Abstract
Paired helical filaments (PHF) is an aberrant structure present in the brain of Alzheimer's disease patients which has been correlated with their degree of dementia. In order to determine the structure of PHF, several studies have been performed using atomic force microscopy (AFM). However, those studies have the limitation that they have not been done in solution and the sample could be far from the real physiological conditions. In this work we present an AFM analysis of PHF in liquid environment and we compare that analysis with that performed in dry conditions. PHF imaging in liquid was only possible by using jumping mode AFM as the imaging technique. Jumping mode AFM images of PHF in solution show first, a notable increase in the absolute values of the height of the filament, and second, a smaller ratio between the height measured at the upper and at the lower part of the PHF. Direct comparison of the experimental data with structural models has been performed. From this we conclude that the PHF structure is compatible with two coupled ribbons with an overall height of 20 nm and a width of 10 nm.
INTRODUCTION
Alzheimer's disease (AD) is a senile dementia. Histopathological analysis of the brains of AD patients have indicated the presence of two aberrant structures: extracellular senile plaques and intracellular neurofibrillary tangles (NFT). The increase in the number of senile plaques appears to correlate with patient age whereas the presence of NFT could correlate with the degree of dementia in AD patients (Arriagada et al., 1992). NFT are composed of bundles of filamentous structures (Terry and Wisniewski, 1972) termed paired helical filaments (PHF) (Kidd, 1963). Several groups have indicated that the microtubule associated protein tau is the major component of PHF (Grundke-Iqbal et al., 1986; Kosic et al., 1986; Wood et al., 1986; Wischik et al., 1988) and that PHF-like polymers could be assembled under different conditions by using tau as the unique protein component (Montejo de Garcini et al., 1988; Wille et al., 1992; Crowther et al., 1994; Goedert et al., 1996; Perez et al., 1996, 2000; Wilson and Binder, 1997; Abraha et al., 2000). Knowledge of the three-dimensional structure of the proteins implicated not only in Alzheimer's disease PHF but in other neurodegenerative disorders is essential for understanding why and how endogenous proteins may adopt a nonnative fold (Temussi et al., 2003).
Different analyses have been carried out to look for PHF structure. The structural characterization of PHF has improved during the last years thanks to the use of techniques such as x-ray diffraction (Wiesniewski et al., 1976) and electron microscopy (Wischik et al., 1985; Montejo de Garcini et al., 1988; Appelt and Balin, 1993; Ruben et al., 1999). Atomic force microscopy (AFM) (Binnig et al., 1986) uses a sharp tip at the end of a flexible cantilever to scan and sense the topography of a sample deposited on a flat surface. AFM was used to confirm the helical nature of PHF and to study its structure (Pollanen et al., 1994, 1995, 1997; Ikonomovic et al., 1995; Moreno-Herrero et al., 2001). Pioneer AFM studies were carried out in air using contact mode AFM (Pollanen et al., 1994; Ikonomovic et al., 1995).
The high resolution and the possibility to study biological systems in their native environment has created an enormous expectation of AFM as an ideal tool for molecular biology (Bustamante et al., 1994). AFM has been used, not only to visualize a wide variety of biological molecules (Bustamante et al., 1997), but also to study the dynamics of proteins (Rief et al., 1997; van Noort et al., 1998); the conformational change of DNA in DNA-protein interactions (Rivetti et al., 1999), and the evolution of complex processes such as transcription (Kasas et al., 1997). However, to date AFM studies on Alzheimer PHF have been done in ambient air and therefore in nonphysiological conditions, and this has raised a main criticism on the published AFM paired helical filaments data.
In this work we present the first AFM images of PHF obtained in solution. AFM imaging in solution is difficult since tip-molecule interaction must be tuned to be weaker than molecule-substrate interaction. In liquids, van der Waals forces are screened and hence molecule-substrate interaction is extremely weak. For this reason, the AFM imaging technique must be chosen in such a way as to minimize intrusiveness. Data obtained in air and in solution have been compared to simulations done using two of the most extended models that attempt to explain the PHF structure: first, the model based on a single twisted ribbon, and second, the model proposed by Crowther and Wischik. From the analysis below we conclude that our AFM data fit better with the latter model.
MATERIALS AND METHODS
PHF isolation
PHF fractions were obtained by using the method of Greenberg and Davies (1990). PHF protein was subjected to sodium dodecyl sulfate acrylamide gel electrophoresis and the fractionated protein was characterized by Western blot using PHF-1 antibody (a kind gift of Dr. P. Davies, Albert Einstein University, Bronx, NY). Additionally, the presence of PHF was determined by electron microscopy as previously described (Nieto et al., 1988).
AFM sample preparation
AFM imaging is very sensitive to the substrate where the biological specimen has to be immobilized. In this work we present data obtained with mica being the deposition substrate. Another important aspect of AFM operation is the interface between substrate and sample. In the case of ambient pressure air, PHF samples were prepared as described in Moreno-Herrero et al. (2001). Data presented in air were obtained using dynamic amplitude constant AFM (DAC-AFM). For the experiments performed in liquid environment, samples were prepared in a similar manner as for air but after a 5-min adsorption time, the mica was placed in a liquid cell and filled with 0.01 M phosphate buffered saline, pH 7.4 (PBS) (Sigma-Aldrich, Alcobendas, Spain). Molecules imaged in liquid were never allowed to dry. Data presented in liquid were obtained using jumping mode AFM (JM-AFM) as the imaging technique.
