Abstract
Opsonization of bacteria by complement proteins is an important component of the immune response. The pathogenic bacterium Streptococcus pyogenes has evolved multiple mechanisms for the evasion of complement-mediated opsonization. One mechanism involves the binding of human regulators of complement activation such as factor H (FH) and FH-like protein 1 (FHL-1). Acquisition of these regulatory proteins can limit deposition of the opsonin C3b on bacteria, thus decreasing the pathogen's susceptibility to phagocytosis. Binding of complement regulatory proteins by S. pyogenes has previously been attributed to the streptococcal M and M-like proteins. Here, we report that the S. pyogenes cell surface protein Fba can mediate binding of FH and FHL-1. We constructed mutant derivatives of S. pyogenes that lack Fba, M1 protein, or both proteins and assayed the strains for FH binding, susceptibility to phagocytosis, and C3 deposition. Fba expression was found to be sufficient for binding of purified FH as well as for binding of FH and FHL-1 from human plasma. Plasma adsorption experiments also revealed that M1+ Fba+ streptococci preferentially bind FHL-1, whereas M1− Fba+ streptococci have similar affinities for FH and FHL-1. Fba was found to contribute to the survival of streptococci incubated with human blood and to inhibit C3 deposition on bacterial cells. Streptococci harvested from log-phase cultures readily bound FH, but binding was greatly reduced for bacteria obtained from stationary-phase cultures. Bacteria cultured in the presence of the protease inhibitor E64 maintained FH binding activity in stationary phase, suggesting that Fba is removed from the cell surface via proteolysis. Western analyses confirmed that E64 stabilizes cell surface expression of Fba. These data indicate that Fba is an antiopsonic, antiphagocytic protein that may be regulated by cell surface proteolysis.
The gram-positive bacterium Streptococcus pyogenes, or group A streptococcus (GAS), is an important human pathogen. Most streptococcal infections (e.g., pharyngitis, impetigo) are relatively mild and readily treated with antibiotics. However, GAS cause a variety of severe and invasive diseases as well (12). For example, rheumatic fever, a poststreptococcal sequela, is endemic in the third world, where it is a major cause of cardiovascular disease and mortality in persons under 50 years of age (36). Also, since the mid-1980s, Western countries have reported significant increases in severe GAS infections (e.g., sepsis, necrotizing fasciitis, streptococcal toxic shock syndrome) with high mortality rates (7, 17, 35).
GAS express an array of cell surface molecules that contribute to pathogenesis. Among the best studied of these are the M proteins (20). M proteins are multifunctional proteins that contribute to GAS pathogenesis in a number of ways, including by adherence to host tissues, intracellular invasion, and autoaggregation of bacterial cells. Perhaps the most important function of M protein, however, is to confer bacterial resistance to phagocytosis. It has been proposed that the resistance of GAS to killing by professional phagocytes is, at least in part, attributable to the binding of human regulators of complement activation (RCAs) by M and M-like proteins (21, 23, 28, 30).
Horstmann et al. (23) first described the role of RCAs in GAS resistance to phagocytosis. They demonstrated that M6 protein is capable of binding an RCA, factor H (FH), and that FH binding resulted in decreased deposition of C3b on streptococci. A second RCA, FH-like protein 1 (FHL-1), has been found to bind the hypervariable region of M6 protein (28). Type 5 M protein binds both FH and FHL-1 as well (28, 32), but many GAS isolates bind neither RCA (39).
FH and FHL-1 are encoded by the same gene. Expression of the two proteins results from the regulation of transcript elongation and processing (22, 37, 52). FH is a 150-kDa protein comprised of 20 repeat elements known as short consensus repeats (SCRs). Each SCR constitutes an independently folded domain of approximately 60 amino acid residues. FHL-1 is a 42-kDa protein comprised of seven SCRs that are identical, with the exception of 4 amino acids at the C terminus, to SCR1 through SCR7 of FH. FH and FHL-1 each regulate complement activity by at least three mechanisms. First, the regulatory proteins can bind the opsonin C3b, thereby blocking the interaction of C3b with complement factor B and with C3b receptors on phagocytes. Secondly, the RCAs function as cofactors in the factor I-mediated cleavage of C3b. C3b cleavage blocks formation of the C3 convertase C3bBb, thereby blocking the amplification mechanism of the alternative complement pathway. Thirdly, both RCAs promote the decay acceleration of C3bBb (22, 25, 38). The predicted effect of GAS binding of RCAs is a reduction in the amount of C3b deposited on bacterial cells with a concomitant decrease in ingestion and killing of bacteria by phagocytes.
Although the model outlined above is consistent with many observations, it does not fully account for the antiphagocytic activity of all M proteins. For example, many GAS strains appear not to bind RCAs (39) and phagocytosis resistance for some serotypes is dependent on fibrinogen binding (13, 24). Furthermore, recent work by Kotarsky et al. (32) suggests that RCA binding plays a limited role in GAS phagocytosis resistance. These authors identified and deleted the region of M5 protein necessary for binding of FH and FHL-1. Although the M5 variants they constructed no longer bound RCAs, the bacteria remained resistant to phagocytosis, leading the authors to propose that RCA binding may play a more important role in GAS adherence to host tissues than in phagocytosis resistance.
The latter proposal is supported by recent reports demonstrating that FH and FHL-1 can serve as adhesive molecules (15, 22, 52). SCR7 of FH and FHL-1 contains a glycosaminoglycan-binding site, and FH can function as a bridging molecule to facilitate the binding of human polymorphonuclear monocytes (PMNs) to heparin and chondroitin (15). SCR4 contains an RGD motif that is recognized by members of the integrin family of cellular receptors (26). Integrins on fibroblasts, epithelial cells, and melanoma cells can all bind to FHL-1, whereas FH is recognized by integrin CD11b/CD18 on PMNs (15, 22).
