Abstract
Functionally required conformational plasticity of transmembrane proteins implies that specific structural motifs have been integrated in transmembrane helices. Surveying a database of transmembrane helices and the large family of G-protein coupled receptors we identified a series of overrepresented motifs associating Pro with either Ser or Thr. Thus, we have studied the conformation of Pro-kinked transmembrane helices containing Ser or Thr residues, in both g+ and g− rotamers, by molecular dynamics simulations in a hydrophobic environment. Analysis of the simulations shows that Ser or Thr can significantly modulate the deformation of the Pro. A series of motifs, such as (S/T)P and (S/T)AP in the g+ rotamer and the TAP and PAA(S/T) motifs in the g− rotamer, induce an increase in bending angle of the helix compared to a standard Pro-kink, apparently due to the additional hydrogen bond formed between the side chain of Ser/Thr and the backbone carbonyl oxygen. In contrast, (S/T)AAP and PA(S/T) motifs, in both g+ and g−, and PAA(S/T) in g+ rotamers decrease the bending angle of the helix by either reducing the steric clash between the pyrrolidine ring of Pro and the helical backbone, or by adding a constrain in the form of a hydrogen bond in the curved-in face of the helix. Together with a number of available experimental data, our results strongly suggest that association of Ser and Thr with Pro is commonly used in transmembrane helices to accommodate the structural needs of specific functions.
INTRODUCTION
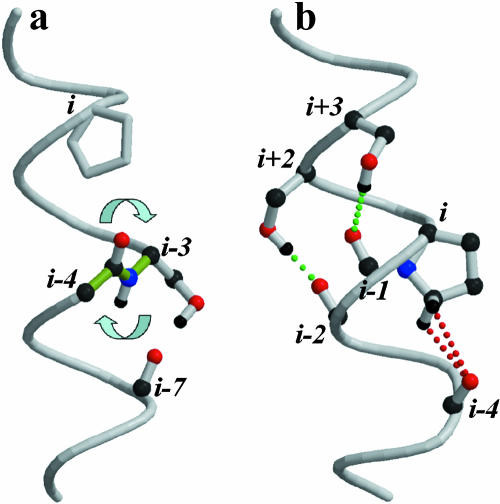
Membrane receptors and channels are integral membrane proteins composed of several transmembrane (TM) α-helices that assemble through tertiary or quaternary structures to form a bundle that crosses the lipid bilayer (Jiang et al., 2003; Palczewski et al., 2000). Biological function of these proteins involves conformational rearrangement of this transmembrane bundle. For example, it is thought that activation of G-protein coupled receptors (GPCRs) requires rigid-body motions of several if not all TM helices (Farrens et al., 1996). Such conformational changes require local flexibility in α-helices, which can, for instance, be provided by proline residues within the helix (Sansom and Weinstein, 2000). Although Pro has the least helix-forming tendency (O'Neil and DeGrado, 1990) and the highest turn-stabilizing tendency in the membrane (Monne et al., 1999) of all naturally occurring amino acids, Pro residues are normally observed in TM helices (Senes et al., 2000) where they usually induce a significant distortion named Pro-kink (Von Heijne, 1991). The break is produced to avoid a steric clash between the pyrrolidine ring of Pro (at position i) and the carbonyl oxygen of the residue in the preceding turn (position i-4) (Chakrabarti and Chakrabarti, 1998) leading to a bending of the helical structure (Cordes et al., 2002). Furthermore, the absence of the backbone N-H group in Pro prevents the formation of the hydrogen bond with the backbone carbonyl oxygen of residue i-4. The role of the N-H group is partly fulfilled by the CδH atoms of Pro, which hydrogen bond a backbone carbonyl oxygen usually located three, four, or five residues preceding the Pro (Chakrabarti and Chakrabarti, 1998). This type of Cδ-H···O=C interaction can help to stabilize the local break. Moreover, due to the distortions of the Pro-kink, the Ni+1-H···O=Ci-3 hydrogen bond is frequently disrupted, and the Ni-1-H···O=Ci-5 hydrogen bond is sometimes replaced by the Ni-1-H···O=Ci-4 hydrogen bond (Chakrabarti and Chakrabarti, 1998). Therefore, in the context of an α-helix, Pro not only induces a break in the helix, but it also destabilizes the hydrogen bond network that normally stabilizes the secondary structure. As a result, Pro introduces a flexible point in the α-helix, which could be of functional importance for membrane proteins (Gether, 2000).
We and others have shown that, in transmembrane proteins, specific sequence motifs preceding the Pro can provide structural properties for α-helices that are significantly different than those of a regular Pro-kink (Govaerts et al., 2001; Ri et al., 1999). These motifs include Ser and Thr residues in the vicinity of the Pro, which induce a structural modification of the Pro-kink through hydrogen bonds of their polar side chains. Experimental studies involving site-directed mutagenesis of the residues forming these motifs have demonstrated the functional importance of the Ser and Thr residues in Pro-containing α-helices (Govaerts et al., 2001; Peralvarez et al., 2001; Ri et al., 1999).
Therefore the question arises as whether combinations of Ser/Thr and Pro are generically used as structural modulators in integral membrane proteins. To address this issue we have systematically surveyed sequences of TM helices to analyze the frequencies of such combinations. Moreover, the conformation of Pro-kinked TM helices containing Ser or Thr residues was assessed by molecular dynamics (MD) simulations in a hydrophobic environment. Our study indicates that membrane proteins, and GPCRs in particular, favor combined motifs of Pro and Ser or Thr in their TM helices and that these motifs have specific structural properties. This suggests that transmembrane proteins have evolved these specific motifs for functional purposes.
METHODS
MD simulations were performed on the model peptides Ace-(Ala)10-X-(Ala)11-NHMe, where X is either AAAP, AASP, AATP, ASAP, ATAP, SAAP, or TAAP; and Ace-(Ala)13-X-(Ala)8-NHMe, where X is either PSAA, PTAA, PASA, PATA, PAAS, or PAAT. These model peptides were built in the standard Pro-kinked α-helix conformation (Table 2 in Olivella et al. (2002)). The side chains of Ser or Thr were built on both the χ1 = g+ and g− rotamer conformations.
TABLE 2.
Percentage of occurrences of the (S/T)xxP, (S/T)xP, (S/T)P, P(S/T), Px(S/T), and Pxx(S/T) motifs in the rhodopsin-like family of G-protein coupled receptors
| Position* | Number of occurrences of Pro | SxxP | TxxP | SxP | TxP | SP | TP | PS | PT | PxS | PxT | PxxS | PxxT | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.36 | 378 | 2 | 4 | 0 | 1 | 5 | 0 | 6 | 12 | 2 | 1 | 3 | 0 | 35 |
| 1.48 | 203 | 0 | 13 | 9 | 0 | 0 | 12 | 2 | 1 | 1 | 0 | 3 | 2 | 44 |
| 2.58 | 546 | 6 | 1 | 10 | 71 | 0 | 1 | 0 | 1 | 1 | 0 | 5 | 2 | 100 |
| 2.59 | 875 | 6 | 16 | 3 | 15 | 5 | 6 | 2 | 3 | 16 | 10 | 1 | 3 | 86 |
| 2.60 | 103 | 5 | 0 | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 8 | 28 | 47 |
| 4.59 | 1139 | 2 | 1 | 42 | 11 | 6 | 4 | 3 | 3 | 0 | 2 | 2 | 0 | 76 |
| 4.60 | 634 | 34 | 12 | 3 | 2 | 1 | 3 | 1 | 1 | 1 | 2 | 5 | 0 | 66 |
| 5.50 | 1588 | 0 | 1 | 3 | 3 | 0 | 3 | 2 | 4 | 11 | 9 | 1 | 5 | 40 |
| 6.50 | 1603 | 8 | 3 | 1 | 0 | 3 | 15 | 1 | 0 | 4 | 4 | 7 | 4 | 51 |
| 6.59 | 97 | 0 | 6 | 1 | 4 | 6 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 21 |
| 7.38 | 235 | 3 | 8 | 9 | 53 | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 3 | 94 |
| 7.45 | 101 | 13 | 4 | 0 | 13 | 0 | 0 | 3 | 0 | 2 | 4 | 0 | 0 | 39 |
| 7.46 | 325 | 1 | 10 | 0 | 0 | 2 | 40 | 4 | 4 | 0 | 0 | 1 | 0 | 62 |
| 7.50 | 1888 | 5 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 9715 | 6 | 5 | 7 | 9 | 2 | 6 | 2 | 2 | 4 | 3 | 2 | 2 | 52 |
Positions in the generalized numbering scheme of (Ballesteros and Weinstein, 1995).