Dynamic amplitude constant AFM
DAC-AFM is characterized by a sinusoidal oscillation with a certain amplitude and a phase lag with respect to the external driving force. In topographic imaging, the amplitude of the oscillation is kept constant by adjusting tip-sample distance. The adjustments of the piezotube that holds the sample are correlated with the height of the particles and stored as height images. In phase imaging, the phase lag of the tip relative to the excitation signal is monitored and recorded while feedback keeps the amplitude at a fixed value. Best results on individual biomolecules in air have been obtained using DAC-AFM since it is possible to operate the microscope without contact between tip and sample (Luna et al., 1998).
For DAC-AFM we have used Olympus type cantilevers with a force constant of 0.75 N/m and resonance frequency of 70 kHz (Olympus Optical, Tokyo, Japan).
Jumping mode AFM
JM-AFM (van der Werf et al., 1994; Rosa-Zeise et al., 1997; de Pablo et al., 1998) works by measuring a sequence of force versus distance curves at each point of the sampled surface with a feedback time in between. First, the tip is in contact with the surface while the feedback is keeping the cantilever deflection at the set point. Then the feedback is turned off and the tip is vertically moved away from the surface. At maximum tip-sample separation, the tip is moved laterally to the next point avoiding lateral forces. Finally, the tip is brought again into contact with the surface.
For JM-AFM in liquid we have used Nanosensors cantilevers with a force constant of 0.02 N/m (Nanosensors, Wetzlar-Blankenfeld, Germany). A comparison between DAC-AFM and JM-AFM and a detailed description of the experimental conditions used in both imaging modes can be found in Moreno-Herrero et al. (2002).
RESULTS
Dynamic amplitude constant AFM of PHF in ambient air
Images were obtained with a commercial microscope (Nanotec Electronica, Madrid, Spain). For imaging in air we have used DAC-AFM. Topographic AFM images of PHF in air show a left-handed helical structure (Fig. 1 i). Pitch value varies from 65 to 80 nm (average pitch: 75 nm) depending on the adsorption conditions. The experimental average height is 16 ± 1 nm at the top part and 10 ± 1 nm at the valley part (Fig. 1 i C). Straight filaments (SF) were also found but in much lower proportion than PHF. We have measured an average height of 8 ± 1 nm for SF particles in air. This height is half of the maximum height measured for PHF. Most of the work has been done using mica as deposition substrate. Mica has been chosen instead of other materials due to its flatness and to the negative charge it exposes when cleaved. Some studies by AFM (Kowalewski and Holtzman, 1999) suggest that for the analysis of other similar polymers like amyloid fibers, graphite seems to be a more suitable substrate. We have found that adsorption of PHF particles to graphite is difficult. This is probably due to the absence of electrostatic charge in the conducting surface of graphite. However, we have performed some experiments on graphite and we have found similar values for the height measurements (data not shown). Averaged measured widths, obtained from several PHF particles, at half-maximum height were 31 ± 3 nm and 32 ± 3 nm at the top and the valley part of the PHF (Fig. 1 i, A and B). All AFM lateral measurements are affected by the dilation phenomena of AFM. This is always present when the dimensions of the object to be imaged are smaller than the tip's dimensions. Typical tip radius is r ≥ 10 nm. Hence, a dilation phenomenon in the AFM images is often present when imaging biological samples.
FIGURE 1.
DAC-AFM data of PHF on mica in air. (i) AFM characterization of one single PHF particle. Profiles taken at top and valley points are shown in (i, A and B). Average height at these points is 16 nm and 10 nm. The effect of the finite size of the tip is demonstrated by a large measured width of ∼30 nm. The characteristic longitudinal profile of the PHF imaged in air is shown in C. The ratio between the height at top and valley points is 1.6. (ii) DAC-AFM topography (ii A) and phase images (ii, B and C) of PHF on mica in air. A detail of the structure of PHF is shown in ii C. The presence of the fuzzy coat around the PHF is clear in the phase images. Phase images also show a substructure composed of circular aggregates of variable sizes varying from 5 nm to 10 nm in diameter.
Phase AFM images of PHF in air are shown in Fig. 1 ii, B and C. The simultaneously recorded topographic data is shown in Fig. 1 ii A. In these images a two-color contrast is clear. The relatively wide topographic structure imaged in Fig. 1 ii A appears as a thin filament surrounded by a dark coat in phase AFM images (Fig. 1 ii, B and C). These findings coincide with the electron microscopy data reported by Crowther (1991). In that work, authors reported that PHF particles were surrounded by a fuzzy coat. This fuzzy coat is claimed to be composed of the N- and C-termini of the tau-protein, the main constituent of PHF (Wischik et al., 1997). Phase images exhibit more contrast than topographic images. An assembly of several structures of variable sizes (5–10 nm) can be distinguished in the high-resolution phase image (Fig. 1 ii C). These molecular assemblies can be attributed to tau-aggregates. The fuzzy coat that surrounds the PHF core has been seen in air and in liquid topographic images. Hence, we believe that the contrast that appears in phase images is not due to air drying artifacts. Indeed, several contributions have investigated the potential of phase imaging to detect variations in material properties such as adhesion, elasticity, and viscoelasticity, or to image samples with large vertical dimensions (Tamayo and García, 1997; Cleveland et al., 1998; Noy et al., 1998). We have also analyzed the error data images but we could not extract any additional information to that provided by the phase images. Error data images were very similar to the derivative of the topographic data.