During the course of our studies of serotype M1 GAS, we found that an M1 mutant bound FH as well as a wild-type (M1+) strain did. In order to identify the factor responsible for FH binding, we searched the M1 GAS genome database (19) for potential FH-binding proteins. This search resulted in the identification of a hypothetical gene, Spy2009, predicted to encode a 40.4-kDa cell wall-anchored protein that had sequence similarity with the FH-binding proteins of other streptococci (14, 27). Here, we report that the gene product of Spy2009 mediates the binding of FH and FHL-1. Moreover, expression of Spy2009 contributes to phagocytosis resistance by GAS. During the course of this work, Terao et al. (48) reported that Spy2009 encodes a fibronectin-binding protein, denominated Fba, which promotes GAS adherence and intracellular invasion.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains used in this study are listed in Table 1. Streptococci were grown in Todd-Hewitt broth supplemented with 1% yeast extract (THY; Difco Laboratories, Detroit, Mich.). Solid media for streptococci were Todd-Hewitt or sheep blood agar. Escherichia coli was grown in Luria-Bertani broth. The solid medium contained 1.5% agar. Antibiotics were used at the following concentrations: 100 μg of spectinomycin/ml for GAS and E. coli, 1 and 350 μg of erythromycin/ml for GAS and E. coli, respectively, and 100 μg of ampicillin/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli XL-1 Blue | recA1 | Stratagene |
| S. pyogenes | ||
| 90-226 | M1 serotype | 16 |
| 90-226 emm1::Km | emm1::Km | 16 |
| DC276 | emm1::Km, fba::pFW5 | This study |
| DC283 | fba::pFW5 | This study |
| DC294 | DC276 (pYT1143) | This study |
| DC297 | DC283 (pYT1143) | This study |
| Plasmids | ||
| pFW5 | GAS suicide vector | 42 |
| pYT1143 | Plasmid for expression of fba in GAS | 48 |
| pSportI | Cloning vector | Invitrogen |
| pSport-fba | fba gene cloned into pSportI | This study |
| pFW5Δfba | 730-bp internal fragment of fba cloned into pFW5 | This study |
DNA techniques.
Isolations of plasmid DNA from E. coli and genomic DNA from GAS were performed using reagents purchased from Promega Corp., Madison, Wis. DNA sequencing was performed by the Kansas University Medical Center Biotechnology Support Facility. PCR was performed by following standard procedures (46). Plasmid transformations of GAS were performed as described by Caparon and Scott (4).
Cloning and inactivation of Spy2009/orfX/fba.
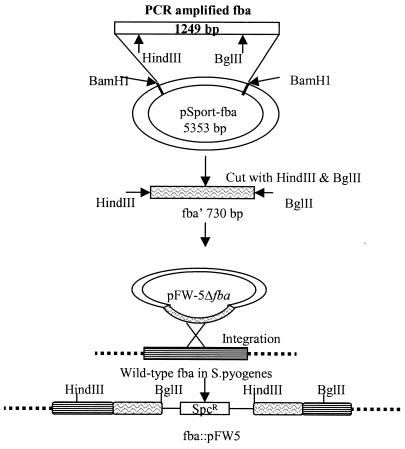
The fba gene (GenBank accession number AB040536) was amplified via PCR from genomic DNA isolated from GAS strain 90-226. The oligonucleotide primer sequences used were ATATGGATCCTTTTTGATGAGGCAGCACATC and TTAAGGATCCAGGAGGACAATATGCGTAGAGC (boldface letters indicate BamHI restriction sites). The 1,265-bp product of the reaction was digested with BamHI, purified from agarose, ligated with BamHI-digested pSportI, and used for transformation of E. coli (Fig. 1). The cloned gene was sequenced and determined to be identical to fba of GAS strain SSI-9 (48). Plasmid pFW5Δfba was constructed by subcloning the 730-bp HindIII-to-BglII fragment of fba into HindIII- and BglII-digested pFW5 (42). pFW5 encodes spectinomycin resistance and does not replicate in GAS. The fba gene in GAS strains 90-226 and 90-226 emm1::Km was inactivated by transformation with pFW5Δfba. Genomic DNA was isolated from the resulting transformants and subjected to Southern and PCR analyses to confirm insertion of the plasmid into fba. For Southern analysis, PCR-amplified fba was labeled with digoxigenin-dUTP by using reagents purchased from Roche Diagnostics, Mannheim, Germany. Genomic DNAs isolated from GAS were digested separately with HindIII and Bsu36I, electrophoresed through 0.8% agarose, transferred to a nylon membrane, and hybridized with the digoxigenin-labeled probe.
FIG. 1.
Construction of fba mutants. The fba gene was amplified via PCR with genomic DNA isolated from GAS strain 90-226 as the template. The oligonucleotide primers were designed to create BamHI restriction sites at the ends of the amplified fragment. The amplicon was digested with BamHI, gel purified, and ligated to BamHI-digested pSportI to create the plasmid pSport-fba. A 730-bp HindIII-to-BglII fragment containing an internal fragment of fba was then subcloned into the suicide vector pFW5 (42). pFW5 carries the aad9 (spectinomycin resistance) gene and does not replicate in GAS. Plasmid pFW5Δfba was introduced into GAS strains 90-226 and 90-226 emm1::Km by electroporation. Southern blot and PCR analyses were performed to verify integration of pFW5Δfba into the chromosomal fba gene.
Proteins, antibodies, and sera.
Human FH was obtained from Quidel Corp., Santa Clara, Calif., and from Calbiochem, La Jolla, Calif. Goat anti-human FH and anti-human C3d monoclonal antibody were purchased from Quidel Corp. Donkey anti-goat immunoglobulin G (IgG) labeled with alkaline phosphatase was from Chemicon International, Temucula, Calif. Mouse anti-rabbit IgG conjugated with alkaline phosphatase was obtained from Sigma-Aldrich, St. Louis, Mo. Rabbit anti-Fba serum was generously provided by Schigetada Kawabata (48). Human plasma and blood were obtained from healthy adult volunteers in accordance with a protocol approved by the Kansas University Medical Center Human Subjects Institutional Review Board. Human serum was purchased from BioWhittaker, Walkersville, Md. Protein mass standards were purchased from Bio-Rad Laboratories, Hercules, Calif.
Plasma adsorption experiments.
Plasma adsorption experiments were performed essentially as described previously (27). GAS strains were grown to an optical density at 560 nm (OD 560) of 0.5. Bacteria were isolated by centrifugation, washed twice with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST), and then suspended in PBST to approximately 1010 CFU/ml. One- hundred-microliter portions of the cell suspensions were mixed with 100 μl of human plasma and incubated with gentle rocking at room temperature for 1 h. The mixtures were then centrifuged for 10 min at 2,000 × g at room temperature. The resulting pellets were washed five times with 500 μl of PBST containing 20 μM E64 and 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich). The cell pellets were then suspended in 100 μl of 0.1 M glycine, pH 2.0, and incubated at room temperature for 10 min. The bacteria were pelleted via centrifugation, and the resulting supernatants were transferred to new tubes. The supernatants were neutralized with NaOH and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were either stained with Coomassie blue or transferred to nitrocellulose membranes. To detect FH, nitrocellulose membranes were successively incubated with TBST (20 mM Tris [pH 7.5], 0.5 M NaCl, 0.05% Tween 20) containing 0.5% gelatin, TBST containing 0.5% gelatin and FH antiserum, and donkey anti-goat IgG conjugated with alkaline phosphatase. Blots were developed with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (XP) p-toluidine (Invitrogen, Carlbad, Calif.).
