Abstract
NADH enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP) was evaluated for studying enzyme kinetics in vitro and in isolated mitochondria. Mass, optical, and nuclear magnetic resonance spectroscopy data were consistent with the UV NADH photolysis reaction being NADH → NAD· + H+ + e−. The overall net reaction was O2 + 2NADH + 2H+ → 2NAD+ + 2H2O, or in the presence of other competing electron acceptors such as cytochrome c, NADH + 2Cytox → NAD+ + H+ + 2Cytred. Solution pH could differentiate between these free-radical scavenging pathways. These net reactions represent the photooxidation of NADH to NAD+. Kinetic models and acquisition schemes were developed, varying [NADH] and [NAD] by altering NADH photolysis levels, for extracting kinetic parameters. UV irradiation levels used did not damage mitochondrial function or enzymatic activity. In mitochondria, [NADH] is a high affinity product inhibitor that significantly reduced the NADH regeneration rate. Matrix NADH regeneration only slightly exceeded the net rate of NADH consumption, suggesting that the NADH regeneration process is far from equilibrium. Evaluation of NADH regeneration in active mitochondria, in comparison to rotenone-treated preparations, revealed other regulatory elements in addition to matrix [NADH] and [NAD] that have yet to be fully characterized. These studies demonstrate that the rapid UV photolysis of NADH to NAD is an effective tool in evaluating the steady-state kinetic properties of enzyme systems. Initial data support the notion that the NADH regeneration process is far from equilibrium in mitochondria and is potentially controlled by NADH levels as well as several other matrix factors.
INTRODUCTION
Enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP) has recently been described as a method of monitoring enzymatic activities in microsamples in vitro or cells using confocal microscopy techniques (Combs and Balaban, 2001). This approach relies on the transient photobleaching or photolysis of a fluorophore product of an enzymatic reaction and observing its enzyme-dependent replenishment. The initial description of this technique was using NADH as the fluorophore in both in vitro samples as well as in imaging experiments in intact cells. Due to the key role that NADH plays in cellular energy metabolism as well as redox signaling and other cellular regulatory processes the development of this tool as a method of determining the cellular distribution of its production rate would, predictably, be very useful (for early examples of NADH detection and imaging studies, see Chance et al., 1962, 1965; Barlow and Chance, 1976). From the original description of this technique, several important issues remain with regard to the use of ED-FRAP for studying NADH metabolism. These include 1), determining the photolysis products of UV excitation of NADH; 2), the generation of a comprehensive kinetic model of NADH ED-FRAP in isolated enzyme systems; 3), the evaluation of nonspecific effects of photobleaching effects on mitochondria, in vitro; and 4), initiation of NADH ED-FRAP studies on intact mitochondria under a variety of conditions.
The common hypothesis is that NADH molecules are destroyed in the photobleaching process in cellular microscopy (see Combs and Balaban, 2001). However, in the photochemistry literature, the photolysis of NADH has been shown to potentially lead to the generation of the free radical of NAD (NAD*) together with a free electron. Both of these free radicals can reduce oxygen or any protein acceptor like oxidized cytochrome or myoglobin resulting in the production of NAD (Czochralska and Lindqvist, 1983; Cunningham et al., 1985; Orii, 1993; Boldridge et al., 1984). This approach has been used to good effect to study free-radical reaction kinetics of cytochrome c and competition with oxygen (Orii, 1993). The net product of NADH photolysis are NAD and reduced oxygen, proteins, or other free-radical products, with little change in the total NADH:NAD metabolite pool. Some evidence also exists for the irreversible generation of NAD dimers by photolysis that might occur due to cross-reaction between the NAD* and other molecules (Umrikhina et al., 1990).
Once the products of NADH photolysis have been confirmed, a kinetic model needs to be generated to analyze the ED-FRAP results. ED-FRAP is fundamentally different from other rapid kinetic approaches, such as stop-flow, temperature jump, NMR saturation transfer, or lineshape analysis, since it displaces a steady-state or equilibrium system by exchanging product for substrate, nearly instantaneously, providing unique insights as well as interpretation problems. Using the information obtained on the photolysis products of NADH, a kinetic model was developed and tested on equilibrium as well as essentially irreversible reaction systems.
Mitochondria play a key role in the regulation of energy in the myocardium and are the major sites of NADH generation as well as the source of most of the NADH fluorescence detected in the intact myocyte (Eng et al., 1989). Thus, NADH ED-FRAP studies in cardiac myocytes are likely dominated by the mitochondria activity. To evaluate the effects of UV irradiation and NADH photolysis on mitochondria, isolated mitochondria were studied where measurements on the metabolic rate, energy coupling, and function could be made in conjunction with the NADH ED-FRAP measurements. Finally, the isolated mitochondria provided an initial biological preparation to evaluate several quantitative aspects of NADH ED-FRAP experiments. Of initial primary interest was the flux capacity of the mitochondria to generate NADH relative to the net rate of consumption by oxidative phosphorylation under a variety of conditions.
To accomplish these aims, a specialized photolysis device was constructed to homogeneously irradiate relatively large samples for biochemical and functional analysis. Using this system, NADH ED-FRAP studies were conducted on several dehydrogenase systems and quantitatively analyzed. A system for performing serial NADH ED-FRAP studies on functioning isolated mitochondria was developed and NADH ED-FRAP studies conducted under a variety of conditions.
MATERIALS AND METHODS
Laser photolysis system
To generate large amounts of NADH photolysis products a relative large sample photolysis system was developed to generate 50-μl UV-irradiated samples. A pulsed laser was used in this study providing an ∼6-ns, 355-nM pulse (Continuum Surelite 1; Continuum, Santa Clara, CA). The peak power generated in this system was measured to be 80 mW over ∼0.2 cm2. The amount of photolysis was varied by changing the power of the laser pulse or the number of pulses impinged on a sample. The laser beam was equally split to impinge on both sides of the cuvette simultaneously to avoid inner filter effects generating unequal irradiation distribution within the reaction chamber. This was especially important in scattering samples such as mitochondria. A 3 × 3 mm quartz cuvette was used as the reaction chamber (model RF-303/45, Spectrocell, Oreland, PA) holding 50 μl solutions. NADH fluorescence was monitored using a fiber-optic-based spectrofluorometer previously described (Territo et al., 2000). Constant amplitude excitation was provided with a 500-W Hg/Xe arc lamp (Oriel, Stratford, CT) fitted with a 360-nm bandpass filter (model 46085, 360 ± 10 nM; Edmund Scientific, Tonawanda, NY). This low-power, focused, excitation source did not lead to measurable bleaching of the NADH fluorescence signal. Fluorescence was detected with a linear CCD array (model PC2000; Ocean Optics, Dunedin, FL). Data were digitally sampled over the spectral bandwidth of 335–1064 nM at 10 Hz with a 12-bit A/D converter.
Absorbance experiments were also conducted to evaluate different chromophores and light scattering variations after photobleaching. White light was transmitted through the reaction chamber using a 150-W tungsten-halogen source (model F0-150; Titan Tool, Buffalo, NY) coupled to the chamber via a single fiber optic. The transmitted light was sampled with the same detector as used for fluorescence. Light scattering was followed using six isosbestic wavelengths as described previously (Territo et al., 2001). Hemoglobin absorbance was followed as a measure of oxygen tension (Territo and Balaban, 2000), and cytochrome c (cytoc) absorbance change as an indicator of free-radical production.
Due to the complex optics used in this system, we did not attempt to regulate the temperature of the reaction chamber and all experiments were conducted at room temperature. To evaluate the effect of the laser pulses on sample temperature, a microtemperature probe was inserted into the chamber during the irradiation. A small transient (<2 s) increase of temperature of <0.1°C was only detected at the highest power (80 mW) in the presence or absence of NADH or mitochondria. No temperature changes were detected at lower powers. It was possible that even this small transient increase in temperature was due to direct absorbance of the UV light by the temperature probe and not true sample heating.
Optical monitoring of free-radical generation
To follow the generation of free radials and oxygen consumption, NADH solutions were prepared containing 200 μM of NADH in a 50-mM sodium phosphate buffer (pH = 7.4) at room temperature, with 20 mM oxidized cytoc to monitor free-radical production, or with 250 μM purified hemoglobin to optically follow oxygen tension. In some experiments the phosphate buffer was removed to aid in the observation of pH changes after the photolysis. The pH was measured either with a micro pH electrode (Yellow Springs Instruments, Yellow Springs, OH) or optically with SNARF (Molecular Probes, Eugene, OR), using the ratiometric evaluation of 580 vs. 638 nm emission with a 532-nm excitation.
Enzyme solutions
Three enzyme solutions were studied, as follows.
Alcohol dehydrogenase (ADH), which catalyzes the following reaction: Ethanol + NAD+ ↔ acetaldehyde + NADH + H+ (Kapp = Keq/[H+] = 10−2 at 25°C and at pH = 8.8). ADH was used as an enzyme model that strongly favors NADH formation. ADH was kept far from equilibrium by using a high-ethanol, alkaline medium, and trapping acetaldehyde with semicarbazide. The reaction media was 0.5 M ethanol, [NADH] (20–400 μM),
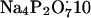 H2O (0.1 M), and semicarbazide (0.1 M).
H2O (0.1 M), and semicarbazide (0.1 M).Glutamate dehydrogenase (GDH): Glutamate + H2O + NAD+ ↔ α-ketoglutarate + NH4+ + NADH + H+ (Keq = 4.5 × 10−14 M2), was used as an equilibrium enzyme that favors NAD production. The reaction media was 5 mM α-ketoglutarate, 5 mM NAD+, 1 mM glutamate, and 300 μM NH4+, with a 50 mM Tris buffer at pH 8.