Jumping mode AFM of PHF in physiological conditions
The main drawback of the data reported in the previous section is that the structure of the PHF can be modified or distorted by the fact that images were obtained in an ambient air environment. In order to overcome this criticism we have performed similar experiments in physiological conditions.
We have first used DAC-AFM for imaging PHF in liquid. When DAC-AFM is used in liquids, the high viscosity of the medium dramatically reduces the quality factor of the cantilever (∼5) with the consequent sensitivity lost. In addition, the resonance frequency of the cantilever is reduced by a factor of 3 since the effective mass of the cantilever increases due to drag of the liquid. Finally, van der Waals forces are screened by the surrounding ions so that operation in the noncontact regime in liquids is difficult, if not impossible. Contact between tip and sample often takes place in liquids when using DAC-AFM (Putman et al., 1994). Therefore, soft materials can be modified by the tapping of the tip on the molecule. Indeed, PHF particles imaged using DAC-AFM in liquids showed progressive damage in consecutive images (data not shown). This is why we decided to use JM-AFM since in this mode the maximum applied force is well established and directly measured.
Here, we report the first AFM images of PHF obtained in liquid environment (Fig. 2). Fig. 2 i shows a typical set of PHF particles at different magnifications. Apparently, data obtained in solution are similar to data collected in air. A detailed analysis of one PHF particle is shown in Fig. 2 ii. Again, a left-handed helical structure is obtained with pitch varying from 65 nm to 80 nm, but in PBS buffer the average pitch is closer to 70 nm instead of the 75 nm obtained in air (Fig. 2 ii C). PHF structure, as imaged with the AFM, appears more compact in liquid than in air. However, there are slight but important differences comparing the data obtained in both environments. The main difference concerns the absolute values of height measured in PBS buffer. We measure an average height of 20 ± 1 nm at the top part and 15 ± 1 nm at the valley part (Fig. 2 ii, A and B). We attribute this difference to the full hydration of PHF in solution. In any helical structure, the maximum height coincides with the maximum width. The average measured height at the top part is in agreement with the maximum width from electron microscopy data reported by Crowther et al. (1991). Filaments with low marked periodicity, which can be considered SF, show an average height of 11 ± 2 nm.
FIGURE 2.
JM-AFM data of PHF on mica in PBS buffer. (i) JM-AFM gallery images of PHF. (i A) A long-range topographic image where several PHF particles can be seen. (i, B and C) Two high-magnification images. The left-handed helical structure is clear from the AFM images. (ii) AFM characterization of one single particle. Profiles taken at top and valley areas are shown in ii, A and B. Average heights at these points are 20 nm and 15 nm, respectively. The characteristic longitudinal profile of the PHF imaged in buffer is shown in ii C. The ratio between the height at top and valley points is 1.3.
Lateral dimensions of PHF particles measured in PBS buffer are slightly larger than those measured in air. Average widths obtained from several PHF particles are 32 ± 3 nm and 37 ± 3 nm at the top and valley parts, respectively. The fuzzy coat shown in phase images obtained in air also appears in JM-AFM topographic images. AFM images show blurred boundaries of the PHF particles. JM-AFM can, simultaneously with the topography, also acquire adhesion maps. The adhesion data showed a negligible adhesion force over the PHF and forces up to 450 pN over clean surface areas (data not shown). AFM is able to measure electrostatic forces in aqueous solution (Butt, 1991). It has also been proved that electrostatic forces are correlated with adhesion forces (Rotsch and Radmacher, 1997). Hence, we believe that the adhesion force contrast measured is due to the difference of electrostatic potential between PHF and clean surface areas.
DATA ANALYSIS, MODELS, AND SIMULATIONS
Structural models
The PHF structure is still under debate. Briefly, two different models have been proposed. One is based on a twisted ribbon (TR) model (Pollanen et al., 1994, 1995), and the other consists of a double-helical stack of transversely oriented subunits, each having three domains (CW model) (Crowther and Wischik, 1985).
A simple way to describe a helical periodic structure is to repeat a two-dimensional architecture of H × W (height × width) while rotating it along the longitudinal direction Z. The helical structure generated by performing this operation will have an oscillating height varying between H and H/2 + W/2. We can use the experimental data to find the best values of H and W that generate a model which fits with the data. For this purpose we must use the images obtained under physiological conditions. These data lead to H = 20 nm and W = 10 nm since the measured height at valley areas is 15 nm. From these numbers we can build up the two models that are under discussion.