These initial structures were placed in a rectangular box (∼62 Å × 55 Å × 55 Å in size) containing methane molecules (∼4200 molecules in addition to the TM α-helix) to mimic the membrane environment, at a density approaching half of hydrocarbons in lipid bilayer. This is due to the different equilibrium distance between carbons in the methane box and in the polycarbon chain of the lipid (Olivella et al., 2002). The peptide-methane systems were subjected to 500 iterations of energy minimization, and then heated to 300 K in 15 ps. This was followed by an equilibration period (15–500 ps), and a production run (500–1500 ps). The simulations were carried out at constant volume and constant temperature (300 K), with the latter maintained through coupling to a heat bath. The particle mesh Ewald method was employed to compute electrostatic interactions (Darden et al., 1993). Structures were collected for analysis every 10 ps during the last 1000 ps of simulation (100 structures). Bending angles of these structures were calculated as the angle between the axes computed as the least square lines through the backbone atoms (N, Cα, C) of the residues before (from 2 to 11) and after (from 16 to 24) the Pro-kink, using the InsightII software (Accelrys, San Diego, CA). One-way analysis of variance plus a posteriori two-sided Dunnett's t-tests was employed, with the SPSS 11 program (SPSS Inc. Chicago, IL), to contrast if the bend angle of the model peptides differs from the control. It is important to remark that this statistical test can only be applied to independent samples. To achieve this prerequisite, structures were collected for analysis every 10 ps so that a given structure is not related to the previous and succeeding structures (Lyapunov instability) (Frenkel and Smit, 1996). The unit twist angles of these structures were calculated, for each set of four contiguous Cα atoms along the α-helix, using the program HELANAL (Bansal et al., 2000). Representative structures for each trajectory were selected by automatically clustering the collected geometries into conformationally related subfamilies with the program NMRCLUST (Kelley et al., 1996). These structures can be downloaded from http://lmc.uab.es/SerThrPro/structures.htm. The MD simulations were run with the Sander module of AMBER 5 (Case et al., 1997), using an all-atom force field (Cornell et al., 1995), the SHAKE bond constraints on all bonds, and a 2-fs integration time step.
RESULTS
Statistical analysis of the presence of Ser/Thr residues in Pro-kinked transmembrane helices
Table 1 lists the observed number of occurrences of the (S/T)xxP, (S/T)xP, (S/T)P, P(S/T), Px(S/T), and Pxx(S/T) patterns in a nonhomologous database of sequences of TM helices (Senes et al., 2000). To find overrepresented and underrepresented patterns we calculated the expected number of occurrences with the TMSTAT formalism that accounts for both the relative frequency of the amino acids and length of the sequence (Senes et al., 2000) (see http://bioinfo.mbb.yale.edu/tmstat/). We used a p-value cutoff of 0.10 to select statistically significant overrepresented (odds ratio >1) and underrepresented (odds ratio <1) patterns. Both TxP (odds ratio of 1.10, p-value of 0.07) and SxP (1.12, 0.02) motifs are overrepresented pairs, suggesting that (S/T)xP is a common pattern in TM helices. In contrast, the SP (0.87, 0.01), PS (0.84, 0.001), and PxxS (0.84, 0.004) are underrepresented pairs.
TABLE 1.
Observed and expected, calculated with the TMSTAT formalism, number of occurrences of the (S/T)xxP, (S/T)xP, (S/T)P, P(S/T), Px(S/T), and Pxx(S/T) motifs in a nonhomologous database of transmembrane helices; and the odds ratio and its significance
| Pair | Observed | Expected | Odds ratio | Significance |
|---|---|---|---|---|
| SxxP | 292 | 294.9 | 0.99 | 0.88 |
| TxxP | 267 | 268.7 | 0.99 | 0.95 |
| SxP | 353 | 314.6 | 1.12 | 0.02 |
| TxP | 316 | 286.6 | 1.10 | 0.07 |
| SP | 291 | 334.2 | 0.87 | 0.01 |
| TP | 325 | 304.6 | 1.07 | 0.21 |
| PS | 280 | 334.2 | 0.84 | 0.001 |
| PT | 308 | 304.6 | 1.01 | 0.83 |
| PxS | 297 | 314.6 | 0.94 | 0.31 |
| PxT | 283 | 286.6 | 0.99 | 0.85 |
| PxxS | 249 | 294.9 | 0.84 | 0.004 |
| PxxT | 250 | 268.7 | 0.93 | 0.23 |
Statistical analysis of the presence of Ser/Thr residues in Pro-kinked transmembrane helices of the rhodopsin-like family of GPCRs
GPCRs, or 7TM receptors, are one of the largest families of membrane proteins with more than 800 human sequences (Venter et al., 2001). These receptors have been classified into five main families: glutamate, rhodopsin, adhesion, frizzled/taste2, and secretin (Fredriksson et al., 2003). The members of the rhodopsin family (the largest subgroup) and bovine rhodopsin, for which the structure is known (Palczewski et al., 2000), share a large number of conserved sequence patterns in the TM segments. This allows for a straightforward identification of TM helices and provides a large set to survey the association of Ser/Thr and Pro residues. Table 2 shows the positions, in the generalized numbering scheme of Ballesteros & Weinstein (Ballesteros and Weinstein, 1995), and the number of occurrences of Pro residues within the TM helices of the 1948 GPCR sequences denoted as Class A rhodopsin-like in GPCR database (Horn et al., 2003), as of September, 2002 release (6.1). We found a total of 9715 Pro residues, providing a very large statistical sample of Pro-kinked TM helices. Whereas Pro are found at many locations throughout the family, a typical TM helix does not contain more than one Pro-kink (except in TM 4). Partly or highly conserved prolines (>5% of the sequences) can be found in TM 1 (positions 1.36 and 1.48), TM 2 (2.58, 2.59, and 2.60), TM 4 (4.59 and 4.60) TM 5 (highly conserved 5.50), TM 6 (highly conserved at position 6.50 and occasionally at 6.59) and TM 7 (7.38, 7.45, 7.46, and 7.50, which belongs to the highly conserved NPxxY motif). Table 2 also shows the probability of finding a Ser or Thr residue in the direct vicinity of the Pro, namely at positions i-3 (S/T)xxP, i-2 (S/T)xP, i-1 (S/T)P, i+1 P(S/T), i+2 Px(S/T), and i+3 Pxx(S/T) relative to Pro.
