Abstract
We have used RNA-RNA in situ hybridization to detect the expression of several Mycobacterium tuberculosis genes in tuberculous granulomas in lung tissue sections from tuberculosis patients. The M. tuberculosis genes chosen fall into two classes. Four genes (icl, narX, and Rv2557 and Rv2558) have been implicated in the persistence of the bacterium in the host, and two genes (iniB and kasA) are upregulated in response to isoniazid exposure. Both necrotic and nonnecrotic granulomas were identified in all of the patients. Necrotic granulomas were divided into three zones: an outer lymphocyte cuff containing lymphocytes and macrophages, a transition zone consisting of necrotic material interspersed with macrophages, and a central acellular necrotic region. Transcripts of all of the genes studied were found in nonnecrotic granulomas and in the lymphocyte cuff of necrotic granulomas. Mycobacterial gene expression was associated with CD68-positive myeloid cells. Rv2557 and/or its homologue Rv2558, kasA, and iniB were expressed within the transition zone of necrotic granulomas, whereas icl and narX transcripts were absent from this area. There was no evidence of transcription of any of the genes examined in the central necrotic region, although mycobacterial DNA was present. The differential expression of genes within granulomas demonstrates that M. tuberculosis exists in a variety of metabolic states and may be indicative of the response to different microenvironments. These observations confirm that genes identified in models of persistence or in response to drug treatment in vitro are expressed in the human host.
Patients with active tuberculosis (TB) are thought to harbor Mycobacterium tuberculosis bacilli in various metabolic states ranging from actively growing bacilli to those in a state of persistence (23, 24). Persistent organisms are phenotypically resistant to chemotherapeutic agents and require at least six months of chemotherapy to achieve sterilization. This extended duration of treatment results in poor patient compliance and increased risk of the emergence of drug resistance. TB therapy would be improved with new drugs that eradicate persistent organisms, thus shortening the duration of chemotherapy.
It is not known how M. tuberculosis survives the immune onslaught from the host, the hostile environment within the granuloma, or long-term antimycobacterial therapy. The Th1 immune response (interleukin-12 and gamma interferon) drives the formation of granulomas, which consist of macrophages and lymphocytes surrounding a central area of necrosis. In animal models, tumor necrosis factor alpha (TNF-α) has been shown to play a central role in the host immune response to TB, including granuloma formation and containment of disease (11, 27). There is no direct evidence of a protective role for TNF-α in patients with TB; however, treating rheumatoid arthritis patients with Infliximab, a TNF-α-neutralizing agent, resulted in reactivation of latent TB (21, 28). As the granuloma becomes necrotic, a crossroads is reached whereby the lesion may resolve (by fibrosis and calcification) or it may lead to dissemination via liquefaction (4). Therefore, the development of necrosis does not necessarily lead to dissemination and can also provide an environment that prevents transmission and spread of disease, but the bacilli remain viable and evade the immune system (26). Therefore, the environment within the granuloma may fail to kill the bacilli while inducing them to alter their metabolic state and enter a state of nonreplicating persistence (16).
A number of in vitro studies have been based on the assumption that granulomas provide a microaerophilic environment. In the Wayne model, oxygen is gradually depleted from mycobacterial cultures and a metabolic “shiftdown” is observed, coinciding with the changeover of the bacilli from rapid to slow metabolism and ultimately into a state of nonreplicating persistence (32). During this shiftdown, isocitrate lyase and nitrate reductase activities increase (33, 34). Isocitrate lyase is an enzyme of the glyoxylate pathway, which M. tuberculosis and other bacteria utilize when growing on acetate and certain fatty acids to replenish intermediates of the tricarboxylic acid cycle (23). This is important when fatty acids are the main source of carbon and energy, as has been suggested for intracellular M. tuberculosis and M. leprae (16). The icl gene is upregulated upon macrophage infection (13, 15) and is also essential for the persistence of M. tuberculosis in mice during the chronic phase of infection (23).
The narX gene, which encodes a putative fused nitrate reductase, was found to be upregulated in anaerobic cultures of M. bovis BCG (20) and in a study of the hypoxic response of M. tuberculosis (29). This gene is not present in other bacteria but shows homology to various subunits of the multimeric nitrate reductase NarGHIJ found in many bacteria, including M. tuberculosis.
A recent study utilizing nutrient starvation to drive M. tuberculosis into a persistent state found that Rv2557 and Rv2558 mRNAs and corresponding proteins were upregulated (3). The RV2557 and RV2558 gene products are highly homologous to each other (69% protein and 81% DNA identities) but not to any other known protein. Their function is unknown. We are unable to distinguish expression of the Rv2557 and Rv2558 genes by hybridization, but there is evidence from proteome studies that both proteins are expressed (3).
In vitro studies have revealed that treatment of M. tuberculosis with isoniazid induces expression of a number of genes (36). These include the iniBAC operon (1), which is proposed to provide a protective response to cell wall damage (2), and kasA, a β-ketoacyl-ACP synthase component of the fatty acid synthase II complex (36). We chose to study expression of iniB and kasA in human tissue in order to confirm that M. tuberculosis gene expression responds to drug treatment in a similar way in the host.
Reverse transcription-PCR techniques have been utilized to detect mycobacterial RNA (5, 14, 17, 25). Recently, we and others have reported the successful use of in situ hybridization to detect mycobacterial DNA and also mycobacterial RNA in patients with Crohn's disease and TB (8, 18). The advantage of in situ hybridization is that the location of the bacilli and spatial variations in gene expression can be visualized.