3-Hydroxybutyrate dehydrogenase (HDH): β-Hydroxybutyrate + NAD+ ↔ acetoacetate + NADH + H+ (K [H+] = 1.45 × 10−9 at 25°C and pH = 7) was used as another equilibrium system with similar kinetics as GDH. The initial reaction media used for HDH was 0.1 M KPO4 buffer, 12.5 mM Hydroxybutyric acid, and 2.5 mM NAD+.
Mitochondria preparation
Porcine heart mitochondria were prepared according to Territo et al. (2001), and were within the guidelines listed in the Animal Care and Welfare Act (7 USC 2142§13). Briefly, the mitochondria were isolated from the left ventricular myocardium, where the final mitochondrial suspension was maintained in Buffer A (137 mM KCl, 10 mM HEPES, 2 mM Pi, 2.5 mM MgCl2, and 0.5 mM EDTA at pH 7.2) at 4°C. Cytochrome a,a3 (cytoa) content was determined spectrophotometrically (Balaban et al., 1996). The experimental medium (Buffer B) contained: 125 mM KCl, 20 mM HEPES, 15 mM NaCl, 5 mM MgCl2, 1 mM K2EDTA, 1 mM EGTA, 2 mM Pi, 0.1 mM malate, and 4 mM TPP+. The pH was titrated to 7.1 at room temperature with KOH and HCl.
Mitochondria (1–6 nM/cytoa per ml) were preincubated while following oxygen consumption in a water jacket reaction chamber with a Clark oxygen electrode at room temperature. After a steady state had been established, a 50-μl sample was rapidly removed and placed in the cuvette for ED-FRAP analysis. Care was taken to assure that the entire ED-FRAP study was accomplished during the steady state of a metabolic perturbation based on the oxygen consumption data.
The concentration of the mitochondria NADH-NAD pool was estimated spectrophotometrically in Triton X-100 solubilized mitochondria. Mitochondria were first incubated at room temperature in the absence of any metabolic substrate in the presence of 300 μM ADP and 3 mM Pi. Under these conditions, the respiratory rate quickly approached zero and it was assumed that the NADH-NAD pool was essentially 100% NAD. The Triton X-100 extraction was then performed by diluting the mitochondria suspension 1:1 in 2% Triton X-100 in 100 mM KPO4 at pH 7.1. The absorbance at 340 nM was determined and then compared with the addition of 5 mM glutamate-malate to the extract that maximally reduced the NADH-NAD pool. The concentration of NADH-NAD pool was then determined using an extinction coefficient of 6510 M and a mitochondria matrix volume of 2 μl/nmol cytoa. Using this approach the total NADH-NAD pool was estimated to be 3.4 ± 0.2 mM (n = 7).
The fluorescence signal was assumed to originate mostly from the relatively immobilized NADH pool in the mitochondria matrix with an extended NADH fluorescence lifetime (Wakita et al., 1995). Previous studies have established that the binding of NADH enhances the lifetime and net efficiency of NADH fluorescence emission (Estabrook, 1962). NADPH is not a significant contaminant in these studies due to its lower concentration, four- to sevenfold lower than NADH (Livingston et al., 1996; Klingenberg et al., 1959; Chance et al., 1965) and fourfold-lower fluorescence enhancement in the matrix (Estabrook, 1962).
Mass spectrometry
NADH, NAD, and photolyzed NADH (1 mM) samples were prepared for mass spectroscopy. Mass spectra were run on a TSQ-700 spectrometer (ThermoFinnegan, San Jose, CA) at 0.5 μl/min at 3 kV. Different levels of photobleaching were obtained by varying the power and number of laser pulses providing a range of photolysis between 0 and 100%. Experiments were run on samples in pure distilled water to simplify the mass spectra.
NMR spectroscopy
NADH, NAD, and photolyzed NADH (400 μM) samples were prepared in 100-mM phosphate-buffered solutions (pH 7.0) made up in 99.8% D2O. 50-μl samples of NADH were irradiated with one or two laser pulses of identical power to photolyze NADH 25% or 50%. Ten 50-μl samples were pooled and placed into a 5-mm NMR tube. Spectra were collected at 300 MHz (Bruker Instruments, Billerica, MA) with no water suppression techniques. Spectra were processed using the MestRe-2.3 software (Santiago de Compostela, Spain) running on a standard PC. The residual water proton signal was used as a chemical shift reference at 4.697 ppm.
Data and statistical analysis
All of the optical spectroscopy data were collected and analyzed with custom programs written in IDL (Research Systems, Ver. 5.4, Boulder, CO) unless stated otherwise. Specific analysis methods are reported in the Results section. Where appropriate, values were expressed as mean ± SE, with a level of significance taken at p < 0.05.
RESULTS
Chamber characterization
To prove that the reaction chamber was homogenously irradiated, critical for the interpretation of the fluorescence recovery, FRAP studies were conducted on pure NADH solutions. The NADH fluorescence signal was immediately decreased by the laser pulse but remained constant for minutes after the irradiation. Similar results were obtained from studies using a nonmetabolized form of NADPH (α-NADPH) added to mitochondria suspensions to assure that the significant scattering properties of the mitochondria did not influence the irradiation pattern. Though the efficiency of a given laser power to photolyze NADH was reduced by the mitochondrial scattering, the homogeneous excitation patterns remained intact, based on the lack of any change in fluorescence intensity after the initial bleaching. These data are consistent with a homogenous irradiation of the reaction chamber without the generation of significant gradients of NADH in the chamber using this double illumination scheme. Thus, no correction for sample diffusion was necessary in these studies.
NADH photolysis products
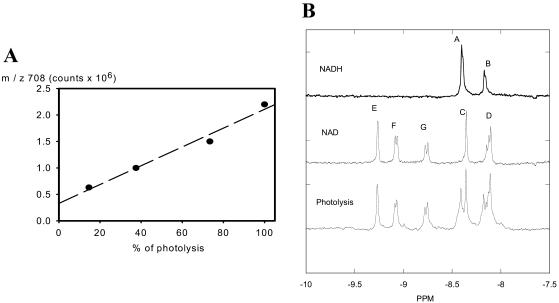
To evaluate the products of NADH photolysis, samples of NADH were photolyzed to different extents by varying the power and number of UV irradiation pulses. Electrospray mass spectrometry and proton NMR were used to evaluate the reaction products. In the former, NADH is detected as its disodium salt (mol wt 709). A 1-mM solution in pure water provides a very low abundance (M + H)+ ion at m/z 710 along with an even less abundant (M + Na)+ ion at m/z 732 (data not shown). Photolysis of this solution produces an abundant ion at m/z 708 corresponding to the (M + H)+ ion of the disodium salt of NAD. This peak increased with the degree of NADH photolysis, finally reaching a value (2.2 × 106 counts) close to that found in a 1-mM solution of NAD, to which two equivalents of sodium bicarbonate had been added (Fig. 1 A). It was not possible to similarly monitor the disappearance of NADH because of the very low abundance of m/z 710 and the fact that it was obscured by the second 13C satellite of m/z 708. In addition, its (M + Na)+ could be detected only until 30% photolysis. The enhanced intensity of the NAD ion relative to the NADH ion in these experiments is undoubtedly due to the presence of its permanently charged quaternary pyridinium group compared to the tertiary dihydropyridine present in NADH. The mass spectrometry data confirms that most of the reaction products from the photolysis of NADH is NAD, although species may be present that were not detected by mass spectrometry due to their failure to charge, or simply because they were below the limits of detection.
FIGURE 1.
Mass spectroscopy and NMR data on NADH photolysis. (A) Mass spectroscopy data. The relationship between the degree of NADH photolysis and the abundance of m/z 708 (disodium salt of NAD). Degree of photolysis was determined by 100 × (1−(NADHphotolysis/NADHc)) where NADHc is control fluorescence and NADHphotolysis is fluorescence after photolysis. (B) 300 MHz proton spectrums of NAD, NADH, and photolysis products of NADH. All samples were initially 0.4 mM in 100 mM phosphate buffer pH 7.0. Each spectrum is the average of 600 transients collected in 2-s intervals. Resonance assignments: NADH is A, C8 of adenosine, and B, C2 of adenosine. NAD is C, C8 of adenine, D, C2 of adenine, E, C2 of nicotinamide, F, C6 of nicotinamide, and G, C4 of nicotinamide. Assignments based on Sarma et al. (1968), Meyer et al. (1962), and Jardetzky and Wade-Jardetzky (1966).
The NMR data was consistent with the mass spectroscopy results. In Fig. 1 B, the proton spectra for a selected region of the proton spectrum is presented. A dose-dependent increase in NAD was associated with a proportional decrease in NADH. This is best illustrated from the NAD specific resonances labeled E–G. Specific peak assignments are presented in the figure legend. The generation of NAD was found to quantitatively match the loss of NADH in two irradiation powers analyzed by integrating these quantitative spectra. The signal-to-noise of these measurements suggested sensitivity on the order of 10%. Other than NAD+, no evidence of another significant product from NADH irradiation was found in these studies.
