Abstract
A degP mutant of Shigella flexneri was identified in a screen for insertion mutants that invaded cultured cells but did not form wild-type plaques in monolayers. The degP mutant SM1100 invaded Henle cells at wild-type levels and induced apoptosis in macrophages but formed smaller plaques than those formed by wild-type S. flexneri in confluent monolayers of Henle and Caco-2 cells. The proportion of SM1100 bacteria with IcsA localized to the bacterial pole, a process required for actin polymerization into actin “tails,” was reduced compared to results with wild-type bacteria. The reduction in proper IcsA localization may account for the reduced plaque size of the degP mutant. Although DegP is a protease, the protease activity of S. flexneri DegP was not required for IcsA localization or the formation of plaques in Henle cell monolayers. DegP was also required for efficient polar IcsA localization in E. coli expressing icsA. In addition, the growth or survival of SM1100 was compromised compared to that of the wild type at elevated temperatures and in acidic conditions.
Shigella flexneri is a gram-negative facultative intracellular pathogen that causes bacillary dysentery. Shigella invades colonic epithelial cells by bacterium-induced phagocytosis, lyses the phagocytic vesicle, multiplies within the cytosol, and subsequently spreads to adjacent cells (37). Most of the genes required for invasion of epithelial cells and intercellular spread are located on the 220-kb virulence plasmid, although full virulence also requires the expression of chromosomally encoded genes. A 31-kb region of the S. flexneri virulence plasmid confers an invasive phenotype on Escherichia coli K-12 (29). This 31-kb invasion locus encodes the Mxi-Spa type III secretion system (TTSS), the secreted invasion plasmid antigens (IpaA, -B, -C, and -D), IpgD, the cytoplasmic chaperone IpgC, and the positive regulator VirB. The ability of Shigella to move within the host cytosol and into adjacent cells by polymerizing host actin requires the expression and proper localization of IcsA (VirG), a virulence plasmid-encoded protein located outside of the 31-kb region (3, 25, 28).
IcsA is a 120-kDa autotransporter protein whose carboxy terminus forms a β-barrel in the outer membrane, through which the amino-terminal portion of IcsA is transported and exposed on the bacterial surface (14, 15). On the bacterial surface, the outer membrane protease SopA (IcsP) can cleave IcsA at Arg758-Arg759 to release a 95-kDa fragment, IcsA* (10, 14, 40). IcsA is the major target of SopA proteolytic activity, but the biological role of this cleavage is unclear (45). SopA has 60% amino acid identity to E. coli OmpT, which is also able to cleave IcsA on the surface (30). In actively dividing bacteria, IcsA is localized to the old pole of the bacillus, although the mechanism for this localization is unclear (15, 16). Inside the host cell, IcsA at the bacterial pole interacts with the eukaryotic proteins vinculin and neural Wiskott-Aldrich syndrome protein (N-WASP) to promote assembly of F-actin tails that propel the bacterium through the host cytosol (46, 47). Defects in polar IcsA localization reduce the efficiency with which Shigella moves into adjacent cells, resulting in either reduced plaque size or the inability to form plaques in Henle cell monolayers.
To identify additional chromosomal and virulence plasmid genes required for S. flexneri to spread intercellularly, a genetic screen was performed using a library of random TnphoA mutants (19, 20). Mutants were identified that invaded Henle cells but did not form wild-type plaques in Henle cell monolayers, an indication of failure of the mutant to either multiply within the Henle cell cytosol or spread to adjacent cells. We report here the analysis of a mutant in which TnphoA was inserted into degP (htrA).
DegP is a periplasmic serine protease that is highly conserved among bacteria, and DegP homologues have been identified in higher organisms including yeast and humans (32). In E. coli, DegP functions as a protease at elevated temperatures and as a chaperone at low temperatures (44). Native substrates of DegP in vitro and in vivo include MalS (44), the colicin A lysis protein (6), and the PapA pilin (C. H. Jones, unpublished data cited in reference 22). For E. coli, degP expression is under the control of the alternative sigma factor σE and the Cpx two-component signal transduction system, both of which respond to extracytoplasmic stress (8, 9, 13). E. coli degP mutants are sensitive to elevated temperatures (27) and oxidative stress (42). Salmonella enterica serovar Typhimurium, Brucella abortus, and Yersinia enterocolitica degP mutants are also sensitive to oxidative stress and show reduced survival either within macrophages or in animal models. This suggests that DegP may be a virulence factor in these pathogens (26, 34, 51). Because the S. flexneri degP mutant was isolated in a screen for mutants with defects in intercellular spread, and since DegP is essential for virulence in other bacterial pathogens, the role of DegP in S. flexneri virulence was investigated further.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. S. flexneri strains were plated on tryptic soy broth agar plates containing 0.01% Congo red. E. coli and S. flexneri strains were grown in Luria broth. When necessary, antibiotics were added at the following concentrations: 125 μg of carbenicillin/ml, 30 μg of chloramphenicol/ml, 50 μg of kanamycin/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristic(s) | Reference |
|---|---|---|
| Shigella strains | ||
| SA100 | S. flexneri serotype 2a | 33 |
| SA101 | Noninvasive virulence plasmid mutant of SA100 | 24 |
| SA511 | Chloramphenicol-resistant SA100 derivative | 20 |
| SA222-7 | SA511 icsA::TnphoA | This work |
| SA222-89 | SA511 degP::TnphoA | This work |
| SM100 | Streptomycin-resistant SA100 derivative | S. Seliger |
| SM1100 | SM100 degP::cm | This work |
| E. coli strains | ||
| AB1515 | E. coli K-12 | C. F. Earhart |
| UT4400 | AB1515 ΔompT | 11 |
| KS474 | degP::kan | 2 |
| GPE100 | AB1515 degP::kan | This work |
| GPE101 | UT4400 degP::kan | This work |
| BL21(DE3)/plysS | E. coli host strain for protein expression | Novagen |
| Plasmids | ||
| pMTLcam | Chloramphenicol resistance gene | 50 |
| pWKS30 | Low-copy-number cloning vector | 48 |
| pHM5 | Suicide vector pGP704 containing sacB, Cbr | 35 |
| pGP25.1 | degP PCR product cloned into pGEM-T-easy (Promega) | This work |
| pGP25.2 | degP from pGP25.1 cloned into the NotI site of pWKS30 | This work |
| pGP25.5 | degP::cam cloned as a blunt fragment into the EcoRV site of pHM5 | This work |
| pGP25.6 | degP from pGP25.2 cloned as a SalI-SphI fragment into pACYC184 | This work |
| pGP38.1 | pWKS30 carrying icsA | This work |
| pGP43.4 | degPS210A cloned into pWKS30 | This work |
| pET21b | T7 RNA polymerase expression vector | Novagen |
| pGP44.5 | DegPS210A-HIS derivative of pET21b | This work |
| pGP44.7 | DegP-HIS derivative of pET21b | This work |
Nucleotide sequence analysis.