The identification of genes essential for M. tuberculosis persistence is the first step in the design of new drugs that specifically target this population. In order to confirm the importance of mycobacterial genes identified in in vitro models, we used in situ hybridization to analyze the expression of these genes in human tuberculous lung granulomas. The use of human tuberculous tissue and the in situ technique should help us to build a realistic picture of what happens at the site of human disease.
MATERIALS AND METHODS
Tissue specimens.
Adult lung tissue was obtained from seven patients undergoing surgery for hemoptysis, a life-threatening bleeding caused by liquefied granulomas rupturing into the blood vessels in the infected lung(s). The tissue was collected from five male and two female patients with active TB ranging from ages 14 to 49 years (Table 1). All seven patients were culture positive but only five were positive for acid-fast bacilli as determined by Ziehl-Neelsen (ZN) staining on the resected lung tissue obtained at the time of surgery. All of the patients were human immunodeficiency virus negative, and all material was obtained from Tygerberg Hospital, Cape Town, South Africa. Ethical approval for the study was obtained from the Ethics Committee of the University of Stellenbosch.
TABLE 1.
Clinical information for hemoptysis patients
| Patient | Age/sexa | Treatment duration | CXR typeb | ZN status | DNA statusc |
|---|---|---|---|---|---|
| TB7 | 16/F | 1 yr | RUL | + | + |
| TB8 | 41/M | 1 yr | LUL | + | + |
| TB9 | 23/M | 2 yr | RUL | + | + |
| TB10 | 18/M | 2 mo | LUL | + | + |
| TB11 | 36/M | 3 mo | LUL | + | + |
| TB12 | 49/M | 6 mo | RUL | − | + |
| TB13 | 14/F | 8 mo | RLL | − | + |
M, male; F, female.
CXR, chest radiograph; RUL, LUL, and RLL, right or left upper lobe and right lower lobe cavitation, respectively.
M. tuberculosis DNA was shown by DNA-DNA in situ hybridization.
Preparation of the probes.
Preparation of the MTB484 probe for the detection of M. tuberculosis DNA has been described elsewhere (8). Regions of the icl, narX, Rv2557, kasA, and iniB genes (Table 2) were PCR amplified from H37Rv DNA, purified by using a PCR purification kit (Promega), and cloned into the pGEMTeasy vector (Promega). The primer sequences and annealing temperatures are shown in Table 2. The high sequence homology between the Rv2557 and Rv2558 genes made it difficult to design a riboprobe that would distinguish between them in in situ hybridization. Therefore, a region of Rv2557 was chosen that we expect will detect transcripts from both genes, and the resulting probe was designated Rv2557*. Plasmids containing the icl, narX, Rv2557*, kasA, and iniB inserts were isolated, restriction mapped, and sequenced. Antisense and sense biotin-labeled riboprobes were transcribed, and confirmation of biotinylation was performed as described previously (9).
TABLE 2.
Primer information for each probe used
| Probe | Rv no. | Primer sequences | Probe length (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| icl | Rv0467 | 5′-aagacgtagccgccactcag-3′ and 5′-tgataaacgggcggaagtc-3′ | 466 | 55 |
| narX | Rv1736C | 5′-agttcgcccatcatgtcacc-3′ and 5′-aactcacggcaactcggcac-3′ | 452 | 55 |
| Rv2557* | Rv2557 and Rv2558 | 5′-gcggaccaagtagatcaagg-3′ and 5′-actcgcattcatcgagttcc-3′ | 234 | 60 |
| iniB | Rv0341 | 5′-gctagccagatcggtgtctc-3′ and 5′-cgacagatgaggcatagcag-3′ | 170 | 60 |
| kasA | Rv2245 | 5′-ccgttgggcatgatcatctg-3′ and 5′-gtcagccttccaccgctaatg-3′ | 442 | 55 |
In situ hybridization.
Consecutive 5-μm sections of paraffin-embedded lung tissue were applied to RNase-free slides coated with 5 μg of aminopropyltriethoxysilane (Sigma Aldrich)/ml. Dewaxing, rehydration through graded ethanols, and the proteinase K treatment, prehybridization, and hybridization steps were performed as previously described (8). Controls for DNA-DNA and RNA-RNA hybridization were as described earlier (8). Hybridization with the antisense or sense probes, respectively, was performed as described previously (8). After the addition of the substrate nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate)-iodonitrotetrazolium violet, a brown color appeared on the sections, which were then counterstained with hematoxylin (Vector USA) for 10 s, rinsed in distilled water, and mounted with Dako Faramount.
Double labeling.
A combination of immunohistochemistry with an anti-CD68 antibody (Dako) and in situ hybridization with the icl, narX, Rv2557*, iniB, and kasA riboprobes was performed as described earlier (8). The sections were not counterstained after the double-labeling technique. As a control, immunohistochemistry for CD68 was performed in the absence of the DNA or RNA probes, and the detection of the red color without counterstaining was as described previously (8).
Photography.
The images were captured by using a Zeiss microscope (Axioskop 2) fitted with a Sony 3CCV video camera. In order to maintain comparability between slides, the light parameters were optimized for the DNA-probed slides and then kept constant for all subsequent slides. The images were saved by using Axiovision from Zeiss (Germany).
Assessment of slides.
In situ hybridization is an empirical staining technique and cannot be accurately quantitated. Granulomas were therefore scored either negative or positive for the presence of DNA or mRNA. It is impossible to determine accurately the number of bacilli present in one granuloma, since one brown spot does not necessarily equate to one bacillus. Two observers assessed duplicate slides; the numbers of granulomas positive for M. tuberculosis DNA and mRNA for each patient were determined at a 5× magnification.