Fig. 3 A shows the TR model with these parameters. Fig. 3 B shows the CW model with D (diameter of the domain) = W/2. Topographic representation of these structures is shown in Fig. 3, C and D, as well as three-dimensional representations (see insets). According to these parameters, longitudinal profiles for both models are shown in Fig. 3, E and F.
FIGURE 3.
Structural models for the structure of PHF in liquids. The model shown in A is based on a twisted ribbon which is obtained by rotating the structure of dimensions (H × W) along the z axis (TR model). The topography image of this model, as well as the three-dimensional representation (inset) is shown in C. The longitudinal profile of this model would have a top height of H and a valley height of H/2 + W/2 (see panel E). The other model under debate is composed of two C-shape regions with three domains of diameter D (CW model). The overall dimensions of this structure are H × W and are shown in B. Topography and three-dimensional images (inset) of the CW model are shown in D and the longitudinal profile is shown in F. Note that the longitudinal profile of the CW model exhibits an asymmetry on the right part of the loop. Model parameters are H = 20 nm and W = 10 nm.
Models presented in Fig. 3 show two ideal structures with no deformation. This is the situation that, presumably, happens in liquids. In contrast, in ambient air, biological samples suffer two processes: dehydration and deformation. Dehydration results in a severe loss of volume since the main component of biomolecules is water. Indeed, PHF and SF heights measured in air are roughly 20% lower than those measured in liquid. Deformation of the structure is due to capillary, electrostatic, and van der Waals forces. This deformation, as mentioned above, will be especially relevant for areas not in contact with the surface of the mica substrate. Deformation and dehydration occur when the polymer is adsorbed to mica. This implies that the axial periodicity of the polymer would not be altered. We have confirmed this point since measured pitch is similar in both environments.
The next step is to introduce dehydration and deformation processes in the models (Fig. 4). Dehydration is easily implemented by reducing model dimensions by means of a reduction factor. Deformation of the filament is stronger in valley areas than in top areas. Thus, a reasonable way to simulate deformation is by subtracting a periodic function, 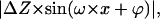 from the original structure. ΔZ is the amplitude of the deformation and ω is the axial frequency of the PHF (
from the original structure. ΔZ is the amplitude of the deformation and ω is the axial frequency of the PHF ( with
with 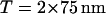 ). The phase, ϕ, is an adjustable parameter to have zero deformation at points of maximum height (top points). For the models presented in Fig. 4, dimensions have been reduced by 20% and ΔZ was adjusted to 2 nm. Fig. 4, A and B, shows, respectively, the modified TR model and CW model. Fig. 4, C and D, shows the corresponding longitudinal profiles.
). The phase, ϕ, is an adjustable parameter to have zero deformation at points of maximum height (top points). For the models presented in Fig. 4, dimensions have been reduced by 20% and ΔZ was adjusted to 2 nm. Fig. 4, A and B, shows, respectively, the modified TR model and CW model. Fig. 4, C and D, shows the corresponding longitudinal profiles.
FIGURE 4.
Structural models for the structure of PHF in air. The structure of the polymer is affected by dehydration and deformation due to molecule-surface interaction forces. The effect of dehydration is simulated by applying a reduction factor to the dimensions of the structure. Hence, new dimensions are H′ × W′. A is the modified TR model and B is the modified CW model. The effect of the deformation (see arrows) is bigger in valley areas and it has been simulated by subtracting, from the unmodified structure, a periodic function with maximum amplitude (ΔZ) at these areas. According to experimental data we have performed a reduction of 20% to the dimensions of the model. Thus, H′ = 16 nm and W′ = 8 nm and ΔZ = 2 nm. Longitudinal profiles for the modified models are shown in C and D.
AFM dilation
Dilation phenomena are always present in AFM when objects are smaller than the tip's dimensions. In order to compare experimental data with models (Figs. 3 and 4) we must perform the convolution of the AFM tip and the model. The simulations below (based on the algorithm of Villarrubia, 1996) assume a parabolic shape for the tip. Model parameters were adjusted to obtain the best fit with the experimental data.
Fig. 5 i, A–C, shows the simulations for the TR model in liquids with H = 20 nm and W = 10 nm. The model is presented in Fig. 5 i A; the AFM simulated image in Fig. 5 i B, and the experimental data in Fig. 5 i C. Tip dimensions were adjusted to obtain similar lateral profiles for the simulated and experimental images. This leads to a tip radius of 8 nm. The corresponding longitudinal profiles are shown below the topographic images. Simulations for the TR model in air are shown in Fig. 5 ii. For these simulations H = 16 nm, W = 8 nm, and ΔZ = 2 nm.
FIGURE 5.
AFM simulations using the TR model. (i) AFM simulations for liquids. The structural model is shown in i A. The simulated AFM image using a tip with a radius of 8 nm is shown in i B and the experimental image in i C. Tip radius was adjusted to obtain a similar value for the lateral dimension of the simulated and the experimental image. Longitudinal profiles for i, A–C can be directly compared. Dimensions of the model are H = 20 nm and W = 10 nm, chosen according to the experimental data. (ii) AFM simulations for air using the modified TR model. The structural model is shown in ii A. The simulated AFM image using an 8-nm tip is shown in ii B and the experimental image in ii C. Longitudinal profiles for ii, A–C can be directly compared. Dimensions of the model are H′ = 16 nm, W′ = 8 nm, and ΔZ = 2 nm, chosen according to the experimental data.
