Abstract
The conduction properties of ClC-0 and ClC-1 chloride channels are examined using electrostatic calculations and three-dimensional Brownian dynamics simulations. We create an open-state configuration of the prokaryotic ClC Cl− channel using its known crystallographic structure as a basis. Two residues that are occluding the channel are slowly pushed outward with molecular dynamics to create a continuous ion-conducting path with the minimum radius of 2.5 Å. Then, retaining the same pore shape, the prokaryotic ClC channel is converted to either ClC-0 or ClC-1 by replacing all the nonconserved dipole-containing and charged amino acid residues. Employing open-state ClC-0 and ClC-1 channel models, current-voltage curves consistent with experimental measurements are obtained. We find that conduction in these pores involves three ions. We locate the binding sites, as well as pinpointing the rate-limiting steps in conduction, and make testable predictions about how the single channel current across ClC-0 and ClC-1 will vary as the ionic concentrations are increased. Finally, we demonstrate that a ClC-0 homology model created from an alternative sequence alignment fails to replicate any of the experimental observations.
INTRODUCTION
Anionic channels are essential in maintaining the integrity of synaptic physiology and perform a diverse range of physiological functions, yet they have been largely neglected in theoretical investigations. Here we focus our attention on a subclass of channels that are selectively permeable to anions: the voltage-gated ClC family of chloride channels, present in the cell membranes of every living organism. ClC Cl− channels perform diverse roles, such as the control of cellular excitability, acidification of intracellular vesicles, and cell volume regulation (see, for recent reviews, Jentsch et al., 1999, 2002; Maduke et al., 2000; Fahlke, 2001). The prototype channel ClC-0, from the Torpedo electroplax, was first discovered and characterized by Miller (1982). Since then, nine ClC channel isoforms have been identified in humans alone, each with a slightly different tissue distribution, but the precise physiological role of several of these isoforms remains unknown. The biggest clue in determining the primary role of each isoform is obtained by examining the diseases induced by ClC channel mutations. Genetic mutations of ClC channels are known to be associated with myotonia congenita, a muscle disease characterized by stiffness on sudden movement (ClC-1); Dent's disease, an inherited kidney disorder (ClC-5); and Bartter's syndrome, a salt-wasting renal tubular disorder (ClC-K). It will be a challenge to uncover how and why mutations of these genes alter the permeation dynamics of Cl− ions.
Over the past decade, many salient properties of ClC-type channels have been uncovered using the techniques of molecular cloning and subsequent heterologous expression (Jentsch et al., 1990). First among these properties is the fast gating mechanism. ClC channels undergo voltage-dependent transitions between open and closed states (Pusch et al., 1995; Chen and Miller, 1996; Rychkov et al., 2001), which are facilitated by Cl− ions in the extracellular solutions. Thus, unlike the cationic voltage-gated channels, a permeating Cl− ion itself appears to be acting as a ligand. Secondly, conduction properties differ among the isoforms. The current-voltage relationships measured from ClC-0 and ClC-2 are linear (Miller, 1982; Lorenz et al., 1996), whereas those measured from other isoforms are either inwardly rectifying (ClC-1; Rychkov et al., 2001) or outwardly rectifying (ClC-3, ClC-4, and ClC-5; Duan et al., 1999; Kawasaki et al., 1995; Steinmeyer et al., 1995; Friedrich et al., 1999; Vonoye and George, 2002). Thirdly, ClC-0, and perhaps other ClC-type channels, show an anomalous mole fraction behavior in a mixed solution of Cl− and NO3 ions (Pusch et al., 1995), thus suggesting that conduction across the pore is a multi-ion process. Because the conductance of all ClC-type channels is low, ranging from 10 pS to <1 pS, detailed characterizations of single channel properties have not yet been carried out.
Despite the availability of x-ray structures of two prokaryotic ClC Cl− channels and their mutations (Dutzler et al., 2002, 2003), as yet there has been no theoretical study that attempts to relate the atomic structure of a ClC channel to the macroscopic properties. One of the difficulties in utilizing the newly unveiled information is that all of the published crystallographic structures, including the E148A mutant channel in the postulated open state, have atoms occluding the pore and obstructing Cl− permeation. In the wild-type E. coli ClC (EcClC) channel structure, residues from the N-termini of the D, F, and N α-helices are constricting the channel and two of these residues are completely blocking the conduction pore. As we follow the EcClC pore from either the extracellular or intracellular opening toward the middle of the pore, it abruptly tapers and vanishes. Investigation of the postulated open state configuration of the E. coli E148A mutant reveals that two residues are still partially occluding the channel, preventing Cl− permeation. It is likely that the conduction path of ions in these channels has become distorted in the process of crystallization or that they represent the channel in a closed state. Thus, before the crystal structure coordinates can be used to investigate the permeation of ions through the channel, a completely open state structure needs to be created by using molecular dynamics or other means. Moreover, the ion-conducting path of the EcClC channel, unlike that of the KcsA channel (Doyle et al., 1998), takes a tortuous course through the protein, instead of being straight and perpendicular to the plane of the membrane. The meandering nature of the ClC pore complicates calculations of the force an ion experiences as it moves through the pore.
Here we examine the dynamics of ion permeation in ClC-0 and ClC-1, the two most thoroughly studied ClC channels, using electrostatic calculations and three-dimensional Brownian dynamics simulations. We first build an open state configuration of the channel with molecular dynamics simulations, by pushing the pore-lining residues located near the middle of the channel outward until the segment attains the desired interior radius of ≈2.5 Å, the smallest radius that allows conduction of both Cl− and 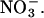 Then, we convert the bacterial EcClC channel into ClC-0 and ClC-1 by replacing all the charged and dipole-containing amino acid residues that are not conserved. Utilizing the full atomic models of the channels so constructed, we carry out electrostatic calculations and Brownian dynamics simulations to elucidate the permeation dynamics across the pores and investigate the effect of several point mutations.
Then, we convert the bacterial EcClC channel into ClC-0 and ClC-1 by replacing all the charged and dipole-containing amino acid residues that are not conserved. Utilizing the full atomic models of the channels so constructed, we carry out electrostatic calculations and Brownian dynamics simulations to elucidate the permeation dynamics across the pores and investigate the effect of several point mutations.
METHODS
Building open-state models of ClC-0 and ClC-1
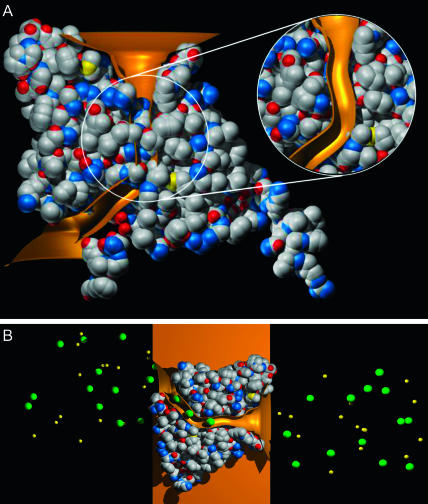
Although the ClC protein is a homodimer, forming two identical pores (Middleton et al., 1996; Ludewig et al., 1996), we simplify our model by dealing with only one of the two pores throughout this study. In Fig. 1 A, we show a view of the pore from the plane of the membrane, created from the crystallographic protein structure of the Escherichia coli ClC channel (EcClC) reported by Dutzler et al. (2002) (Protein Data Bank accession code 1KPK). Despite the slightly higher resolution of the E148A crystal structure (Dutzler et al., 2003), we choose the wild-type EcClC as our template for this study, due to the presence of the key gating residue Glu-148 in its native state. Later we demonstrate that the results we obtain with the model based on the low-resolution x-ray structure do not differ from those obtained with the model based on the high-resolution structure. To reveal the ion-conducting path, the front half of the protein is removed in the figure. There is no continuous course which ions can traverse from the intracellular to extracellular sides of the protein, as shown in Fig. 1 A. There are deep depressions on each side of the protein, but atoms from two of the central residues in the EcClC structure, Ser-107 and Glu-148, occlude the pore. Although the pathway to the external vestibule can be deduced from the wild-type structure, the task of locating the ion-conducting pathway is simplified by overlaying the E148A postulated open structure. Here, three Cl− ions sit in between the channel entrances, two of which are trapped in the protein between the intracellular and extracellular depressions by Ser-107 and Lys-147, suggesting that the two depressions join to form a conducting conduit for Cl− ions. Although the width of the pathway is not large enough to allow a Cl− to pass, the likely location of the pore axis can be determined.