Another series of experiments were performed to show that the NAD+ formed from the NADH photolysis is active for enzymatic activity and further estimate the efficiency of the NADH to NAD+ photolysis process. A cuvette containing the reaction mixture for ADH (50 μl) with NADH (160 μM) was photolyzed with varying laser pulse power or number of pulses. ADH (20 U in 2 μl) was then injected into the cuvette to resynthesize NADH from the generated NAD. Data was corrected for the 4% dilution caused by the addition of ADH. NADH fluorescence was monitored during the entire process (see Fig. 2, insert). After photolysis, NADH fluorescence recovered rapidly with the injection of ADH (see Fig. 2, insert). These data are summarized in Fig. 2. The percentage recovery of NADH decreases roughly proportionally with the amount of NADH photolysis, i.e., (% recovery) = 100 − 0.22 (% photolysis), (R = 0.9). The decrease in efficiency with increasing NADH photolysis is likely due to the increase in free-radical generation along with the reduction of [O2] available to scavenge free radicals. Even with 80% photolysis of NADH, the efficiency of conversion to NAD+ was still ∼85%. Lower photolysis levels lead to a higher efficiency of conversion to NAD+. These data confirm the notion that the photolysis of NADH leads primarily to NAD+ and that this NAD+ is still enzymatically active. The small fraction of NADH not recovered in this process might be due to irreversible products generated in this reaction that were not detected in the mass spectroscopy or NMR experiments, especially if these products were different populations of small molecules.
FIGURE 2.
Enzymatic reversal of NADH photolysis. Insert represents a time course of a typical experiment. After NADH photolysis, ADH was injected into the cuvette and the NADH recovery determined. The photolysis percentage was calculated as outlined in Fig. 1, and recovery percentage was determined as 100*(NADHc–NADHADH)/(NADHc–NADHphotolysis), where NADHADH is the fluorescence level after the addition of ADH.
Free-radical generation
Previous studies have suggested that two free radicals are generated during the photolysis of NADH, ·NAD* and a free hydrated electron, that must be eliminated:
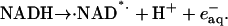 |
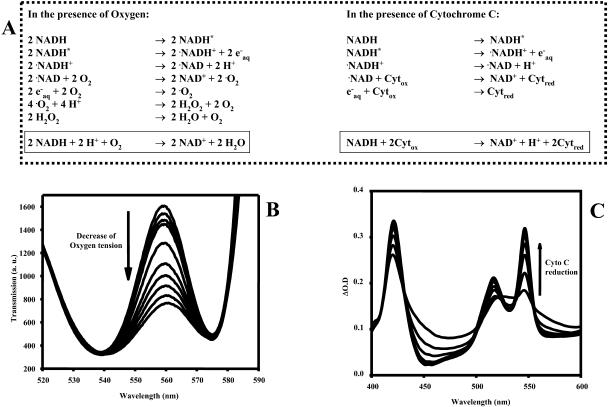
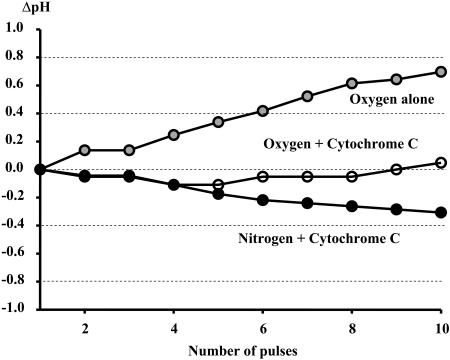
Two basic mechanisms for free-radical scavenging were evaluated that are relevant to cellular studies. The free radicals could be eliminated with reaction with oxygen or by any electron acceptor as outlined in the top of Fig. 3 (adapted from Orii, 1993). These pathways were experimentally verified as illustrated below each pathway. With regard to oxygen scavenging, the photolysis of NADH was found to decrease the oxygen tension, as determined from hemoglobin absorbance, and alkalize the medium (Fig. 4) as predicted for the oxygen free-radical scavenging pathway. With regard to electron acceptor pathways, the photolysis of NADH was associated with a reduction of added oxidized cytoc and an acidification of the medium in the absence of oxygen in the medium (Fig. 4). A “crossover” between these two pathways is possible through the slow reaction of the superoxide anion directly with oxidized cytoc generating oxygen and reduced cytoc (Orii, 1993); however, this reaction also proceeds as a net acidifying reaction since the protein is the final electron acceptor. The results not only confirm the free-radical scavenging pathways but also suggest that pH could be used to differentiate between these different pathways with regard to the final electron acceptor. When both oxygen and oxidized cytoc were present in the medium the pH remained neutral, suggesting these reactions competed equally for free-radical electrons (Fig. 4).
FIGURE 3.
Proposed mechanisms and products of NADH photolysis in the presence of O2 or oxidized cytoc. (A) Proposed reaction pathways for oxygen or oxidized cytoc for scavenging photolysis-generated free radicals. (B) Effect of NADH photolysis on hemoglobin (Hb) oxygenation. A series of UV pulses was applied to a 200-μM NADH sample including 250 μM of hemoglobin. The absorption spectrum of the hemoglobin is presented in a stack plot. The oxygenation of the sample decreased with each irradiation pulse as evidenced by the decrease in absorbance at ∼560 nm. Note that the nonlinear dependence of hemoglobin oxygenation on oxygen tension contributed to the nonlinear response of absorbance to the photolysis steps. (C) Reduction of cytoc during the photolysis of NADH. A series of UV pulse as applied to the sample of NADH including 20 mM of oxidized cytoc. An increase in cytoc reduction was observed by the absorbance changes at ∼550 nm. Absorbance changes at ∼520 and ∼416 nm were also characteristic of cytoc.
FIGURE 4.
Effect of NADH photolysis on pH. The solution pH was followed from SNARF fluorescence. Experiments were conducted on 1 mM in pure water in the absence of additional pH buffers. Experiments were conducted in the presence of oxygen, oxygen and oxidized cytoc (20 mM), or oxidized cytoc alone (nitrogen environment). As predicted from the equations in Fig. 3, the photolysis of NADH was associated with an increase in pH in oxygen, whereas in the presence of cytoc a net decrease in pH was observed.
Summary of NADH photolysis data
These data are consistent with the conversion of NADH to NAD+ as the major reaction in the photolysis of NADH. As a consequence of this reaction, free radicals are generated that can consume oxygen or directly reduce electron acceptors. No evidence for any product other than NAD+ was found in the mass spectroscopy or NMR studies. However, evidence for irreversible products of NADH photolysis was obtained in enzymatic assay for NAD+ where as much as 15% of the NADH was not recovered by ADH. This irreversible product is likely not a single product, since it was not detected in either NMR or mass spectroscopy, and likely is a variety of dimers or, more likely, other small molecules from free-radical reactions.
Effect of NADH photolysis on isolated mitochondria and mitochondria extracts
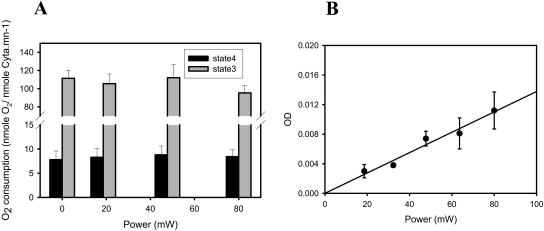
Previously it was demonstrated that ED-FRAP experiments could be conducted on isolated mitochondria in the confocal microscope (Combs and Balaban, 2001). Using the macroscopic technique developed for this study, we can evaluate the net biochemical consequences of the irradiation pulses on isolated mitochondria systems. Mitochondria suspensions (2–6 nmol cytoa/ml) were subjected to NADH photolysis from 20% to 60% using a laser power from 18.6 to 80 mW. The resting oxygen consumption (State 4), maximum rate of ADP-Pi driven oxygen consumption (State 3), and light scattering at 540 nm (at State 4) were evaluated before and after the irradiation (Fig. 5). The irradiation resulted in no significant change in State 4 or State 3 respiratory rates. A small proportional change in light scattering was observed with irradiation at State 4 consistent with small increases in matrix volume, however, these changes were very small in comparison to physiological manipulations of Ca2+ or carbon substrates (Territo et al., 2001). These results suggest that the respiration chain and oxidative phosphorylation are minimally affected at low UV power, or low free-radical loads induced by NADH photolysis. These data suggest that the mitochondria can tolerate the UV irradiation pulses used in these studies and have adequate mechanisms to eliminate the free radicals generated from NADH without significant damage to the oxidative phosphorylation process.
FIGURE 5.
Effect of NADH photolysis on isolated mitochondria respiration and light scattering. (A) Mitochondria respiratory rate at States 3 and 4 as a function of UV irradiation power. (B) Effect of UV irradiation on mitochondria light scattering at 540 nm.
We can estimate the amount of free-radical generation in mitochondria by NADH photolysis by assuming that matrix [NADH] is 3.4 mM in these quiescent mitochondria (i.e., the entire NAD-NADH pool is in its reduced form), thus a 30% photolysis of NADH results in the generation of >2 mM free radicals (two free radicals/NADH photolyzed) in a few nanoseconds, which require 0.5 mM oxygen to eliminate. Assuming a mitochondrial volume of 2 μl/nmol cytoa, this amount of oxygen corresponds to 1 nmol O2/nmol cytoa, which is insignificant with regard to the background State 4 oxygen consumption rate of ∼20 nmol O2/min per nmol cytoa and well below the maximum rate approaching 150 nmol O2/min per nmol cytoa at room temperature. Since the oxygen tension is only 0.2 mM, it is unlikely that all of the free radicals are consumed by oxygen even when supplemented by diffusion from the extramitochondrial space. For example, assuming that the mitochondrion is 1 μm in diameter and an oxygen diffusion coefficient of 3 × 10−3 mm2/s, the average time for diffusion of oxygen into the center of the matrix is ∼0.5 ms, relatively long for a free radical's lifetime. These calculations also suggest that the oxygen tension might transiently (microseconds) drop during the photolysis. However, as demonstrated in the experiments with oxygen and cytoc, other electron acceptors could contribute to the elimination of the free radicals. Thus, it is likely that the extensive free-radical scavenging system within the mitochondria, including the oxidized cytochromes, contribute to the scavenging process, in addition to molecular oxygen.