Binding of FH by GAS and enzyme-linked immunosorbent assay with Fba antiserum.
Binding of FH to immobilized GAS was performed, with modifications, as previously described (10, 11). Unless stated otherwise, bacteria were harvested from log-phase (OD560 of approximately 0.5) cultures grown in THY. Bacterial cells were harvested by centrifugation, washed once with PBS, and suspended to an OD560 of 0.05 in 50 mM carbonate buffer, pH 9.6. Wells of microtiter plates were coated with suspensions of bacterial cells overnight at 5°C. After removal of unbound cells, wells were incubated with wash buffer (PBS containing 0.5% gelatin and 0.05% Tween 20) for 60 min at 37°C. Except where stated otherwise, 100 μl of wash buffer containing 10 μg of purified FH/ml was then added to the wells and the plates were incubated for 2 h at room temperature. As controls, buffer only was added to wells coated with each bacterial strain. After washing to remove unbound FH, 100 μl of wash buffer containing FH antibody was added to the wells and the plates were incubated for 60 min at 37°C. After removing unbound antibody, wash buffer containing an alkaline phosphatase-labeled secondary antibody was added and the incubation was repeated. Finally, wells were washed and 200 μl of 0.1 M glycine (pH 10.5) containing 1.5 mg of p-nitrophenylphosphate/ml, 1 mM CaCl2, and 1 mM ZnCl2 was added to each well. Plates were incubated at 37°C, and absorbance at 405 nm was determined. For each individual strain, absorbance values from wells not incubated with FH were subtracted as background. The subtracted absorbance values typically ranged from 0.025 to 0.1. Data are from three independent experiments in which each strain was assayed in triplicate. Assays of bacterial binding of Fba antiserum were performed similarly. For some experiments, bacteria were isolated from stationary-phase cultures grown in either THY or THY supplemented with 25 μM E64.
Extraction and analysis of cell surface proteins.
GAS were isolated via centrifugation from stationary-phase cultures grown in either THY or THY supplemented with 25 μM E64. Harvested cells were washed twice with TES (10 mM Tris [pH 8], 1 mM EDTA, 25% sucrose) (40) and then suspended in a 1/4 volume of TES containing 1 mg of lysozyme/ml, 500 U of mutanolysin/ml, 100 μg of RNase A/ml, 25 μM E64, and 1 mM PMSF. The suspensions were incubated at 37°C for 30 min and were then centrifuged at 2,500 × g for 10 min at 4°C. Trichloroacetic acid was added to the resulting supernatants to a final concentration of 16% (vol/vol), and the mixtures were incubated on ice for 20 min. The mixtures were then centrifuged at 11,500 × g for 10 min at 4°C. The pellets were washed with acetone and suspended in 50 mM NaOH. The preparations were fractionated by SDS-PAGE and either stained with Coomassie blue or transferred to nitrocellulose membranes. To detect Fba, the membranes were blocked with TBST containing 0.5% gelatin and then incubated with Fba antiserum and a labeled secondary antibody. To detect FH binding, membranes were successively incubated with TBST containing 3% bovine serum albumin (BSA), TBST containing 3% BSA and 10 μg of FH/ml, FH antiserum, and a labeled secondary antibody.
Peptide sequencing.
N-terminal sequencing of FHL-1 blotted onto polyvinylidene difluoride membrane (Bio-Rad) was performed by Midwest Analytical Inc. (St. Louis, Mo.).
Measurement of C3 deposition on streptococci.
Streptococci were harvested from log-phase cultures by centrifugation. The cell pellets were washed with 1 volume of veronal-buffered saline (VBS; Sigma-Aldrich), pH 7.4, and then suspended to an OD560 of 1.0 in VBS containing 10 mM EGTA and 5 mM MgCl2. One milliliter of each bacterial suspension was then mixed with 1 ml of human serum, and the mixtures were incubated at room temperature for 30 min with gentle rocking. The bacteria were then harvested by centrifugation at 4,000 × g for 10 min. The resulting pellets were washed three times with VBS containing 10 mM EDTA, 20 μM E64, and 1 mM PMSF and finally suspended in 50 mM carbonate buffer, pH 9.6. The bacterial suspensions were then diluted in carbonate buffer to OD560s of 0.1, 0.05, and 0.025. One hundred microliters of each dilution was then applied to wells of microtiter plates in quadruplicate. As a control, some wells were mock coated with carbonate buffer. Bacteria were absorbed to the plates by overnight incubation at 5°C. The plates were then washed and blocked as described above. C3 deposition was detected using an anti-human C3d monoclonal antibody that recognizes all forms of C3. For each individual strain, absorbance values (typically <0.1) from wells not incubated with the primary antibody were subtracted as background. For each experiment, the mean absorbance values for strain 90-226 (M1+ Fba+) were assigned a value of 1. Values for the mutant strains are expressed relative to that for strain 90-226. Data are from three independent experiments performed with the same serum. Additional experiments were performed with pooled human plasma samples that yielded similar results.
Bactericidal assays.
The ability of GAS to survive in human blood was measured as previously described (16). Briefly, log-phase cultures of streptococci were diluted in PBS and 100 μl (102 CFU) of each bacterial suspension was added to 1.25 ml of heparinized blood and to 1.25 ml of plasma derived from the same blood sample. A portion of each culture was plated in sheep blood agar to determine the input CFU. Tubes were incubated at 37°C with gentle rocking for 3 h. Portions of each culture were again plated to determine the number of surviving CFU. Plates were incubated overnight at 37°C before the counting of colonies. The growth index of each strain was calculated by dividing the number of surviving CFU by the number of input CFU. For each experiment, the growth index for strain 90-226 (M1+ Fba+) was assigned a value of 1. Growth indices for the mutant strains are expressed relative to that of strain 90-226. The growth indices for strain 90-226 ranged from 46 to 139 in blood and 84 to 160 in plasma. Statistical significance of the data was determined by Student's t test by using Microsoft Excel 2000 software. P values of <0.05 were considered significant.
RESULTS
Generation and characterization of fba mutants.