The statistical analysis shows that 52% of the Pro-containing TM helices contain either Ser or Thr in the vicinity of the proline. The most common combinations are the (S/T)xP motifs, found in 16% of all Pro-kinks in GPCRs (9% for TxP, 7% for SxP). Notably, the distribution of TxP and SxP is not uniform over the different Pro-kinks, as some specific locations, such as 2.58, 4.59, and 7.38, show much higher frequencies than the others. Moreover, Ser and Thr are not equally found. For example, Pro2.58 predominantly associates with a Thr (forming a TxP2.58 motif) whereas Pro4.59 displays four times more SxP4.59 than TxP4.59 motifs. This indicates that Ser and Thr are not equivalent and already suggests, because their hydrogen-bonding propensities are similar, that the methyl group differentiating their side chains may have a defined structural or functional role (see below). Additionally, SP motifs (2%) are observed at lower frequency than TP motifs (6%) further suggesting different roles for these two side chains. Finally, the frequency of Ser/Thr residues at positions preceding Pro (35% of the Pro-kinks) is larger than at positions after Pro (16% of the Pro-kinks).
Conformation of Pro-kinked transmembrane helices containing Ser or Thr residues at positions from i-3 to i+3 relative to Pro
The strong association of Ser/Thr residues with Pro in TM helices, described above, suggests a possible structural role. Due to the paucity of membrane protein structures, it is not possible to investigate such structural role by surveying TM helices in known crystal structures. We have previously shown that structural characteristics of TM helices (including Pro-containing helices) can be accurately reproduced by MD simulations (Olivella et al., 2002). Thus, we have used a similar approach here to assess the conformation of model peptides containing the (S/T)AAP, (S/T)AP, (S/T)P, P(S/T), PA(S/T), and PAA(S/T) patterns, with Ser and Thr side chains built in both χ1 = gauche+ (g+) or gauche− (g−) conformation (see Methods). In α-helices, the side chains of Ser and Thr adopt primarily the g+ or g− where the hydrogen-bonding capacity of their OγH side chain can be satisfied by interacting with the backbone carbonyl oxygen in the preceding turn of the helix (Gray and Matthews, 1984). In contrast, the trans (t) conformation is not stable because it cannot form such hydrogen bond as it points the OγH group away from the backbone. The control of these simulations was the simulation of a Pro-containing polyAla α-helix (see Methods). We monitored the distortions caused by these patterns in the α-helix by the hydrogen bond interactions in the Pro-kink turn (Figs. 1 and 2), the bending angle (Table 3), and the unit twist angles (Fig. 3) (see Methods). The results of the simulations are presented separately for each motif. The notation employed in the manuscript assigns the i position to Pro and the j position to Ser/Thr, and all other amino acids are named relative to these reference points. Moreover, the distribution, in percentage, of the intrahelical hydrogen bond between the OγH side chain of Ser/Thr and the carbonyl oxygen at positions j-4 and j-3 as observed during the MD simulations, is also shown in Table 3.
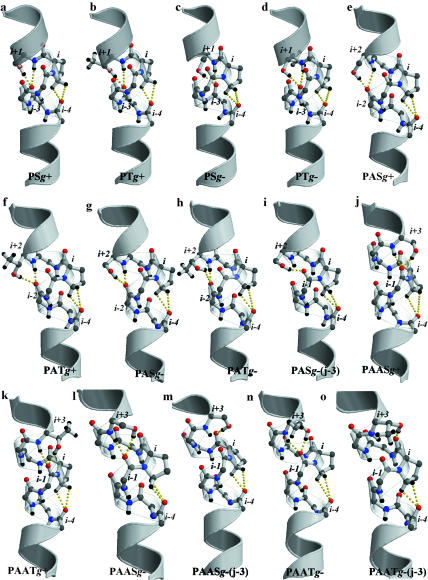
FIGURE 1.
Detailed view of the hydrogen bond pattern, in the Pro-kink turn, of model peptides containing Pro and the (S/T)P, (S/T)AP, and (S/T)AAP patterns, with Ser and Thr side chains built in both g+ and g− conformations. Only polar hydrogens are depicted to offer a better view. The OγH group of Ser/Thr is interacting with the carbonyl oxygen at position j-4, relative to Ser/Thr, except when noted. Figures were created using MolScript v2.1.1 (Kraulis, 1991) and Raster3D v2.5 (Merritt and Bacon, 1997).
FIGURE 2.
Detailed view of the hydrogen bond pattern, in the Pro-kink turn, of model peptides containing the P(S/T), PA(S/T), and PAA(S/T) patterns, with Ser and Thr side chains built in both g+ and g− conformations. Only polar hydrogens are depicted to offer a better view. The OγH group of Ser/Thr is interacting with the carbonyl oxygen at position j-4, relative to Ser/Thr, except when noted.
TABLE 3.
Percentage (%) of the intrahelical hydrogen bond between the OγH side chain of Ser/Thr and the O=Cj-4 and O=Cj-3 carbonyl, and bend angle (mean ± SD) as observed during the molecular dynamics simulations of polyAla α-helices (see Methods)
|
gauche+
|
gauche−
|
|||||||
|---|---|---|---|---|---|---|---|---|
| O=Cj-4
|
O=Cj-3
|
O=Cj-4
|
O=Cj-3
|
|||||
| % | Bend | % | Bend | % | Bend | % | Bend | |
| SAAP | 100 | 15.8 ± 6.4‡ | 0 | – | 94 | 14.2 ± 5.4§ | 6 | – |
| TAAP | 100 | 15.1 ± 7.3§ | 0 | – | 89 | 13.5 ± 4.7§ | 11 | – |
| SAP | 100 | 30.3 ± 11.6§ | 0 | – | 2 | – | 98 | 34.8 ± 9.6§ |
| TAP | 98 | 27.2 ± 7.6§ | 2 | – | 88 | 26.5 ± 7.4§ | 12 | – |
| SP | 99 | 30.5 ± 8.2§ | 1 | – | 67 | 19.6 ± 4.8 | 33 | 49.8 ± 10.1§ |
| TP | 99 | 26.9 ± 6.9§ | 1 | – | 92 | 20.9 ± 6.1 | 8 | – |
| P* | 19.7 ± 7.3 | |||||||
| PS | 100 | 18.9 ± 6.4 | 0 | – | 83 | 17.5 ± 5.8 | 17 | – |
| PT | 100 | 18.9 ± 6.8 | 0 | – | 98 | 19.7 ± 6.5 | 2 | – |
| PAS | 100 | 16.0 ± 6.5‡ | 0 | – | 65 | 14.2 ± 5.6§ | 35 | 14.6 ± 6.6‡ |
| PAT | 100 | 14.1 ± 6.8§ | 0 | – | 93 | 16.4 ± 5.1† | 7 | – |
| PAAS | 100 | 14.0 ± 6.2§ | 0 | – | 35 | 29.6 ± 8.6§ | 65 | 30.0 ± 10.9§ |
| PAAT | 100 | 15.6 ± 6.6‡ | 0 | – | 74 | 24.7 ± 8.8‡ | 26 | 32.4 ± 10.0§ |
A Pro-containing polyAla α-helix is used as a control. One-way analysis of variance plus a posteriori two-sided Dunnett's t-tests was employed to contrast if the bend angle of the model peptides differs from the control simulation (†p < 0.05; ‡p < 0.01; §p < 0.001).
FIGURE 3.