RESULTS
Histopathology.
All seven patients were found to have both necrotic as well as nonnecrotic granulomas. Five of the seven patients were positive for acid-fast bacilli upon ZN staining, and these bacilli were detected within giant cells and nonnecrotic granulomas and sometimes within the necrotic regions of granulomas (data not shown). The M. tuberculosis strains isolated from the seven patients were found to be sensitive to both isoniazid and rifampin. Granulomas were surrounded by well-defined lymphocyte cuffs consisting predominantly of cells with the morphology of lymphocytes and macrophages. The center of nonnecrotic granulomas consisted of macrophages and giant cells. In comparison, the centers of granulomas with caseous necrosis were devoid of cells. The transition zone is situated between the lymphocyte cuff and the central necrotic area. It exhibits features of both regions with some intact macrophages interspersed with cell debris.
Expression of mycobacterial genes within granulomas at low-power magnification.
Consecutive sections of lung tissue from the seven TB patients (Table 1) were subjected to RNA-RNA in situ hybridization with the riboprobes for icl, narX, and kasA. Sections from only four of the patients were probed with Rv2557* and iniB because there was insufficient material available from patients TB7, TB8, and TB9 (Table 3).
TABLE 3.
Summary of data obtained from seven hemoptysis patients after analysis for the presence of M. tuberculosis DNA and narX, icl, Rv2557 and/or Rv2558, iniB, and kasA mRNAs by in situ hybridization
| Patient | Total no. of granulomas | No. of granulomas positive for:
|
|||||
|---|---|---|---|---|---|---|---|
| DNA | narX mRNA | icl mRNA | Rv2557∗ mRNA | iniB mRNA | kasA mRNA | ||
| TB7 | 32 | 30 | 2 | 15 | NTb | NT | 28 |
| TB8 | 18 | 18 | 1 | 8 | NT | NT | 15 |
| TB9 | 10 | 9 | 1 | 5 | NT | NT | 7 |
| TB10 | 9 | 9 | 1 | 5 | 9 | 9 | 7 |
| TB11 | 9 | 9 | 0 | 6 | 9 | 9 | 6 |
| TB12 | 25 | 25 | 2 | 19 | 22 | 22 | 21 |
| TB13 | 19 | 19 | 1 | 16 | 19 | 19 | 16 |
DNA, Mycobacterium tuberculosis DNA.
NT, not tested.
A total of 122 granulomas were analyzed for the presence of M. tuberculosis DNA, and 119 (98%) were found to be positive (Table 3). Both necrotic and nonnecrotic granulomas from all seven patients were positive for M. tuberculosis DNA. The M. tuberculosis DNA was widespread, being found within cells with the morphology of macrophages and giant cells in the lymphocyte cuff, in the transition zone as well in the regions of necrosis in necrotic granulomas (data not shown).
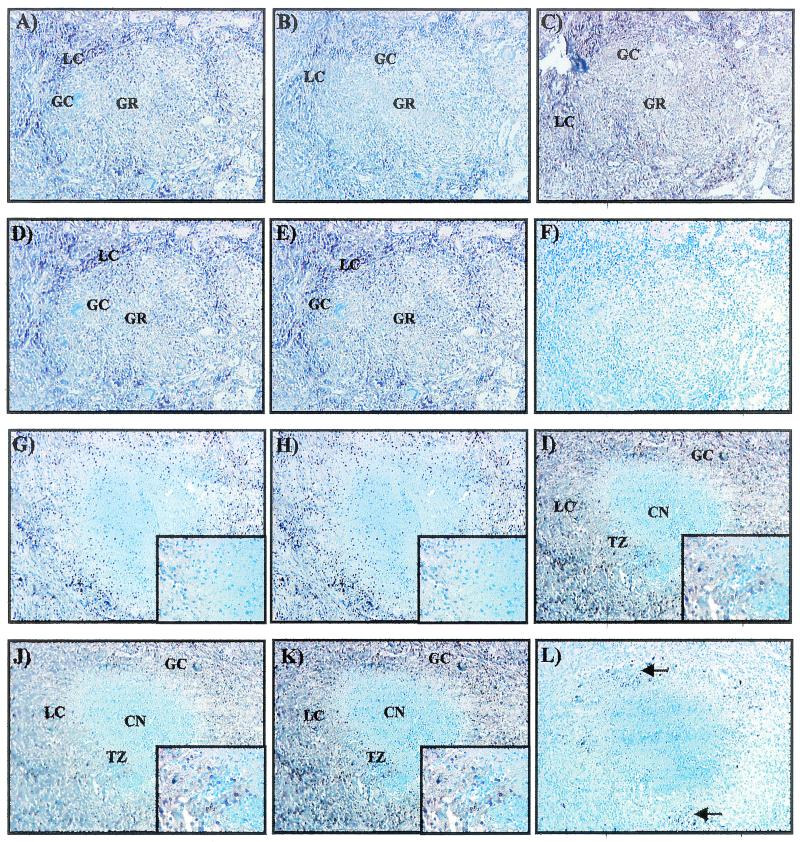
A representative nonnecrotic granuloma from patient TB11 is shown in Fig. 1A to E. On the outer edges is the lymphocyte cuff, rich in cells with the morphology of lymphocytes and macrophages. In the center are giant cells interspersed with numerous cells with the morphology of macrophages and lymphocytes. This granuloma was positive for icl, narX, Rv2557*, kasA, and iniB mRNAs as visualized by the brown signal (Fig. 1A, B, C, D, and E, respectively).