FH binding experiments were performed with GAS strain 90-226 and an isogenic M1− mutant derivative, 90-266 emm1::Km (Fig. 2) (9, 16). It was found that both GAS strains readily bound FH, indicating that a factor other than M1 protein mediated binding. Therefore, the genome sequence of the serotype M1 strain SF370 (19) was searched for genes that encoded a protein with similarity to previously characterized FH-binding proteins (14, 27). The best candidate gene was Spy2009. Spy2009 encoded a hypothetical 40.4-kDa protein with an N-terminal signal sequence and a C-terminal cell wall-anchoring motif. An allelic variant of Spy2009, designated orfX, was previously described by Podbielski et al. (41). Since the initiation of this study, Terao et al. (48) reported that Spy2009 encodes a fibronectin-binding protein that they designated Fba. The latter designation will be used throughout the remainder of this report.
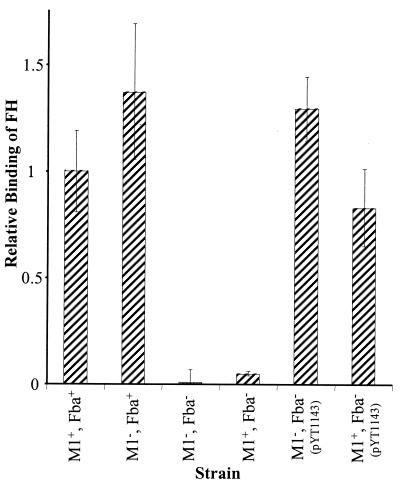
FIG. 2.
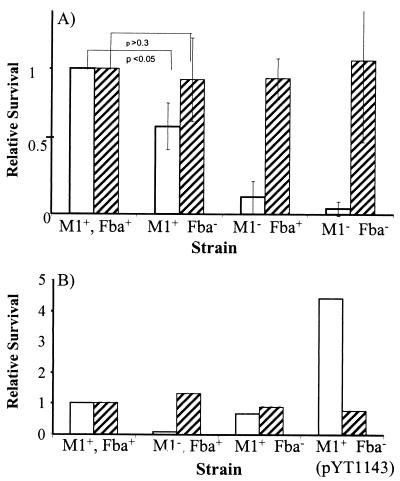
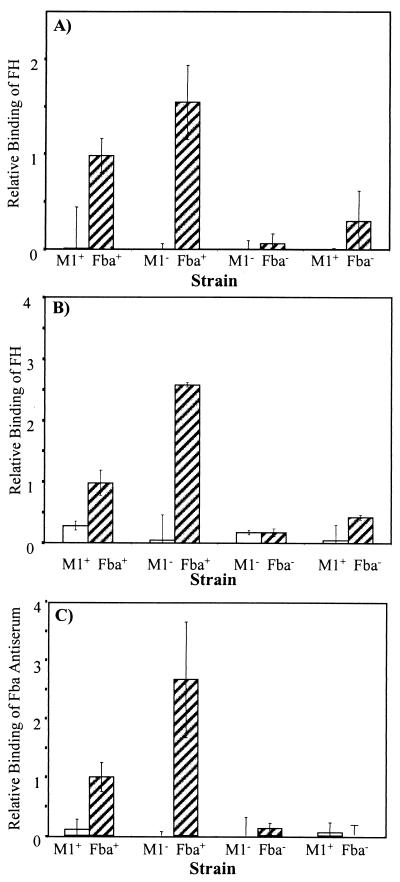
Inactivation of fba inhibits FH binding. Bacterial cells were harvested from exponential-phase cultures and adsorbed to wells of microtiter plates. The immobilized cells were incubated with FH followed by goat anti-human FH serum and an alkaline phosphatase-labeled secondary antibody. FH binding is expressed relative to that of the wild-type strain 90-226. Data are from three independent experiments, wherein each assay was performed in triplicate. Error bars represent standard deviations. Strains: M1+ Fba+, 90-226; M1− Fba+, 90-226 emm1::Km; M1− Fba−, DC276; M1+ Fba−, DC283.
The fba gene from strain 90-226 was cloned and sequenced and determined to be identical to fba of GAS strain SS-9 described by Terao et al. (48). The fba gene in strains 90-226 and SS-9 (48) is 72 bp shorter than the strain SF370 gene and is predicted to encode a 37.8-kDa protein. To inactivate fba, a 730-bp internal fragment of the gene was cloned into the suicide vector pFW5 (Fig. 1). The resulting plasmid was introduced into strains 90-226 and 90-226 emm1::Km. Genomic DNA was isolated from the transformants and the parental strain and used in PCR and Southern analyses to verify the genomic structure of the mutants.
FH binding experiments were then performed. The results, shown in Fig. 2, indicated that there was no appreciable binding of FH by the fba mutants. To verify that the loss of FH binding activity was due to the inactivation of fba and not due to an effect of the plasmid insertion on downstream genes, pYT1143 was introduced into the fba mutant strains. Plasmid pYT1143 carries the intact fba gene under the control of the GAS recA promoter (48). PYT1143 completely restored FH binding by the Fba− strains (Fig. 2).
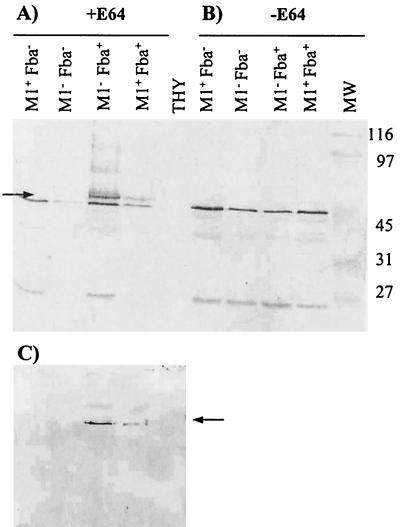
Cell surface proteins were extracted from the Fba+ and Fba− strains and subjected to Western analysis using Fba antiserum (Fig. 3). The results revealed that the Fba+ strains expressed a protein with an apparent molecular mass of 58 kDa. The 58-kDa protein was not expressed by the Fba− mutants. The predicted mass of the cell surface form of Fba (33.8 kDa) is considerably lower than 58 kDa. Terao et al. (48) reported, however, that Fba migrates anomalously on SDS-PAGE gels. This is probably due to the high proline content of the protein. To confirm that the 58-kDa band represented Fba, cell surface proteins were transferred to nitrocellulose membranes and the membranes were successively incubated with FH, FH antiserum, and a labeled secondary antibody (Fig. 3C). As controls, some membranes were incubated with only the primary and secondary antibodies to verify that the antibodies did not bind directly to the blotted proteins. FH was determined to bind to the same 58-kDa band that reacted with Fba antiserum.