Inverse PCR was performed on SA222-89 using primer 1 (5′-ATATCGCCCTGAGCAG-3′) and primer 2 (5′-CAACCGGTGTCAAAACC-3′), which are located within TnphoA. DNA sequencing was performed using an ABI Prism 377 automatic sequencer. The obtained DNA sequence was compared to the GenBank database using the BLASTN algorithm (1).
Construction of degP plasmids and strains.
S. flexneri degP was amplified by PCR using primer 3 (5′-TCATCATTTGCACGAAGC-3′) and primer 4 (5′-CACCACCATTTCGGTTAG-3′) designed using the E. coli degP sequence, GenBank accession number AE000125. The PCR product was cloned into the pGEM-T-easy vector (Promega), generating pGP25.1.
SM1100 was constructed by allelic exchange as follows: the chloramphenicol cassette from pMTL24cam was inserted into the BclI site of degP, and degP::cm was cloned as a blunt fragment into the EcoRV site of the suicide vector pHM5 containing the sacB gene for counterselection. This recombinant plasmid, pGP25.5, was delivered by conjugation from SM10λpir to S. flexneri SM100. Double-crossover events were identified as colonies that were both sucrose and chloramphenicol resistant but carbenicillin sensitive. The mutant SM1100 was confirmed by PCR. The strains GPE100 and GPE101 were constructed by P1 transduction of degP::kan from KS474 into AB1515 and UT4400, respectively, and confirmed by PCR.
To introduce a mutation changing the putative active site Ser-210 to a codon for Ala, splice-overlap PCR was performed (18). PCR was performed by using a 10:1 mixture of Taq and Pfu DNA polymerases with the TaqI0 X buffer and by using the primers degPSer-Ala1 (5′-CACCACCGGCGTTACCAC-3′) and degPSer-Ala2 (5′-GTGGTAACGCCGGTGGTG-3′) with primer 3 and primer 4, respectively, to generate two overlapping fragments each containing the mutation encoding the Ala codon. The resulting PCR products then were used as template for a second PCR using primer 3 and primer 4 to amplify the entire degPSer210Ala gene. The second PCR product was digested with SalI and KpnI and cloned into pWKS30, generating pGP43.4. pGP43.4 sequence analysis indicated that the Ser-210 codon had been changed to one encoding Ala, but no other nucleotide changes had occurred.
Tissue culture, cell invasion, and plaque assays.
The ability of S. flexneri to invade Henle cells (Intestinal 407, American Type Culture Collection [ATCC]) was determined by the procedure of Hale and Formal (17). Infections of the macrophage cell line J774A.1 (ATCC) were performed in the same manner, and macrophages were tested for the onset of apoptosis using the ApoAlert Annexin V-FITC apoptosis kit (Clontech). Plaque assays on Henle and Caco-2 (ATCC) cells were performed as described by Oaks et al. (31).
SDS-PAGE and Western blot analysis.
Whole-cell and secreted proteins were normalized to the number of bacterial cells and analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). Gels were stained with Coomassie brilliant blue, and proteins were transferred to nitrocellulose for Western analysis. Immunoblotting procedures were carried out using mouse monoclonal antibodies recognizing IpaB or IpaC and rabbit polyclonal antibodies against IpaD and IcsA. All S. flexneri antisera were provided by Edwin Oaks (Walter Reed Army Institute of Research, Washington, D.C.).
Immunofluorescence labeling.
To label bacteria grown in vitro, indirect immunofluorescence labeling was performed as described previously (19). For double labeling, bacteria were labeled with primary antibody to IcsA and a fluorescein isothiocyanate-conjugated secondary antibody, followed by labeling with primary antibody to PhoA and a tetramethyl rhodamine isocyanate (TRITC)-conjugated secondary antibody. IcsA labeling of bacteria growing inside Henle cells was performed as follows: Henle cell invasions were performed using the standard protocol (above), and after 2 h, the Henle cells were fixed with 1% paraformaldehyde and permeabilized with 0.2% (vol/vol) Triton X-100. The intracellular bacteria were then labeled with polyclonal IcsA antibodies and fluorescein isothiocyanate-conjugated secondary antibody. For labeling of actin, infected Henle cells were fixed and permeabilized as described above and then stained with 50 μg of TRITC-phalloidin (Sigma)/ml for 40 min at room temperature. For IcsA localization in E. coli strains, IcsA was expressed from the plasmid pGP38.1, which was constructed as follows. icsA was amplified by PCR using primer 5 (5′-GGAATTCCTTGCGGTTTGAAGCAGAC-3′) and primer 6 (5′-GCGGATCCGCTTCCTGTGTAACGCCAAG-3′) designed using the S. flexneri virulence plasmid sequence, GenBank accession number SFPWR100. The PCR product was digested with EcoRI-BamHI and cloned into pWKS30 (48), generating pGP38.1. Fluorescence microscopy was performed using an Olympus BX41 microscope and a Leica TCS 4D confocal laser scanning microscope.
Protein purification.
To construct the expression vectors pGP44.5 and pGP44.7, degP and degPSer210Ala were each amplified using primer 7 (5′-GGAGATATACATATGAAAAAAACCACATTAGCA-3′) and primer 8 (5′-GCTTCAGATCTTGCATTAACAGGTAGATGG-3′) using a 10:1 ratio of Taq and Pfu DNA polymerases. PCR products were digested with NdeI and BglII and cloned into the NdeI and BamHI site of pET21b. The sequences of cloned degP and degPSer210Ala were confirmed by nucleotide sequencing. DegP and DegPSer210Ala were purified from E. coli BL21(DE3)/plysS under native conditions using nickel-nitrilotriacetic acid resin (Qiagen) according to the manufacturer's protocol. Proteins were eluted at an imidizole concentration of 100 mM.