FIG. 1.
Low-power examination of a nonnecrotic and a necrotic granuloma for the detection of icl, narX, Rv2557 and/or Rv2558, kasA, and iniB mRNAs by RNA-RNA in situ hybridization. The nonnecrotic granuloma contains a number of giant cells (GC) and a lymphocyte cuff (LC) consisting of lymphocytes and macrophages. This nonnecrotic granuloma was positive for icl (A), narX (B), Rv2557* (C), kasA (D), and iniB (E) mRNAs. (F) Hybridization with the kasA sense probe produced no signal. Positive signals are brown, whereas negative signals are blue. A representative necrotic granuloma positive for icl (G), narX (H), Rv2557* (I), kasA (J), and iniB (K) mRNAs can be seen, as evidenced by the brown signal. The center of this granuloma consists of a region of caseous necrosis (CN) and is surrounded by a region called the transition zone (TZ), which is surrounded by a lymphocyte cuff (LC). icl and narX mRNAs localized only to the lymphocyte cuff and were absent from the transition zone (insets of panels G and H); Rv2557*, kasA, and iniB mRNAs were detected within the transition zone as shown in the insets of panels I, J, and K (insets, total magnification of ×360). Hybridization with the kasA sense riboprobe produced no positive signal, as evidenced by the blue color (L). Black spots indicate carbon deposits and are shown with arrows on the edges of the granuloma. Total magnification for each section, ×90.
A representative necrotic granuloma from patient TB10 consisting of a central region of caseous necrosis surrounded by a lymphocyte cuff is shown in Fig. 1G to L. Between the region of caseous necrosis and the lymphocyte cuff is the transition zone. This granuloma was positive for icl and narX mRNAs only in the lymphocyte cuff (Fig. 1G and H, respectively). No signal was detected for icl or narX mRNA within the transition zone or within the region of necrosis (Fig. 1G and H, inset). The same granuloma was also positive for Rv2557*, kasA, and iniB mRNAs, with the brown signal being present in the lymphocyte cuff and within the transition zone of the granuloma (Fig. 1I, J, and K, as shown in the inset). No Rv2557*, kasA, or iniB mRNA signal was detected within the central necrotic area of granulomas. icl, narX, Rv2557*, kasA, and iniB mRNAs were always associated with granulomas positive for M. tuberculosis DNA. No signal was obtained when the sense riboprobes were used; the kasA sense probe is shown in Fig. 1F and L, thus proving the specificity of the binding of the probes.
Expression of mycobacterial genes within granulomas at high-power magnification.
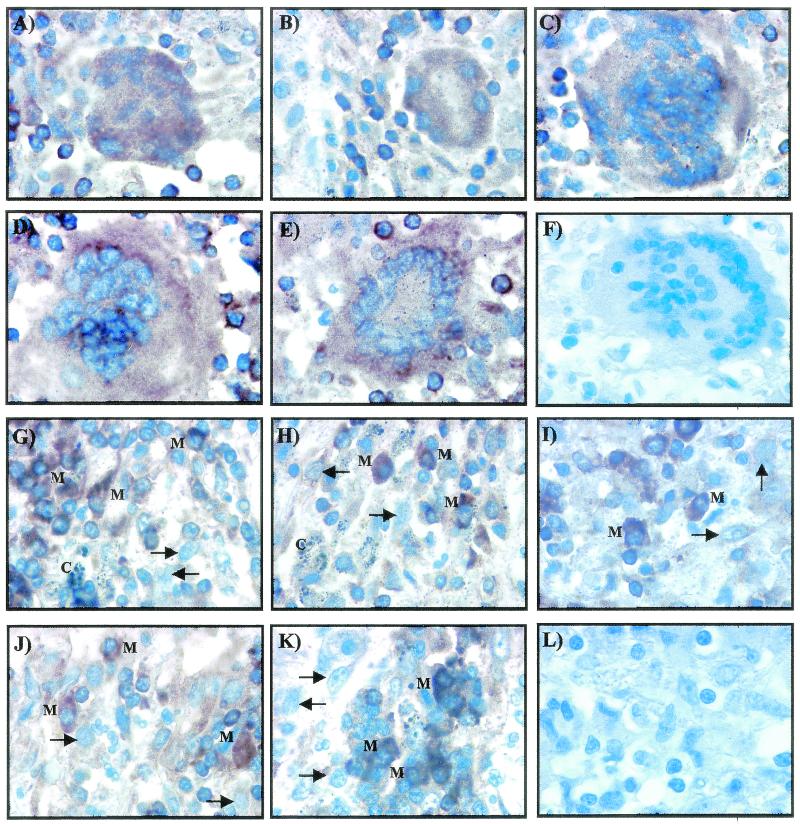
Cells present within the boundaries of the lymphocyte cuff were also found to contain transcripts of icl, narX, Rv2557*, kasA, and iniB mRNAs as visualized by the brown signal (Fig. 2). Giant cells (Fig. 2A to E) and isolated cells with the morphology of macrophages (Fig. 2G to K) were positive for icl, narX, Rv2557*, kasA, and iniB mRNAs. Not all cells with the morphology of macrophages were positive for the different mycobacterial mRNAs, and these stained blue. The sense probes were negative for any hybridization signals, as shown by the signal obtained with the sense riboprobe in Fig. 2F and L.
FIG. 2.