FIGURE 1.
The shape of the model channel and Brownian dynamics simulation system. The atomic detail of the crystal structure of the E. coli channel, EcClC, taken from Dutzler et al. (2002) (A) and our open model of the channel (inset) are shown with the front half of the atoms removed to reveal the pore. The surface of the open pore in gold is superimposed on each diagram. (B) A cylindrical reservoir containing a fixed number of Na+ (shown in yellow) and Cl− (green) is attached at each end of the channel for Brownian dynamics simulation. The channel protein is imbedded in a low dielectric region representing the membrane (gold).
For a Cl− to navigate across either the wild-type or the E148A EcClC channel, the blocking residues must move sideways so as to create a pore with radius >1.81 Å, that of a chloride ion. To allow conduction of larger ionic species, such as I− and 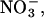 and reproduction of other experimental results, the minimum pore radius must be increased closer to 2.5 Å. We create an open state shape in as simple a way as possible, by gently pushing the occluding residues outward and allowing them to find their own orientation.
and reproduction of other experimental results, the minimum pore radius must be increased closer to 2.5 Å. We create an open state shape in as simple a way as possible, by gently pushing the occluding residues outward and allowing them to find their own orientation.
Expansion of the EcClC channel, using the x-ray structure reported by Dutzler et al. (2002), to an open state is carried out with CHARMM (Brooks et al., 1983), utilizing Version 19 extended-atom parameters for protein. Atoms blocking the pore are pushed aside using a CHARMM MMFP cylindrical repulsive force. Because the region where the pore is blocked is curved, six short MMFP cylinders are placed along the channel center line, with each cylinder axis aligned tangential to the axis of the pore. Since the surrounding atoms have radii of ∼1.5 Å, the radius of the MMFP cylinders (measured to the atom centers) is set to 5 Å. By placing these cylinders, starting from z = −12.5 Å, to follow the curvature of the pore, all the atoms protruding into the pore can be contained within their interiors. Any atoms inside the cylinders are then pushed outward with harmonic force of 85 kT/Å2 (or 50 kcal/mol/Å2) relative to the surface. All atoms outside the cylinders are held near their initial positions using comparatively weak harmonic constraints of 1.6 kT/Å2. The expansion is carried out in two stages of 500 steps of CHARMM dynamics at 298 K, with the final atom positions from the first stage used as new initial positions for constraints during the second stage.
The open structure of the pore thus created is shown in the inset of Fig. 1 A. The shape of the open pore surface used for our calculations is indicated by the gold-colored surface superimposed on the closed and open structures. The ClC pore takes a tortuous course through the protein, unlike the KcsA potassium channel, which is straight and lies perpendicular to the plane of the membrane. We have imposed the condition that the cross-section of the channel perpendicular to the channel axis must be circular, to simplify the computational steps involved in determining and storing the electric field inside the pore. The channel remains quite narrow, having a minimum radius of r ≈ 2.5 Å near the center, but opens up quite rapidly at each end. Notably, the orientation of the Glu-148 residue is similar to that seen in the E148A mutant.
We make an assumption that the overall shape of the open-state pore for EcClC that we created is approximately the same as for ClC-0 and ClC-1. Thus, we hypothesize that it is the differing amino acid sequence that yields the different conductance properties of these channel isoforms. This is a plausible assumption and is supported by a recent study that examines the location of inhibitor binding sites to suggest a high degree of structural similarity between bacterial and mammalian ClC channels (Estévez et al., 2003).
Next we convert the open-state bacterial ClC channel into ClC-0 and ClC-1 using primarily the sequence alignment of Dutzler et al. (2002). There is a limited sequence similarity between the bacterial and prokaryotic homologs, and the alignment in many regions is dubious. For example, the residue population between the various domains in ClC-0 is far denser than in the bacterial channel. Thus, for ClC-0, we also generated two additional sequence alignments using ClustalW (Thompson et al., 1994), one without and the other one with manual adjustment. To generate a sequence alignment between ClC-0 and EcClC, we use the ClC-0 amino acid sequence from Torpedo california (Genebank number: P35522). To overcome the large difference in the length of the sequence, we crop the nonaligned C-terminus residues of the ClC-0 isoform, leaving a tail of ∼20 nonaligned residues. This truncated ClC-0 sequence is then aligned with the EcClC primary sequence with ClustalW. This alignment without any further manual adjustment is one of the alternative sequences we use for building a homology model.
The ClustalW alignment is further adjusted manually to obtain an alternative sequence alignment. For manual adjustments, we search the EcClC primary sequence for patterns of conserved residues from the ClC-0 isoforms. If these conserved sequences are misaligned with the EcClC sequence, we manually adjust the alignment where possible, using the scattered regions of unmatched residues as buffers that mark each end of the misaligned regions. We then shift the nonaligned regions up or down the sequence alignment by the required number of residues until all the sequences are aligned. The sequence alignment of ClC-0 adjusted manually in this way is identical to that of the alignment reported by Dutzler et al. (2002), except for four positions. These are positions 73, 142, 310, and 459. Dutzler et al. (2002) identify the first three of these positions as glutamine, whereas in our alignment they are glycine. The last position, identified as arginine by Dutzler et al. (2002), is glutamate according to our alignment. Because this residue occupies a strategic position in the conducting pathway, the charge it carries will have a pronounced effect on the permeation dynamics. We first characterize ClC-0 assuming that the residue at position 459 is glutamate and then examine the effects of changing this residue to arginine in a subsequent section.
To make the conversions, the bacterial ClC coordinates are aligned with the primary sequences of ClC-0 and ClC-1 and all nonconserved pore-lining, charged, and dipole-containing residues are replaced. If the side chain of an existing amino acid is larger than the one with which it is to be replaced, the extra atoms are cropped, the residue is renamed and, unless the vacant space is accessible to the bulk solution it is filled with a dielectric material of ɛ = 2 . If the existing side-chain is smaller than the one with which it is to be replaced, we build onto the original coordinates to form a new functional group. In this way, we ensure that the mutated residue lies in the same plane as the original residue. Although a slight rearrangement of the atoms to accommodate the new residues is likely, provided they stay close to the initial positions, the electrostatic forces and thus our simulations will not be affected. Using these procedures, we have made 173 substitutions, three insertions, and five deletions to convert the prokaryotic ClC channel to ClC-0. The corresponding numbers to mutate from the prokaryotic ClC channel to ClC-1 are 179, 3, and 6. Because we are interested in the permeation of ions through the transmembrane pore, extramembrane regions of ClC-0 and ClC-1 are ignored, as their structures remain unknown.
To avoid confusion, the EcClC numbering scheme is used for all the aligned sequences throughout this study. It is difficult to determine the charge state of the acidic and basic residues in the protein, as traditional approaches require the use of the Poisson-Boltzmann theory, whose validity inside narrow channels is questionable (Corry et al., 2003). In previous simulation studies on the KcsA potassium channel, Chung et al. (2002) reduced the charge on the Glu-118 and Arg-117 residues to 0.7e or 0.3e so as to obtain the best agreement with experimental data. When these residues are fully charged, the outward current decreases, and the inward current increases by ∼20% (see Fig. 3 of Chung et al., 2002). Here, all basic and acidic residues in the protein are kept fully charged. We have not examined the effects of reducing the charge states of these residues on the conductance properties of ClC-0 and ClC-1.
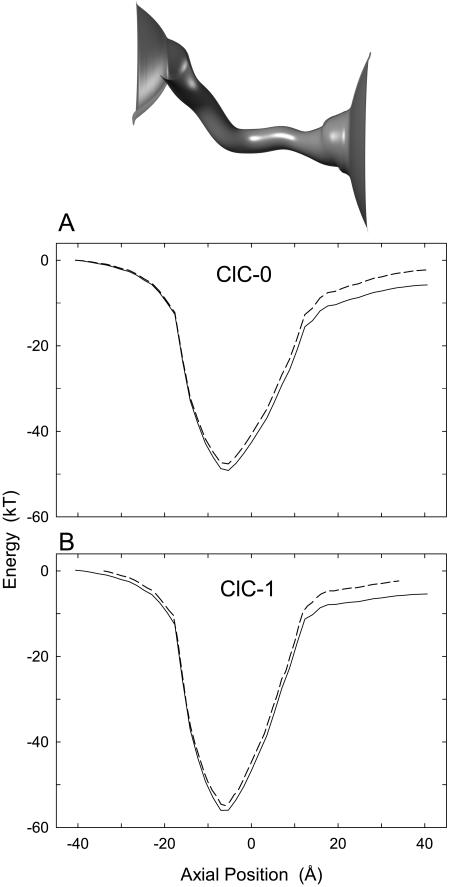
FIGURE 3.