Proteolytic activity of purified DegP.
Proteolytic activities of DegP and DegPSer210Ala were determined using resorufin-labeled casein (0.4% [wt/vol] in water) following the manufacturers instructions (Boehringer Mannheim).
CAT assays.
The chloramphenicol acetyltransferase (CAT) protein was quantitated as nanograms/milliliter/optical density at 600 nm using the CAT enzyme-linked immunosorbent assay kit (Roche Molecular Biochemical) following the manufacturer's instructions.
Environmental stress assays.
Unless indicated otherwise, all cultures were stationary-phase cultures grown in Luria broth and plated to determine viability before and after exposure to the indicated condition. To determine whether strains were sensitive to acid, cultures were diluted 1:50 in Luria broth, adjusted to pH 2.5 with HCl, and incubated for 2 h. Sensitivity to oxidative stress was determined by the addition of hydrogen peroxide (H2O2) to a final concentration of 80 mM or of cumene hydroperoxide to a final concentration of 2 mM. In an additional assay for sensitivity to oxidative stress, L agar plates were overlaid with soft L agar containing 100 μl of an overnight culture upon which disks containing 32.6 mg of H2O2 or 34.5 mg of cumene hydroperoxide were placed. Zones of inhibition were measured following overnight incubation at 37°C. To test the response to reducing conditions, the growth of cultures in the presence of 25 mM β-mercaptoethanol was determined. For low-osmolarity conditions and high-osmolarity conditions, the growth of cultures in Luria broth without NaCl or in Luria broth with 0.464 M sucrose, respectively, was determined. Growth in high salt conditions was measured using Luria broth containing 0.34 M NaCl. Sensitivities to the detergents SDS and sodium deoxycholate, dyes or antibiotics, and serum were determined as described previously (19).
RESULTS
Characterization of the degP mutant in tissue culture assays.
S. flexneri TnphoA mutants that invaded Henle cells but did not form wild-type plaques in Henle cell monolayers were isolated as described previously (19, 20). To amplify the gene into which TnphoA was inserted, inverse PCR was performed using primers within the transposon. Comparison of the inverse PCR fragment sequence from one of these mutants, S. flexneri SA222-89, with the GenBank database indicated that the interrupted gene shared 98% nucleotide identity with the E. coli K-12 degP (htrA) gene. An additional degP mutant, SM1100, was constructed in S. flexneri SM100 by replacing the wild-type gene with one containing a chloramphenicol cassette. The ability of SM1100 to invade cultured Henle cells and spread from cell to cell was then tested.
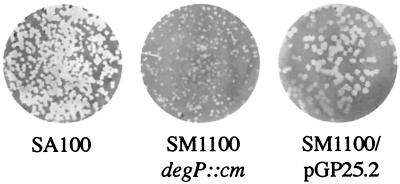
In a standard invasion assay, 94% of Henle cells were infected with the SM1100 degP mutant, comparable to the 97% invasion obtained with wild-type bacteria (Table 2). SM1100 secreted Ipa proteins at levels equivalent to those for the wild-type strain, as determined by SDS-PAGE and Western blot analysis of culture supernatants (data not shown). When tested for plaque formation on either Henle or Caco-2 cell monolayers, the plaque size of SM1100 was less than half that of the wild-type strain SM100 (Fig. 1 and Table 2). The small-plaque phenotype was complemented by supplying the low-copy-number plasmid pGP25.2, which encodes S. flexneri degP.
TABLE 2.
Characteristics of the degP mutant SM1100 in tissue culture assays
| Strain (description) | % Invasiona | Intracellular doubling time (min)b | Plaque size (mm)c | Apoptosisd |
|---|---|---|---|---|
| SA100 (WT) | 97 ± 4 | 30 | 1.4 ± 0.29 | + (80.15) |
| SM1100 (degP::cm) | 94 ± 8 | 30.5 | 0.65 ± 0.24 | + (96.22) |
| SA101 (noninvasive) | 0 | NDe | ND | − (34.30) |
Percent invasion is calculated as the percentage of Henle cells that contain three or more bacteria 30 min following invasion.
The intracellular doubling time was determined by calculating the number of bacteria per infected Henle cells every 30 min. Henle cells were counted, and the percentage of infected cells was determined. The number of intracellular bacteria was obtained by lysing infected Henle cells and plating the contents.
The average diameter of plaques formed in Henle cell monolayers is reported.
Shigella-infected macrophages undergoing apoptosis were detected using the ApoAlert Annexin V-FITC assay, and the mean fluorescence of cells was determined by fluorescence-activates cell sorting The ability of strains to induce apoptosis is indicated as + or −, and the mean fluorescence of 10,000 cells is reported.
ND, not determined.
FIG. 1.
The S. flexneri degP mutant forms small plaques in Henle cell monolayers. Plaque formation by SA100, SM1100 (degP::cm), and SM1100/pGP25.2 (DegP+) on confluent Henle cell monolayers is shown.
The inability of the degP mutant to form wild-type plaques could result either from a growth defect in the intracellular environment or from a reduced ability to spread to adjacent cells. As shown in Table 2, SM1100 multiplied within the Henle cell cytoplasm at the same rate as the wild-type parent strain, indicating that failure to form full-size plaques was not due to an inability to grow within the Henle cell environment.
Proper expression and secretion of the virulence plasmid-encoded Ipa proteins by the TTSS are essential for invasion as well as cell-to-cell spread (39). The ability of SM1100 to induce apoptosis in macrophages, an IpaB-mediated effect (52), was examined to determine whether intracellular secretion of Ipa proteins was affected by the degP mutation. The J774A.1 macrophage cell line was infected with SM1100, wild-type SM100 cells, and SA101, a mutant that does not secrete Ipa proteins. SM1100-infected macrophages showed signs of apoptosis that were similar to those for macrophages infected with wild-type SM100 and clearly distinguishable from results with SA101-infected macrophages not undergoing apoptosis (Table 2). The induction of apoptosis in SM1100-infected macrophages indicates that the TTSS functions normally for the secretion of IpaB into the intracellular environment. Thus, DegP is not required for invasion, intracellular multiplication, or induction of apoptosis in macrophages but is required for maximal cell-to-cell spread in S. flexneri.