FIG. 3.
Western analysis of cell surface proteins from strain 90-226 and its isogenic M1− and Fba− derivatives. Cell surface proteins were extracted from stationary-phase cultures of GAS, fractionated by SDS-PAGE, and transferred to nitrocellulose membranes. The phenotypes of the strains are listed above the blots. Proteins used in blots A and C were extracted from streptococci cultured to stationary phase in THY containing 25 μM of the cysteine protease inhibitor E64. Proteins for blot B were extracted from THY-grown cultures. (A and B) Blots were incubated with Fba antiserum. The arrow in panel A indicates the position of the 58-kDa band present in extracts from Fba+ strains that was not present in extracts from Fba− strains. Cell surface Fba was not detectable in cultures grown in the absence of E64. Lanes: THY, mock extraction performed with sterile culture medium; MW, molecular mass standards, with masses indicated in kilodaltons. (C) FH binding to Fba is shown. The same extracts used for blot A were transferred to a membrane, blocked, and successively incubated with 10 μg of FH/ml, FH antiserum, and a labeled secondary antibody. The arrow indicates the position of the same protein band indicated by the arrow in panel A.
Adsorption of FH and FHL-1 from human plasma.
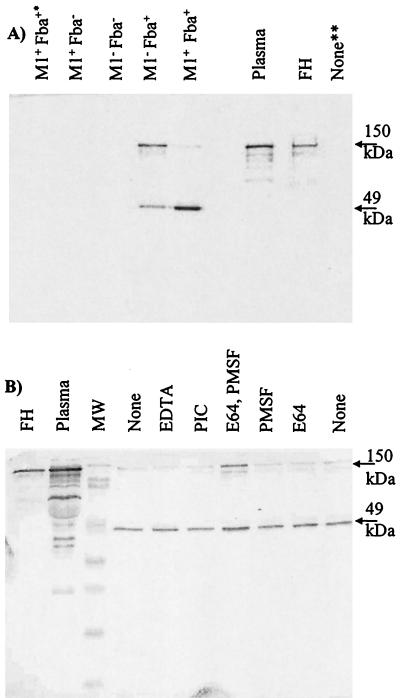
It was next determined whether GAS could bind the FH present in human plasma. Bacteria were incubated with plasma and washed extensively, and the bound proteins were eluted and subjected to Western analyses with FH antiserum (Fig. 4). No FH was recovered from the incubation of the fba mutants with plasma. Conversely, both Fba+ strains bound FH as well as a second, approximately 49-kDa, protein. The size of the latter protein and its reactivity with FH antiserum suggested it was FHL-1. Amino-terminal sequencing of the 49-kDa protein was performed. The sequence XDCNELPPRRN was obtained, indicating that the protein was either FH or FHL-1 (18, 47). It seemed possible that the 49-kDa band could have been derived from the proteolytic cleavage of FH. To test this possibility, plasma adsorption experiments were performed in the presence of various protease inhibitors (Fig. 4B). The addition of inhibitors had no effect on the pattern of eluted proteins. These results support the conclusion that the 49-kDa protein was FHL-1.
FIG. 4.
Adsorption of FH and FHL-1 from human plasma. (A) GAS strain 90-226 and its isogenic M1− and Fba− derivatives were incubated with human plasma, harvested by centrifugation, and washed extensively. Bound plasma proteins were eluted and subjected to Western analysis with FH antiserum. The phenotypes of the strains are listed above the blot. Adsorption experiments were performed with plasma or serum from four different sources. The topmost arrow to the right of the figure indicates the 150-kDa FH band. The lower arrow indicates a 49-kDa band determined by N-terminal sequencing to be FH or FHL-1. Lanes: Plasma, loaded with human plasma; FH, loaded with purified FH; M+ Fba+∗, mock adsorption performed with bacteria in the absence of plasma; None∗∗, mock adsorption performed with plasma in the absence of bacteria. (B) Plasma adsorptions were performed in the presence of protease inhibitors. Plasma adsorptions were performed with wild-type GAS in the absence of protease inhibitors (None) or in the presence of 25 μM E64, 1 mM PMSF, 25 μM E64 and 1 mM PMSF, 25 μM EDTA, or a cocktail of protease inhibitors (PIC; Sigma-Aldrich) as indicated.
The wild-type strain was found to preferentially bind FHL-1, whereas the M1− Fba+ strain appeared to bind FH and FHL-1 equally well. As binding of either plasma protein was dependent on Fba expression, these results suggested that M1 protein influenced the binding specificity of Fba. Experiments like those shown in Fig. 4 were performed with plasma or serum from four different sources. Similar results were obtained from all experiments. Therefore, binding of FHL-1 is reflective of the ligand specificity of the Fba expressed by the wild-type organism.
Survival of streptococci in human blood and plasma.
To determine if Fba contributes to phagocytosis resistance, we compared survival of the wild-type and Fba− strains in human blood and plasma. As anticipated, the wild-type strain survived and multiplied in blood whereas the M1− strains did not (Fig. 5A). Survival of the M1+ Fba− mutant was intermediate to that of the M1+ Fba+ and the M1− strains, indicating that Fba contributes to phagocytosis resistance. Moreover, expression of Fba in trans increased survival of the M1+ Fba− strain (Fig. 5B). There was no significant impact of the Fba mutations on bacterial survival in human plasma.
FIG. 5.
(A) Survival of GAS in human blood and plasma. GAS strains were grown to early- to mid-log phase, and 102 CFU of each strain were added to heparinized human blood (open bars) or human plasma (hatched bars). A portion of each culture was plated to determine the input CFU. Cultures were incubated at 37 C for 3 h. Portions of each culture were again plated to determine the number of surviving CFU. The growth index of each culture was calculated by dividing the number of surviving CFU by the number of input CFU. For each experiment, the growth index for strain 90-226 (M1+ Fba+) was assigned a value of 1. Growth indices for the mutant strains are expressed relative to strain 90-226. The data in the figure were obtained from four independent experiments. Error bars represent standard deviations. (B) Complementation of the fba mutation. Experiments were performed as described for panel A. The M1+ Fba− strain is DC283. M1+ Fba− (pYT1143) is DC283 carrying the Fba expression plasmid pYT1143 (48). Values are the averages from two experiments.
Measurements of C3 deposition on streptococci.