Electrostatic energy profiles encountered by a single Cl− ion moving across ClC-0 (A) and ClC-1 (B). A Cl− is moved across the pore, 1 Å at a time, and the potential energy at each point is calculated by solving Poisson's equation. The calculations are carried out in the absence of an applied potential (broken lines) and in the presence of −80 mV, inside negative with respect to outside (solid lines). The outline of the pore is shown in the inset. The channel extends from −27 to +27 Å.
Solution of Poisson's equation
To calculate the electric forces acting on ions in or around the channel, we solve Poisson's equation using a finite difference technique (Moy et al., 2000). We represent the channel and aqueous solution as continuous dielectric regions with constants ɛp = 2 for the protein, ɛw = 80 for the bulk water outside of the channel, and ɛc = 60 for the interior of the pore. Using this technique, the bent path of the pore can be easily modeled by determining which grid points are in the protein and which are not. The calculated potential inside the channel converges rapidly as the grid size is reduced (Edwards et al., 2002). No improvement in solution is obtained using a grid size of <0.6 Å.
The potential energy profile encountered by a single ion moving along the pore is obtained by moving an ion along the center of the pore in 1 Å steps and calculating the potential energy at each position. As will be shown later, the ClC channel is usually occupied by two ions. To visualize the shape of the energy profile a Cl− ion encounters as it attempts to enter a pore that is already occupied by two or more resident ions, we construct multi-ion energy profiles. We move one of the ions from the extracellular space into the channel in 1 Å steps, holding it fixed at each step. We then allow the resident ions, placed initially at the binding sites, to adjust their positions so that the force on them will be 0, thus minimizing the total energy of the system. The minimization is performed at each step as we bring the third ion into the channel and the positions of the ions and the total energy are recorded. This corresponds to the total electrostatic energy required to bring in the charge on the ions from an infinite distance in infinitesimal amounts. Energy minimizations are carried out using a modified version of the steepest descent algorithm (Press et al., 1989; Chung et al., 1999).
Brownian dynamics simulations
The energy profiles provide only a qualitative picture of the permeation dynamics, since the random motion of ions is not taken into account. To deduce the conductance of ions through the channel, we carry out three-dimensional Brownian dynamics simulations.
In these simulations, we place 15 Cl− ions and 15 Na+ ions in cylindrical reservoirs of radius 30 Å at each end of the channel to mimic the extracellular or intracellular space (Fig. 1 B). We adjust the height of the cylinder to 61.2 Å to bring the solution to 150 mM. We then trace the motion of these ions under the influence of electric and random forces using the Langevin equation:
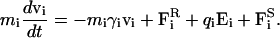 |
(1) |
Here, mi, vi, γi, and qi are the mass, velocity, friction coefficient, and charge on an ion with index i, whereas 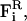 Ei, and
Ei, and 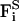 are the random stochastic force, systematic electric field, and short range forces experienced by the ion, respectively. We calculate the total force acting on each and every ion in the assembly and then calculate new positions for the ions a short time later. A multiple time step algorithm is used, where a time step of Δt = 100 fs is employed in the reservoirs and 2 fs in the channel where the forces change more rapidly.
are the random stochastic force, systematic electric field, and short range forces experienced by the ion, respectively. We calculate the total force acting on each and every ion in the assembly and then calculate new positions for the ions a short time later. A multiple time step algorithm is used, where a time step of Δt = 100 fs is employed in the reservoirs and 2 fs in the channel where the forces change more rapidly.
Since calculating the electric forces at every step in the simulation is very time-consuming, we store precalculated electric fields and potentials due to one- and two-ion configurations in a system of lookup tables (Hoyles et al., 1998). To do this, the electric potential is broken into four components,
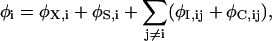 |
(2) |
where the sum over j runs over all the other ions in the system. The symbols in Eq. 2 assume the following significance: φX,i is the external potential due to the applied field, fixed charges in the protein wall, and charges induced by these; φS,i is the self-potential due to the surface charges induced by the ion i on the channel boundary; φI,ij is the image potential felt by ion i due to the charges induced by ion j; and φC,ij is the direct interaction between ions i and j. The first three potential terms in Eq. 2 are calculated using a finite difference solution of Poisson's equation as described above. The first term is stored in a three-dimensional table to save time and storage space. The second and third terms are stored in two- and five-dimensional tables utilizing the symmetry developed in the construction of the pore. As the cross-section of the pore is circular, the potential and field is only calculated at one azimuthal angle in this plane (but still using a three-dimensional solution to Poisson's equation) and the values at an arbitrary point in the plane are interpolated from these. The ion-ion interactions include the Coulomb term and an oscillating short-range potential derived from molecular dynamics simulations as described previously (Corry et al., 2001), and are calculated on the fly during the simulation. The short-range forces include these short-range ion-ion interactions as well as those between ions and the channel walls.
The Langevin equation is solved with the algorithm of van Gunsteren and Berendsen (1982), using the techniques described by Li et al. (1998). Bulk ionic diffusion coefficients of 1.33 × 10−9 m2 s−1 for Na+ and 2.03 × 10−9 m2 s−1 for Cl− ions are employed in the reservoirs and vestibules. These values are reduced to 50% of the bulk values in the pore, as determined with molecular dynamics studies (Allen et al., 2000). Simulations under various conditions, each lasting usually 10–20 μs, are performed with symmetric ionic concentrations in the two reservoirs. The current is computed from the number of ions that pass through an imaginary plane near the end of the channel during a simulation period. For further technical details of the Brownian dynamics simulation method, see Chung et al. (1998, 1999, 2002).
RESULTS
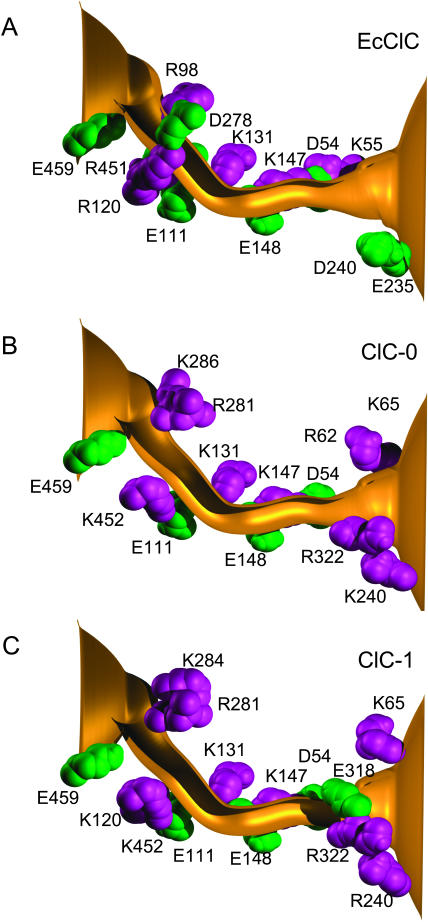
The channel wall of an open-state ClC channel is lined with many charged amino acids, both basic and acidic. In Fig. 2, we show the charged residues that are lining the protein wall of the prokaryotic ClC channel, EcClC (Fig. 2 A), ClC-0 (Fig. 2 B), and ClC-1 (Fig. 2 C). Here, positively-charged arginine and lysine residues are shown in purple and negatively-charged glutamate and aspartate residues are shown in green. There is an additional positively-charged residue (Arg-126) in ClC-0 that is not shown in Fig. 2 B, as it lies behind the pore, between Arg-281 and Lys-131. A cursory inspection of the figure reveals that EcClC, ClC-1, and ClC-0 contain, respectively, seven, five, and four glutamate and aspartate residues lining the protein wall, and six, nine, and 10 arginine and lysine residues. Thus, EcClC has one net negative charge lining the pore, whereas ClC-1 and ClC-0 have, respectively, four and six net positive charges. By counting the number of charged residues, one can reasonably deduce that the current of anions across EcClC will be substantially smaller compared to ClC-0 or ClC-1. This agrees with the experimental findings (C. Miller, personal communication). Also, we notice that there is an additional glutamate residue, namely, Glu-318, guarding the extracellular mouth of ClC-1, which is absent in ClC-0. The presence of this negatively-charged residue would make the entry of a Cl− ion from the extracellular space more difficult. Also, this residue will most likely make it harder for ions to move from the center of the channel to the extracellular space. Thus, we can predict that the current in ClC-1 will be smaller than in ClC-0. Moreover, if the channel holds multiple ions, we can expect that the current-voltage relationship of ClC-1 will show rectification. This follows as an ion moving toward the extracellular space will be aided past the Glu-318 residue by the Coulomb repulsion of the other Cl− ions behind it. A lone ion entering from the extracellular space would find passing this residue much more difficult.