Comparison of the evolution of the unit twist angles (°) along the α-helix, from turns (i-11, i-8) to (i+4, i+7), between a Pro-containing polyAla α-helix (control, solid black line) and (a) (S/T)P, (b) (S/T)AP, (c) (S/T)AAP, (d) P(S/T), (e) PA(S/T), and (f) PAA(S/T) motifs. The color and style codes are: Ser (red), Thr (blue), g+ rotamer (solid line), g− rotamer interacting with the carbonyl oxygen at position j-4 (dotted line), and g− rotamer interacting with the carbonyl oxygen at position j-3 (dashed line).
Pro
The bending angle of 20° we have obtained by MD simulations (Table 3) coincides with the reported bending angle of 21° measured from a database of Pro-containing TM helices (Cordes et al., 2002), whose structures are known at a resolution of 4 Å or better. Fig. 3 shows the evolution of the unit twist angles along the α-helix, from turns (i-11, i-8) to (i+4, i+7), as observed during the MD simulations. The helical distortion induced by the Pro residue, in the control simulation, is reflected at the level of twist angles from approximately turn (i-5, i-2) up to (i-1, i+2). Fig. 1 a shows a detailed view of the Pro-kink. Both CδH atoms of Pro act as hydrogen bond donors in the hydrogen bond interaction with the carbonyl oxygen at position i-4 (Chakrabarti and Chakrabarti, 1998).
(S/T)P
gauche+
The side chain of both Ser and Thr residues always interact with the backbone carbonyl oxygen at position j-4 (Table 3). Thus, the OγH side chain of Ser/Thr interacts, in addition to the Ni-1-H amide, with the carbonyl oxygen at position i-5 (Fig. 1, b and c). As a result, these helices are strongly bent, with average bending angles of 31° for SP and 27° for TP (∼7–11° larger than the 20° of the reference Pro-kinked α-helix) (Table 3). Moreover, there is an increase of 13° in SP and 4° in TP in the twist angle (i.e., a closing of the helix) at the (i-4, i-1) (Fig. 3 a).
gauche−
Notably, this rotamer differentiates Ser and Thr. Although Ser can form a hydrogen bond with either the carbonyl oxygen at position j-3 (33%, see Table 3) or j-4 (67%), Thr is almost only hydrogen bonding the carbonyl oxygen at position j-4 (92%) due to steric restriction of the additional methyl group (see Discussion). When the OγH side chain of Ser/Thr interacts with the carbonyl oxygen at position j-4, there is little or no structural effect on the Pro-kink, as measured by the bending angle (20° for SP or 21° for TP versus 20° for the reference Pro-kink) or the twist angle profile (Fig. 3 a) of the helix. Each CδH group of Pro acts as a hydrogen donor, either with the O=Ci-4 carbonyl oxygen or the Oγ atom of Ser/Thr (Fig. 1, d and e).
In contrast, when the OγH side chain of Ser interacts with the carbonyl oxygen at position j-3 (Oγ-H···O=Cj-3), there are significant changes in the hydrogen bond network of the Pro-kink turn, inducing a strong structural effect. The CδH atoms of Pro interact with both carbonyl oxygens at positions i-3 and i-4, causing the intrahelical Ni+1-H···O=Ci-3 to be disrupted, and the Ni-1-H amide to relocate between O=Ci-4 and O=Ci-5 (Fig. 1 f). This induces an increase in the bending angle (50° vs. 20° of Pro, see Table 3) and a closing of the helix at the Pro-kink, as reflected by an increase of the twist angle at the (i-4, i-1) turn (121° vs. 101° for the reference Pro-kink simulation, see Fig. 3 a). To corroborate these results, we performed additional MD simulations on the SP motif, with the side chain built in the g− conformation, and restraining the intrahelical hydrogen bond of the Hγ atom of Ser to the carbonyl oxygen at positions j-3 and j-4. The analysis of these independent restrained MD trajectories confirms our initial conclusions obtained from unrestrained MD trajectories. The hydrogen bond patterns are similar to the ones depicted in Fig. 1, e and f: the bending angles are 16° and 48° and the twist angles at the (i-4, i-1) turn are 99° and 120°, for the SP motif with the Oγ-H···O=Cj-4 and Oγ-H···O=Cj-3 hydrogen bond, respectively.
(S/T)AP
gauche+
TAP and SAP motifs induce an important bending of the α-helix (27° and 30°, respectively, ∼7–10° more than a regular Pro-kink). However, in contrast to (S/T)P in g+, there are no large changes in the unit twist angle profile relative to the reference Pro-kinked α-helix (Fig. 3 b). These effects are due to the specific hydrogen bond network, in the Pro-kink turn, as depicted in Fig. 1, g and h. The hydroxyl moiety of both Ser and Thr interacts, in addition to the Ni-2-H amide, with the carbonyl oxygen at the i-6 position (j-4 to Ser/Thr) (Table 3). Moreover, the CδH groups of Pro act as hydrogen bond donors in the hydrogen bond interaction with both lone pairs of the carbonyl oxygen at i-4.
gauche−
As for (S/T)P, the g− rotamer shows significant differences between SAP and TAP motifs. The OγH side chain of Thr hydrogen bonds preferably the carbonyl oxygen at the j-4 position (88%, Table 3) and only rarely to j-3, due to steric hindrance between the methyl moiety of the side chain and the carbonyl oxygen of residue j-4 (see Discussion). In contrast, the hydroxyl group of Ser hydrogen bonds almost exclusively the carbonyl oxygen at position j-3 (i-5 to Pro) (98%, Table 3). This leads to a very different hydrogen bond pattern and conformation of the Pro-kinked α-helix (Fig. 1, i and j). When both the OγH of Thr and the Ni-2-H amide are interacting with the i-6 carbonyl oxygen, the CδH group of Pro acts as a hydrogen bond donor in the hydrogen bond interaction with the carbonyl oxygen at position i-4. In this case, the average bend angle of the α-helix is 27° (∼7° larger than the reference Pro-kinked α-helix, Table 3); and the unit twist angle profile is not strongly modified relative to the reference system (Fig. 3 b). In other words, the TAP motif presents a similar structural effect in both g+ and g− rotamers.
However, in the SAP g− motif, the OγH of the Ser residue is interacting with the carbonyl oxygen at position j-3 (i-5 relative to the Pro), and while the CδH group of Pro keeps interacting with the carbonyl oxygen at position i-4 and the Ni+1-H···O=Ci-3 hydrogen bond is preserved, the Ni-1-H···O=Ci-5 hydrogen bond is disrupted (the Ni-1-H group seems to interact with the Oγ atom of Ser) and the Ni-2-H amide reorients between O=Ci-5 and O=Ci-6 (Fig. 1 i). This results in an average bend angle of 35° (∼15° larger than the reference Pro-kinked α-helix, Table 3), and an increase in the twist angle at the (i-5, i-2) turn of 17° relative to the Pro-kinked helix (Fig. 3 b).
