixed ligand concentration (in this example 250 nM), each of the binding sites at the single DNA molecule limit (Dtot → 0) will be maximally occupied for a range of relevant Kds. Above approximately a concentration of binding sites of 100 nM the fractional occupation becomes lowered dramatically.
It is likely that the relatively high degree of fractional occupation of DNA binding sites in the single-molecule experiment is a factor that prevents the previously described formation of intramolecular bridges by H-NS. First of all, a high fractional occupation of binding sites on the DNA and the consequent higher effective persistence length (by local rigidification of the DNA as suggested by Fig. 6 in Amit et al., 2003) should result in a lower probability of intrastrand contacts. Second, under such conditions, the probability that DNA bound H-NS molecules will encounter naked (rather than a tract already covered with H-NS) is lowered.
Generally, the interpretation of single-molecule data of architectural proteins without sequence specificity is faced with similar complexity as described for the situation with H-NS. Other such proteins have not yet been studied, but, for instance, Lrp-type proteins—like H-NS—are known from bulk experiments to have the ability to bridge DNA strands (Beloin et al., 2003; Tapias et al., 2000) and analysis of Lrp-DNA interactions at the single-molecule level should therefore be interpreted in a similar fashion. Another class of important architectural proteins is thought to organize DNA by the (dynamic) induction of local DNA bends. Examples include the prokaryotic HU (Dame and Goosen, 2002) and the eukaryotic HMG proteins (Thomas and Travers, 2001). Such proteins are expected to reduce the apparent length of a DNA molecule (when analyzed in a tweezers set-up) by transient binding and bending at random positions. It is likely that such an effect only takes place if relatively few molecules are bound, since bending by many such proteins involves folding around the protein of free DNA adjacent to a minimal binding site. If the level of saturation of the binding sites increases, the effective ability of each bound protein to bend may be reduced. In this case, since a considerable level of saturation will be reached at relatively low protein concentrations, the bending regime might be easily overlooked in single-molecule observations. These theoretical considerations indicate that for this type of experiments the interpretation of results is not straightforward, and comparison between those experiments performed in bulk and those performed at the single-molecule level requires caution.
In previous tweezers studies, differences between measurements in bulk and at the single-molecule level have hardly been addressed. Yet our considerations have general implications, reaching far beyond the example of H-NS binding described here. The difference between bulk and single-molecule experiments will be most evident for ligands (like H-NS), which bind DNA nonspecifically and at many sites simultaneously. There may also be important implications for sequence-specific proteins, which may display significant levels of nonspecific binding at random sites. Such proteins have been studied relatively often using single-molecule techniques. It is therefore important to always be aware of a possible “single-molecule effect.” If one intends to properly compare data from both kinds of experiments, it may be important to perform the described type of single-molecule studies in a bulk context as well, i.e., in the presence of other DNA molecules free in solution. Understanding single-molecule behavior requires more than single molecules.
References
- Ali, B. M., R. Amit, I. Braslavsky, A. B. Oppenheim, O. Gileadi, and J. Stavans. 2001. Compaction of single DNA molecules induced by binding of integration host factor (IHF). Proc. Natl. Acad. Sci. USA. 98:10658–10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit, R., A. B. Oppenheim, and J. Stavans. 2003. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 84:2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin, C., J. Jeusset, B. Revet, G. Mirambeau, F. Le Hegarat, and E. Le Cam. 2003. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 278:5333–5342. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., and N. Goosen. 2002. HU: promoting or counteracting DNA compaction? FEBS Lett. 529(2–3):151–156. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2000. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28:3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie. 83:231–234. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, R. Wurm, R. Wagner, and N. Goosen. 2002. Structural basis for H-NS–mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146–2150. [DOI] [PubMed] [Google Scholar]
- Esposito, D., A. Petrovic, R. Harris, S. Ono, J. F. Eccleston, A. Mbabaali, I. Haq, C. F. Higgins, J. C. Hinton, P. C. Driscoll, and J. E. Ladbury. 2002. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324(4):841–850. [DOI] [PubMed] [Google Scholar]
- Le Chatelier, H. L. 1888. Recherches expérimentales et théoriques sur les équilibres chimiques. Annales des Mines. 13:157–182. [Google Scholar]
- McGhee, J. D., and P. H. von Hippel. 1974. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86:469–489. [DOI] [PubMed] [Google Scholar]
- Rippe, K. 1997. Analysis of protein-DNA binding at equilibrium. BIF Futura. 12:20–26. [Google Scholar]
- Schneider, R., R. Lurz, G. Luder, C. Tolksdorf, A. Travers, and G. Muskhelishvili. 2001. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 29:5107–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, H., T. Iwaki, R. Ieda, H. Kurumizaka, C. Ueguchi, T. Mizuno, S. Morikawa, H. Nakamura, and H. Kuboniwa. 1995. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 360:125–131. [DOI] [PubMed] [Google Scholar]
- Shindo, H., A. Ohnuki, H. Ginba, E. Katoh, C. Ueguchi, T. Mizuno, and T. Yamazaki. 1999. Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Lett. 455:63–69. [DOI] [PubMed] [Google Scholar]
- Smyth, C. P., T. Lundback, D. Renzoni, G. Siligardi, R. Beavil, M. Layton, J. M. Sidebotham, J. C. Hinton, P. C. Driscoll, C. F. Higgins, and J. E. Ladbury. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36 (4):962–72. [DOI] [PubMed] [Google Scholar]
- Tapias, A., G. Lopez, and S. Ayora. 2000. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 28:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related “architectural” DNA-binding proteins. Trends Biochem. Sci. 26:167–174. [DOI] [PubMed] [Google Scholar]
- Tupper, A. E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. Ferguson, J. M. Sidebotham, J. C. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]