The degP mutant shows altered IcsA localization.
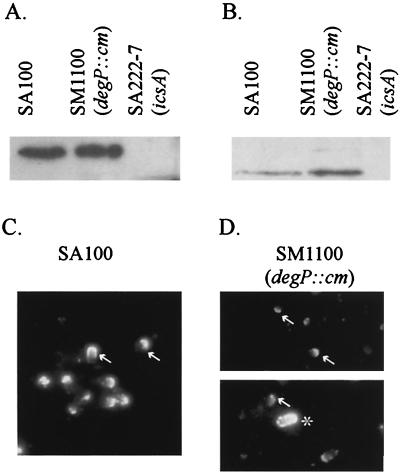
To determine if SM1100 expresses IcsA and delivers it to the bacterial surface, whole cells and culture supernatants were analyzed by Western blotting. Equal amounts of IcsA were present in SA100 and SM1100 cells (Fig. 2A), and IcsA* was detected in the supernatant of the degP mutant, indicating that delivery of IcsA to the bacterial surface and its subsequent cleavage were not eliminated by the degP mutation (Fig. 2B). To determine if the small-plaque phenotype of SM1100 was due to aberrant localization of IcsA on the cell surface, indirect immunofluorescence with antibody to IcsA was performed on SA100 and SM1100 cells from logarithmic-phase cultures. IcsA was present at the bacterial surface and localized to the pole of 55% of wild-type SA100, whereas IcsA was on the surface and localized to the pole of only 25.4% of SM1100 bacteria (Fig. 2C and D). When the intensities of staining for the wild type and the mutant were compared, there also appeared to be a reduction in the amount of IcsA on the surface of the mutant. A small proportion of the SM1100 cells (<1%) were stained with α-IcsA antibody over the entire surface. Double-labeling analysis indicated that this population of SM1100 also stained with primary antibody to a periplasmic protein (data not shown). Thus, those bacteria that appear to have IcsA over the entire surface are most likely permeable to the antibody and may be dead or dying.
FIG. 2.
IcsA expression and localization in the S. flexneri degP mutant. Western analysis with IcsA antibodies of SA100, SM1100, and SA222-7 bacteria (A) and culture supernatants (B) is shown. IcsA localization in broth-grown cultures of SA100 (C) and SM1100 (D) was observed by indirect immunofluorescence. IcsA “caps” are denoted by arrows. An asterisk indicates a bacterium representative of the minority population of SM1100 that appears to have IcsA over the entire surface.
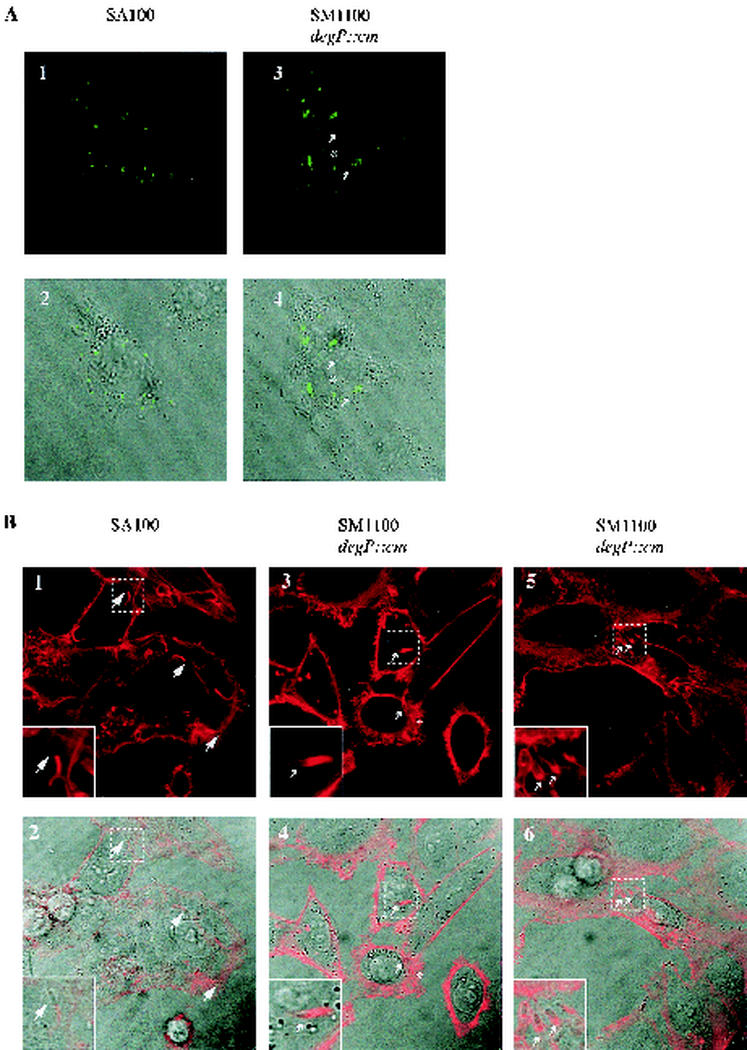
IcsA surface presentation and localization were also examined in SA100- or SM1100-infected Henle cells (Fig. 3A). Inside Henle cells, for roughly 50% of the wild-type bacteria, IcsA was on the surface of the bacterium and the IcsA was localized exclusively at the pole. On average, in the SM1100-infected Henle cells, only 18% of the bacteria had IcsA on the surface. Within that population, IcsA was localized at the pole with the rare exception of IcsA staining over the entire surface as before. These observations indicate that the degP mutant SM1100 may have a defect in secretion of IcsA to the bacterial pole.
FIG.3.