(S/T)AAP
gauche+
The effect of (S/T)AAP motifs in the helix is radically different from (S/T)P or (S/T)AP. SAAP (16°) and TAAP (15°) significantly decrease the bending angle of the helix relative to the reference Pro-kink (20°, Table 3). The decrease in bending is due to specific hydrogen bond networks that reduce the steric clash between the pyrrolidine ring of Pro and the O=Ci-4 carbonyl (see Discussion). Fig. 1, k and l show how, in both cases, the Ni-3-H amide (N-H group of Ser/Thr) is located between the O=Ci-6 and O=Ci-7 carbonyl oxygens. Adaptation of the standard intrahelical Ni-3-H···O=Ci-7 hydrogen bond toward the Ni-3-H···O=Ci-6 hydrogen bond, modifies the orientation of the O=Ci-4 carbonyl. Notably, this relocation of the O=Ci-4 carbonyl allows the CγH group of Pro to interact with the carbonyl oxygen instead of the standard CδH atoms.
gauche−
There is also a straightening of the helix for both SAAP (14°) and TAAP (14°) relative to the standard Pro-kink (20°, Table 3). As in the g+ conformation, both Ser and Thr residues establish preferably the hydrogen bond with the backbone carbonyl at position j-4 (i-7 to Pro) (Table 3). The hydrogen bond network at the Pro-kink turn is also very similar: the Ni-3-H amide is located between the O=Ci-6 and O=Ci-7 carbonyl oxygens, the relocation of O=Ci-4 reduces the steric clash with the Pro side chain, and the CδH and CγH groups of Pro interact with the O=Ci-4 carbonyl oxygen (Fig. 1, m and n).
P(S/T)
gauche+, gauche−
PS and PT motifs in both g+ and g− rotamers show no significant changes in either the bending angle (Table 3) or the twist profile (Fig. 3 d) relative to the control simulation. The side chain of both Ser and Thr residues forms the hydrogen bond, in addition to the Ni+1-H amide, with the carbonyl oxygen at position j-4 (i-3 to the Pro) (Table 3). The CδH group of Pro acts as a hydrogen bond donor in the hydrogen bond interaction with the carbonyl oxygen at the i-4 position (Fig. 2, a–d).
PA(S/T)
gauche+
The average bending angles of the α-helices for PAS (16°) and PAT (14°) are slightly decreased relative to the reference Pro-kink (Table 3), whereas the twist angle profile remains unchanged (Fig. 3 e). The fact that the steric clash between the CδH atoms of Pro and the O=Ci-4 carbonyl group and the hydrogen bond between the OγH side chain of Ser/Thr and the O=Ci-2 carbonyl group, are in opposite sides of the α-helix could be responsible for this effect (see Discussion). Although the CδH···O=Ci-4 clash tends to bend the helix toward the opposite side of the Pro residue, the additional OγH···O=Ci-2 hydrogen bond partly counters the bending of the helix in this direction. The detailed view of the hydrogen bond network depicted in Fig. 2, e and f shows that the hydroxyl group of either Ser or Thr side chains is interacting with carbonyl oxygen at the j-4 position (i-2 to the Pro) (Table 3).
gauche−
Ser and Thr side chains behave differently in this g− conformation. Although Ser can form a hydrogen bond with the carbonyl oxygen at position j-3 (35%, Fig. 2 i) or at position j-4 (65%, Fig. 2 g), the methyl group of Thr forces the OγH side chain to hydrogen bond the carbonyl oxygen at position j-4 (93%, Fig. 2 h) (Table 3). All these conformations, despite these variations in the hydrogen bond pattern, reduce the bending angle of the helix relative to the standard Pro-kink (14° or 15° for PAS hydrogen bonding the j-4 or the j-3 carbonyl oxygen, respectively; and 16° for PAT, Table 3). Similarly to the g+ conformation, the formation of either the OγH···O=Ci-2 or OγH···O=Ci-1 hydrogen bond in the reverse side of the helix, from where Pro is located, impedes its bending. There are no significant changes in the twist angles profile (Fig. 3 e).
PAA(S/T)
gauche+
These motifs also reduce the bending of the helix relative to the control (14° for PAAS and 16° for PAAT, Table 3). The side chain of both Ser and Thr residues is hydrogen bonding the O=Ci-1 carbonyl oxygen (O=Cj-4 relative to Ser/Thr) (Table 3) (Fig. 2, j and k). As previously observed for the PAS motif in the g− rotamer conformation, the formation of this additional OγH···O=Ci-1 hydrogen bond impedes the bending of the helix. There are no significant changes in the twist angles profile (Fig. 3 f).
gauche−
PAAS and PAAT motifs in g− are the only patterns of all Ser/Thr residues following Pro in which the bending of the helix increases relative to the control Pro-kinked helix (see Table 3). Moreover, the Thr side chain in PAAT is the only case of all patterns that contain Thr in which the OγH side chain is able to interact, in a significant amount of conformations (26%, Table 3), with the O=Cj-3 carbonyl (O=Ci relative to Pro). In contrast to the other motifs, the formation of this hydrogen bond is feasible due to an increase of 6° in the unit twist angle (i.e., a closing of the helix) at the (i-1, i+2) helical turn (see Fig. 3 f), which relocates the position of the O=Ci-1 and O=Ci carbonyls. In all cases, the CδH groups of Pro hydrogen bond the O=Ci-4 carbonyl (Fig. 2, l–o), leading to similar bending angles (30° for PAAS hydrogen bonding either O=Cj-4 or O=Cj-3; and 25° or 32° for PAAT hydrogen bonding O=Cj-4 or O=Cj-3, respectively). Although the side chain of Ser/Thr in both g+ and g− can interact with the same O=Ci-1 carbonyl, the effect on the bending of the helix is opposite, decreasing in g+ and increasing in g− (Table 3). Although in the g+ conformation the OγH···O=Ci-1 hydrogen bond is in the opposite face of the helix that the steric CδH···O=Ci-4 clash (Fig. 2, j and k; see Discussion), in the g− conformation both interactions occur in the same face (Fig. 2, l and n).
Due to the different behavior of the PAA(S/T) motifs in g−, these results were further investigated by additional MD simulations restraining the intrahelical hydrogen bond of the Hγ atom of Ser/Thr to the carbonyl oxygen at positions j-3 and j-4. Analysis of these new trajectories leads to bend angles similar to those previously obtained (28° or 31° for PAAS hydrogen bonding O=Cj-4 or O=Cj-3, respectively; and 27° or 29° for PAAT hydrogen bonding O=Cj-4 or O=Cj-3, respectively), thus confirming our results.
DISCUSSION
Signaling membrane proteins achieve structural diversity within a constraining environment. Specific TM helices must have included specific structural motifs, adapted to the functional role of each of these proteins. Deciphering the structure-function relationship in membrane proteins will therefore require an understanding of the structural motifs that govern the conformational diversity. Besides the important role of Pro residues (Sansom and Weinstein, 2000), Ser and Thr residues also induce helical distortions of possible functional importance (Ballesteros et al., 2000). In this study we have shown that combinations of these residues can produce dramatic effects on the conformation of TM α-helices, whereas their unusual distribution in TM segments further suggests a possible functional role as structural adapters.
The (S/T)P and (S/T)AP motifs in the gauche+ rotamer and the TAP and PAA(S/T) motifs in the gauche− rotamer increase the bending of Pro-kinked transmembrane helices
Fig. 4 a shows the conformation of Pro-kinked (control, yellow), SP (red), and SAP (dark green) in the g+ rotamer, and TAP (light green) and PAAS (purple) in the g− rotamer TM α-helices. The OγH group of Ser/Thr is hydrogen bonding the peptide carbonyl oxygen at position j-4, relative to Ser/Thr, in all the cases. The helix conformations of TP and TAP in g+ and PAAT in g− are similar to SP and SAP in g+ and PAAS in g−, respectively, and are omitted for clarity. Clearly, all these motifs and rotamer conformations induce a bending angle in the helix that is ∼7–12° larger than the control Pro-kinked helix (see Results and Table 3). Incorporation into the α-helix of these modified conformations at one side of the cell membrane results in a significant displacement of the residues located at the other side of the membrane. The magnitude of the relocation might be estimated from the models depicted in Fig. 4 a. Thus, the distances between the α-carbon positions between the control Pro-kinked helix (yellow) and the other modified helices are in the 2–4 Å range for an amino acid located 10 residues away from Pro. The additional hydrogen bond formed between the OγH group of Ser or Thr and the backbone carbonyl oxygen in the preceding turn of the helix, together with the additional flexibility provided by the adjacent Pro, seems to cause the observed conformation of the helices.