The Crowther and Wischik model is more complex to simulate than the twisted-ribbon model. In the CW-model the fiber is composed of small domains of 5 nm. According to the model shown in Fig. 4 B, this implies small grooves of 2.5 nm between domains. Fig. 6 i shows the simulations for the CW model in liquids. Model, AFM image simulation, and experimental data are respectively presented in Fig. 6, A–C. Longitudinal profiles are shown below the topographic images. The effect of the size of the tip is especially relevant for the AFM simulation of the CW model since nanoscopic features coming from the subunits are rounded due to tip dimensions (see profiles). These features originate an asymmetry on the right part of the loop (see arrows) that is reproduced in the experimental data. Fig. 7 shows some simulated and experimental profiles of the CW model and PHF in liquids. Simulation leads to three different profiles with one, two, and three bulbs obtained at different parts of the simulated PHF structure (profiles type A, B, and C with a resolution of 0.68 nm/pixel). Profiles with one and two bulbs are also obtained from the experimental data. Profiles of type C were not obtained from the PHF images. This can be attributed to the limited resolution of the images obtained in liquids (3.9 nm/pixel).
FIGURE 6.
AFM simulations using the CW model. (i) AFM simulations for liquids. The structural model is shown in i A. The simulated AFM image is shown in i B and the experimental image in i C. Longitudinal profiles for i, A–C can be directly compared. Dimensions of the model are H = 20 nm, W = 10 nm, and D = 5 nm, chosen according to the experimental data. The tip radius for the AFM simulation was 8 nm. (ii) AFM simulations for air using the modified CW model. The structural model is shown in ii A. The simulated AFM image using an 8-nm tip is shown in ii B and the experimental image in ii C. Longitudinal profiles are shown in ii, A–C. Dimensions of the model are H′ = 16 nm, W′ = 8 nm, and ΔZ = 2 nm. The direction of the helix (see arrow), predicted by the model, is reproduced in the experimental data.
FIGURE 7.
(A–C) Characteristic profiles of the AFM simulation of the CW model obtained with a lateral resolution of 0.68 nm/point. Profiles with one and two bulbs are experimentally reproduced and shown in D and E. The C profile is not experimentally reproduced due to the low lateral resolution of the experimental image (3.9 nm/point).
Simulations for the CW model in air are shown in Fig. 6 ii. In this case, the asymmetry at the right part of the loop, although it is clear in the simulated image, is not reproduced in the experimental image, probably due to deformation of the PHF. However there is a high agreement between the simulated and the experimental direction of the helix (see arrows).
DISCUSSION
We have carried out a structural analysis of the paired helical filaments in air and in liquid environment using AFM. AFM appears to be a unique tool for imaging biological assemblies in its native media with high resolution (Engel et al., 1997). Imaging individual biomolecules in liquids is challenging because tip-molecule interaction is similar to molecule-substrate interaction. Studies in air were done with DAC-AFM (Fig. 1) and those performed in liquids with JM-AFM (Fig. 2). These imaging modes were chosen in order to minimize sample damage and intrusiveness. The helical nature of PHF was confirmed in air and in solution as it has been already described in air and in vacuum conditions (Wischik et al., 1985, 1997; Appelt and Balin, 1993; Pollanen et al., 1997; Ruben et al., 1999). Phase AFM images obtained in air confirm the presence of a fuzzy coat that surrounds the core structure of the PHF particles. Moreover, these images reveal that the polymer is made of structures of nanometric size placed along the longitudinal axis of the filament. Images obtained in buffer complement and extend the information deduced from the images acquired in air. From these images we have deduced that PHF are filamentous structures of variable heights ranging from 15 nm to 20 nm.
Direct comparison of data obtained in air and in liquids shows two major differences. First is the value of the averaged maximum height at the top and valley areas. We measure 16 nm at the top part in air and 20 nm in solution. At the valley part these values are 10 nm in air and 15 nm in liquid. These data suggest that JM-AFM is a nonintrusive AFM technique for imaging biomolecules in liquids and showing the native structure of PHF. The nonintrusive behavior of JM-AFM has been also demonstrated by imaging microtubules (de Pablo et al., 2003) and DNA (Moreno-Herrero et al., 2003) in solution.
The second important difference of the data obtained in solution with respect to the data measured in air is the ratio between top and valley heights. In air this ratio is ∼1.6 and in liquids ∼1.3. This suggests that the structure of the PHF is well preserved in liquids and not modified when imaged with the AFM. If the structure of the PHF is similar to a twisted ribbon, then the parts situated between two consecutive maximums would not make contact with the surface of mica where the molecule is adsorbed. This would explain the ratio between top and valley heights measured in PBS buffer. When imaging in air, electrostatic, capillary, and van der Waals forces strongly squeeze the molecule into the substrate modifying its structure. This deformation is dramatic in the valley parts since these areas are free standing in the air in a similar way as a bridge is held by two adjacent pillars. This deformation would explain the value of the top/valley height ratio in air. The fact that the measured pitch is less disperse in PBS buffer than in air also proves that deformation of the PHF due to surface forces is minimized in liquids.