IcsA localization and actin staining inside Shigella-infected Henle cells. (A) IcsA localization observed by indirect immunofluorescence. In the SM1100 panels A3 and A4, arrows point to representative bacteria with polar IcsA localization and an asterisk indicates IcsA localization over the entire bacterium. (B) Actin was stained with TRITC-phalloidin, and representative bacteria have been labeled as follows: a large arrowhead points to normal actin tail formation (B1 and B2), small arrowheads point to a tail descending on the bacterium (B3 to B6), and an asterisk indicates actin cloud formation (B3 and B4). Fluorescence images (A1, A3, B1, B3, and B5) and differential interference contrast overlay images (A2, A4, B2, B4, and B6) are shown.
The degP mutant forms both actin tails and actin “clouds.”
To determine whether the degP mutant was impaired in its ability to form actin tails, the actin of Shigella-infected Henle cells was stained with TRITC-phalloidin. In infections with wild-type SA100, long actin tails focused on the pole of the bacteria were formed (Fig. 3B). The actin polymerized strictly at the bacterial pole, and there was no staining along the sides of the bacterium. In contrast, some of the SM1100 degP bacteria showed an altered phenotype. A small population of SM1100 appeared to be surrounded by actin clouds, possibly because IcsA was distributed on the entire surface of the bacterium (Fig. 3B). Another subpopulation of SM1100 did form actin tails, but compared to the actin tails on wild-type SA100, these tails were not as focused and covered more than the pole of the bacterium. In some of these, the outline of the entire bacterium could be seen (Fig. 3, panels B3 to B6). The actin outline may result from bacteria that targeted IcsA to the pole but did not maintain it there, resulting in a gradient of IcsA that continues over the entire bacterial surface. Altered actin polymerization, especially by those degP mutant bacteria forming actin clouds, may reduce the efficiency of intercellular spread for the degP mutant population.
DegP protease activity is not essential for IcsA localization.
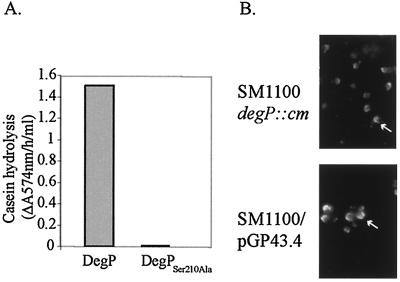
In the deduced amino acid sequence of SA100 DegP, two important regions of the DegP protein are conserved: the serine-protease catalytic triad and the PDZ domains, which are necessary for the formation of proteolytically active hexamer (38). In E. coli, DegP functions as a periplasmic chaperone at low temperatures and its proteolytic activity increases when the temperature rises from 30 to 42°C, presumably by undergoing a conformational change that makes the active-site serine (Ser210) more accessible (44). At 37°C, DegP may act as a chaperone or a protease, and Ser210Ala point mutations of E. coli DegP abolish protease function without changing chaperone activity (41, 44). The high sequence similarity between E. coli DegP and S. flexneri DegP suggested that Shigella DegP also could have both protease and chaperone activity. To determine if the protease activity was necessary for normal plaque formation by Shigella, an S. flexneri DegPSer210Ala point mutant was generated. DegP and DegPSer210Ala proteins were purified and assayed for proteolytic ability at 37°C using resorufin-labeled casein to confirm loss of protease activity by the mutant. Wild-type S. flexneri DegP was proteolytically active, but S. flexneri DegPSer210Ala was not (Fig. 4A). The plasmid pGP43.4, encoding the S. flexneri DegPSer210Ala point mutant, was tested for its ability to complement the SM1100 plaque defect in Henle cell monolayers. The SM1100 plaque size in Henle cell monolayers was restored by the plasmid-encoded DegPSer210Ala to 1.71 (±0.12) mm, comparable to the 1.69 (±0.09)-mm plaque size of SM1100 complemented with wild-type DegP encoded by pGP25.2. Restoration of wild-type IcsA localization in SM1100/pGP43.4 also was confirmed by indirect immunofluorescence (Fig. 4B). Thus, the protease activity of DegP is not essential for proper IcsA secretion and localization or for wild-type plaque formation by S. flexneri, suggesting that the role of DegP in IcsA secretion at the bacterial “cap” is associated with DegP chaperone activity.
FIG. 4.
Characterization of S. flexneri DegPSer210Ala activity. (A) Proteolytic activity of DegPSer210Ala using the substrate resorufin-labeled casein. (B) IcsA localization in broth-grown SM1100 and SM1100/pGP43.4 bacteria. IcsA “caps” are denoted by arrows.
DegP contributes to proper IcsA localization in E. coli.
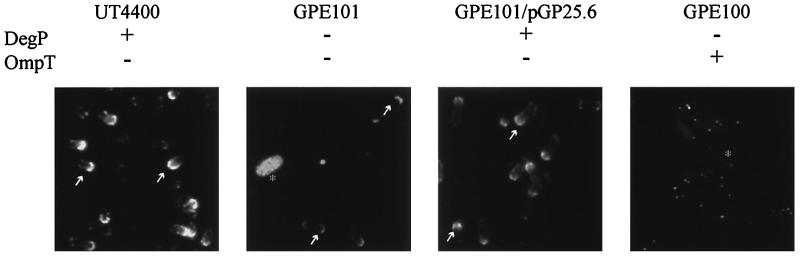
In wild-type E. coli, the OmpT protease cleaves IcsA, preventing the establishment of IcsA on the surface (30). However, IcsA is properly localized to the polar cap of E. coli ompT mutants, suggesting that IcsA localization is independent of Shigella-specific determinants (7, 36). In addition, DegP protease and possibly chaperone activity are conserved between Shigella and E. coli. To determine whether DegP is part of a general IcsA localization mechanism, IcsA was examined by indirect immunofluorescence in E. coli ompT and degP mutants where icsA was expressed from the plasmid pGP38.1. In the wild-type parent strain, E. coli AB1515, IcsA was not present on the surface (data not shown), presumably because the strain contains a functional OmpT. In the ompT deletion mutant UT4400, IcsA was delivered to the surface and localized to the bacterial pole of roughly 70% of the bacterium (Fig. 5). In GPE101, an E. coli degP ompT double mutant, only 30% of the bacteria had IcsA on the surface. Similar to the Shigella degP mutant, there was a reduction in the intensity of IcsA staining observed for the E. coli degP ompT mutant GPE101 compared to the DegP+ OmpT− strain UT4400 (Fig. 5). Interestingly, when IcsA localization was observed for GPE100, the E. coli degP mutant that contains a functional OmpT protease, 100% of the bacteria were expressing IcsA diffusely over the entire surface and had some punctate fluorescence. Since the degP mutant GPE100 is OmpT+, we had anticipated that OmpT would rapidly remove IcsA from the surface. IcsA localization in both E. coli degP mutants GPE100 and GPE101 could be restored to the respective DegP+ phenotype by the addition of the plasmid pGP25.6, which encodes S. flexneri DegP (Fig. 5 and data not shown).