FIGURE 4.
Comparison of helix bending between a Pro-containing polyAla α-helix (control, yellow) and (a) SP (red) and SAP (dark green) in the g+ rotamer, and TAP (light green) and PAAS (purple) in the g− rotamer transmembrane α-helices (the OγH group of S/T is interacting with the carbonyl oxygen at position j-4, relative to S/T, in all the cases); and (b, c) SP (orange) and SAP (light green) in the g− rotamer interacting with the carbonyl oxygen at position j-3 relative to Ser (different views rotated by 90° along the helical axis). The side chains of Pro and Ser/Thr are shown.
The (S/T)P motifs in the gauche− rotamer do not modify the bending of Pro-kinked transmembrane helices
SP and TP motifs in the g− rotamer can also hydrogen bond the i-5 carbonyl oxygen (j-4 relative to Ser/Thr), but do not change the bending of the Pro-kinked TM helix (see Results and Table 3). In contrast to other motifs and conformations, in this g− rotamer the Oγ atom of Ser or Thr hydrogen bonds the CδH group of Pro (see Fig. 1, d and e). This new intrahelical hydrogen bond adds an additional constrain that counterbalances the increase of bending angle caused by the additional hydrogen bond formed between the Ser/Thr side chain and the i-5 peptide carbonyl oxygen. The CδH···Oγ hydrogen bond is only feasible when Ser/Thr is preceding Pro and Ser/Thr are in the g− rotamer. In the g+ rotamer the lone pairs of the Oγ atom of Ser/Thr are pointing away from the helix (see Fig. 1, b and c).
The SP and SAP motifs in the gauche− rotamer interacting with the carbonyl oxygen at position i-3 increase the bending and alter the direction of Pro-kinked transmembrane helices
SP (33%) and SAP (98%) motifs in the g− rotamer can hydrogen bond the j-3 carbonyl oxygen relative to Ser (Table 3). The formation of this interaction, rather than the standard with the j-4 carbonyl oxygen relative to Ser, induces a striking modification in the conformation of the helix, as illustrated in Fig. 4, b and c. Notably, the bend angle of the helix increased significantly from 20° for Pro to 50° for SP or 35° for SAP α-helices (Table 3). Moreover, the helices point to a completely different direction in space (Fig. 4 c): toward the center for Pro (control, yellow), left for SAP (light green), or right for SP (orange) α-helices. This change in direction is attributed to the closing of the helix at the Pro-kink, as illustrated by the increase, relative to the Pro-kinked helix, of the twist angle at the (i-4, i-1) turn for SP (20°), and at the (i-5, i-2) turn for SAP (17°) (see Fig. 3). It is probable that the closing of the helix at different turns, (i-4, i-1) for SP or (i-5, i-2) for SAP, makes both helices to point toward opposite directions in space.
The methyl group of Thr prevents the interaction of OγH with the carbonyl oxygen at position j-3
Fig. 5 shows the evolution of the distance between the Hγ atom of Ser and the oxygen of the O=Cj-4 (solid line) or O=Cj-3 (broken line) carbonyl, relative to Ser, and the values of χ1 of Ser obtained during the production run of the MD simulations of SP (left panel) and SAP (right panel) in the g− rotamer. There is a clear correlation between the type of side-chain interaction and χ1. When OγH is hydrogen bonding Oj-4 (from ∼850 ps to 1500 ps in SP) χ1 stays in the ∼35–55° range (average value of 46° for SP). The value of χ1 goes up to the ∼55–75° range (average values of 68° for SP and 65° for SAP) if OγH is hydrogen bonding Oj-3 (from 500 ps to ∼850 ps in SP and from ∼550 ps to 1500 ps in SAP). Thus, the formation of the hydrogen bond with the j-3 carbonyl oxygen, relative to Ser, requires an increase of the χ1 dihedral. These values of χ1 are unfavorable for Thr because of the steric hindrance between its methyl group and the O=Cj-4 carbonyl. Thus, Thr residues rarely (see Table 3) form the interaction with the O=Cj-3 carbonyl. In these conformations local rearrangement of the backbone is necessary to accommodate the methyl group (results not shown). The side chain of Thr in the PAAT motif interacts with the O=Cj-3 carbonyl (O=Ci relative to Pro) in a larger number of structures (26%) than the other Thr-containing motifs (Table 3). The closing of the helix at the (i-1, i+2) helical turn (see Fig. 3 f) relocates the position of the O=Ci-1 (the O=Cj-4 carbonyl that would clash with the methyl group of Thr) and O=Ci (interacting with the OγH side chain of Thr) carbonyls, thus facilitating the accommodation of the methyl group.
FIGURE 5.
Evolution of the distance (bottom panels) between the Hγ atom of Ser and the oxygen of the O=Cj-4 (solid line) or O=Cj-3 (broken line) carbonyl, relative to Ser, and the values of χ1 (top panels) of Ser obtained during the production run of the molecular dynamics simulations of SP (left panels) and SAP (right panels) in the g− rotamer.
Molecular mechanisms of decreasing the bending of Pro-kinked transmembrane helices
(S/T)AAP and PA(S/T) motifs, in both g+ and g−, and PAA(S/T) in g+ rotamers decrease the bending of Pro-kinked TM helices (see Table 3). However, the mechanisms by which these motifs modify the conformation of the helix differ.
(S/T)AAP motifs moderate the steric clash between the pyrrolidine ring and the O=Ci-4 carbonyl oxygen by modifying the conformation of the Ni-3-H amide group. The additional hydrogen bond formed between OγH of Ser/Thri-3 and the O=Ci-7 carbonyl oxygen (j-4 relative to Ser/Thr) modifies the intrahelical Ni-3-H···O=Ci-7 hydrogen bond. Adjustment of the Ni-3-H amide group (see bottom arrow in Fig. 6 a) alters the orientation of the O=Ci-4 carbonyl (see top arrow in Fig. 6 a), due to the planarity of the OCi-4-Ni-3H peptide bond (in green), away from the helix; reducing the steric clash between the CδH atoms of Pro and the O=Ci-4 carbonyl; and decreasing the bending angle of the helix (Table 3).
FIGURE 6.
Molecular mechanisms of decreasing the bending of Pro-kinked transmembrane helices. (a) (S/T)AAP motifs in both g+ and g− rotamers moderate the steric clash between the pyrrolidine ring and the O=Ci-4 carbonyl oxygen by modifying the conformation of the Ni-3-H amide group, throughout the additional hydrogen bond formed between OγH of S/Ti-3 and the O=Ci-7 carbonyl oxygen. (b) PA(S/T), in both g+ and g−, and PAA(S/T) in g+ rotamers obstruct the bending of the helix by adding a constrain in the form of a hydrogen bond (in green) in the other side of the helix where the steric CδH···O=Ci-4 clash (in red) occurs.