From the data obtained in air and in liquid we have roughly constructed two of the most extended and discussed models that are under debate. First, the model based on a twisted ribbon and second, the model based on two coupled ribbons with two subunits and several domains. Both models are characterized by a height (H) and a width (W) dimension. The latter model is also characterized by the diameter (D) of the domain (Fig. 3). To elaborate and simulate a model that fits the experimental data obtained in air and in liquids is difficult. Dimensions of both models were adjusted from the data obtained in solution. In order to fit these models to the data obtained in air, two modifications were applied (Fig. 4). First, model dimensions were 20% reduced in order to simulate the effect of the absence of water that occurs in air. Second, we have simulated the deformation due to sample-surface interaction. This has been done by subtracting a periodic function from the data. Once the models for air and for liquids were established, AFM simulations were carried out by taking into account the effect of tip finite size.
At first glance, simulated images and experimental data obtained in both environments agree fairly well when using either of the presented models (Figs. 5 and 6). The helical structure and the heights at top and valley areas coincide in both the experimental and simulated images. However, although both models fit for the general shape of PHF, a detailed analysis of the data shows some differences. The direction of the helix (Fig. 6 ii) and the substructure detected in liquids (Fig. 7) are better predicted with the CW model than with the TR model. The dimensions of the CW model show a polymer with a maximum height of 20 nm and a thickness in the central part of 10 nm with a domain of 5 nm. Data obtained on SF in air and in buffer suggest that PHF and SF are related structural variants of the two coupled ribbons described in the CW model as described by Crowther (1991). The thin structure reported in air when using ultrasharp tips (Pollanen et al., 1994; Moreno-Herrero et al., 2001) would be also compatible with the CW model since this model has a relatively thick nucleus (composed of four domains) and a thin tail composed of one domain. The thickness of the polymer measured when using sharp tips is ∼5 nm, in good coincidence with the size of the domain. Ultrasharp tips could not be used in a liquid environment because of its relatively high force constant. The development of soft and sharp tips for imaging in liquid would be very helpful for this type of structural analysis.
In summary, we have used AFM to study PHF structure in ambient pressure air and in solution. The experiments in solution are especially relevant since molecular structure is only preserved in liquid environment. PHF imaging in solution is extremely difficult due to the small surface contact area of the molecules to the substrate. Only the JM-AFM method is successful in obtaining repetitive PHF images in liquid showing the native structure of the polymer. PHF particles imaged in solution show 1), larger absolute filament heights; and 2), a smaller top/valley height ratio than those particles imaged in air. The data have been compared to AFM simulated images of the TR model and of the CW model. A detailed analysis of the images shows that PHF structure is compatible with the model proposed by Crowther and Wischik consisting of two coupled ribbons with an overall height of 20 nm and a width of 10 nm.
Acknowledgments
This work was supported by a grant from Comunidad Autónoma de Madrid (to A.M.B.) and by grants from Centro de Investigacion Científica y Tecnologica (Spain), Fundación Lilly, and by an institutional grant from Fundación Ramón Areces (to J.A.). F.M.-H. is a recipient of a fellowship from Comunidad Autónoma de Madrid.
References
- Abraha, A., N. Ghoshal, T. C. Gamblin, V. Cryns, R. W. Berry, J. Kuret, and L. I. Binder. 2000. C-terminal inhibition of tau assembly in vitro and in Alzheimer's disease. J. Cell Sci. 113:3737–3745. [DOI] [PubMed] [Google Scholar]
- Appelt, D. M., and B. J. Balin. 1993. Analisys of paired helical filaments (PHFs) found in Alzheimer's disease using freeze-drying/rotary shadowing. J. Struct. Biol. 111:85–95. [DOI] [PubMed] [Google Scholar]
- Arriagada, P. V., J. H. Growdon, E. T. Hedley-Whyte, and B. T. Hyman. 1992. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology. 42:631–639. [DOI] [PubMed] [Google Scholar]
- Binnig, G., C. F. Quate, and C. Gerber. 1986. Atomic force microscope. Phys. Rev. Lett. 56:930–933. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., D. A. Erie, and D. Keller. 1994. Biochemical and structural applications of scanning force microscopy. Curr. Opin. Struct. Biol. 4:750–760. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., C. Rivetti, and D. J. Keller. 1997. Scanning force microscopy under aqueous solutions. Curr. Opin. Struct. Biol. 7:709–716. [DOI] [PubMed] [Google Scholar]