FIGURE 2.
Comparison of EcClC, ClC-0 and ClC-1. The locations of the charged residues lining the pores are illustrated. Arginine and lysine residues are shown in purple and aspartate and glutamate in green. Here and throughout the article, the intracellular aspect of the channel is on the left-hand side.
The results of simulations reported here are in accord with these conjectures. Because no experimental data on the prokaryotic ClC channels are available in the literature, here we focus our attention on ClC-0 and ClC-1. We first describe the results obtained with the open-state configuration of EcClC based on the x-ray structure of Dutzler et al. (2002) and the homology models constructed from the sequence alignments given also by Dutzler et al. (2002). Then, we compare these results with those obtained with an alternative ionic pathway created from the high-resolution x-ray structure (Dutzler et al., 2003). We also demonstrate that a homology model of ClC-0 constructed from a different sequence alignment fails to replicate any of the experimental observations.
Energy landscapes of ClC-0 and ClC-1
As a Cl− ion navigates across the pore, it encounters not only charged amino acid residues but also many dipole-containing residues that are lining the protein wall. In general, the positive poles or the NH backbones of these residues are pointing toward the water-filled pore. Some among these residues are Gln-103, Ser-107, Tyr-445, and His-120 (and for ClC-1, Thr-348). The arrangements of these residues relative to the ion-conducting path and the presence of four more pore-lining, positively-charged arginine and lysine residues than aspartate and glutamate residues in ClC-0 and ClC-1 indicates that the channels will permit anions to pass across, while effectively blocking cations from entering it.
The electrostatic potential energy profiles for various ion configurations in ClC-0 and ClC-1 are given in Figs. 3 and 4. A Cl− ion entering an empty ClC-0 channel encounters a deep energy well of ∼47 kT (ClC-0), or 52 kT for ClC-1 (1 kT = 4.11 × 10−21 J) created by the positively-charged and polar residues in the protein wall (broken lines). The energy profiles obtained from ClC-0 (Fig. 3 A) and ClC-1 (Fig. 3 B) in the absence of an applied potential are broadly similar, except that the well is slightly narrower and shallower for ClC-0. For both profiles, the nadir of the well for a Cl− ion occurs at −7 Å, a little left of the position where Glu-148 is situated. In contrast, for a Na+ or K+ ion attempting to enter the pore, the surplus of positively-charged residues produce an energy barrier of approximately equal magnitude to the well seen by a Cl− ion, that effectively excludes cations from the channel. When a potential of −80 mV (inside negative with respect to outside) is applied, the potentials on the right-hand side of the profiles become lower than those at the left-hand side (Fig. 3, solid lines). Under the influence of the applied potential, Cl− ions tend to drift from inside to outside the cell.
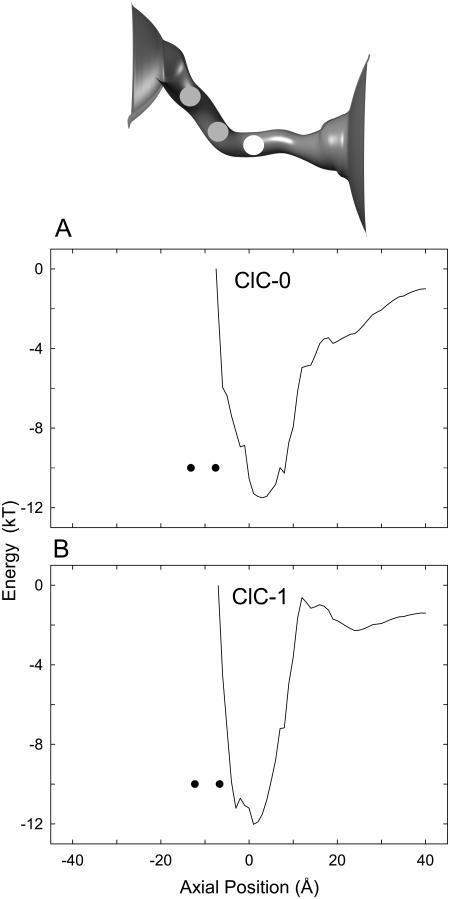
FIGURE 4.
Electrostatic energy profiles encountered by an outermost Cl− ion while two resident ions are at the equilibrium positions. With two ions placed in the binding site, a third ion is moved toward the extracellular space, 1 Å at a time, and the electrostatic potential energy at each position is calculated. At each fixed position of the third ion, the stable configuration of the first two ions is determined iteratively, and the total energy of the assembly is computed. The locations of the two resident ions, when the test ion is at z = 0 Å, are indicated in solid circles in A and B and as dark spheres in the inset.
The wells in both ClC-0 and ClC-1 are deep enough to accommodate two Cl− ions and enables them to exist in a stable equilibrium. The two-ion equilibrium is disrupted when a third ion enters the channel. The energy landscapes seen by the outermost ion with three ions in the channel, in the presence of an applied potential of −80 mV, are illustrated in Fig. 4 A (ClC-0) and 4 B (ClC-1). The positions of the two Cl− ions in the pore, when the outermost ion is at z = 0 Å, are indicated in solid circles. The outermost ion in ClC-0 encounters an energy barrier of ∼11 kT to climb out of the well and conduct into the intracellular space. The ion can occasionally surmount this residual barrier through its random motions aided by Coulomb repulsion exerted by the other ions, allowing it to move into the extracellular space. Thus, from these profiles we deduce that the channel normally appears to cradle two Cl− ions and conduction across the ClC pore takes place when a third ion stumbles into the channel. In contrast to ClC-0, the exiting Cl− ion experiences a sharper energy barrier at the extracellular side of ClC-1, which it must overcome before leaving the channel. This barrier is due to the effects of the pore-lining residue 318, which is a negatively-charged glutamate residue in ClC-1 but neutral in ClC-0.
Current-voltage-concentration profiles
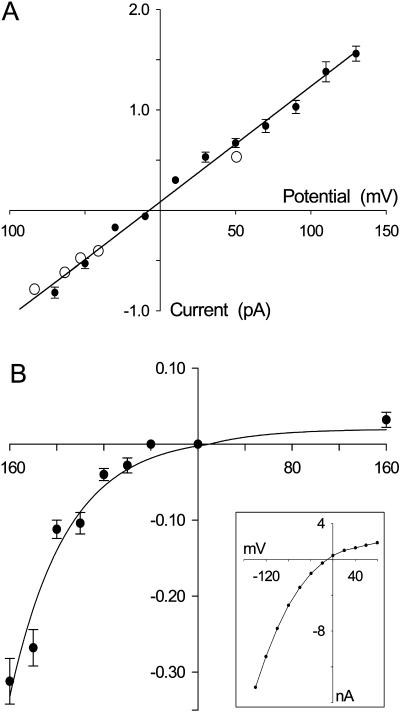
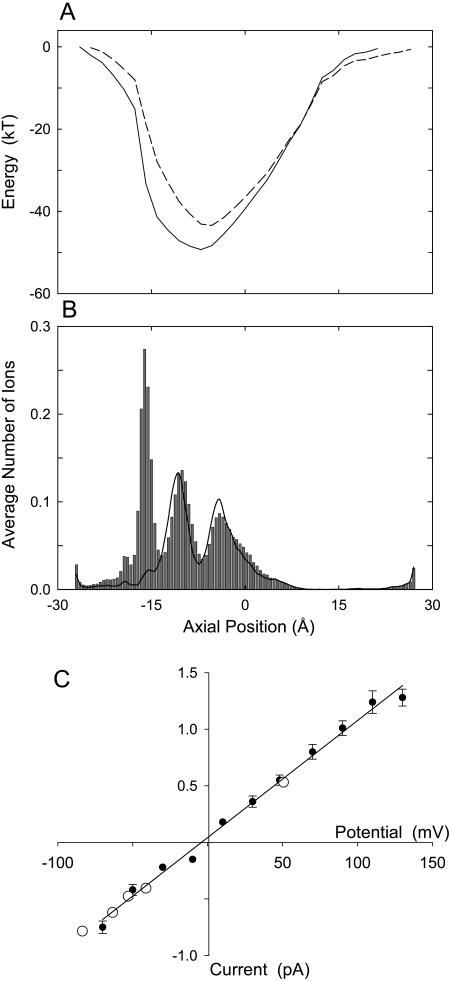
We study the conductance properties of ClC-0 and ClC-1 under various conditions by performing BD simulations. The current-voltage relationships of ClC-0 and ClC-1, shown in Fig. 5, are obtained with symmetrical solutions of 150 mM in both reservoirs. The relationship for ClC-0 obtained from our simulations (solid circles in Fig. 5 A) is linear through the origin when the applied potential is between −70 and +140 mV. The experimental measurements reported by Miller (1982) are superimposed (open circles). The core conductance, derived by fitting a linear regression through the data points, is 11.3 ± 0.5 pS, compared to the experimental value of 9.4 ± 0.1 pS. The relationship, however, abruptly deviates from Ohm's law with further decreases in the membrane potential <−70 mV. The nonlinear section of the data is not shown in the figure. This discontinuity results from a sudden change in the permeation dynamics. A strong negative applied potential, when it exceeds a certain critical value, forces an additional Cl− ion into the channel, thus enhancing the likelihood of the outermost ion exiting the pore.