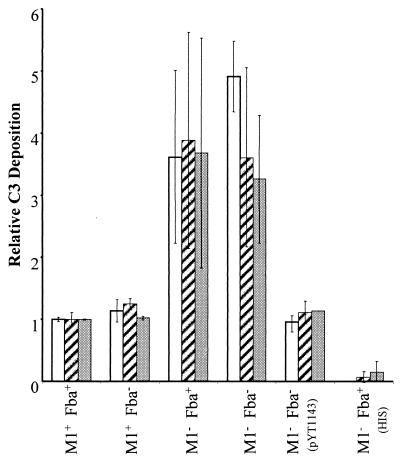
RCA binding could decrease the susceptibility of GAS to phagocytosis by limiting the amount of C3b deposited onto bacterial cells. To determine whether M1 protein and/or Fba affected C3 deposition, GAS were incubated with human serum, washed, and immobilized in microtiter plates. C3 deposition was determined with a monoclonal antibody that reacts with all forms of C3 (Fig. 6). Approximately four times more C3 was deposited on M1− streptococci than on the wild-type strain. In contrast, the fba mutation did not affect C3 deposition. Moreover, C3 deposition was equivalent for the M1− Fba+ and M1− Fba− strains. Thus, fba in single copy had no significant impact on C3 deposition in these assays. However, the presence of multiple copies of fba significantly reduced C3 deposition on M1− Fba− streptococci (Fig. 6). No C3 deposition was detected when bacteria were incubated with heat-inactivated serum.
FIG. 6.
C3 deposition on streptococci. Bacterial cells were suspended in VBS containing 10 mM EGTA and 5 mM MgCl2. The suspensions were mixed with an equal volume of human serum or heat-inactivated human serum (HIS) and incubated at room temperature for 30 min. Bacteria were harvested, washed, and suspended in carbonate buffer. The cells were then diluted in carbonate buffer to final OD560s of 0.025 (open bars), 0.05 (hatched bars), and 0.1 (filled bars) and applied to wells of microtiter plates. C3 deposition was detected with a monoclonal anti-human C3d antibody.
Evidence that the cell surface activity of Fba is regulated by proteolysis.
Our early attempts to assay FH binding by streptococci yielded inconsistent results, especially with regard to strain 90-226, which frequently failed to bind FH. It was suspected that the inconsistencies might have been due to the use of early, stationary-phase cultures in the experiments. Therefore, the assay procedure was modified to use bacterial cells harvested in exponential phase. This change resulted in obtaining readily reproducible results for FH binding by Fba+ GAS.
To confirm that FH binding activity was lost in stationary phase, we compared the binding by mid-log and late-stationary phase (i.e., 14 h) cultures. FH binding was greatly reduced for stationary-phase cells (Fig. 7A). SpeB is an extracellular cysteine protease of GAS that reportedly can cleave proteins from the cell surface (2, 5, 43). To determine if SpeB might be involved in the loss of FH binding activity, FH binding and reactivity with Fba antiserum were compared for bacteria grown to stationary phase in the presence or absence of the cysteine protease inhibitor E64. Growth of GAS in the presence of E64 resulted in the retention of FH binding activity in stationary-phase cultures (Fig. 7B). Accordingly, GAS grown in the presence of E64 reacted strongly with Fba antiserum whereas cells grown without the inhibitor reacted weakly (Fig. 7C).
FIG. 7.
Fba is removed from the cell surface in stationary phase. (A) FH binding activity is lost in stationary phase. Wells of microtiter plates were coated with GAS grown to mid-exponential phase (hatched bars) or stationary phase (open bars). FH binding was assayed and is presented as described in the legend to Fig. 2. (B) E64 stabilizes FH binding activity. Microtiter plates were coated with GAS grown to stationary phase in THY medium (open bars) or THY medium containing 25 μM of E64 (hatched bars). (C) E64 stabilizes Fba expression. Wells were coated with GAS grown to stationary phase in THY medium (open bars) or THY medium containing 25 μM of E64 (hatched bars). Fba expression was assayed with Fba antiserum. All data are from two independent experiments in which assays were performed in triplicate (panels A and B) or duplicate (panel C).
Western analyses of cell surface proteins extracted from stationary-phase cultures were also performed. No Fba was detected in extracts from any strain grown in the absence of E64 (Fig. 3B). Fba was detectable, however, in proteins extracted from the Fba+ strains cultured with E64 (Fig. 3A). These results are consistent with the proteolytic removal of Fba from the cell surface in stationary-phase cultures.
DISCUSSION
Opsonization of pathogenic bacteria by complement proteins or antibodies represents a major component of the immune response. GAS have evolved a number of mechanisms to limit opsonization and avoid ingestion and killing by phagocytes. The hyaluronic acid capsule of GAS can serve as a physical barrier to the interaction of phagocytes with bacterial-bound opsonins (13). M and M-like proteins can bind the Fc regions of immunoglobulins, thereby inhibiting activation of the classical complement pathway (3). M protein binding of the plasma protein fibrinogen can also inhibit complement activation and may alter the interaction between GAS and phagocytes (13, 45). The streptococcal C5a peptidase can inactivate C5a, a chemoattractant for neutrophils (51). The recently described Mac protein can inhibit phagocytosis and killing of GAS by binding to CD16 on PMNs (33).
Bacterial binding of RCAs has also been proposed as a mechanism for GAS evasion of the innate immune response. Several members of the M protein family can bind the RCA C4b-binding protein (29, 49). M5 and M6 proteins can bind both FH and FHL-1 (28, 32). FH and FHL-1 both function as cofactors in factor I-mediate cleavage of C3b to iC3b, thereby inhibiting the amplification mechanism of the alternative complement pathway. FH and FHL-1 also promote decay acceleration of the C3 convertase C3bBb and can sequester surface-bound C3b (37). Previously, binding of human RCAs by GAS has been attributed to M and M-like proteins. The data presented here demonstrate that Fba, a protein unrelated to M, can also mediate binding of FH and FHL-1.
Insertional inactivation of fba inhibited FH binding in vitro, and expression of Fba in trans restored FH binding to the Fba− strains. Plasma adsorption experiments were performed to determine if Fba could mediate FH binding in a complex mixture of proteins. The results corroborated the results of direct binding assays performed with purified FH; i.e., Fba expression is sufficient for FH binding.