- Butt, H. J. 1991. Measuring electrostatic, van der Waals, and hydration forces in electrolyte solutions with an atomic force microscope. Biophys. J. 60:1438–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, J. P., B. Anczykowski, A. E. Schmid, and V. B. Elings. 1998. Energy dissipation in tapping-mode atomic force microscopy. Appl. Phys. Lett. 72:2613–2615. [Google Scholar]
- Crowther, R. A. 1991. Straight and paired helical filaments in Alzheimer's disease have a common structural unit. Proc. Natl. Acad. Sci. USA. 88:2288–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther, R. A., O. F. Olesen, M. J. Smith, R. Jakes, and M. Goedert. 1994. Assembly of Alzheimer-like filaments from full length tau protein. FEBS Lett. 337:135–138. [DOI] [PubMed] [Google Scholar]
- Crowther, R. A., and C. M. Wischik. 1985. Image reconstruction of the Alzheimer paired helical filament. EMBO J. 4:3661–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pablo, P. J., J. Colchero, J. Gómez-Herrero, and A. M. Baro. 1998. Jumping mode scanning force microscopy. Appl. Phys. Lett. 73:3300–3302. [Google Scholar]
- de Pablo, P. J., I. A. T. Schaap, and C. F. Schmidt. 2003. Observation of microtubules with scanning force microscopy in liquid. Nanotechnology. 14:143–146. [Google Scholar]
- Engel, A., C. A. Schoenenberger, and D. J. Muller. 1997. High resolution imaging of native biological sample surfaces using scanning probe microscopy. Curr. Opin. Struct. Biol. 7:279–284. [DOI] [PubMed] [Google Scholar]
- Goedert, M., R. Jakes, M. G. Spillantini, M. Hasegawa, M. J. Smith, and R. A. Crowther. 1996. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 383:550–553. [DOI] [PubMed] [Google Scholar]
- Greenberg, S. G., and P. Davies. 1990. A preparation of Alzheimer paired helical filaments that displays distinct tau proteins by polyacrylamide gel electrophoresis. Proc. Natl. Acad. Sci. USA. 87:5827–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal, I., K. Iqbal, Y. C. Tung, M. Quinlan, H. M. Wisniewski, and L. I. Binder. 1986. Abnormal phosphorylation of microtubule-associated protein tau in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA. 83:4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomovic, M. D., D. M. Armstrong, S. H. Yen, C. Obcemea, and B. Vidic. 1995. Atomic force microscopy of paired helical filaments isolated from the autopsied brains of patients with Alzheimer's disease and immunolabeled against microtubule-associated protein tau. Am. J. Pathol. 147:516–528. [PMC free article] [PubMed] [Google Scholar]
- Kasas, S., N. H. Thomson, B. L. Smith, H. G. Hansma, X. Zhu, M. Guthold, C. Bustamante, E. T. Kool, M. Kashlev, and P. K. Hansma. 1997. Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry. 36:461–468. [DOI] [PubMed] [Google Scholar]
- Kidd, M. 1963. Paired helical filaments in electron microscopy of Alzheimer's disease. Nature. 197:192–193. [DOI] [PubMed] [Google Scholar]
- Kosic, K. S., S. F. Bakalis, D. J. Selkoe, M. W. Pierce, and L. K. Duffy. 1986. High molecular weight microtubule-associated proteins: purification by electro-elution and amino acid components. J. Neurosci. Res. 15:543–551. [DOI] [PubMed] [Google Scholar]
- Kowalewski, T., and D. M. Holtzman. 1999. In situ atomic force microscopy study of Alzheimer's beta-amyloid peptide on different substrates: new insights into mechanism of beta- sheet formation. Proc. Natl. Acad. Sci. USA. 96:3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, M., J. Colchero, and A. M. Baro. 1998. Intermittent contact scanning force microscopy: the role of liquid necks. Appl. Phys. Lett. 72:3461–3463. [Google Scholar]
- Montejo de Garcini, E., J. L. Carrascosa, I. Correas, A. Nieto, and J. Avila. 1988. Tau factor polymers are similar to paired helical filaments of Alzheimer's disease. FEBS Lett. 236:150–154. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero, F., P. J. de Pablo, M. Álvarez, J. Colchero, J. Gómez-Herrero, and A. M. Baró. 2003. Jumping mode scanning force microscopy: a suitable technique for imaging DNA in liquids. Appl. Surf. Sci. 210:22–26. [Google Scholar]
- Moreno-Herrero, F., P. J. de Pablo, R. Fernández-Sánchez, J. Colchero, J. Gómez-Herrero, and A. M. Baró. 2002. Scanning force microscopy jumping and tapping modes in liquids. Appl. Phys. Lett. 81:2620–2623. [Google Scholar]
- Moreno-Herrero, F., J. M. Valpuesta, M. Perez, J. Colchero, A. M. Baro, J. Avila, and E. Montejo de Garcini. 2001. Characterization by atomic force microscopy and cryoelectron microscopy of tau polymers assembled in Alzheimer's disease. J. Alzheimers Dis. 3:443–451. [DOI] [PubMed] [Google Scholar]
- Nieto, A., I. Correas, E. Montejo de Garcini, and J. Avila. 1988. A modified form of microtubule-associated tau protein is the main component of paired helical filaments. Biochem. Biophys. Res. Commun. 154:660–667. [DOI] [PubMed] [Google Scholar]
- Noy, A., C. H. Sanders, D. V. Vezenov, S. S. Wong, and C. M. Lieber. 1998. Chemically-sensitive imaging in tapping mode by chemical force microscopy: relationship between phase lag and adhesion. Langmuir. 14:1508–1511. [Google Scholar]