FIG. 5.
Indirect immunofluorescence of E. coli strains expressing IcsA. The relevant phenotype is given for each strain. Arrows point to proper IcsA localization to the bacterial pole.
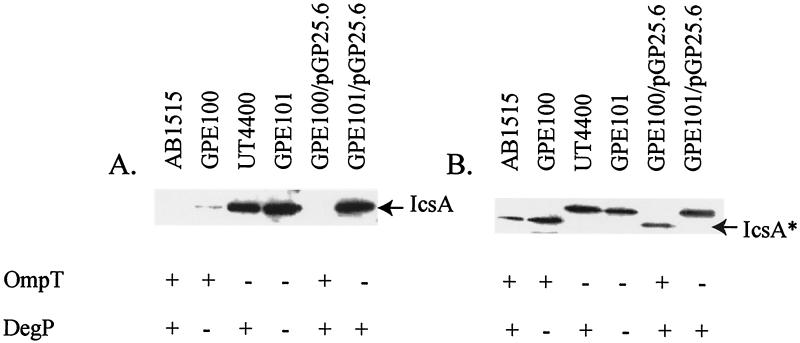
The reduced percentage of cells with IcsA on the bacterial surface for the E. coli degP ompT mutant GPE101 relative to the ompT mutant UT4400 could be due to either reduced synthesis or reduced secretion. Western blot analysis of E. coli mutants expressing IcsA was performed to distinguish between these two possibilities. We found that in the degP ompT mutant GPE101, the IcsA protein level was not reduced from that for the parent strain AB1515 or UT4400 (Fig. 6). Also, we found that the degP mutant GPE100 had a small amount of full-length IcsA associated with the bacterial cells, while the parent strain, AB1515, did not. These data suggest that for E. coli, as for S. flexneri, DegP is required for proper IcsA delivery to the surface and possibly for IcsA maintenance at the bacterial pole.
FIG. 6.
Western analysis of E. coli strains expressing IcsA. Whole-cell proteins (A) and proteins from culture supernatants (B) of E. coli strains expressing IcsA from pGP38.1 were analyzed with IcsA antibodies. The location of IcsA and the IcsA* fragment are indicated. The presence (+) or absence (−) of OmpT and DegP in each strain is indicated.
The degP mutant releases cellular proteins into the culture supernatant when grown at 37 or 43°C.
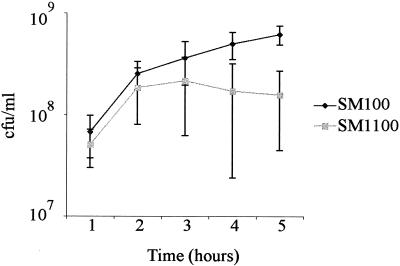
The ability of tissue culture assays to mimic all conditions encountered in the infection process is limited, so we investigated survival or growth of the degP mutant in other environmental conditions. E. coli degP mutants were described originally as temperature sensitive for growth (27). The potential temperature sensitivity of SM1100 relative to wild-type SM100 was examined by comparing survival at elevated temperatures. Bacteria were grown to mid-logarithmic phase at 30°C and then shifted to either 37 or 44°C. The number of viable cells was measured over time by plate counts. The shift from 30 to 37°C did not affect the viability of SM1100 (data not shown). After the shift to 44°C, SM1100 did not increase in number of CFU/ml from 2 to 5 h, whereas the number of CFU of the wild-type strain/ml increased over time (Fig. 7). This may be due to either lack of growth or lysis of growing SM1100 bacteria. When SM1100 was transferred from 30 to 43°C instead of 44°C, it grew at the same rate as SM100 (data not shown).
FIG. 7.
Survival of S. flexneri degP mutant at 44°C. SM100 and SM1100 cultures were grown for 2 h at 30°C until early log phase and then transferred to a 44°C environment. Standard deviation for three independent experiments is shown.
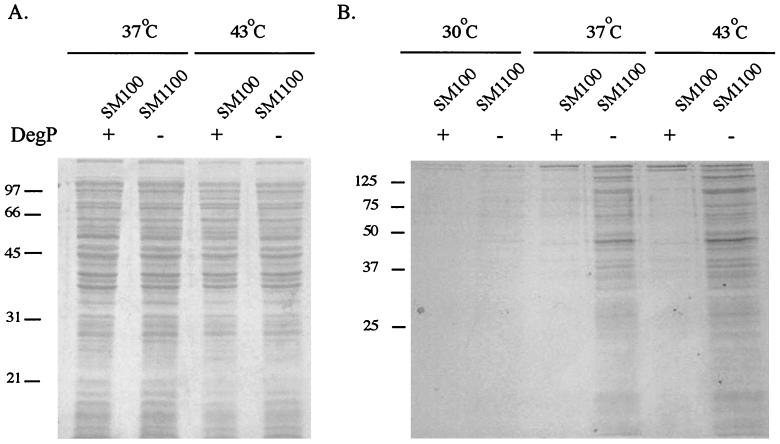
To further characterize the response of the degP mutant to high temperatures, strains were grown at the threshold temperature of 43°C. To determine if SM1100 lysed at elevated temperatures, SDS-PAGE analysis of culture supernatants was performed to look for proteins released into the supernatant. At both 37 and 43°C, the whole-cell protein profile of SM1100 was indistinguishable from that of SM100 (Fig. 8A). In contrast, there was more protein in the culture supernatant of the S. flexneri degP mutant than in that of wild-type SM100, especially at 43°C (Fig. 8B). The profile of the proteins present in the SM1100 supernatant more closely resembled that of the total cell protein than that of the subset of proteins secreted by wild-type SM100. This suggested that these proteins were not those normally secreted by the TTSS but could be indicative of bacterial lysis. To determine whether the proteins were a result of cell lysis, an enzyme-linked immunosorbent assay was performed to determine the amount of the cytoplasmic protein CAT present in the SM1100 bacteria and culture supernatant. The degP mutant SM1100 has a chloramphenicol resistance cassette encoding CAT inserted into the chromosomal degP gene. SA511, a chloramphenicol derivative of SA100 that contains the chloramphenicol resistance cassette on the pHS2 plasmid, was used as a DegP+ control. SM1100 and SA511 were grown at 30°C and then shifted to 37 or 43°C. Two hours after the temperature shift, the amounts of CAT protein in the bacterial cells and in the cell supernatant were determined. Our findings agree with those described previously for an E. coli degP mutant that released increasing amounts of CAT protein into the supernatant at elevated temperatures (23). When the mutant was shifted to 37 and 43°C, the cell supernatant contained 6 and 15% of the total CAT protein, respectively. Only 1% of the total CAT protein was found in the supernatant of the DegP+ strain with a shift to either 37 or 43°C. Release of cytoplasmic proteins into the supernatant by the degP mutant SM1100 but not SA511 indicated lysis or reduced integrity of the cell wall of the degP mutant at 43°C and to a lesser extent at 37°C.