High-power magnification of nonnecrotic and necrotic granulomas demonstrate that giant cells and individual cells with the morphology of macrophages contain transcripts detected by icl, narX, Rv2557*, kasA, and iniB probes. RNA-RNA in situ hybridization shows that giant cells are positive for icl (A), narX (B), Rv2557* (C), kasA (D), and iniB (E). Cells with the morphology of macrophages are also positive for icl (G), narX (H), Rv2557* (I), kasA (J), and iniB (K). Positive macrophages are indicated with an “M,” and negative macrophages are indicated with an arrow. The positive signal is indicated by the brown color; the negative signal is blue. (F and L) No signal was observed with the sense probes, and a representative giant cell and macrophages are shown. Total magnification for each section, ×900.
CD68-positive cells colocalize to icl, narX, Rv2557*, kasA, and iniB mRNAs.
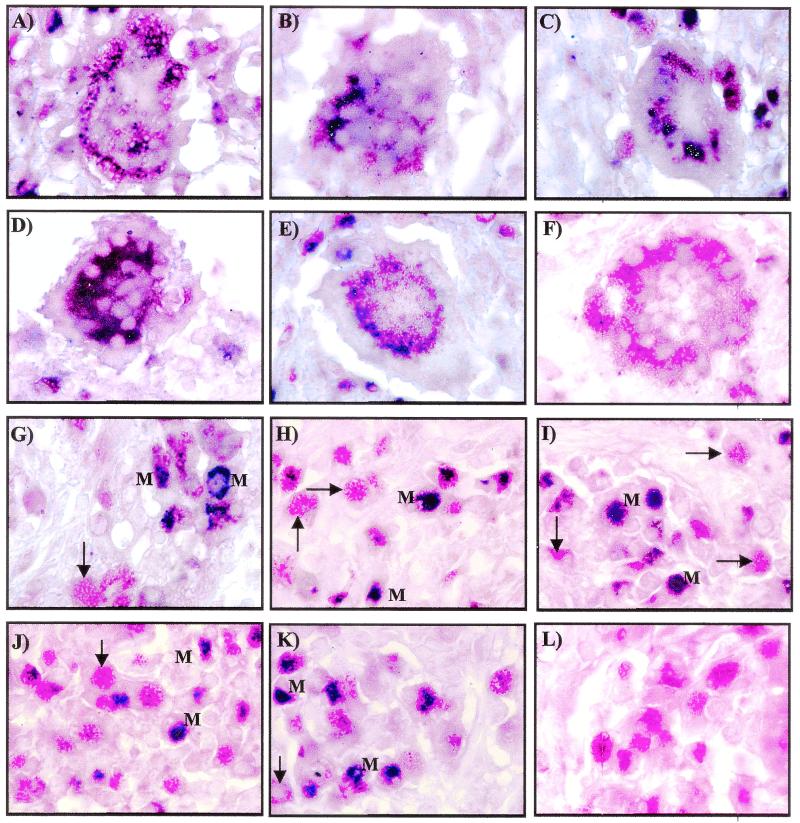
A dual-labeling technique combining immunohistochemistry for CD68 and in situ hybridization for icl, narX, Rv2557*, kasA, and iniB mRNAs was used to immunologically identify the cell types containing the mycobacterial mRNA. Giant cells (Fig. 3A to E) and individual macrophages (Fig. 3G to K) show colocalization of the macrophage cell marker CD68 (red) with icl, narX, Rv2557*, kasA, and iniB mRNAs (blue). All of the mycobacterial mRNA species investigated in the present study colocalized to CD68-positive cells as visualized by the blue and red signals hybridizing to the same cells. Not all CD68-positive cells contained mycobacterial mRNA, and these stained red only. Immunohistochemistry for CD68 performed in the absence of any of the different riboprobes produced a red color only (Fig. 3F and L).
FIG. 3.
RNA-RNA in situ hybridization (blue color) and immunohistochemistry for CD68 (red color) shows that CD68-positive cells colocalize to the different mycobacterial mRNAs. CD68-positive giant cells colocalize to icl (A), narX (B), Rv2557/8 (C), kasA (D), and iniB (E). (F) A giant cell staining red for CD68 only is shown. CD68-positive cells with the morphology of macrophages were also positive for icl (G), narX (H), Rv2557/8 (I), kasA (J), and iniB (K). Not all CD68-positive cells were positive for the various mycobacterial mRNAs, as shown with arrows; those that were positive are indicated with an “M.” (L) Immunohistochemistry performed in the absence of in situ hybridization produced CD68-positive cells which stained red only. Total magnification for each section, ×900.
DISCUSSION
In our previous study, we demonstrated that in situ hybridization could be used to detect mycobacterial DNA and rpoB mRNA in paraffin-embedded lung tissue from patients with TB (8). We extended the present study to monitor the expression of genes proposed to be important for the survival of mycobacteria.
M. tuberculosis DNA and mRNA were detected in all seven study patients, whereas M. tuberculosis bacteria were seen in only five patients by ZN staining. M. tuberculosis DNA was associated with CD68-positive macrophages and giant cells within all types of granulomas as well as in the transition zone of necrotic granulomas. M. tuberculosis DNA was also localized to the center of necrotic granulomas where no cells could be identified. These results are in agreement with previous findings that have demonstrated the presence of mycobacterial proteins or DNA in the absence of acid-fast bacilli (12, 18, 19).