PA(S/T), in both g+ and g−, and PAA(S/T) in g+ rotamers obstruct the bending of the helix by adding a constrain in the form of a hydrogen bond in the opposite side of the Pro residue, the curved-in face of the helix. Thus, the formation of either the OγH···O=Ci-2 or OγH···O=Ci-1 hydrogen bond by Ser/Thri+2 or Ser/Thri+3 side chain (Fig. 6 b) in the other side of the helix where the steric CδH···O=Ci-4 clash occurs, attenuates the bending of the helix. However, PAA(S/T) in the g− rotamer also interact with the O=Ci-1 carbonyl, but in this case there is an increase of the bending angle (Table 3). In this g− conformation the formation of the OγH···O=Ci-1 hydrogen bond by the Ser/Thri+3 side chain occurs on the same side as the steric clash (compare Fig. 2, j and k with Fig. 2, l and n), hence reinforcing the deformation.
P(S/T) motifs in both g+ and g− rotamers do not modify the conformation of the helix relative to the standard Pro-kink. In this case the additional hydrogen bond between the Ser/Thri+1 side chain occur with the O=Ci-3 carbonyl, which is neither at the same nor at the opposite face of the helix than the Pro side chain. We suggest that this particular hydrogen bond network maintains the conformation of the Pro-kinked helix unchanged.
Influence of the Ser/Thr side-chain rotamer in mechanisms of signal transduction
Our simulations suggest that Ser and Thr could act as molecular switches in signal transduction. Although in TM helices these side chains are normally in either g+ or g− conformation, interaction with an external partner (i.e., bound ligand) could induce a change toward the trans rotamer. Our results indicate that such rearrangement of the Ser or Thr can lead to conformational modification of the whole helix, with possible functional importance. Similarly, structural changes could be induced by a rotamer change from a g+ to g− conformation (and vice versa). To illustrate this mechanism, Fig. 7 a shows the conformation of SAP in the g+ rotamer (dark green), t rotamer (yellow, taken as a regular Pro-kinked α-helix), and g− rotamer interacting with the carbonyl oxygen at position j-3 relative to Ser (light green); and Fig. 7 b shows the conformation of SP in the g+ rotamer (red) and g− rotamer interacting with the carbonyl oxygen at position j-4 (dark orange) and j-3 (light orange). Clearly, variation of the side-chain rotamer (Fig. 7 a), or changing the hydrogen bond from j-4 to j-3 or vice versa (Fig. 7 b), covers a wide structural range in the α-helix. Similarly, rotation of Ser/Thr side chain in the PAA(S/T) motif from g+ to g− would modify the bend angle of the helix from ∼15° to ∼28° (see Table 3).
FIGURE 7.
Comparison of helix bending between (a) SAP in the g+ rotamer (dark green), t rotamer (yellow), and g− rotamer interacting with the carbonyl oxygen at position j-3 relative to Ser (light green); and (b) SP in the g+ rotamer (red) and g− rotamer interacting with the carbonyl oxygen at position j-4 (dark orange) and j-3 (light orange). The conformation of the t rotamer was taken as a regular Pro-kinked α-helix. The side chains of Pro and Ser/Thr are shown.
Functional relevance of Ser and Thr as Pro-kinked modulators of transmembrane α-helices
The statistical analysis of the presence of these motifs in the rhodopsin-like family of GPCRs (Table 2) shows that, among all the occurrences found in the search, the predominant sequences are TxP (9%), SxP (7%), TP (6%), SxxP (6%), TxxP (5%). Thus, there is a clear tendency of Ser/Thr residues to be found at positions preceding Pro. Among all the possibilities of Ser/Thr preceding Pro, only the SP motif is not often observed in the database (2%). This might be attributed to the extreme conformation this motif achieves in the g− rotamer interacting with the carbonyl oxygen at position j-3.
Clearly, (S/T)xP motifs are highly observed in both a nonhomologous database of sequences of TM helices and a database of sequences of rhodopsin-like GPCRs. The functional role of Ser/Thr residues in the highly conserved Thr-x-Pro2.58 motif of the chemokine receptors (TM 2) was probed by site-directed mutagenesis and functional assay of the CCR5 receptor (Govaerts et al., 2001, 2003). Although mutation of Thr2.56 to Ser, Cys, Ala, or Val does not affect chemokine binding, it strongly influences the functional response. The functional impairment is highly dependent on the specific residue substituted for the Thr, and the rank order parallels the structural deformation of the α-helix (Govaerts et al., 2001).
The second TM segment of connexin32 contains the TP87 motif. Mutations of Thr86 to Ser, Ala, Cys, Val, Asn, or Leu shift the conductance-voltage relation of wild type, such that the mutated channels close at smaller transjunctional voltages (Ri et al., 1999). It was proposed that the hydrogen bonding potential of Thr86 mediates the conformational changes between open and closed channel states (Ri et al., 1999).
Bacteriorhodopsin contains TP91 in TM helix C. Mutation of Thr90 to Ala alters the proton pumping efficiency, and suggests that Thr90 has an important structural role in the proton pumping mechanism (Peralvarez et al., 2001). Fig. 8 shows the superimposition of helix C as observed in the crystal structure of bacteriorhodopsin (PDB entry 1AP9) that contains the TP91 motif in the g+ rotamer (red), and the computationally determined conformations of a Pro-kinked (yellow) and TP in the g+ rotamer (orange) polyAla α-helices. Clearly, helix C of bacteriorhodopsin exhibits a larger bending than a regular Pro-kinked helix that matches the computationally determined conformation of TP in the g+ rotamer. The bending angle of 30° obtained around the Pro-kink of helix C is in good agreement with the average value of 27° obtained in computer simulations (see Table 3).
FIGURE 8.

Comparison of helix bending between the computationally determined conformations of Pro (control, yellow) and TP in the g+ rotamer (red) polyAla α-helices; and the experimentally determined conformation of helix C of bacteriorhodopsin that contains TP91 in the g+ rotamer (orange), as observed in the PDB structure 1AP9. The side chains of Pro and Ser/Thr are shown.
CONCLUSIONS
Our simulations on model peptides offer molecular explanations for the high association of Ser and Thr residues with Pro in TM helices. Through specific action of their polar side chains, these residues modulate the structural deformations caused by the pyrrolidine ring of Pro. Such modulations cover a wide structural range, with, for instance, a variation of the bending angle from 14° to 30° by simply rotating the side chain of Ser in the PAAS motif from g+ to g−. These significant conformational changes could obviously have important functional roles, allowing or preventing specific interactions to take place. Therefore, combinations of Ser and Thr residues with Pro appear as a possible mechanism for structural adaptation of membrane proteins. Of course, our single helix simulations cannot take into account helix-helix interactions, which most likely have important structural effects as well. Nevertheless, our findings show how structurally (and hence functionally) important polar residues such as Ser and Thr can modulate the conformation of Pro-kinked TM helices, and therefore provide a structural basis for further understanding the function of membrane proteins.
Acknowledgments
This work has been supported by the Ministerio de Ciencia y Tecnología (SAF2002-01509), and Improving Human Potential of the European Community (HPRI-CT-1999-00071). Computer facilities were provided by the Centre de Computació i Comunicacions de Catalunya.
Cedric Govaerts' present address is Cellular and Molecular Pharmacology, University of California, San Francisco, 600 16th St., San Francisco, CA 94143-2240.
References
- Ballesteros, J. A., X. Deupi, M. Olivella, E. E. J. Haaksma, and L. Pardo. 2000. Serine and threonine residues bend α-helices in the χ1 = g-conformation. Biophys. J. 79:2754–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros, J. A., and H. Weinstein. 1995. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods in Neurosciences. 25:366–428. [Google Scholar]
- Bansal, M., S. Kumar, and R. Velavan. 2000. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 17:811–819. [DOI] [PubMed] [Google Scholar]
- Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vicent, M. Crowley, D. M. Ferguson, R. J. Radmer, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 1997. AMBER 5. University of California, San Francisco, CA.