FIGURE 5.
The current-voltage relationships of ClC-0 (A) and ClC-1 (B). The current measured at various applied potentials is obtained with symmetrical solutions of 150 mM in both reservoirs. In this and all subsequent figures, unless stated otherwise, a data point (solid circle) represents the average of 24–48 sets of simulations, each set lasting 4 × 106 time steps (or 0.4 μs). Error bars in this and following figures have a length of mean ± SE of 1 and are not shown when they are smaller than the data points. Superimposed on the simulated data in A are the experimental measurements obtained by Miller (1982), shown in open circles. The whole-cell current-voltage relationship illustrated in the inset of B is reproduced from Bennetts et al. (2001).
In contrast to ClC-0, the inward and outward currents are both smaller and pronouncedly asymmetrical in ClC-1 (Fig. 5 B). The conductance at −100 mV is 1.0 pS, increasing to 1.9 pS when the applied potential is increased to −160 mV. With the polarity of the applied potential reversed, the current across the channel is <0.03 pA (or no ion crosses the pore in the period of 14.4 μs) until the driving force is increased to +160 mV. The conductance at this potential is ∼0.2 pS. There are several whole-cell current measurements demonstrating that the ClC-1 channel is inwardly rectifying (Rychkov et al., 2001; Bennetts et al., 2001; Fahlke et al., 1995), as shown in the inset of Fig. 5 B. Because of the low conductance, no single channel current-voltage relationship of ClC-1 has been reported. Using low intracellular pH, which causes a slowing of gating kinetics, Saviane et al. (1999) determined the channel conductance to be ∼1.2 pS. This value is in a close agreement to that determined by using concatemeric channels containing one subunit each of ClC-0 and ClC-1 (Weinrich and Jentsch, 2001). Both the shape of the current-voltage relationship and the inward conductance determined from our simulation are in accord with these experimental findings.
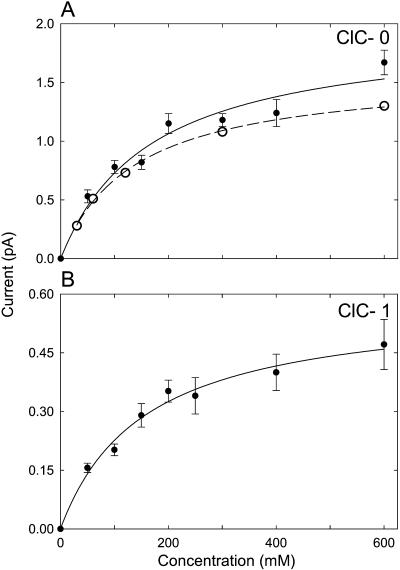
A current-concentration relationship of a single channel reveals important information about the underlying permeation mechanism. Here we provide the results of our simulations on ClC-0 and ClC-1. Experimentally, the current, I, across many channels first increases with an increasing ionic concentration [Cl] and then saturates, leading to a current-concentration relationship of the Michaelis-Menten form: I = Imax/(1 + Cls/[Cl]). Thus, the current approaches the saturation current, Imax when [Cl]≫Cls. Theoretically, the conductance-concentration curve is expected to saturate if the transport of ions across the channel is determined by two independent processes, one of which depends on ionic concentrations on the two sides of the channel and one that does not. In this case we expect that the time spent waiting for a third ion to enter the channel will decrease as concentration is increased, whereas the time for the outermost ion to cross to the extracellular space will not. In Fig. 6, the currents obtained from Brownian dynamics simulations in ClC-0 (Fig. 6 A) and ClC-1 (Fig. 6 B), under the applied potential of −80 mV, are plotted against the concentration of Cl− ions in the reservoirs. The experimental data obtained from ClC-0 with an applied potential of −80 mM are shown in open circles in Fig. 6 A (Tsung-Yu Chen, personal communication) and fitted with a broken line. The solid lines fitted through our data points are calculated from the Michaelis-Menten equation. The half-saturation points Cls determined from the fitted curves are 163 ± 51 mM for ClC-0 and 144 ± 23 mM for ClC-1. The value we obtained from ClC-0 is slightly higher than that reported by Chen (personal communication) and White and Miller (1981). Their half-saturation constant Cls, measured at +40 mV, is ∼85 mM (75 mM Cl− activity). The corresponding value obtained from Chen's measurements is 136 ± 8 mM.
FIGURE 6.
Current-concentration relationships for ClC-0 (A) and ClC-1 (B). The outward currents are obtained with symmetrical solutions of varying concentrations of NaCl in the reservoirs under an applied potential of −80 mV. The data points are fitted by solid lines using the Michaelis-Menten equation. The experimental measurements obtained by Chen (personal communication) are shown in open circles in A.
Ions in the channels
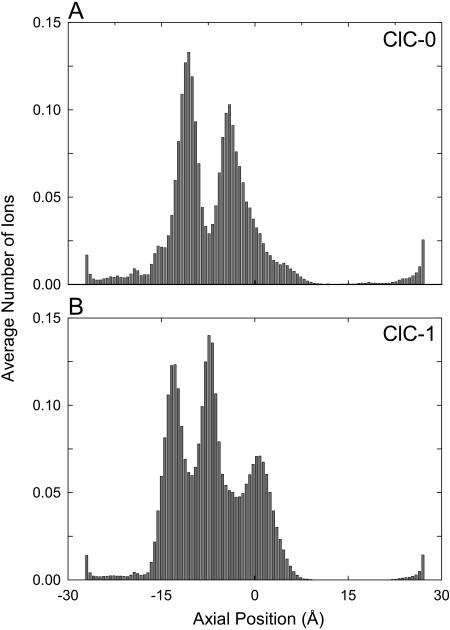
Next we examine where ions reside in ClC-0 and ClC-1 and the steps involved in conduction using three-dimensional Brownian dynamics simulations. In Fig. 7, we illustrate the likelihood of finding ions in different regions of ClC-0 (Fig. 7 A) and ClC-1 (Fig. 7 B), in the presence of an applied potential of −80 mV. To construct the histograms, the channel is divided into 100 thin sections, and we plot the average number of ions in each section during a 0.4-μs simulation. There are two regions in ClC-0 where ions dwell preferentially, one at z = −11.0 and the other at z = −4.4 Å. In a conducting state (with an applied field), there are, on average, 2.3 ions in the channel. In ClC-1, without an applied field, there are on average 2.5 Cl− ions in the pore. Two prominent binding sites are apparent at −10.3 Å and the other at −4.9 Å. In the conducting state, driven by an applied potential of −80 mV, the binding sites shift toward the right by 2.1 Å and a third peak in the histogram appears at the center of the pore near z = 0. The average number of ions in the pore is now increased to 2.9.
FIGURE 7.
The dwell histograms of ClC-0 (A) and ClC-1 (B). The channel is divided into 100 thin sections, and the average number of ions in each section is calculated over a simulation period (0.4 μs) in the presence of an applied potential of −80 mV.