To attempt to evaluate the mechanism of the free-radical elimination process, the matrix pH (SNARF fluorescence) and cytochrome redox state was monitored optically under identical photolysis conditions. No significant change of matrix pH or cytochrome redox state was observed (data not shown). This suggests that either the combined reduction of oxygen and chromophores resulted in a near neutral pH reaction or that the proton buffering capacity of the mitochondria blunted the pH effect. The lack of redox change in the cytochromes suggests that the cytochromes themselves play a small role, despite the fact that cytochrome c and a are mostly oxidized under State 4 conditions (Chance and Williams, 1956), or that the redox effects are very transient and missed in the rather slow ∼100-ms temporal resolution of the current experiments. This latter conclusion is supported by the notion that the overall delivery of reducing equivalents by the photolysis is small relative to the normal flux through the cytochrome chain.
Since the free radicals from NADH might be generated in the immediate vicinity of the DH active sites, we also conducted a series of experiments evaluating the effects of irradiation on the NADH generating capacity of mitochondria extracts. After irradiating the intact mitochondria (2 nnol/nmol cytoa per ml) at different powers, an extract was made by diluting the mitochondria in a equal volume of Triton X-100 (2%). The total DH activity of this extract was estimated by monitoring the rate of NADH formation in a standard spectrofluorometer (PerkinElmer, Boston, MA) after adding 5 mM glutamate and malate along with varying amounts of NAD. No significant effect of the preirradiation of the mitochondria could be detected in the extracts (Fig. 6). Despite the fact that an extract is an imperfect model of the matrix conditions (see following kinetic studies), these paired experiments suggest that UV irradiation, under these experimental conditions, do not irreversibly damage the DH activity of the enzymes represented in this crude extract. Similar results were obtained for ADH, GDH, and HDH with powers up to 80 mW studied as outlined above in the absence of Triton X-100 (data not shown). These results are consistent with the notion that neither the direct UV illumination nor free-radical generation during NADH photolysis has significant adverse effects on isolated mitochondria function or NADH-generating enzyme complexes.
FIGURE 6.
Effect of pre-NADH photolysis on mitochondria extract DH enzymatic activity. The production NADH rate was monitored on extracts after preirradiated intact mitochondria with different UV power levels from 18.6 to 80 mW. Extract reaction was initiated by the addition of 5 mM G/M along with varying amounts of NAD. NADH levels were monitored spectrophotometrically in a standard cuvette instrument (PerkinElmer).
Kinetic modeling
The determination of NAD as a major product of NADH photolysis significantly changes the original kinetic interpretation of ED-FRAP (Combs and Balaban, 2001). Instead of extensively destroying NADH, the NADH pool is directly converted to NAD, the substrate for the formation of NADH, significantly altering the thermodynamics as well as [substrate] dependent kinetics. To our knowledge, this is a unique condition in the evaluation of enzyme kinetic reactions. Also important is the fact that it is applicable to the study of a key metabolic intermediate, NADH, both in vitro and in vivo. Since the degree of perturbation can be modulated via the irradiation power, the concentration dependence of many of these processes can also be directly evaluated. Due to this unique kinetic condition, we attempted to generate a theoretical model of NADH ED-FRAP to aid in the interpretation of the kinetic information obtained both in vitro and in vivo.
Since the analysis of NADH ED-FRAP depends on the temporal recovery of NADH after photolysis to NAD, most applications will involve reactions at steady state or near equilibrium. On considering the different approaches for extracting kinetic information, we concluded that the analysis of the initial rate of recovery was most advantageous since the concentration of other reactants, both known and unknown, can be assumed to remain constant over the initial time period. Examples of the initial rate analysis of an NADH ED-FRAP study are presented in Fig. 7. The main figure simply shows the time course of NADH fluorescence in a reaction mixture for HDH. Photolysis was initiated at the arrow. NADH recovered in a monotonic fashion to a near complete recovery. This initial rate was calculated from the first 0.5–1 s of the data maintaining a linear correlation coefficient of >0.99. The timing interval for the data collection was always evaluated to assure that the initial rate was not limited by the acquisition speed. The insert shows the effect of increasing the irradiation power on the recovery process. Increasing the degree of NADH photolysis increased the initial rate of NADH regeneration. This is not surprising since the initial concentration of the product (NADH) decreased whereas the reactant (NAD) increases with NADH photolysis.
FIGURE 7.
Time course of NADH fluorescence in an ED-FRAP experiment. Experiment was performed on the HDH system outlined in Methods. Analysis focused on the initial rate of recovery that was linear for the first 1-s measurement. Different levels of NADH photolysis (shown in small panel) essentially simultaneously varied the [NADH] and [NAD] aiding in the extraction of kinetic parameters for the reaction. Note that in this specific case, the full recovery can be considered as a simple monoexponential process.
For in vitro experiments, where little or no net flux occurs (i.e., reaction at equilibrium or steady state), the initial rate for the recovery can be analyzed as a Bi-Bi kinetic mechanism, as below, that is commonly used for many dehydrogenase reactions.
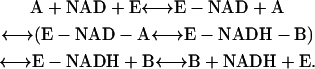 |
The initial velocity expression for this mechanism is complex, but some rearrangements can be performed to express it as a function of [NADH] and [NAD]:
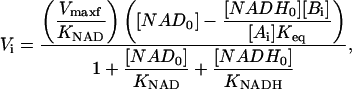 |
(1) |
where the parameters Vmaxf, KNAD, and KNADH are complicated aggregates of rate constants and [Ai] and [Bi], but are proportional to the forward velocity, the affinity constant for [NAD], and the inhibition constant of [NADH] respectively, and thus will be called apparent constants. The subscripts i and 0 refer to initial values before the photobleaching and just after the photobleaching, respectively, and 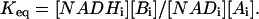
To estimate these apparent kinetics parameters the following strategy was derived. The extent of the photolysis was expressed as shown below for [NADH0] and [NAD0].
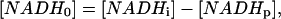 |
(2) |
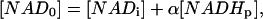 |
(3) |
where [NADHp] is the amount of NADH photolyzed and α is the fraction of photolyzed NADH that was converted to NAD. This later term was to take into account any irreversible changes in the metabolite pool caused by the irradiation. The α-value was determined by comparing the initial value of NADH with the final value after complete recovery. In each experimental condition, a characteristic α-value was found that was constant over the entire range of [NADHp] generated and was in the range of 0.75 to 1 in the isolated enzyme systems.
Combining Eqs. 1, 2, and 3 results in a characteristic equation of the initial velocity in an ED-FRAP experiment using the model
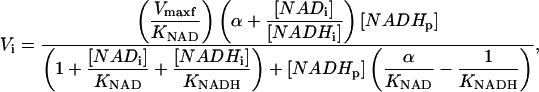 |
(4a) |
or in a simplified form, as
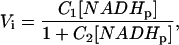 |
(4b) |
with
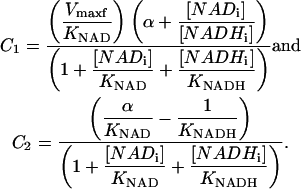 |
Thus, in the Lineweaver-Burk form, a linear relationship between 1/Vi and 1/[NADHp] should be observed as
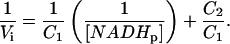 |
(5) |
Using this approach, two simple situations can be evaluated to extract the kinetic parameters, Case 1 and Case 2, as follows.
Case 1
[NADi]/[NADHi] ≫ 1 and [NADi] ≫ KNAD, Eq. 4a) simplifies to
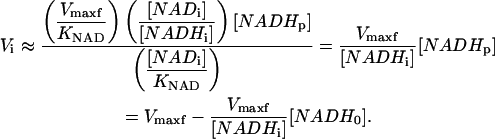 |
(6) |
In this case, the initial slope analysis allows the direct determination of Vmaxf from an ED-FRAP experiment by varying [NADHp] (by extrapolating to [NADHp] = [NADHi], or by plotting Vi versus [NADH0], and extrapolating to [NADH0] = 0). Note that in this case, the full recovery is monoexponential, and a rate constant k = Vmaxf/[NADHi] can be extracted again assuming that any changes in [Ai] and [Bi] are negligible.
Case 2
[NADi]/[NADHi] ≪ 1, and [NADi] ≪ KNAD (NADi small), Vi is given by
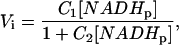 |
(7) |
with
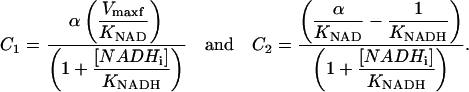 |
Thus, by varying [NADHp], it is possible to extract C1 and C2. Additional experiments with different [NADHi] values permit extraction of all kinetic parameters.