Fba contributes to the survival of GAS in human blood, but Fba expression is neither sufficient nor absolutely essential for resistance to phagocytosis. Accordingly, a single copy of fba did not significantly affect C3 deposition on streptococci. Interestingly, fba carried on a multicopy plasmid did significantly inhibit C3 deposition and enhance phagocytosis resistance. The latter results suggest that upregulation of fba expression could significantly enhance GAS survival in vivo. M1 protein limits C3 deposition to a greater extent and is necessary for bacterial survival in blood. It is important to note that M1 protein inhibits C3 deposition and resistance to phagocytosis in the absence of detectable FH or FHL-1 binding. Thus, results reported here are similar to those of Kotarsky et al. (32), who reported that the ability of M5 protein to confer phagocytosis resistance is independent of FH and FHL-1 binding. Even though we have not detected FH binding that could be attributable to M1 protein, we cannot exclude the possibility that M1 does bind FH. In fact, Kihlberg et al. (31) demonstrated that purified M1 protein can bind purified FH. It is clear, however, that Fba can bind purified FH as well as FH and FHL-1 in plasma.
In this study, M1 protein was found to influence the ligand binding specificity of Fba. In plasma adsorption experiments, the M1− Fba+ strain bound FH as well as FHL-1. The wild-type strain also bound both proteins, though it preferentially bound FHL-1. The preferential binding of FHL-1 by M1+ Fba+ streptococci is especially intriguing, as the concentration of FH in human blood is 10 to 40 times higher than that of FHL-1 (22, 52). The mechanism whereby M1 affects ligand binding by Fba is at present unknown. An obvious possibility is that M1 and Fba physically interact when coexpressed on the cell surface. M1 protein does bind a number of human plasma proteins, including fibrinogen, albumin, IgG, and fibronectin (1, 9, 11). It is possible that binding of one or more of these factors influences the binding specificity of Fba. Because FHL-1 can function as an adhesion molecule for a variety of human cells (22, 52), the preferential binding of FHL-1 by M1 GAS could be highly significant with regard to streptococcal-host interactions.
Data presented here also show that Fba is removed from the surface of streptococci in stationary phase. Cells harvested from exponential-phase cultures reproducibly bound FH, whereas FH binding by cells from stationary-phase cultures was weak or absent. The addition of E64 to culture media resulted in retention of FH binding activity and detectable Fba expression by stationary-phase cells. The fba gene is regulated by the positive transcriptional regulator Mga (41, 48). Mga-regulated genes, which include emm1, sic, and scpA, as well as fba, are transcribed in exponential phase and shut off in stationary phase (34). Our results are consistent with Fba being expressed on the cell surface in log phase and removal of the protein in stationary phase. The fact that E64 can stabilize Fba expression suggests involvement of the cysteine protease SpeB in the stationary-phase removal of Fba. The speB gene is present in all strains of GAS, and SpeB is produced in stationary phase (6). The addition of exogenous SpeB to GAS cultures can result in cleavage of biological active proteins from the cell surface (2, 43). Endogenous SpeB can also alter the interaction between GAS and host cells (5, 43, 44, 50). The latter phenomenon is due, at least in part, to SpeB-mediate cleavage of fibronectin-binding protein(s) (5). Although our results are consistent with the direct involvement of SpeB in the removal of Fba, it would be premature to draw this conclusion. Collin and Olsen (8) reported that SpeB is an inactive zymogen in cultures of strain 90-226 emm1::Km, the M1− Fba+ strain used in this study. Thus, it is possible that E64 inhibits the activity of another streptococcal protease or that E64 has effects on the physiology of GAS other than protease inhibition.
It has been proposed that the proteolytic cleavage of bacterial cell surface proteins could promote dissemination of bacteria under conditions of high population density and nutrient limitation. Furthermore, proteins released from the cell surface could interact with host cells and macromolecules, thereby affecting the interaction between pathogen and host (2, 5, 44). It remains to be determined whether removal of Fba from the cell surface releases a biologically active protein into the environment. This matter, and the mechanism of Fba removal from the cell surface, warrants further investigation.
Acknowledgments
This work was supported by National Institutes of Health grant RR16443-01 and a Lied Basic Science Research Grant from the Kansas University Medical Center Research Institute.
We thank Shigetada Kawabata and Yutaka Terao for Fba antiserum and pYT1143, Darlene Sheffer for help with the phagocytosis assays, Hildegard Bell for assistance with the C3 deposition assays, Bala Chandran for advice and encouragement, and Chia Lee for advice and critical reading of the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Akesson, P., K. H. Schmidt, J. Cooney, and L. Bjorck. 1994. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem. J. 300:877-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berge, A., and L. Bjorck. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270:9862-9867. [DOI] [PubMed] [Google Scholar]
- 3.Berge, A., B. M. Kihlberg, A. G. Sjoholm, and L. Bjorck. 1997. Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J. Biol. Chem. 272:20774-20781. [DOI] [PubMed] [Google Scholar]
- 4.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., R. L. Cole, and J. P. M. van Putten. 2000. Streptococcal erythrogenic toxin B abrogates fibronectin-dependent internalization of Streptococcus pyogenes by cultured mammalian cells. Infect. Immun. 68:3226-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Hauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518-521. [DOI] [PubMed] [Google Scholar]
- 8.Collin, M., and A. Olsen. 2000. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36:1306-1318. [DOI] [PubMed] [Google Scholar]
- 9.Cue, D., P. E. Dombek, H. Lam, and P. P. Cleary. 1998. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect. Immun. 66:4593-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cue, D., H. Lam, and P. P. Cleary. 2001. Genetic dissection of the Streptococcus pyogenes M1 protein: regions involved in fibronectin binding and intracellular invasion. Microb. Pathog. 31:231-242. [DOI] [PubMed] [Google Scholar]