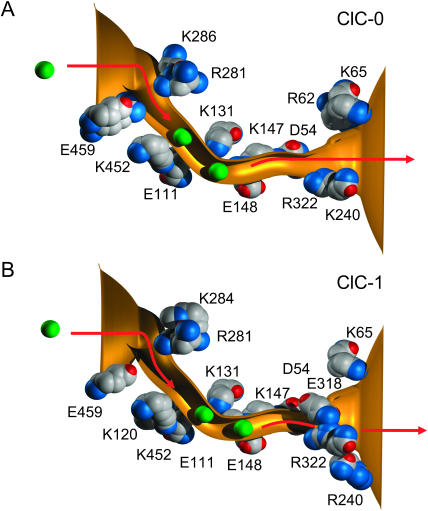
The locations of ions in the channel, together with the main charged amino acid residues lining the pores, are illustrated in Fig. 8. The two resident ions in ClC-0 (Fig. 8 A) stay very close to the positively-charged residues, Lys-452 and Lys-131. Comparing the position of these ions to the innermost two ions trapped in the crystallized E148A EcClC channel, the positions of the ions in our simulation are shifted slightly toward the extracellular side of the channel. This shift reflects the presence of an additional positively-charged residue, Arg-62, lining the extracellular end of the ClC-0 pore and attracting Cl− ions deeper into the channel. For conduction to take place, a third ion must enter the pore from the intracellular side of the channel. This disrupts the stable equilibrium of the two resident ions. With an applied potential of −70 mV, it takes on average 125 ns for a third ion to stumble into ClC-0 from the intracellular reservoir. Once three ions are in the pore, the outermost ion near the Lys-131 residue manages to climb out of the energy well in ∼70 ns. Thus, there are two rate-limiting steps in the outward conduction process. When the polarity of the applied field is reversed, the locations of the binding sites remain unaltered. Here, the step that limits the number of ions the channel can process is the entrance of a third ion from the extracellular space into the pore, which on average takes nearly 200 ns. Once three ions are in the pore, the innermost ion, located near the Lys-452 residue, is expelled from the pore almost instantaneously.
FIGURE 8.
Permeation dynamics of ClC-0 (A) and ClC-1 (B). The locations of two Cl− ions occupying the binding sites are indicated with green spheres. The figures show the time it takes for a third ion to enter the pore (step 1) that is already occupied by the two resident ions, and the time it takes for the outermost ion to exit the pore (step 2), given that the third ion has entered the pore. The average times are calculated over a simulation period of 0.4 μs in the presence of −70 mV for ClC-0 and −80 mV for ClC-1.
The dynamics of ion permeation in ClC-1 are similar to that in ClC-0 (see Fig. 8 B). The two resident ions are situated near the Lys-131 and Lys-147 residues, shifted slightly toward the extracellular side compared to those in ClC-0. The dwell histogram obtained from ClC-1 shows the presence of a third peak, in the extracellular side of the channel at z = −13.4 Å. This indicates that a third ion spends a considerable amount of time in the channel before a successful conduction event occurs. The location of these three peaks in ClC-1 corresponds almost exactly to the position of the three chloride ions in the crystal structure of E148A EcClC (Dutzler et al., 2003). Under the influence of an applied potential of −80 mV, a third ion enters the pore, this process taking on average 270 ns. With three ions in the pore, it takes >1 μs for the outermost ion to exit the pore. The number of ions the channel can process is thus limited by the energy barrier created by the additional negatively-charged residue Glu-318.
The conduction process across ClC-0 and ClC-1 is broadly similar to that in the KcsA potassium channel, where a third K+ ion entering the central cavity accelerates toward the selectivity filter and effects conduction by knock-off (Allen and Chung, 2001). Pusch et al. (1995) showed that current through the ClC-0 channel is reduced substantially when two permeating ionic species, Cl− and 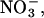 are mixed compared to that observed when only one of the two ions is present in the solution. This so-called “anomalous mole fraction” behavior can only occur if the pore has to be occupied by two or more ions for conduction to take place. The results of our simulation are consistent with this experimental deduction.
are mixed compared to that observed when only one of the two ions is present in the solution. This so-called “anomalous mole fraction” behavior can only occur if the pore has to be occupied by two or more ions for conduction to take place. The results of our simulation are consistent with this experimental deduction.
Ionic pathways and sequence alignments
There are two caveats to the approach we use in constructing the models. These are to do with, firstly, the method of creating an ionic path across the EcClC protein and, secondly, the sequence alignments used for creating the homology models of ClC-0 and ClC-1. We address these issues in this section.
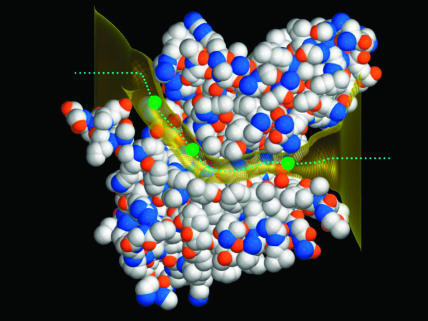
To ascertain the reliability of the open-state EcClC configuration that we use, we compare the pathway to open structures created using the high-resolution x-ray structures of the wild-type EcClC and E148A EcClC reported by Dutzler et al. (2003) (Protein Data Bank accession code 1OTS and 1OTT, respectively). When the E148A EcClC structure is aligned with the low-resolution structure, the three embedded Cl− ions in the E148A structure pass through the center of the ionic pathway created using the crystal coordinates reported in Dutzler et al. (2002). The expansion of both high-resolution structures are carried out using the same procedures detailed previously, except that the trapped Cl− ions are not moved during the expansion process. In Fig. 9, we show the open-state configurations created with the low-resolution wild-type EcClC (gold surface) and high-resolution E148A EcClC crystal structures. Three Cl− ions in the x-ray structure (Protein Data Bank accession code: 1OTT) are indicated as green spheres, and the central axis of the ionic path we created is shown as a dotted green line. The two pore shapes are virtually indistinguishable. The high-resolution structure has seven residues less than that of the low-resolution structure: it is five residues shorter at the N-terminus and two at the C-terminus. When we select atoms in the vicinity of the pore, ∼25% of all atoms, and compare the open-state configuration derived from the E148A high-resolution structure with that derived from the wild-type low-resolution structure, we find that the root-mean square average difference in atom position is 1.01 Å. We then used the axial coordinates of the open state mapped through the E148A EcClC to create an open-state configuration for the high-resolution wild-type EcClC structure. The equivalent root-mean square deviation of atom position is 0.95 Å. The results of these tests show that the structure of the open pore we construct is independent of the original structure used to create it. We demonstrate later that the shape of the energy profiles and the conduction properties obtained from two different open-state models, one obtained from the low-resolution x-ray structure (Dutzler et al., 2002) and the other obtained from the WT high-resolution structure (Dutzler et al., 2003), are virtually identical.
FIGURE 9.
The ionic pathway created from the high-resolution x-ray structures. The shape of the pore is virtually indistinguishable from that created from the low-resolution x-ray structure, illustrated as a gold surface. Three Cl− ions are indicated as green spheres, and the central axis of the pore created from the high-resolution structures is shown as a dotted line.
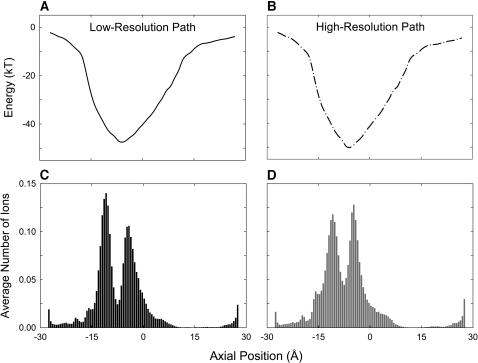
In Fig. 10, we compare the energy profiles and dwell histograms obtained from the ClC-0 models based on the low-resolution (left-hand column) and high-resolution (right-hand column) x-ray structures. The sequence alignment used to create the homology models is the same as the one we used throughout, except the glutamine residues at positions 73, 142, and 310 are replaced by glycine (i.e., we use the ClustalW alignment with manual adjustment). In the ClC-0 notation, these residue numbers correspond to 89, 160, and 344. The energy landscapes encountered by a single Cl− ion traversing channel show a deep well centered at −8 Å. The two profiles, in the presence of an applied potential of +20 mV, are virtually identical. The maximum depth of the well obtained from the low-resolution structure is 2.1 kT shallower than that obtained from the high-resolution structure (47.4 kT vs. 49.5 kT). The two dwell histograms illustrated in Fig. 10 are similar. Both reveal two prominent binding sites in the channel, centered at z = −11 and −4.4 Å. The average number of ions in the channel in the conducting state for the low-resolution homology model (C) is 2.31, compared to 2.45 for the high-resolution homology model (D). The difference is not statistically significant. There is no significant difference in the magnitudes of current flowing across the two models, determined by using Brownian dynamics. With an applied potential of −40 mV, the currents we observe, during the simulation period of 17.2 μs, are −0.35 ± 0.08 and −0.33 ± 0.08 pA for the low-resolution and high-resolution models, respectively. From these results, we conclude that the open-state configurations created from the x-ray structures of Dutzler et al. (2002, 2003) are almost identical, and the conductance properties deduced from the two models do not differ appreciably.