Kinetic measurements on isolated dehydrogenases in vitro
This approach was first tested in vitro on the three enzyme systems presented in the Methods section. GDH and HDH are Case 1 systems with the reaction favoring the production of NAD, and [NADi] ≫ KNAD. As expected, a monoexponential recovery of NADH was obtained for both systems, with a rate constant proportional to [E] (not shown) (Combs and Balaban, 2001). The initial slope analysis is shown for the GDH experiments in Fig. 8. An increase of the initial slope is observed when the amount of NADH photolysis to NAD is increased. This increase is due to an increase of [NAD] as shown by the linear relationship between initial slope and initial NADH0 as expected. Extrapolating to [NADH0] permits the estimation of Vmaxf. A linear relationship was established between this extrapolated Vmaxf value and [E] as predicted. Similar qualitative results were obtained for the HDH system with different kinetic constants being extracted (raw data for HDH shown in Fig. 7).
FIGURE 8.
Initial slope analysis of GDH NADH ED-FRAP experiments. (A) Linear regression of initial NADH recovery after photobleaching for different levels of NADH photolysis. Note the increase of the slope when the amount of [NADH] is decreased and [NAD] is increased. (B) Linear relationship between initial slope and initial [NADH0] level. To extrapolate to [NADH0] = 0 allows obtaining an estimation of Vmaxf; see text. (C) Estimated Vmaxf vs. [GDH] concentration. The linear relationship shows that the estimated Vmaxf is well related to enzyme concentration.
ADH is an example of Case 2 with a high NADH/NAD with the Vi described by Eq. 7. The Vi increased with [NADHp] with a Michaelis-Menten dependence on [NADHp]. Thus, for each initial value of [NADHi], an estimation of C1 and C2 can be obtained by performing a fit using Eq. 7 (Fig. 9). Then, by plotting 1/C1 vs. [NADHi] or 1/C2, it is possible to extract all the parameters. We found KNADH = 325 μM, KNAD = 40 μM, and Vmaxf = 19 μM/s. Finally, by varying the concentration of [ADH], we confirmed that the extrapolated Vmaxf (or the term C1) is linearly related to the [ADH].
FIGURE 9.
Initial slope analysis of ADH NADH ED-FRAP experiments. (Top left) Relation between initial slope and amount of photolyzed NADH. A classical Michaelis-Menten relationship is observed as expected. (Top right) Linear relationship observed between the parameters 1/C1 and [NADH] consistent with Eq. 7. (Bottom left) Effect of [ADH] variation on the relation between the initial recovery and the amount of photolyzed NADH. (Bottom right) Demonstration of the dependence of the parameter C1 of Eq. 7 with [ADH].
NADH ED-FRAP in isolated mitochondria
We evaluated the use of NADH ED-FRAP to measure the NADH generating capacity of isolated mitochondria relative to the net utilization of NADH by oxidative phosphorylation. There are several consequences of attempting this analysis in the rather fixed environment of the mitochondria matrix. The total NAD-NADH pool will remain constant at ∼3.4 mM and is difficult to manipulate as an independent variable. All mitochondria dehydrogenase reactions will contribute to NADH formation in the NADH ED-FRAP experiment potentially leading to highly complex kinetics. Logically, the reactions with the highest activity will dominate the global NADH ED-FRAP measurement. However, NADH ED-FRAP will accurately reveal the net capacity of intermediary metabolism to generate NADH from all of these sources. Finally, unlike the isolated enzyme systems, a net flux through the reaction sequence occurs due to oxidative phosphorylation and/or proton leaks in the inner membrane. This later aspect will require modifications to the models presented above.
No general information on the relative kinetic constants for NAD or NADH is available for the mitochondria matrix and some information on these parameters will be useful in simplifying the kinetic analysis of the data. This is especially the case in mitochondria where the absolute concentration of NADH-NAD cannot be changed as in the isolated enzyme system. To estimate the kinetic constants for the mitochondria dehydrogenase system, we used a Triton X-100 extract of the mitochondria preparation as described in the Methods section. NADH ED-FRAP experiments in Triton X-100 mitochondria extracts demonstrated a robust NADH recovery demonstrating that active matrix DH was present and that the NADH ED-FRAP effects were not dependent on an intact mitochondrial membrane system (data not shown). Classical nonequilibrium methods were used to determine the net apparent KNAD, KNADH, and Vmaxf of the DH reactions in the Triton extracts. The extract KNAD was found to be 107 ± 17 μM (n = 5). The KNADH was very low (<15 μM) as estimated from the concentration dependence of NADH inhibition on the forward reaction. The Vmaxf was only 26 nmol NADH/nmol cytoa per min in this system, well below the maximum rate of NADH consumption at State 3. These later data suggest that suboptimal conditions for NADH generation were present in Triton extracts that include an ∼1000-fold dilution of matrix reactants and a direct inhibition of some enzyme activity. For example, succinate dehydrogenase was found to be inactive in Triton extracts (i.e., no reduction of NADH by succinate). This low activity suggests that some caution must be taken in extrapolating these results to the intact mitochondria conditions, but should provide some guidelines for the kinetic constants involved.
For our initial experiments, the kinetic analysis was simplified by providing the mitochondria in Buffer B with NADH-linked substrates (5 mM glutamate and malate) in the presence of the site 2 inhibitor rotenone (40 μM) with ATP (5 mM), Pi (5 mM), and free Ca2+ (500 nM). The latter agents were added to optimize the NADH recovery rate as would be discussed below. Rotenone was added to minimize the net flux of NADH utilization by respiration or FADH-linked reverse electron flow (Combs and Balaban, 2001) but maintain oxygen high and cytochromes oxidized for free-radical scavenging. An example of a mitochondrial NADH ED-FRAP experiment in the presence of rotenone is shown in Fig. 10. As in the enzyme systems, the rate of NADH recovery increased with increasing NADH photolysis similar to that observed with in vitro enzymes (Fig. 7) and within intact cells (Combs and Balaban, 2001). The incomplete recovery of the NADH after bleaching is partially due to the bleaching of background, non-NADH fluorophores in the mitochondria that accounted for almost 15% of the fluorescence signal, though some lack of recovery of the NADH pool may be occurring. To convert initial rates to a quantitative measure of NADH production, it was assumed that the total NAD-NADH matrix concentration was 3.4 mM. Fully reduced NADH was established with rotenone or anoxia that provided a very reproducible value. The fully oxidized state was estimated from the signal level in the presence of ADP and Pi in the absence of carbon substrates. This oxidized value was less reliable due to the aforementioned non-NADH fluorescence from the mitochondria and the effect of photobleaching. This led to as much as an ∼15% variability in this parameter when normalized to cytoa content. The amount of NADH was determined from the cytoa content by assuming 2 μl of matrix volume/nmol cytoa. This resulted in a final factor of 6.8 nmol NAD-NADH/nmol cytoa.
FIGURE 10.
NADH ED-FRAP experiments on isolated porcine heart mitochondria. Mitochondria were pretreated with 40 μM of rotenone to block any net flux through the system. (A) Time courses of NADH recovery after NADH photolysis. Variable degrees of NADH photolysis was accomplished by varying UV laser power from 18.6 to 80 mW. (B) Observed relationship between initial slope and the amount of NADH photolyzed, consistent with Eq. 7.
For kinetic analysis, we assumed that rotenone-treated mitochondria with no net flux is similar to Case 2 presented above with NAD/NADH ≪ 1. With in vitro enzymes it was demonstrated that the initial velocity in ED-FRAP experiments could be expressed as
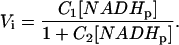 |
This provides a simple two-parameters equation to fit the initial rate data as a function of [NADHp]. An example of such a fit is presented in Fig. 10 along with the Vi vs. [NADHp] data from mitochondria. To estimate KNAD/KNADH and Vmaxf from C1 and C2, the following approximations were made based on the Triton extracts and measured quantities in the rotenone study: KNAD = 0.1 mM; KNAD/KNADH (>6); and NADHi = 6.8 nmol/nmol cytoa. These assumptions reduce C1 and C2 to
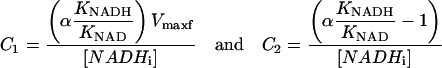 |
Using the data presented in Fig. 11, we obtained C1 = 6.57 (0.79) and C2 = −0.14 (0.01) (SD). If we assumed that [NADHi] in the presence of rotenone = 6.8 nmol/nmol cytoa and α = 1, it gives an estimation of Vmaxf ∼1400 nmol NADH/nmol cytoa per min and (KNAD/KNADH) ∼30. Decreasing 1 to 0.5, to account for any irreversible modification of NADH, only reduced the ratio of KNAD/KNADH to 15 while not affecting Vfmax. Since no direct evidence for an α-irreversible product of NADH photolysis was found in vitro, we assumed the value of α = 1 for these studies. These Vmaxf values can be compared to the net utilization of NADH under State 4 and State 3 conditions in mitochondria under identical conditions in the absence of rotenone by measuring the oxygen consumption and assuming 2 NADH/O2. The NADH consumption rate was 20 nmol NADH/nmol cytoa per min at State 4 and 160 nmol NADH/nmol cytoa per min at State 3. The calculated Vmaxf value is ∼70× higher than State 4 rate and 10× the State 3. However, Vmaxf represents the forward rate in the absence of NADH, not a realistic condition. We can estimate the unidirectional fluxes for the observed DH reaction knowing the NADH/NAD ratio under different experimental conditions. Taking the State 3 and 4 conditions with the NADH/NAD ratio of 0.4 and 4, respectively, we can estimate the forward (Ff) and back (Fb) fluxes in this model. Using the following approximations of apparent equilibrium constant = 20, and KNAD = 100 μM, and KNAD/KNADH = 30, State 4 is Ff = 23 nmol NADH/nmol cytoa per min and Fb = 5 nmol NADH/nmol cytoa per min, and State 3 is Ff = 187 nmol NADH/nmol cytoa per min and Fb = 4 nmol NADH/nmol cytoa per min. The forward rates in these comparisons are very close to the estimated NADH consumption rate under these conditions, and the reverse flux was exceedingly small due to the inhibitory action of NADH on this process. The implications of these observations will be reviewed in the Discussion section.