Using in situ hybridization, we identified icl mRNA in 61% of the granulomas studied. No icl mRNA was detected in the transition zone or the central necrotic region of necrotic granulomas. By combining immunohistochemistry for the macrophage marker CD68 and in situ hybridization for icl mRNA, icl mRNA was shown to colocalize to CD68-positive cells within the lymphocyte cuff, on the outer edges of necrotic granulomas and within nonnecrotic granulomas. icl is upregulated during in vitro infection of macrophages and is important in promoting bacterial survival in activated macrophages (23). The present study confirms that icl is expressed within macrophages at the site of pathology in human tuberculous lungs. icl-deficient M. tuberculosis has been studied in activated macrophages from gamma interferon and TNF-α knockout mice, and the requirement for icl has been shown to depend on the immune status of the host cell (23). We have previously demonstrated that each granuloma is a microenvironment with expression of a unique set of cytokines (9, 10). The expression of icl in some granulomas and not others may reflect the heterogeneity of the immune response. Studies are under way to explore this hypothesis. In an in vitro model of persistence involving oxygen deprivation, isocitrate lyase activity increases as oxygen becomes limiting (35). However, in the granuloma itself, icl is expressed in association with CD68-positive myeloid cells throughout nonnecrotic granulomas and also in the lymphocyte cuff of necrotic granulomas. It is assumed that oxygen availability decreases from the outer edge of the granuloma toward the center, with the central region of necrosis being the most severely depleted of oxygen. The icl gene is not expressed in the central necrotic area or in the transition zone, implying that oxygen starvation may not be the primary inducer of icl expression. It seems likely that the factors causing upregulation of icl are very complex and oxygen tension may not be the only signal.
The narX gene localized to CD68-positive cells and giant cells within nonnecrotic granulomas and in the lymphocyte cuff of necrotic granulomas. No narX mRNA was detected in the transition zone or within the central necrotic region of necrotic granulomas, despite the presence of mycobacterial DNA. This expression pattern is similar to that observed for icl and also rpoB, which we had previously shown to be absent from this region (8). However, narX was present in only 7% of granulomas, whereas icl expression was observed in 61% of granulomas. The narX gene was upregulated in anaerobic cultures of BCG (20), and therefore we expected it to be expressed in the areas of the granuloma most inaccessible to oxygen. Failure to observe narX implies that the gene is not expressed or that its expression is below the detection limit of this technique. Alternatively, the mycobacterial DNA that is present is debris from bacteria that have been killed by the hostile necrotic environment.
We cannot distinguish between Rv2557 and Rv2558 expression due to their high degree of homology. We have shown that one or both genes are expressed in necrotic and nonnecrotic granulomas and also colocalize to CD68-positive cells and giant cells. Rv2557 and/or Rv2558 mRNA was associated with 95% of granulomas, but its distribution followed a granuloma- specific pattern. In nonnecrotic granulomas, the Rv2557* probe detected transcripts in cells throughout the granuloma- whereas in necrotic granulomas, cells containing Rv2557 and/or Rv2558 transcripts were found in both the lymphocyte cuff and the transition zone. This may indicate that Rv2557and/or Rv2558 expression is required during the adaptive response of the bacillus from an intracellular to a more hostile extracellular necrotic environment. Since neither icl, narX, nor rpoB was expressed in this region, Rv2557 and/or Rv2558 expression may be indicative of a further step in the metabolic shutdown required for persistence.
All of the patients included in the present study were receiving TB treatment at the time of their lung resections, and therefore it was decided to investigate the expression of kasA and iniB, both of which have been shown to be induced by isoniazid in vitro (1, 30, 36). The genes were expressed widely, and their expression patterns were similar in the granulomas. kasA mRNA was present in 81% of granulomas, and iniB was present in 95%. Transcripts of both genes were observed in CD68-positive cells and in giant cells within nonnecrotic granulomas, whereas in necrotic granulomas kasA and iniB mRNAs were detected both in the lymphocyte cuff and in the transition zone within CD68-positive macrophages. Transcripts were not detected in the regions of necrosis, where no intact host cells could be discerned. The presence of kasA and iniB transcripts within the transition zone of necrotic granulomas may imply that these genes are upregulated in these isoniazid-treated patients in agreement with the in vitro observations of Wilson et al. (36).
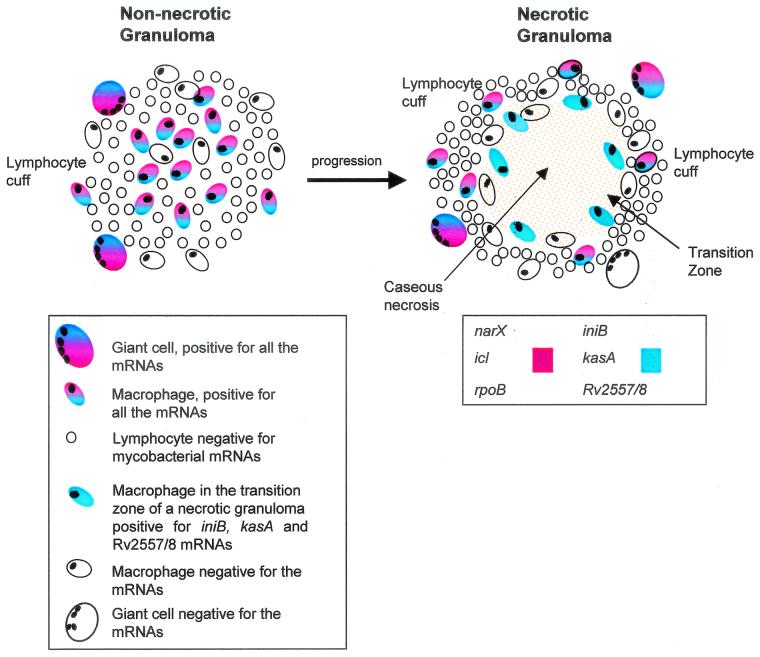
It is now possible to define spatially distinct regions of granulomas in terms of mycobacterial gene expression (Fig. 4). Nonnecrotic granulomas have a lymphocyte cuff that is interspersed with cells with the morphology of macrophages and giant cells containing mycobacteria. Not all macrophages and giant cells were positive for mycobacterial transcripts. However, mycobacterial mRNA always localized to CD68-positive cells, unlike the mycobacterial DNA that was also found in the acellular region of the necrosis. The reason why certain CD68-positive cells are negative for mycobacterial expression is unknown, but this may reflect the activation state of the macrophage and the surrounding lymphocytes. There was no “zonation” of mycobacterial gene expression in this type of granuloma.