FIGURE 10.
Energy profiles and dwell histograms obtained from the models based on the pathways created from the low-resolution (A and C) and high-resolution x-ray structures (B and D).
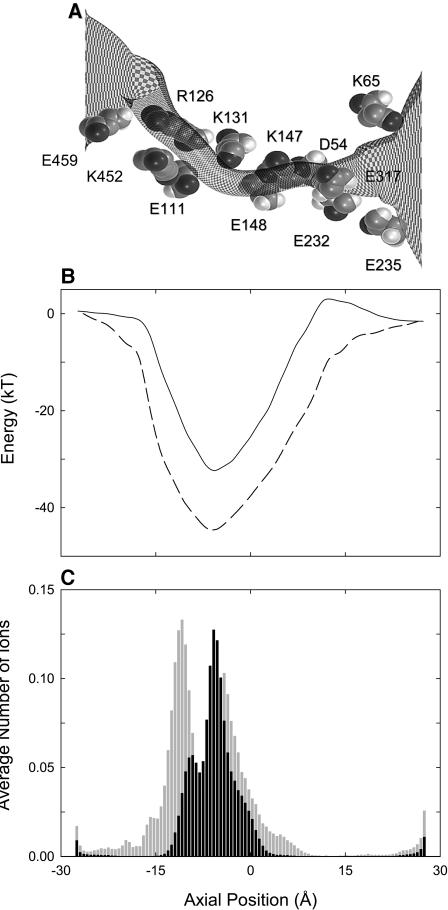
We next examine whether or not the ClustalW alignment of ClC-0 without manual adjustment (see the Method section) can also reproduce any of the experimental observations. The locations of the charged amino acid residues along the pore are shown in Fig. 11 A. Two positively-charged residues guarding the external gate of the channel in the manually adjusted alignment, namely Arg-322 and Lys-240, are now replaced with glutamate residues, Glu-232 and Glu-235 (see Fig. 2 B). There are two net negatively-charged residues lining the pore, compared to six net positive charges in the alignment of Dutzler et al. (2002). The energy landscape encountered by a Cl− ion, shown in Fig. 11 B, reveals several features that are not present in the profile illustrated in Fig. 3 A, reproduced here in dashed line. The energy well is narrower and shallower than that obtained from the homology model constructed from the alignment using Dutzler et al. (2002). The depth of the well at z = −7 Å is 32 kT, compared to 47 kT in the profile shown in Fig. 3 A. Also, there is a prominent energy barrier of ∼3.5 kT located at z = +12 Å. The dwell histogram obtained in the presence of an applied potential of −80 mV is illustrated in Fig. 11 C. Instead of having two prominent peaks, the histogram shows one main peak centered at z = −5.6 Å. For comparison, the histogram shown in Fig. 7 A is superimposed on this figure. There are, on average, 1.4 ions in the channel, compared to 2.4 ions in the histogram illustrated in Fig. 7 A. A resident ion is permanently trapped at the binding site, unable to exist outside the pore. As a second ion enters from the intracellular reservoir, it is forced out of the channel by the Coulomb repulsion of the trapped ion. We measure the current across the model channel using Brownian dynamics. In the simulation period of 9.6 μs with an applied potential of −80 mV, the observed current is −0.09 pA. No ions traverse in the opposite direction during the same simulation period, even at an applied potential of −160 mV. We thus conclude that the homology model constructed with the ClustalW alignment with no manual adjustment is incapable of replicating the experimental observations.
FIGURE 11.
Homology model constructed from the ClustalW alignment with no manual adjustment. The locations of the charged residues lining the pore are indicated in A. In B, the energy profile encountered by a single ion traversing the pore (solid line) is compared to that obtained from the model based on the manually adjusted alignment (broken line). The dwell histogram in C shows one prominent peak. For comparison, the histogram illustrated in Fig. 7 A is reproduced as light shade.
Finally, we examine the effects of performing a point mutation on the native ClC-0 model, mutating position 459 from glutamate to arginine. This residue is identified as arginine in the alignment given by Dutzler et al. (2002), whereas our ClustalW alignment identifies it as glutamate. We demonstrate that the permeation dynamics of the E459R ClC-0 mutant involve an extra ion, but is otherwise similar to that of ClC-0. Harking back to Fig. 8, we note that the Glu-459 residue is located near the entrance of the intracellular vestibule. Fig. 12 A compares the energy wells encountered by a Cl− ion moving through the mutant channel (solid line) and ClC-0 (broken line, reproduced from Fig. 3 A). Changing a negative residue to a positive residue is reflected in the depth and the width of the energy well: it is 8.3 kT deeper and slightly broader than that of the native ClC-0. Now three Cl− ions occupy the pore, oscillating from their equilibrium positions at z = −16.3, −10.7, and −4.7 Å (Fig. 12 B). These positions are close to three positively-charged residues, Arg-459, Arg-281, and Lys-131. In the presence of an applied potential of −80 mV, we find that there are almost 3.5 ions on average in the mutant channel, indicating that the presence of the fourth ion destabilizes the three-ion equilibrium and causes conduction. The entry of the fourth Cl− ion in the pore from the intracellular reservoir is the rate-limit step in conduction in this mutant channel, taking on average 270 ns at −80 mV. The three resident ions shuffle toward the extracellular side of the pore to accommodate the additional ion that enters. Once the outermost ion reaches the position of Lys-147, it dwells there temporarily, but usually exits within a few nanoseconds. Although conduction across the E459R mutant pore becomes a four-ion process, instead of being a three-ion process, the magnitude of currents across the pore remains unaltered. In Fig. 12 C, the current-voltage curve for the mutant pore is illustrated. The curve is linear through the origin, with the core conductance of 10.3 ± 0.3 pS. Our simulated data shows excellent agreement with corresponding measurements reported by Miller (1982), which are superimposed (open circles).
FIGURE 12.
Mutant ClC-0 channel. The Glu-459 residue guarding the intracellular gate of ClC-0 is changed to arginine and then the mutant channel is characterized. The potential energy profile in the absence of an applied potential (A) obtained from the mutant channel (solid line) is compared with the unmutated channel (broken line), reproduced from Fig. 3 A. In the dwell histogram, an additional binding site appears near the intracellular entrance of the pore (B). The current-voltage relationship (C) remains virtually unchanged from that obtained from the unmutated ClC-0 channel. The experimental measurements (open circles) obtained by Miller (1982) is superimposed on the simulated data (solid circles).
DISCUSSION
In this article, we have attempted to relate the molecular structure of the ClC chloride channels to some of their macroscopically observable properties using several different computational approaches. Using molecular dynamics, an open-state structure of the prokaryotic ClC channel is first created by moving the atoms that are occluding the ion-conducting path. The x-ray structures of Dutzler et al. (2002, 2003) correspond to a closed state, although whether the pore is closed by a slow or fast gating mechanism or through a structural change in the process of crystallization is not known. In creating an open conformation, we have made the minimum possible adjustments to the original crystallographic structure. Only those atoms that sterically impede ionic passage are slowly pushed outward, leaving all other residues largely unperturbed. The narrowest segment of the pore in the open-state conformation we create is just under 2.5 Å. We have ascertained in a preliminary study that the conductance is not appreciably affected when the radius of this segment is reduced to 2.3 or 2.1 Å. When the channel is in a conducting state, the constricted segment of the pore near E148 must be wider than the van der Waals radii of Cl− and I− ions, which are 1.80 and 2.15 Å, respectively, and also must accommodate 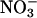 ions. Thus, the minimum pore radius of 2.5 Å we adopted in this study is likely to be a good approximation.
ions. Thus, the minimum pore radius of 2.5 Å we adopted in this study is likely to be a good approximation.