FIGURE 11.
Comparison of rotenone, State 4, and State 3 mitochondrial NADH ED-FRAP initial rate data. The initial recovery rate is plotted versus the [NADHo] in nM NADH/nM cytoa. (Rotenone, solid circles; State 4, unfilled circles, and State 3, solid triangles.) The shift in the curves is due to the differences in the starting [NADH] under the three different conditions.
To assure that the mitochondria NADH recovery rate measured in the presence of rotenone was monitoring a relevant process, we evaluated the effect of Ca2+ on the NADH recovery process. Ca2+ has been well established to activate the net DH activity of these mitochondria (Balaban, 2002). In a series of studies, mitochondria were initially incubated in the standard media in the presence of rotenone without the addition of Ca2+. NADH ED-FRAP measurements were conducted and then the free Ca2+ was raised to 500 nM and experiments repeated. From NADHo levels of 50–75% of control the recovery rate was enhanced 30 ± 4% (n = 5, p < 0.0001) in the presence of Ca2+ consistent with the activation of several DH reactions by Ca2+ (Denton and McCormack, 1986). Similar effects were obtained by the removal and reintroduction of Pi (3 mM) or ATP (3 mM) to the mitochondria medium. The recovery rate was insensitive to additions of ADP (up to 1 mM) or free-radical scavengers, including butylated hydroxytoluene (BHT) (up to 3 mM). These later experiments resulted in the incubation medium containing Ca2+, ATP, and Pi to optimize the NADH recovery rate. Subsequent studies will be required to establish the mechanism of these molecules on the NADH recovery process.
State 4 and State 3 conditions
Rotenone was used to eliminate any net flux through the NADH to simplify the analysis. However, in cells and under most physiologically interesting conditions, a net flux of reducing equivalents through the NADH pool will occur. A net flux can be envisioned as a competing oxidation of NADH as schematically presented below,
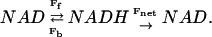 |
The NAD and NADH is exchanged through the mechanisms probed in the rotenone data whereas the Fnet flux is a new reaction corresponding to the utilization of NADH to reduce oxygen. Intermediate exchange reactions such as, e.g., FAD or cytochromes, were ignored in this simple model. For fitting purposes the kinetic constants (e.g., affinity) for supporting Fnet was considered to be independent from the constants involved in Ff and Fb.
This simple model was tested under two conditions, State 4 (low net flux) and State 3 (maximum net flux) (Chance and Williams, 1956). State 4 mitochondria were prepared by simply excluding rotenone from the Buffer B solution containing Pi, ATP, and Ca2+. Experiments were conducted while the oxygen tension was well above 10% saturation. For State 3 mitochondria, 1 mM ADP was added to the mitochondria on entry to the reaction cuvette. This was to assure that anoxia was not reached before the NADH ED-FRAP experiment could be completed. Anoxia was easy to detect, and therefore avoid, due to the large increase in NADH fluorescence that occurs in comparison to the State 3 levels. The initial rates as a function of [NADHo] from the NADH ED-FRAP experiments on State 4 and State 3 mitochondria are presented in Fig. 11 together with the rotenone data, for comparison purposes. The offset in the data is due to the differences in the control NADH/NAD levels under the different conditions with NADH being the highest in rotenone followed by State 4 and then State 3. The variable photolysis levels generated by either laser power or number of pulses (rapidly applied at <50 ms between pulses) permitted the direct comparison of the rate at identical NADH/NAD ratios. Clearly both State 3 and State 4 conditions resulted in a lower recovery rate than in rotenone. In addition, State 3 was slower than State 4 for a given NADH/NAD ratio. Attempting to model this process using the model presented above failed. The addition of Fnet, with independent kinetic parameters, to the kinetic parameters of the rotenone studies for Ff and Fb, either using a numerical simulation or the linearization strategy outlined above, failed to converge on reasonable kinetic values. This is likely due to the several differences in the matrix environment in the presence of rotenone when compared to State 4 or State 3 conditions that affect the kinetic aspects of this process in addition to Fnet. These include an increase in mitochondria membrane potential and pH gradient, higher reduction level of FADH, and cytochromes in States 3 and 4 when compared to the rotenone condition. Just the back flux from FADH could result in a significant complication of the interpretation of the NADH initial rate data (see Combs and Balaban, 2001). More experiments will be required to characterize the reverse electron flow contribution as well as the potential role of DY and cytochrome redox state. However, these data are consistent with the notion that the rate of NADH regeneration is inhibited in the presence of net flux when compared to the rotenone conditions.
DISCUSSION
This study demonstrates that NADH ED-FRAP can be a useful tool to study enzymatic processes involving NAD and NADH redox reactions in vitro and, predictably, in vivo. These studies confirmed that the major product of photolysis of NADH by UV light is NAD. Thus, NADH itself can be used as a endogenous form of “caged” NAD (Cohen et al., 1997). The photolysis of NADH was shown to generate free radicals that can be neutralized with either oxygen or protein electron acceptors (oxidized cytoc in this case) consistent with previous studies. Evidence is presented for limited damage by these free radicals in isolated enzyme or mitochondria preparations undergoing the NADH-EDFRAP experiment. A kinetic model was derived to aid in the interpretation of the complex NADH recovery kinetics in NADH ED-FRAP of enzyme systems. This approach was evaluated for different enzyme model systems, in vitro. On isolated mitochondria, it was shown that the NADH generation capacity of the mitochondria could be directly observed but revealed very complex regulatory processes.
NADH photolysis
The NADH ED-FRAP experiment was initially presented as an approach where NADH is destroyed in the photobleaching process (Combs and Balaban, 2001). In the current study, more defined systems were used to characterize some of the biochemical and kinetic properties of this approach. The first part of this study was focused on the mechanism of UV photobleaching/photolysis of NADH. It has been previously shown that UV irradiation of NADH can produce a superoxide anion, NAD* free radical, and hydrated electron (Czochralska and Lindqvist, 1983; Orii, 1993; Cunningham et al., 1985; Umrikhina et al., 1990; Boldridge et al., 1984). These free radicals can further react with electron acceptors like oxygen, cytochrome c, or myoglobin to generate reduced products and NAD+. We confirmed that the major product of NADH photolysis is NAD+ under our experimental conditions via mass spectrometry and NMR spectroscopy as well as enzymatic methodology. The photolysis of NADH can be associated with a reduction of molecular oxygen or cytoc. The relative contribution of oxygen or other proteins as free-radical scavengers can be determined from the change in pH. The reaction with oxygen results in an increase in pH while a reaction with protein results in a net decrease in pH. When oxidized cytoc and oxygen were mixed, no change in pH was detected, suggesting that oxygen and cytoc are equally contributing to free-radical scavenging.
Two issues are of some concern with this approach. First, two moles of free radicals are generated per NADH photolyzed. This is a considerable free-radical load in the mitochondria where a 30% photolysis of NADH should result in ≈1 mM of free radicals in the matrix (assuming [NADH] ∼3.4 mM). However, these radicals can be scavenged by a variety of mechanisms and no evidence for adverse effects of these free radicals were found in intact mitochondria, mitochondria extracts, or isolated enzymes. In intact cells or mitochondria, a considerable defense mechanism is present that reduces the lifetime of free radicals and this will likely contribute to the lack of biochemical effects observed in those systems. The lack of pH change in the matrix with NADH photolysis suggests that both oxygen and proteins are equally contributing to free-radical scavenging. However, the matrix proton buffering capacity might dampen any changes in pH even though mM [H+] will be created or removed if one mechanism dominated.
The second issue was the lack of complete recovery of NADH after photolysis in some systems, suggesting an irreversible conversion of NADH. This was most notable in situations where the NAD pool, that could be used to replenish “damaged” NADH, was relatively small (i.e., Case 2 enzyme systems and highly reduced mitochondria, i.e., State 4 and rotenone). Some contribution of the small NAD(P)H pool in the mitochondria studies also cannot be ruled out in this process. The source of this irreversible behavior is still unclear. The lack of a large single component other than NAD observed by mass spectrometry or NMR spectroscopy during the photolysis of NADH in simple solutions suggest that some of these effects might only occur in the presence of more complex solutions. NMR studies on photolyzed NADH in the ADH reaction mixture failed to reveal any new products or significant discrepancy between the ratio of NADH disappearance or NAD production (not shown). However, some small fraction of NADH covalently binding to ADH (via a free-radical reaction) or a wide dispersion of different smaller molecules would not have been observed. In any event, no adverse effects of these photolysis products were detected in the enzyme or mitochondria systems studied. Specifically, more studies are required on the photolysis products of NADH in the mitochondria matrix.