- Chakrabarti, P., and S. Chakrabarti. 1998. C–H.O hydrogen bond involving proline residues in alpha-helices. J. Mol. Biol. 284:867–873. [DOI] [PubMed] [Google Scholar]
- Cordes, F. S., J. N. Bright, and M. S. Sansom. 2002. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323:951–960. [DOI] [PubMed] [Google Scholar]
- Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, Jr., D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- Darden, T. A., D. York, and L. Pedersen. 1993. Particle mesh Ewald: aAn N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- Farrens, D. L., C. Altenbach, K. Yang, W. L. Hubbell, and H. G. Khorana. 1996. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 274:768–770. [DOI] [PubMed] [Google Scholar]
- Fredriksson, R., M. C. Lagerstrom, L. G. Lundin, and H. B. Schioth. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63:1256–1272. [DOI] [PubMed] [Google Scholar]
- Frenkel, D., and B. Smit. 1996. Understanding Molecular Simulation: From Algorithms to Applications. Academic Press, San Diego, CA.
- Gether, U. 2000. Uncovering molecular mechanisms involved in activation of G protein- coupled receptors. Endocr. Rev. 21:90–113. [DOI] [PubMed] [Google Scholar]
- Govaerts, C., C. Blanpain, X. Deupi, S. Ballet, J. A. Ballesteros, S. J. Wodak, G. Vassart, L. Pardo, and M. Parmentier. 2001. The TxP motif in the second transmembrane helix of CCR5: a structural determinant in chemokine-induced activation. J. Biol. Chem. 276:13217–13225. [DOI] [PubMed] [Google Scholar]
- Govaerts, C., A. Bondue, J. Y. Springael, M. Olivella, X. Deupi, E. Le Poul, S. J. Wodak, M. Parmentier, L. Pardo, and C. Blanpain. 2003. Activation of CCR5 by chemokines involves an aromatic cluster between transmembrane helices 2 and 3. J. Biol. Chem. 278:1892–1903. [DOI] [PubMed] [Google Scholar]
- Gray, T. M., and B. W. Matthews. 1984. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J. Mol. Biol. 175:75–81. [DOI] [PubMed] [Google Scholar]
- Horn, F., E. Bettler, L. Oliveira, F. Campagne, F. E. Cohen, and G. Vriend. 2003. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 31:294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K(+) channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Kelley, L. A., S. P. Gardner, and M. J. Sutcliffe. 1996. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng. 9:1063–1065. [DOI] [PubMed] [Google Scholar]
- Kraulis, J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 24:946–950. [Google Scholar]
- Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505–524. [DOI] [PubMed] [Google Scholar]
- Monne, M., M. Hermansson, and G. von Heijne. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288:141–145. [DOI] [PubMed] [Google Scholar]
- O'Neil, K. T., and W. F. DeGrado. 1990. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 250:646–651. [DOI] [PubMed] [Google Scholar]
- Olivella, M., X. Deupi, C. Govaerts, and L. Pardo. 2002. Influence of the environment in the conformation of alpha-helices studied by protein database search and molecular dynamics simulations. Biophys. J. 82:3207–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. L. Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto, and M. Miyano. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 289:739–745. [DOI] [PubMed] [Google Scholar]
- Peralvarez, A., R. Barnadas, M. Sabes, E. Querol, and E. Padros. 2001. Thr90 is a key residue of the bacteriorhodopsin proton pumping mechanism. FEBS Lett. 508:399–402. [DOI] [PubMed] [Google Scholar]
- Ri, Y., J. A. Ballesteros, C. K. Abrams, S. Oh, V. K. Verselis, H. Weinstein, and T. A. Bargiello. 1999. The role of a conserved proline residue in mediating conformational changes associated with voltage gating of Cx32 gap junctions. Biophys. J. 76:2887–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom, M. S. P., and H. Weinstein. 2000. Hinges, swivels and switches: the role of prolines in signalling via transmembrane α-helices. Trends Pharmacol. Sci. 21:445–451. [DOI] [PubMed] [Google Scholar]
- Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol. 296:921–936. [DOI] [PubMed] [Google Scholar]
- Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, A. Yao, J. Ye, M. Zhan, W. Zhang, H. Zhang, Q. Zhao, L. Zheng, F. Zhong, W. Zhong, S. Zhu, S. Zhao, D. Gilbert, S. Baumhueter, G. Spier, C. Carter, A. Cravchik, T. Woodage, F. Ali, H. An, A. Awe, D. Baldwin, H. Baden, M. Barnstead, I. Barrow, K. Beeson, D. Busam, A. Carver, A. Center, M. L. Cheng, L. Curry, S. Danaher, L. Davenport, R. Desilets, S. Dietz, K. Dodson, L. Doup, S. Ferriera, N. Garg, A. Gluecksmann, B. Hart, J. Haynes, C. Haynes, C. Heiner, S. Hladun, D. Hostin, J. Houck, T. Howland, C. Ibegwam, J. Johnson, F. Kalush, L. Kline, S. Koduru, A. Love, F. Mann, D. May, S. McCawley, T. McIntosh, I. McMullen, M. Moy, L. Moy, B. Murphy, K. Nelson, C. Pfannkoch, E. Pratts, V. Puri, H. Qureshi, M. Reardon, R. Rodriguez, Y. H. Rogers, D. Romblad, B. Ruhfel, R. Scott, C. Sitter, M. Smallwood, E. Stewart, R. Strong, E. Suh, R. Thomas, N. N. Tint, S. Tse, C. Vech, G. Wang, J. Wetter, S. Williams, M. Williams, S. Windsor, E. Winn-Deen, K. Wolfe, J. Zaveri, K. Zaveri, J. F. Abril, R. Guigo, M. J. Campbell, K. V. Sjolander, B. Karlak, A. Kejariwal, H. Mi, B. Lazareva, T. Hatton, A. Narechania, K. Diemer, A. Muruganujan, N. Guo, S. Sato, V. Bafna, S. Istrail, R. Lippert, R. Schwartz, B. Walenz, S. Yooseph, D. Allen, A. Basu, J. Baxendale, L. Blick, M. Caminha, J. Carnes-Stine, P. Caulk, Y. H. Chiang, M. Coyne, C. Dahlke, A. Mays, M. Dombroski, M. Donnelly, D. Ely, S. Esparham, C. Fosler, H. Gire, S. Glanowski, K. Glasser, A. Glodek, M. Gorokhov, K. Graham, B. Gropman, M. Harris, J. Heil, S. Henderson, J. Hoover, D. Jennings, C. Jordan, J. Jordan, J. Kasha, L. Kagan, C. Kraft, A. Levitsky, M. Lewis, X. Liu, J. Lopez, D. Ma, W. Majoros, J. McDaniel, S. Murphy, M. Newman, T. Nguyen, N. Nguyen, M. Nodell, S. Pan, J. Peck, M. Peterson, W. Rowe, R. Sanders, J. Scott, M. Simpson, T. Smith, A. Sprague, T. Stockwell, R. Turner, E. Venter, M. Wang, M. Wen, D. Wu, M. Wu, A. Xia, A. Zandieh, and X. Zhu. 2001. The sequence of the human genome. Science. 291:1304–1351. [DOI] [PubMed] [Google Scholar]
- Von Heijne, G. 1991. Proline kinks in transmembrane alpha-helices. J. Mol. Biol. 218:499–503. [DOI] [PubMed] [Google Scholar]