FIG. 8.
Cells and culture supernatants of S. flexneri SM100 and degP mutant SM1100. (A) Whole cells from SM100 and SM1100 at 37 and 43°C analyzed by SDS-PAGE. (B) Culture supernatants from equal numbers of bacteria grown at 30, 37, and 43°C were precipitated and analyzed by SDS-PAGE. The gels are stained with Coomassie blue.
DegP is important for S. flexneri acid resistance in vitro.
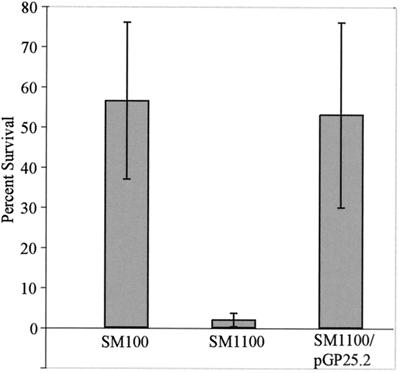
degP mutants in other organisms are sensitive to a variety of conditions other than temperature, including oxidative stress and osmotic stress. SM1100 had the same sensitivity profile as the wild type for a number of conditions, including oxidative and reducing stress, low osmolarity, high osmolarity, high salt, the detergents SDS and sodium deoxycholate, serum, and various dyes and antibiotics (data not shown). However, the survival of stationary-phase SM1100 was less than that of SM100 upon exposure to acidic conditions. The number of viable SM1100 cells decreased 10-fold relative to viable SM100 cells in Luria broth at pH 2.5, and this defect was complemented by degP expressed from the low-copy-number plasmid pGP25.2 (Fig. 9). Thus, DegP plays a role in survival in acidic conditions.
FIG. 9.
Survival of S. flexneri in acid. Percent survival of SM100, SM1100 (degP::cm), and SM1100/pGP25.2 (DegP+) after exposure to Luria broth (pH 2.5) for 2 h is shown.
DISCUSSION
DegP may play a role in S. flexneri virulence, because the S. flexneri degP mutant is impaired in the ability to spread intercellularly. We have shown that the S. flexneri degP mutant formed small plaques on Henle and Caco-2 monolayers, although SM1100 invaded Henle cells, grew within the cytosol, and induced apoptosis in macrophages. There were fewer SM1100 bacteria properly expressing IcsA on the surface compared to the wild type, and therefore only a small percentage of degP mutant bacteria are capable of polymerizing actin in a focused unipolar tail that can propel the bacteria within the infected cell and into adjacent cells. The reduced population of SM1100 bacteria that is able to spread from cell to cell most likely accounts for the small-plaque phenotype of the degP mutant.
Our characterization of IcsA localization in S. flexneri and E. coli degP mutants suggests that DegP may be important in a protease-independent manner for the efficient delivery of IcsA to the bacterial surface. As a member of the autotransporter family of proteins, IcsA is thought to be transported across the inner membrane in a Sec-mediated process but then translocates itself across the outer membrane (14, 15). Prior to this translocation, IcsA is present transiently in the periplasm in a protease-resistant folded conformation (5). As a chaperone, DegP may facilitate the folding of IcsA in the periplasm or the rapid transit of IcsA to the outer membrane. A defect in IcsA secretion in the Shigella degP mutant is suggested by the reduced percentage of SM1100 bacteria with IcsA on the surface, although the amount of IcsA made by the mutant was the same as for the wild type. The same defect was observed in the E. coli degP ompT mutant GPE101. In the E. coli degP mutant GPE100, which possesses functional OmpT capable of rapidly cleaving IcsA, a small amount of full-length IcsA was associated with the bacterial cells and a diffuse amount of IcsA was observed on the bacterial surface. A possibility exists that this small amount of IcsA was inappropriately delivered to the bacterial surface, and its cleavage site may be inaccessible to the outer membrane protease OmpT. Alternatively, it may be that the delivery of both IcsA and OmpT to the outer membrane is altered in the degP mutant GPE100, because IcsA was less efficiently cleaved from the bacterial surface than in the parental E. coli strain AB1515. The lack of proper IcsA localization in E. coli degP ompT mutants and in the Shigella degP mutant suggests that the proposed defect in delivery of IcsA to the outer membrane observed in GPE100 also occurs in these strains.