Kinetic models
We confirmed previous studies that demonstrated that the rate of NADH recovery was proportional to [enzyme] concentration (Combs and Balaban, 2001) for two other enzyme systems, in vitro. Using the biochemical information that NAD is being generated from NADH, we created a simple kinetic model to estimate useful parameters from these data. Since NADH photolysis simultaneously reduces product (NADH) while increasing a reactant (NAD), both forward and reverse flux rates need to be considered. We reasoned that the initial slope measurement was the most robust form of analysis since it permitted the assumption that the metabolite concentrations were constant and in some conditions permitted the extrapolation to the true initial rate condition with [NADH] = 0 (Vmaxf and KNAD/KNADH). Using the simplest kinetic model of this process outlined in Eq. 1, we were able to extract basic kinetic characteristics from model systems poised at very different equilibrium conditions for the NADH:NAD redox couples. Important in this analysis was the ability to alter the concentration of NADH and NAD by varying the extent of conversion of NADH to NAD with the laser power. This permitted determination of the concentration dependence of the reactant and product in a very similar manner that occurs in the intact cell where the NADH:NAD pool is essentially constant during acute perturbations. The extracted kinetic values were in good agreement with literature values for these enzymes using more conventional approaches (for example, KNAD ∼ 40 μM for ADH; Wratten and Cleland, 1963). These data suggest that this approach of varying the concentration of NADH and NAD by varying the degree of NADH photolysis in an NADH ED-FRAP experiment should provide a useful tool in evaluating the kinetics of enzymatic processes both in vitro and in vivo.
Isolated mitochondria studies
NADH ED-FRAP was capable of monitoring the rate of NADH regeneration in intact isolated mitochondria. Like most measurements, there is some perturbation of the system to make the measurement. The greatest concern with this approach is the relatively high power UV pulse used in photolysis of NADH. The pulsed UV irradiation used in these studies did not affect the net rates of respiration in State 3 and State 4 or significant changes in light scattering. These data suggest that any adverse effects of the irradiation were not significant on the overall membrane integrity or oxidative phosphorylation. Due to the considerable amount of free radicals generated by the NADH photolysis, we also evaluated the effect of a free-radical scavenger, BHT, on the NADH recovery rates. No effect of 3 mM BHT was found on the NADH recovery rates, suggesting that the inherent free-radical scavenging system of the mitochondria was adequate to handle the free-radical load introduced. These data are consistent with the notion that the perturbation to the system due to the UV light pulse was minimal to the overall function of the mitochondria.
In the first series of studies the net flux through the NADH pool was inhibited at site 2 with rotenone. This permitted the simple analysis of the NADH regenerating capacity in a similar manner as used for the isolated enzyme systems with no net flux. The extent of conversion of NADH to NAD was varied with laser power or the number of irradiation pulses, and resulted in an increasing rate of NADH regeneration with the NAD/NADH ratio as observed in isolated enzyme systems as well as intact cells (Combs and Balaban, 2001). An adequate range of NADH:NAD values were obtained to fit these data to the simple kinetic model of this process. Using this approach, we estimated that the apparent affinity for NADH is much higher than NAD ∼1:30, consistent with more conventional measurements in Triton X 100 extracts. Taking KNAD as 100 μM from the extract data projects the KNADH as ∼4 μM. This high affinity for NADH results in a strong product inhibition of NADH generation in the matrix where the [NADH] rarely drops below the mM range. These data suggest that the product inhibitory effect of NADH seems to be the largest factor in the observed increase in NADH recovery rate with increasing level of NADH photolysis.
Using these kinetic parameters, estimated in the absence of net flux, we can estimate the unidirectional fluxes that occur under conditions of net flux, assuming the only regulatory process involved is the concentration of NAD and NADH. Taking the NADH/NAD ratio determined in State 4 (low net flux) and State 3 (high net flux) mitochondria we calculated that the forward flux (Ff) would be ∼5 (State 4) to ∼50 × (State 3) of the reverse flux (Fb). Since the estimated Ff was essentially identical to the net consumption of NADH estimated from the oxygen consumption, these data suggest that the DH reaction is far from being at equilibrium conditions, especially under maximum flux conditions. The similarity of Ff with the estimated net flux at maximum rates is consistent with the notion that the rate of NADH generation is a major determinant of the maximum rate of ATP formation in heart mitochondria as previously suggested by Mootha et al. (1997). Furthermore, these data suggest that the generation of NADH by intermediary metabolism is not occurring under near equilibrium conditions as previously suggested (Nishiki et al., 1978). The apparent lack of evidence for a near equilibrium of the DH reaction also brings into question the use of DH metabolite redox couples to estimate matrix NADH/NAD ratio (Williamson et al., 1967). The use of these redox couples requires that the dehydrogenase is operating near equilibrium to equate the metabolic intermediate couples with the NADH/NAD redox state (Katz, 1988; Williamson et al., 1967). The ability of cardiac mitochondrial dehydrogenase systems to reflect the mitochondrial NADH/NAD ratio in heart has been controversial for some time (see Nuutinen et al., 1981; Opie and Owen, 1975) based mostly on conventional measurements of enzyme activity in vitro, and comparing this potential flux capacity with the rate of NADH utilization in vivo. These NADH ED-FRAP data determined within the mitochondria matrix suggest that it is highly unlikely that any single dehydrogenase system is operating near equilibrium, i.e., with a reverse and forward flux that significantly exceeds the net flux, since even under State 4 conditions the forward flux only slightly exceeded the measured net flux whereas the reverse rate was significantly lower. The relatively modest NADH generating capacity is likely due to inhibitory factors in the matrix that are not adequately reflected in most in vitro assays. These data suggest that the use of dehydrogenase redox couples for estimating cardiac mitochondria NADH/NAD ratios should be applied with extreme caution as previously suggested (Opie and Owen, 1975).
We found that several parameters in the incubation medium enhanced the NADH recovery rate. Ca2+ is known to increase the resting NADH level (Territo et al., 2000) and activate several matrix dehydrogenases (Denton and McCormack, 1986). Thus, the enhancement of the NADH recovery with Ca2+ was not surprising. However, both the addition of Pi and ATP were found to significantly increase the NADH regeneration rate. ADP was found to have no effect. Pi has been shown to activate some dehydrogenases in the matrix (Rodriguez-Zavala et al., 2000) but is usually not considered a major regulatory factor. ATP is often considered an inhibitor of Kreb cycle activity, thus the current results are somewhat unexpected. The mechanism of action of Pi and ATP will need to be further evaluated in future studies.
To evaluate the role of the net consumption of NADH by the formation of water from oxygen by the cytochrome chain, mitochondria were studied at low (State 4) and high (State3) flux conditions. The addition of the net flux resulted in a large decrease in the rate of NADH regeneration in the NADH ED-FRAP experiment during both State 3 and State 4 conditions. The simplest explanation for this would be the competition between the regeneration of NADH by DH with the oxidation of NADH by the cytochrome chain on the way to reduce oxygen. However, based on several modeling approaches, the reduction in the recovery rate is much too large to be explained by this process, especially for the State 4 data where the increase in respiratory rate was only equivalent to 20 nmol NADH/nmol cytoa per min. Furthermore, attempts to globally model all of the rotenone, State 3, and State 4 data using numerical or the linearization approaches discussed above failed to converge using a single set of maximum velocities or affinities. These data suggested that some other factors were inhibiting the rate of NADH generation in the presence of net flux (i.e., State 4/3) that were not present in the rotenone experiments. With rotenone, ΔΨ and the reduction level of the cytochromes would be much lower than with either State 3 or State 4 mitochondria. If an electrogenic transport process is involved in the recycling of NADH even in the presence of rotenone, then the rate of NADH formation might be inhibited at high ΔΨ, contributing to the difference between the rotenone and State 4/3 data. To test this hypothesis, we added succinate to the rotenone-treated mitochondria that reestablished ΔΨ, confirmed by monitoring the TPP+ distribution across the inner membrane in paired experiments (see Territo et al., 2001). The addition of succinate, even though it is “insulated” from the NADH reaction by the addition of rotenone, inhibited the rate of NADH regeneration in an ED-FRAP experiment by 60 ± 8.0% (n = 6, p < 0.0001) at identical NADHo values. The subsequent addition of FCCP, to dissipate ΔΨ, stimulated the NADH regeneration rate back to near control levels. These experiments are consistent with the notion that the generation of NADH, even in the presence of rotenone, is influenced by ΔΨ in addition to the concentrations of NADH and NAD. It should be noted that another parameter that correlates with the inhibition of NADH regeneration is the redox state of cytochromes b and c. These cytochromes are reduced in State 4/3 or rotenone + succinate when the recovery rate is slow, and oxidized with rotenone or rotenone + succinate + FCCP when the recovery rate is stimulated. Thus, at the present time it is still unclear what is apparently inhibiting NADH regeneration in States 4 and 3 when compared to rotenone conditions. The strong product inhibition of NADH generation and the apparent inhibition of NADH generation by ΔΨ suggest that the flow of reducing equivalents is limited to that which is needed for energy conversion with very little exchange flow, that is, the Fb is very small. We speculate that this might be beneficial to the system in minimizing the generation of free radicals within the cytochrome chain by permitting reducing equivalents to enter the cytochrome chain only when they will be constructively used in the reduction of oxygen. For example, under conditions of low energy conversion, high [NADH] and ΔΨ will restrict free-radical generation by inhibiting reducing equivalent entry into the cytochrome chain, with increased oxidative phosphorylation, [NADH] and ΔΨ depolarizes, activating the reducing equivalent flow to support ATP formation. The actual mechanisms for the apparent restriction of NADH generation will require further study.