FIG. 4.
Schematic diagram of mycobacterial gene expression within a nonnecrotic tuberculous granuloma as it progresses and becomes necrotic. In a nonnecrotic granuloma consisting predominantly of macrophages, lymphocytes, and giant cells, mycobacteria expressing rpoB, narX, icl, Rv2557 and/or Rv2558, iniB, and kasA mRNAs were detected by using nonradioactive in situ hybridization. This nonnecrotic granuloma was also positive for M. tuberculosis DNA. As the granuloma progresses from a nonnecrotic to a necrotic state, the microenvironment changes, macrophages apoptose, and the bacteria slow their growth rate. The lymphocyte cuff of the necrotic granuloma contains a number of giant cells and isolated macrophages positive for the various mycobacterial genes. The transition zone of the necrotic granuloma still contains a small number of macrophages positive for Rv2557 and/or Rv2558, iniB, and kasA mRNA. However, the center of the necrosis is devoid of macrophages and mycobacterial gene expression but does contain M. tuberculosis DNA, as shown by DNA-DNA in situ hybridization.
In necrotic granulomas, three distinct regions can be classified. First, there is the lymphocyte cuff, which again is interspersed with macrophages and giant cells containing mycobacteria expressing all of the genes examined in the present study. The transition zone between the lymphocyte cuff and region of the necrosis contains a number of cell types, including macrophages harboring mycobacteria that express Rv2557 and/or Rv2558, kasA, and iniB only. The presence of Rv2557 and/or Rv2558, kasA, and iniB within the transition zone may be indicative of a stress response, since the bacilli sense the more necrotic, cell-deficient areas of the granuloma. Alternatively, these genes may indicate a change in microbial metabolism, allowing survival in the hostile necrotic environment at the center of the granuloma. The transition zone can thus be said to be a restrictive environment in which nutrients and oxygen may be limiting, and this may cause upregulation of Rv2557 and/or Rv2558, kasA, and iniB. These genes are not detected in the region of central necrosis, and this may indicate bacterial death or mRNA levels below the limits of detection of this technology. A different set of genes may be upregulated in the central necrotic region. Macrophages within the transition zone might be undergoing apoptosis (7), possibly due to increased ATP from lysed adjacent cells (6, 22) or the activity of cytotoxic CD8+ T cells (31), and this may influence mycobacterial gene expression. The innermost region, consisting of caseous necrotic areas, is devoid of intact human cells. Although we do observe mycobacterial DNA and in some cases a few acid-fast bacilli in this region, we have not seen any evidence of transcription of the genes we have examined.
In summary, we have demonstrated the presence of mRNA from M. tuberculosis genes thought to be involved in persistence and drug response in human tuberculous granulomas. The specific distribution of gene expression suggests that different microenvironments exist within granulomas to which the bacilli are responsive, adopting differing metabolic states. This ability to adapt to these various microenvironments may enable them to survive the host immune system, as well as the stress of chemotherapeutic agents, and thus persist for long periods of time.
Acknowledgments
This work was funded by the GlaxoSmithKline Action TB Initiative program, the Medical Research Council of South Africa, and the University of Stellenbosch.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Alland, D., I. Kramnik, T. R. Weisbrod, L. Otsubo, R. Cerny, L. P. Miller, W. R. Jacobs, Jr., and B. R. Bloom. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., A. J. Steyn, T. Weisbrod, K. Aldrich, and W. R. Jacobs, Jr. 2000. Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 182:1802-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Dannenberg, A. M., and G. A. Rook. 1994. Pathogenesis of pulmonary tuberculosis: an interplay of tissue-damaging and macrophage-activating immune responses—dual mechanisms that control bacillary multiplication, p. 459-483. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 5.Desjardin, L. E., M. D. Perkins, K. Wolski, S. Haun, L. Teixeira, Y. Chen, J. L. Johnson, J. J. Ellner, R. Dietze, J. Bates, M. D. Cave, and K. D. Eisenach. 1999. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am. J. Respir. Crit. Care Med. 160:203-210. [DOI] [PubMed] [Google Scholar]
- 6.Fairbairn, I. P., C. B. Stober, D. S. Kumararatne, and D. A. Lammas. 2001. ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X(7)-dependent process inducing bacterial death by phagosome-lysosome fusion. J. Immunol. 167:3300-3307. [DOI] [PubMed] [Google Scholar]
- 7.Fayyazi, A., B. Eichmeyer, A. Soruri, S. Schweyer, J. Herms, P. Schwarz, and H. J. Radzun. 2000. Apoptosis of macrophages and T cells in tuberculosis associated caseous necrosis. J. Pathol. 191:417-425. [DOI] [PubMed] [Google Scholar]