- Perez, M., R. Cuadros, M. A. Smith, G. Perry, and J. Avila. 2000. Phosphorylated, but not native, tau protein assembles following reaction with the lipid peroxidation product, 4-hydroxy-2-nonenal. FEBS Lett. 486:270–274. [DOI] [PubMed] [Google Scholar]
- Perez, M., J. M. Valpuesta, M. Medina, E. Montejo de Garcini, and J. Avila. 1996. Polymerization of tau into filaments in the presence of heparin: the minimal sequence required for tau-tau interaction. J. Neurochem. 67:1183–1190. [DOI] [PubMed] [Google Scholar]
- Pollanen, M. S., P. Markiewicz, C. Bergeron, and M. C. Goh. 1994. Twisted ribbon structure of paired helical filaments revealed by atomic force microscopy. Am. J. Pathol. 144:869–873. [PMC free article] [PubMed] [Google Scholar]
- Pollanen, M. S., P. Markiewicz, and M. C. Goh. 1997. Paired helical filaments are twisted ribbons composed of two parallel and aligned components: image reconstruction and modeling of filament structure using atomic force microscopy. J. Neuropathol. Exp. Neurol. 56:79–85. [DOI] [PubMed] [Google Scholar]
- Pollanen, M. S., P. Markiewicz, M. C. Goh, and C. Bergeron. 1995. Alzheimer paired helical filaments: a comparison with the twisted ribbon model. Acta Neuropathol. 90:194–197. [DOI] [PubMed] [Google Scholar]
- Putman, A. J., K. O. van der Werf, B. de Grooth, N. F. van Hulst, and J. Greve. 1994. Tapping mode atomic force microscopy in liquid. Appl. Phys. Lett. 72:2454–2456. [Google Scholar]
- Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- Rivetti, C., M. Guthold, and C. Bustamante. 1999. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 18:4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Zeise, A., E. Weilandt, S. Hild, and O. Marti. 1997. The simultaneous measurement of elastic, electrostatic and adhesive properties by scanning force microscopy: pulsed-force mode operation. Meas. Sci. Technol. 8:1333–1338. [Google Scholar]
- Rotsch, C., and M. Radmacher. 1997. Mapping local electrostatic forces with the atomic force microscopy. Langmuir. 13:2825–2832. [Google Scholar]
- Ruben, G. C., J. Z. Wang, I. Grundke-Iqbal, and K. Iqbal. 1999. Paired helical filaments have a wide range of widths with similar helical periods. In Alzheimer's Disease and Related Disorders. K. Iqbal, D. F. Swaab, B. Winglad, and H. M. Wisniewski, editors. John Wiley & Sons, West Sussex, England. 187–192.
- Tamayo, J., and R. García. 1997. Effects of elastic and inelastic interactions on phase contrast images in tapping-mode scanning force microscopy. Appl. Phys. Lett. 71:2394–2396. [Google Scholar]
- Temussi, P. A., L. Masino, and A. Pastore. 2003. From Alzheimer to Huntington: why is a structural understanding so difficult? EMBO J. 22:355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, R. D., and H. M. Wisniewski. 1972. Ultrastructure of senile dementia and of experimental analogs. In Aging and Brain. C. M. Gaitz, editor. Plenum Publishing, New York. 89–116.
- van der Werf, K. O., C. A. J. Putman, B. G. Grooth, and J. Greve. 1994. Adhesion force imaging in air and liquid by adhesion mode atomic force microscopy. Appl. Phys. Lett. 65:1195–1197. [Google Scholar]
- van Noort, S. J., K. O. van der Werf, A. P. Eker, C. Wyman, B. G. de Grooth, N. F. van Hulst, and J. Greve. 1998. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys. J. 74:2840–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarrubia, J. S. 1996. Scanned probe microscope tip characterization without calibrated characterizers. J. Vac. Sci. Technol. B. 14:1518–1521. [Google Scholar]
- Wiesniewski, H. M., H. K. Narang, and R. D. Terry. 1976. Neurofibrillary tangles of paired helical filaments. J. Neurol. Sci. 27:173–181. [DOI] [PubMed] [Google Scholar]
- Wille, H., G. Drewes, J. Biernat, E. M. Mandelkow, and E. Mandelkow. 1992. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J. Cell Biol. 118:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. M., and L. I. Binder. 1997. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer's disease. Am. J. Pathol. 150:2181–2195. [PMC free article] [PubMed] [Google Scholar]
- Wischik, C. M., R. A. Crowther, M. Stewart, and M. Roth. 1985. Subunit structure of paired helical filaments in Alzheimer's disease. J. Cell Biol. 100:1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischik, C. M., R. Y. K. Lai, and C. R. Harrington. 1997. Modeling prion-like processing of tau protein in Alzheimer's disease for pharmaceutical development. In Brain Microtubule Associated Proteins: Modification in Disease. J. Avila, R. Brandt, and K. S. Kosic, editors. Harwood Academic Publishers, Chur, Switzerland. 185–241.
- Wischik, C. M., M. Novak, P. C. Edwards, A. Klug, W. Tichelaar, and R. A. Crowther. 1988. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. USA. 85:4884–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, J. G., S. S. Mirra, N. J. Pollock, and L. I. Binder. 1986. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau. Proc. Natl. Acad. Sci. USA. 83:4040–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]