The use of NADH ED-FRAP has permitted the direct observation of NADH generating capacity in the isolated enzyme systems, intact mitochondria, and cells. The ability to perform rapid kinetic perturbations of enzyme systems near equilibrium in extremely small volumes inducing the rapid exchange of product for substrate may prove to be a useful tool to the investigator interested in enzyme mechanisms. In the mitochondria and cell, NADH ED-FRAP can provide a new measure of a fundamental process in energetics, the generation of NADH. The rate of recovery of NADH in mitochondria was surprisingly slow, since enzyme activity studies would suggest that the sum total of DH activity would support a near-equilibrium activity resulting in a rapid recovery of the NADH levels after a perturbation. NADH ED-FRAP suggests that the rate of NADH generation is highly regulated by [NADH] and potentially the ΔΨ and/or cytochrome redox state. The product inhibition by NADH was found to be one of the most powerful inhibitors of this activity in extracts and in the intact mitochondria. In some regards, these initial mitochondria NADH EFRAP data have raised more questions with regard to the nature of the NADH generation than they have resolved, especially with regard to the consequences of a net flux in this process. However, NADH ED-FRAP should provide a new useful tool in further studying the regulation of NADH generation in intact mitochondria, cells, and tissues.
Acknowledgments
We thank Drs. Paul Territo and Jay Knutson for helping construct the photolysis system, Dr. James Ferretti for initial help on the NMR interpretations, and Alan Olson at the National Institutes of Health In Vivo NMR Center for collecting the NMR data. Special thanks to Stephanie French for the mitochondria preparation and for the idea of using the nonmetabolized NADPH anomer as fluorescence standard in the mitochondria preparation.
Dr. Joubert's present address is Institut National de la Santé et de la Recherche Médical, U-446, Laboratory of Cellular and Molecular Cardiology, faculté de Pharmacie, Châtenay-Malabry, France.
References
- Balaban, R. S. 2002. Cardiac energy metabolism homeostasis: role of cytosolic calcium. J. Mol. Cell. Cardiol. 34:1259–1271. [DOI] [PubMed] [Google Scholar]
- Balaban, R. S., V. K. Mootha, and A. Arai. 1996. Spectroscopic determination of cytochrome c oxidase content in tissues containing myoglobin or hemoglobin. Anal. Biochem. 237:274–278. [DOI] [PubMed] [Google Scholar]
- Barlow, C. H., and B. Chance. 1976. Ischemic areas in perfused rat hearts: measurement by NADH fluorescence photography. Science. 193:909–910. [DOI] [PubMed] [Google Scholar]
- Boldridge, D. W., T. H. Morton, and G. W. Scott. 1984. Formation kinetics and quantum yield of photon-induced electron ejection from NADH in aqueous solution. Chem. Phys. Lett. 108:461–465. [Google Scholar]
- Chance, B., P. Cohen, F. Jobsis, and B. Schoener. 1962. Intracellular oxidation-reduction states in vivo. Science. 137:499–508. [DOI] [PubMed] [Google Scholar]
- Chance, B., and C. M. Williams. 1956. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. 17:65–134. [DOI] [PubMed] [Google Scholar]
- Chance, B., J. R. Williamson, D. Famieson, and B. Schoener. 1965. Properties and kinetics of reduced pyridine nucleotide fluorescence of the isolated and in vivo rat heart. Biochem. Z. 341:357–377. [Google Scholar]
- Cohen, B. E., B. L. Stoddard, and D. E. Koshland. 1997. Caged NADP and NAD. Synthesis and characterization of functionally distinct caged compounds. Biochemistry. 36:9035–9044. [DOI] [PubMed] [Google Scholar]
- Combs, C. A., and R. S. Balaban. 2001. Direct imaging of dehydrogenase activity within living cells using enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP). Biophys. J. 80:2018–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, M. L., J. S. Johnson, S. M. Giovanazzi, and M. J. Peak. 1985. Photosensitized production of superoxide anion by monochromatic (290–405 nm) ultraviolet-irradiation of NADH and NADPH coenzymes. Photochem. Photobiol. 42:125–128. [DOI] [PubMed] [Google Scholar]
- Czochralska, B., and L. Lindqvist. 1983. Biphotonic one-electron oxidation of NADH on laser excitation at 353 nm. Chem. Phys. Lett. 101:297–299. [Google Scholar]
- Denton, R. M., and J. G. McCormack. 1986. The calcium sensitive dehydrogenases of vertebrate mitochondria. Cell Calcium. 7:377–386. [DOI] [PubMed] [Google Scholar]
- Eng, J., R. M. Lynch, and R. S. Balaban. 1989. NADH fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophys. J. 55:621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook, R. W. 1962. Fluorometric measurements of reduced pyridine nucleotide in cellular and subcellular particles. Anal. Biochem. 4:231–245. [DOI] [PubMed] [Google Scholar]
- Jardetzky, O., and N. G. Wade-Jardetzky. 1966. The conformation of pyridine dinucleotides in solution. J. Biol. Chem. 241:85–91. [PubMed] [Google Scholar]
- Katz, A. 1988. Mitochondrial redox state in skeletal-muscle cannot be estimated with glutamate-dehydrogenase system. Am. J. Physiol. 254:C587–C588. [DOI] [PubMed] [Google Scholar]
- Klingenberg, M., W. Slenchzka, and E. Ritt. 1959. Verfleichende biochemie de pyridinnucleotid-systeme in mitochondrien verschiedener organe. Biochem. Z. 332:47–66. [PubMed] [Google Scholar]
- Livingston, B. E., R. A. Altschuld, and C. M. Hohl. 1996. Metabolic compartmentalization in neonatal swine myocytes. Pediatr. Res. 40:59–65. [DOI] [PubMed] [Google Scholar]
- Meyer, W. L., H. R. Mahler, and R. H. Baker. 1962. Nuclear magnetic resonance spectra and conformation of 1,4-dihyropyridines. Biochim. Biophys. Acta. 64:353–358. [Google Scholar]
- Mootha, V. K., A. E. Arai, and R. S. Balaban. 1997. Maximum oxidative phosphorylation capacity of the mammalian heart. Am. J. Physiol. 272:H769–H775. [DOI] [PubMed] [Google Scholar]
- Nishiki, K., M. Erecinska, and D. F. Wilson. 1978. Energy relationships between cytosolic metabolism and mitochondrial respiration in the rat heart. Am. J. Physiol. 234:C73–C81. [DOI] [PubMed] [Google Scholar]
- Nuutinen, E. M., J. K. Hiltunen, and I. Hassinen. 1981. The glutamate dehydrogenase system and the redox state of mitochondrial free nicotinamide adenine dinucleotide in myocardium. FEBS Lett. 128:356–360. [DOI] [PubMed] [Google Scholar]
- Opie, L. H., and P. Owen. 1975. Effects of increased mechanical work by isolated perfused rat heart during production or uptake of ketone bodies. Assessment of mitochondrial oxidized to reduced free nicotinamide-adenine dinucleotide ratios and oxaloacetate concentrations. Biochem. J. 148:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii, Y. 1993. Immediate reduction of cytochrome-c by photoexcited NADH—reaction mechanism as revealed by flow flash and rapid-scan studies. Biochemistry. 32:11910–11914. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Zavala, J. S., J. P. Pardo, and R. Moreno-Sanchez. 2000. Modulation of 2-oxoglutarate dehydrogenase complex by inorganic phosphate, Mg2+, and other effectors. Arch. Biochem. Biophys. 379:78–84. [DOI] [PubMed] [Google Scholar]
- Sarma, R. H., V. Ross, and N. O. Kaplan. 1968. Investigation of the conformation of b-diphosphopyridine nucleotide (b-nicotinamide-adenine dinucleotide) and pyridine dinucleotide analogs by proton magnetic resonance. Biochemistry. 7:3052–3062. [DOI] [PubMed] [Google Scholar]
- Territo, P. R., and R. S. Balaban. 2000. Rapid spectrophotometric determination of oxygen consumption using hemoglobin, in vitro: light scatter correction and expanded dynamic range. Anal. Biochem. 286:156–163. [DOI] [PubMed] [Google Scholar]
- Territo, P. R., S. A. French, M. C. Dunleavy, F. J. Evans, and R. S. Balaban. 2001. Calcium activation of heart mitochondrial oxidative phosphorylation: rapid kinetics of mVO2, NADH, and light scattering. J. Biol. Chem. 276:2586–2599. [DOI] [PubMed] [Google Scholar]
- Territo, P. R., V. K. Mootha, S. A. French, and R. S. Balaban. 2000. Ca2+ activation of heart mitochondrial oxidative phosphorylation:role of F0/F1ATPase. Am. J. Physiol. 278:c423–c435. [DOI] [PubMed] [Google Scholar]
- Umrikhina, A. V., A. N. Luganskaya, and A. A. Krasnovskii. 1990. ESR signals of NADH and NADPH under illumination. FEBS Lett. 260:294–296. [Google Scholar]
- Wakita, M., G. Nishimura, and M. Tamura. 1995. Some characteristics of the fluorescence lifetime of reduced pyridine nucleotides in isolated mitochondria, isolated hepatocytes, and perfused rat liver in situ. J. Biochem. (Tokyo). 118:1151–1160. [DOI] [PubMed] [Google Scholar]
- Williamson, J. R., P. Lund, and H. A. Krebs. 1967. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratten, C. C., and W. W. Cleland. 1963. Product inhibition studies on yeast and liver alcohol dehydrogenases. Biochemistry. 2:935–941. [DOI] [PubMed] [Google Scholar]