- 11.Cue, D., S. O. Southern, P. J. Southern, J. Prabhakar, W. Lorelli, J. M. Smallheer, S. A. Mousa, and P. P. Cleary. 2000. A nonpeptide integrin antagonist can inhibit epithelial cell ingestion of Streptococcus pyogenes by blocking formation of integrin α 5β 1-fibronectin-M1 protein complexes. Proc. Natl. Acad. Sci. USA 97:2858-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale, J. B., R. G. Washburn, M. B. Marques, and M. R. Wessels. 1996. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect. Immun. 64:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dave, S., A. Brooks-Walter, M. K. Pangburn, and L. S. McDaniel. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect. Immun. 69:3435-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiScipio, R. G., P. J. Daffern, I. U. Schraufstatter, and P. Sriramarao. 1998. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves αMβ2 (CD11b/CD18). J. Immunol. 160:4057-4066. [PubMed] [Google Scholar]
- 16.Dombek, P. E., D. Cue, J. Sedgewick, H. Lam, S. Ruschkowski, B. B. Finlay, and P. P. Cleary. 1999. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol. Microbiol. 31:859-870. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, B. K., J. Andersson, S. E. Holm, and M. Norgren. 1999. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J. Infect. Dis. 180:410-418. [DOI] [PubMed] [Google Scholar]
- 18.Estaller, C., W. Schwaeble, M. Dierich, and E. H. Weiss. 1991. Human complement factor H: two factor H proteins are derived from alternatively spliced transcripts. Eur. J. Immunol. 21:799-802. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischetti, V. A. 2000. Surface proteins on gram-positive bacteria, p. 11-24. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 21.Fischetti, V. A., R. D. Horstmann, and V. Pancholi. 1995. Location of the complement factor H binding site on streptococcal M6 protein. Infect. Immun. 63:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellwage, J., S. Kuhn, and P. F. Zipfel. 1997. The human complement regulatory factor-H-like protein 1, which represents a truncated form of factor H, displays cell-attachment activity. Biochem. J. 326:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horstmann, R. D., H. J. Sievertsen, J. Knobloch, and V. A. Fischetti. 1988. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA 85:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horstmann, R. D., H. J. Sievertsen, M. Leippe, and V. A. Fischetti. 1992. Role of fibrinogen in complement inhibition by streptococcal M protein. Infect. Immun. 60:5036-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hourcade, D. E., L. Mitchell, L. A. Kuttner-Kondo, J. P. Atkinson, and M. E. Medof. 2002. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J. Biol. Chem. 277:1107-1112. [DOI] [PubMed] [Google Scholar]
- 26.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 27.Janulczyk, R., F. Iannelli, A. G. Sjoholm, G. Pozzi, and L. Bjorck. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 275:37257-37263. [DOI] [PubMed] [Google Scholar]
- 28.Johnsson, E., K. Berggard, H. Kotarsky, J. Hellwage, P. F. Zipfel, U. Sjobring, and G. Lindahl. 1998. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J. Immunol. 161:4894-4901. [PubMed] [Google Scholar]
- 29.Johnsson, E., A. Thern, B. Dahlback, L. O. Heden, M. Wikstrom, and G. Lindahl. 1997. Human C4BP binds to the hypervariable N-terminal region of many members in the streptococcal M protein family. Adv. Exp. Med. Biol. 418:505-510. [DOI] [PubMed] [Google Scholar]
- 30.Kihlberg, B. M., M. Collin, A. Olsen, and L. Bjorck. 1999. Protein H, an antiphagocytic surface protein in Streptococcus pyogenes. Infect. Immun. 67:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kihlberg, B. M., J. Cooney, M. G. Caparon, A. Olsen, and L. Bjorck. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299-315. [DOI] [PubMed] [Google Scholar]
- 32.Kotarsky, H., J. Hellwage, E. Johnsson, C. Skerka, H. G. Svensson, G. Lindahl, U. Sjobring, and P. F. Zipfel. 1998. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J. Immunol. 160:3349-3354. [PubMed] [Google Scholar]
- 33.Lei, B., F. R. DeLeo, N. P. Hoe, M. R. Graham, S. M. Mackie, R. L. Cole, M. Liu, H. R. Hill, D. E. Low, M. J. Federle, J. R. Scott, and J. M. Musser. 2001. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat. Med. 7:1298-1305. [DOI] [PubMed] [Google Scholar]
- 34.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser, J. M., V. Kapur, J. Szeto, X. Pan, D. S. Swanson, and D. R. Martin. 1995. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect. Immun. 63:994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olivier, C. 2000. Rheumatic fever—is it still a problem? J. Antimicrob. Chemother. 45:13-21. [DOI] [PubMed] [Google Scholar]
- 37.Pangburn, M. K. 2000. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology 49:149-157. [DOI] [PubMed] [Google Scholar]
- 38.Pangburn, M. K., K. L. Pangburn, V. Koistinen, S. Meri, and A. K. Sharma. 2000. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J. Immunol. 164:4742-4751. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Caballero, D., S. Alberti, F. Vivanco, P. Sanchez-Corral, and S. Rodriguez de Cordoba. 2000. Assessment of the interaction of human complement regulatory proteins with group A Streptococcus. Identification of a high-affinity group A Streptococcus binding site in FHL-1. Eur. J. Immunol. 30:1243-1253. [DOI] [PubMed] [Google Scholar]
- 40.Piard, J. C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podbielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. (Berlin) 185:171-181. [DOI] [PubMed] [Google Scholar]
- 42.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 43.Raeder, R., M. Woischnik, A. Podbielski, and M. D. Boyle. 1998. A secreted streptococcal cysteine protease can cleave a surface-expressed M1 protein and alter the immunoglobulin binding properties. Res. Microbiol. 149:539-548. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen, M., and L. Bjorck. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43:537-544. [DOI] [PubMed] [Google Scholar]
- 45.Ringdahl, U., H. G. Svensson, H. Kotarsky, M. Gustafsson, M. Weineisen, and U. Sjobring. 2000. A role for the fibrinogen-binding regions of streptococcal M proteins in phagocytosis resistance. Mol. Microbiol. 37: 1318-1326. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Skerka, C., R. D. Horstmann, and P. F. Zipfel. 1991. Molecular cloning of a human serum protein structurally related to complement factor H. J. Biol. Chem. 266:12015-12020. [PubMed] [Google Scholar]
- 48.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 49.Thern, A., L. Stenberg, B. Dahlback, and G. Lindahl. 1995. Ig-binding surface proteins of Streptococcus pyogenes also bind human C4b-binding protein (C4BP), a regulatory component of the complement system. J. Immunol. 154:375-386. [PubMed] [Google Scholar]
- 50.Tsai, P.-J., C.-F. Kuo, K.-Y. Lin, Y.-S. Lin, H.-Y. Lei, F.-F. Chen, J.-R. Wang, and J.-J. Wu. 1998. Effect of group A streptococcal cysteine protease on invasion of epithelial cells. Infect. Immun. 66:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wexler, D. E., D. E. Chenoweth, and P. P. Cleary. 1985. Mechanism of action of the group A streptococcal C5a inactivator. Proc. Natl. Acad. Sci. USA 82:8144-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zipfel, P. F., and C. Skerka. 1999. FHL-1/reconectin: a human complement and immune regulator with cell-adhesive activity. Immunol. Today 20:135-140. [DOI] [PubMed] [Google Scholar]