- 8.Fenhalls, G., L. Stevens-Muller, R. Warren, N. Carroll, J. Bezuidenhout, P. van Helden, and P. Bardin. 2002. Localisation of mycobacterial DNA and mRNA in human tuberculous granulomas. J. Microbiol. Methods 51:197-208. [DOI] [PubMed]
- 9.Fenhalls, G., L. Stevens, J. Bezuidenhout, G. E. Amphlett, K. Duncan, P. Bardin, and P. T. Lukey. 2002. Distribution of IFN-γ, IL-4 and TNF-α protein and CD8 T cells producing IL-12p40 mRNA in human lung tuberculous granulomas. Immunology 105:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenhalls, G., A. Wong, J. Bezuidenhout, P. van Helden, P. Bardin, and P. T. Lukey. 2000. In situ production of gamma interferon, interleukin-4, and tumor necrosis factor alpha mRNA in human lung tuberculous granulomas. Infect. Immun. 68:2827-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn, J. L., M. M. Goldstein, J. Chan, K. J. Triebold, K. Pfeffer, C. J. Lowenstein, R. Schreiber, T. W. Mak, and B. R. Bloom. 1995. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2:561-572. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie, J., L. L. Barton, and E. W. Rypka. 1986. Phenotypic changes in mycobacteria grown in oxygen-limited conditions. J. Med. Microbiol. 21:251-255. [DOI] [PubMed] [Google Scholar]
- 13.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellyer, T. J., L. E. Desjardin, L. Teixeira, M. D. Perkins, M. D. Cave, and K. D. Eisenach. 1999. Detection of viable Mycobacterium tuberculosis by reverse transcriptase-strand displacement amplification of mRNA. J. Clin. Microbiol. 37:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höner Zu Bentrup, K., A. Miczak, D. L. Swenson, and D. G. Russell. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181:7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höner Zu Bentrup, K., and D. G. Russell. 2001. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9:597-605. [DOI] [PubMed] [Google Scholar]
- 17.Hu, Y. M., P. D. Butcher, K. Sole, D. A. Mitchison, and A. R. Coates. 1998. Protein synthesis is shutdown in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol. Lett. 158:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Hulten, K., H. M. El Zimaity, T. J. Karttunen, A. Almashhrawi, M. R. Schwartz, D. Y. Graham, and F. A. El Zaatari. 2001. Detection of Mycobacterium avium subspecies paratuberculosis in Crohn's diseased tissues by in situ hybridization. Am. J. Gastroenterol. 96:1529-1535. [DOI] [PubMed] [Google Scholar]
- 19.Humphrey, D. M., and M. H. Weiner. 1987. Mycobacterial antigen detection by immunohistochemistry in pulmonary tuberculosis. Hum. Pathol. 18:701-708. [DOI] [PubMed] [Google Scholar]
- 20.Hutter, B., and T. Dick. 1999. Upregulation of narX, encoding a putative ‘fused nitrate reductase' in anaerobic dormant Mycobacterium bovis BCG. FEMS Microbiol. Lett. 178:63-69. [DOI] [PubMed] [Google Scholar]
- 21.Keane, J., S. Gershon, R. Wise, E. Mirabile-Levens, J. Kasznica, W. D. Schwieterman, J. N. Siegel, and M. M. Braun. 2001. Tuberculosis associated with infliximab, a tumor necrosis factor alpha neutralizing agent. New Engl. J. Med. 345:1098-1104. [DOI] [PubMed] [Google Scholar]
- 22.Kusner, D. J., and J. A. Barton. 2001. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome-lysosome fusion. J. Immunol. 167:3308-3315. [DOI] [PubMed] [Google Scholar]
- 23.McKinney, J. D., K. Höner Zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 24.Parrish, N. M., J. D. Dick, and W. R. Bishai. 1998. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107-112. [DOI] [PubMed] [Google Scholar]
- 25.Patel, B. K., D. K. Banerjee, and P. D. Butcher. 1993. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat-shock protein mRNA. J. Infect. Dis. 168:799-800. [DOI] [PubMed] [Google Scholar]
- 26.Raupach, B., and S. H. E. Kaufmann. 2001. Immune responses to intracellular bacteria. Curr. Opin. Immunol. 13:417-428. [DOI] [PubMed] [Google Scholar]
- 27.Roach, D. R., A. G. Bean, C. Demangel, M. P. France, H. Briscoe, and W. J. Britton. 2002. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J. Immunol. 168:4620-4627. [DOI] [PubMed] [Google Scholar]
- 28.Roth, S., E. Delmont, P. Heudier, R. Kaphan, E. Cua, J. Castela, J. M. Verdier, R. M. Chichmanian, and J. G. Fuzibet. 2002. Anti-TNF alpha monoclonal antibodies (Infliximab) and tuberculosis: apropos of 3 cases. Rev. Med. Interne 23:312-316. [DOI] [PubMed] [Google Scholar]
- 29.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slayden, R. A., and C. E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659-669. [DOI] [PubMed] [Google Scholar]
- 31.Stenger, S., D. A. Hanson, R. Teitelbaum, P. Dewan, K. R. Niazi, C. J. Froelich, T. Ganz, S. Thoma-Uszynski, A. Melian, C. Bogdan, S. A. Porcelli, B. R. Bloom, A. M. Krensky, and R. L. Modlin. 1998. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282:121-125. [DOI] [PubMed] [Google Scholar]
- 32.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wayne, L. G., and L. G. Hayes. 1998. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuberc. Lung Dis. 79:127-132. [DOI] [PubMed] [Google Scholar]
- 34.Wayne, L. G., and K. Y. Lin. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]