In addition to its role in IcsA localization, DegP is required for maximum resistance to acid, encountered by Shigella bacteria passing through the human stomach. S. flexneri survives at pH 2.5 for several hours, which contributes to the low infectious dose of this organism (43, 49). In addition, the degP mutation results in compromised membrane integrity for a fraction of the population, as shown by the increased percentage of CAT in SM1100 culture supernatants, and increased antibody staining of a periplasmic protein. This increased permeability accounts for the small proportion of SM1100 bacteria that were stained over the entire surface with α-IcsA antibody. However, the S. flexneri degP mutant is not sensitive to other environmental conditions, including oxidative stress, for which DegP is required in E. coli, S. enterica, B. abortus, and Y. enterocolitica (12, 21, 51), indicating that S. flexneri may respond to oxidative stress by another mechanism. DegP is required for full virulence of a number of organisms, presumably as a stress response protein that counters the various stresses in the host environment. For example, S. enterica, B. abortus, and Y. enterocolitica encounter oxidative stress while inside macrophages, and the respective degP mutants are at a disadvantage compared to the wild type in macrophage infections. DegP homologues in Pseudomonas aeruginosa, AlgW and MucD, were identified and may play a role in the transition to the mucoid phenotype that is associated with chronic respiratory infections in cystic fibrosis patients (4). In addition, Jones et al. (22) recently showed that degP mutants of the gram-positive pathogen Streptococcus pyogenes are attenuated for virulence in a mouse model of streptococcal infection. Since DegP is important for resistance to acid as well as efficient intercellular spread, it is likely a virulence factor of S. flexneri.
Acknowledgments
We thank E. Oaks for the generous gift of antisera and G. Georgiou for the E. coli ompT and degP mutant strains UT4400 and KS474. We also thank Barbera Goettgens and John Mendenhall for technical assistance with the confocal microscope. We thank Elizabeth Wyckoff and Laura Runyen-Janecky for critical review of the manuscript.
This work was funded by NIH grant AI16935.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx, F., and G. Georgiou. 1990. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 172:491-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandon, L. D., and M. B. Goldberg. 2001. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J. Bacteriol. 183:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavard, D., C. Lazdunski, and S. P. Howard. 1989. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J. Bacteriol. 171:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles, M., M. Perez, J. H. Kobil, and M. B. Goldberg. 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc. Natl. Acad. Sci. USA 98:9871-9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 10.Egile, C., H. d'Hauteville, C. Parsot, and P. J. Sansonetti. 1997. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol. Microbiol. 23:1063-1073. [DOI] [PubMed] [Google Scholar]
- 11.Elish, M. E., J. R. Pierce, and C. F. Earhart. 1988. Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J. Gen. Microbiol. 134:1355-1364. [DOI] [PubMed] [Google Scholar]
- 12.Elzer, P. H., R. W. Phillips, G. T. Robertson, and R. M. Roop II. 1996. The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect. Immun. 64:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda, I., T. Suzuki, H. Munakata, N. Hayashi, E. Katayama, M. Yoshikawa, and C. Sasakawa. 1995. Cleavage of Shigella surface protein VirG occurs at a specific site, but the secretion is not essential for intracellular spreading. J. Bacteriol. 177:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg, M. B., O. Barzu, C. Parsot, and P. J. Sansonetti. 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. Infect. Agents Dis. 2:210-211. [PubMed] [Google Scholar]
- 16.Goldberg, M. B., J. A. Theriot, and P. J. Sansonetti. 1994. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infect. Immun. 62:5664-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, T. L., and S. B. Formal. 1981. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Hong, M., Y. Gleason, E. E. Wyckoff, and S. M. Payne. 1998. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun. 66:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, M., and S. M. Payne. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24:779-791. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 22.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langen, G. R., J. R. Harper, T. J. Silhavy, and S. P. Howard. 2001. Absence of the outer membrane phospholipase A suppresses the temperature-sensitive phenotype of Escherichia coli degP mutants and induces the Cpx and sigma(E) extracytoplasmic stress responses. J. Bacteriol. 183:5230-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawlor, K. M., P. A. Daskaleros, R. E. Robinson, and S. M. Payne. 1987. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 55:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lett, M. C., C. Sasakawa, N. Okada, T. Sakai, S. Makino, M. Yamada, K. Komatsu, and M. Yoshikawa. 1989. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J. Bacteriol. 171:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, S. R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino, S., C. Sasakawa, K. Kamata, T. Kurata, and M. Yoshikawa. 1986. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell 46:551-555. [DOI] [PubMed] [Google Scholar]
- 29.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakata, N., T. Tobe, I. Fukuda, T. Suzuki, K. Komatsu, M. Yoshikawa, and C. Sasakawa. 1993. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 9:459-468. [DOI] [PubMed] [Google Scholar]
- 31.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 33.Payne, S. M., D. W. Niesel, S. S. Peixotto, and K. M. Lawlor. 1983. Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J. Bacteriol. 155:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, R. W., and R. M. Roop II. 2001. Brucella abortus HtrA functions as an authentic stress response protease but is not required for wild-type virulence in BALB/c mice. Infect. Immun. 69:5911-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandlin, R. C., and A. T. Maurelli. 1999. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect. Immun. 67:350-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sansonetti, P. J., C. Egile, and C. Wenneras. 2001. Shigellosis: from disease symptoms to molecular and cellular pathogenesis, p. 335-385. In E. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, New York, N.Y.
- 38.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 39.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying postinvasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 40.Shere, K. D., S. Sallustio, A. Manessis, T. G. D'Aversa, and M. B. Goldberg. 1997. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol. Microbiol. 25:451-462. [DOI] [PubMed] [Google Scholar]
- 41.Skorko-Glonek, J., A. Wawrzynow, K. Krzewski, K. Kurpierz, and B. Lipinska. 1995. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene 163:47-52. [DOI] [PubMed] [Google Scholar]
- 42.Skorko-Glonek, J., D. Zurawa, E. Kuczwara, M. Wozniak, Z. Wypych, and B. Lipinska. 1999. The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol. Gen. Genet. 262:342-350. [DOI] [PubMed] [Google Scholar]
- 43.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 45.Steinhauer, J., R. Agha, T. Pham, A. W. Varga, and M. B. Goldberg. 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol. Microbiol. 32:367-377. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, T., H. Miki, T. Takenawa, and C. Sasakawa. 1998. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 17:2767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, T., S. Saga, and C. Sasakawa. 1996. Functional analysis of Shigella VirG domains essential for interaction with vinculin and actin-based motility. J. Biol. Chem. 271:21878-21885. [DOI] [PubMed] [Google Scholar]
- 48.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 49.Waterman, S. R., and P. L. Small. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]
- 50.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zychlinsky, A., B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti. 1994. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol. 11:619-627. [DOI] [PubMed] [Google Scholar]