In building the models, we assume that the open-state shape of the prokaryotic ClC channel is the same as that for ClC-0 and ClC-1, and that the structural features that confer specific characteristics of each ClC isoform are the polar and charged amino acid residues near the ion-conducting path. Whether this assumption is justified will remain unknown until the structures of these two channels are determined, crystallographically or otherwise, although a recent study suggests our assumption is plausible (Estévez et al., 2003). The three-dimensional atomic models of ClC-0 and ClC-1 we constructed by replacing amino acid residues that are not conserved with EcClC successfully reproduce the experimentally observed conductances and the shape of the current-voltage curves. The accurate replication of the experimental data with our Brownian dynamics simulations is not brought about by judiciously adjusting free parameters. There is one unknown constant that features in each of the Langevin and Poisson's equations. The first one of these is the friction coefficient, γ, which is related to the diffusion coefficient, D, by the Einstein relation, γ = kT/mD, where m is the mass of the ion. Molecular dynamics simulations were carried out by Allen et al. (2000) to obtain estimates of diffusion coefficients of biologically important Na+, K+, and Cl− ions in various segments of the KcsA and schematic channels. In the hydrophobic chamber, their diffusion coefficients are reduced to ∼38% of bulk diffusion on average. In this study, we use 50% of the bulk values in the pore. Unlike in the Poisson-Nernst-Planck theory, where conductance scales linearly with assumed D, conductance deduced from Brownian dynamics simulations is less sensitive to this parameter. The outward and inward currents across the potassium channel are reduced only slightly as DK is reduced from the bulk value to 10% of this (Chung et al., 1999).
In solving Poisson's equation, in this and all our previous studies, we use dielectric constants of 2, 60, and 80 for the protein, channel, and reservoir. Unlike water and lipid, which form homogeneous media, proteins are quite heterogeneous, exhibiting large variations in polarizability depending on whether we are dealing with the interior or exterior of a protein (see Schutz and Warshel, 2001). There are several microscopic investigations of the dielectric constant of proteins from molecular dynamics simulations (Smith et al., 1993; Simonson and Brooks, 1996; Pitera et al., 2001). The dielectric constant for the whole protein, according to these studies, varies between 10 and 40, but when only the interior region of the protein consisting of the backbone and uncharged residues is considered, the value drops to 2 or 4. From a microscopic point of view, this should make assigning a fixed ɛp value to an entire protein in continuum electrostatic calculations problematic. The effects of changing ɛp from 2 to 3.5 and 5 were examined by Chung et al. (2002), using the KcsA potassium channel. They showed that the precise value adopted in solving Poisson's equation has negligible effects on the macroscopic properties derived from Brownian dynamics simulations. For further discussion on this issue, see Burykin et al. (2002, 2003).
Assigning the appropriate value of dielectric constant of water, ɛw, within the ion channel is also nontrivial. In bulk water, molecules polarize so as to shield interactions within the dielectric media by a factor of ∼1/80. However, given the preferential alignment of water in narrow pores, especially in regions of high charge, this shielding is likely to be far less effective. In theory, to determine ɛw, one can either examine the interaction of the fluctuating dipole moment with a reaction field acting at the boundary—the so-called Kirkwood-Frölich technique (Frölich, 1968)—or measure the induced polarization in response to an applied electric field (Heinz et al., 2001; Kusalik et al., 1994). In practice, neither method gives a reliable answer when they are applied to channel-like geometries that contain ions. This issue clearly deserves further investigation. In the meantime, we have been consistently adopting the value of ɛw as 60, under the assumption that the polar residues on the protein wall are acting partially like water molecules in shielding ionic charges. In a number of different types of ion channels that we studied using Brownian dynamics, the use of ɛw = 60 in the narrow pore successfully reproduced many of the experimentally determined properties.
Incorporating the atomic models of ClC-0 and ClC-1 in three-dimensional Brownian dynamics, we are able to make a number of predictions that can be tested experimentally. Among these are the conductance-concentration profiles for ClC-0 and ClC-1. For both, we obtain the half-saturation values of ∼150 mM, in the same range as many cationic channels, such as the potassium channels (Coronado et al., 1980; Chung et al., 2002). Theoretically, the conductance-concentration curve is expected to saturate if the transport through the channel is determined by two independent processes, of which only one depends on ionic concentrations on the two sides of the channel. In ClC-0 and ClC-1, outward conduction involves two such steps as illustrated in Fig. 8. The first is the entry of a third ion into the channel from the intracellular space, which depends on the ionic concentration and the applied potential. The second step is the outermost ion climbing out of the energy well and into the extracellular space, which is independent of the ionic concentration and depends solely on the applied potential. Thus, the current in these channels first increases and then saturates with increasing ionic concentration, following the Michaelis-Menten form derived in Chung et al. (1999). Finally, we show that a single point mutation of the Glu-459 residue guarding the intracellular gate to arginine causes an increase in the number of ion binding sites from two to three (Fig. 12). The current-voltage curve, however, remains virtually unchanged. Although the mutant and wild-type channels will be indistinguishable macroscopically, an additional Cl− ion is predicted to be present in the mutant x-ray structure.
Our studies have shed light on the detailed mechanism of ion permeation in ClC channels. The success of our technique of constructing models of these channels has prompted us to examine how the conduction properties of EcClC, ClC-0, and ClC-1 will be affected following theoretical mutations and to study other ClC isoforms, whose single channel properties have not yet been characterized. It has not been possible to measure currents across some ClC-type channels, such as the prokaryotic ClC channels, probably because currents are too small to be resolved experimentally. For these, we are able to model site-directed mutagenesis and ascertain which residues need to be mutated to enhance the magnitude of currents flowing across the pores. The results of our mutation studies will be reported in detail elsewhere. In studying the family of ClC channels, we can now make testable predictions while refining our models as new experimental data comes to hand. Thus, our understanding of the mechanics of ion channels can progress through a fruitful interaction between theory and experiment.
Acknowledgments
The calculations upon which this work is based were carried out using the Compaq AlphaServer Supercomputer of The Australian National University Supercomputer Facility. We thank Dr. David K. Bisset for creating an open-state structure of the EcClC channel and Dr. Tsung-Yu Chen for making his unpublished data available to us. Dr. Chen's experimental measurements are reproduced in Fig. 6 with his kind permission.
This work was supported by grants from the Australian Research Council, the Australian Partnership of Advanced Computing, and the National Health and Medical Research Council of Australia.
References
- Allen, T., and S. H. Chung. 2001. Brownian dynamics study of an open-state KcsA potassium channel. Biochim. Biophys. Acta. 1515:83–91. [DOI] [PubMed] [Google Scholar]
- Allen, T. W., S. Kuyucak, and S. H. Chung. 2000. Molecular dynamics estimates of ion diffusion in model hydrophobic and KcsA potassium channels. Biophys. Chem. 86:1–14. [DOI] [PubMed] [Google Scholar]
- Bennetts, B., M. L. Roberts, A. H. Bretag, and G. Y. Rychkov. 2001. Temperature dependence of human muscle ClC-1 chloride channel. J. Physiol. 535:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187–217. [Google Scholar]
- Burykin, A., C. N. Schutz, J. Villa, and A. Warshel. 2002. Simulations of ion current in realistic models of ion channels: the KcsA potassium channel. Proteins. 47:265–280. [DOI] [PubMed] [Google Scholar]
- Burykin, A., M. Kato, and A. Warshel. 2003. Exploring the origin of the ion selectivity of the KcsA potassium channel. Proteins. 52:412–426. [DOI] [PubMed] [Google Scholar]
- Chen, Y. T., and C. Miller. 1996. Nonequilibrium gating and voltage dependence of the ClC-0 Cl− channel. J. Gen. Physiol. 108:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. H., M. Hoyles, T. W. Allen, and S. Kuyucak. 1998. Study of ionic currents across a model membrane channel using Brownian dynamics. Biophys. J. 75:793–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. H., T. W. Allen, M. Hoyles, and S. Kuyucak. 1999. Permeation of ions across the potassium channel: Brownian dynamics studies. Biophys. J. 77:2517–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. H., T. W. Allen, and S. Kuyucak. 2002. Conducting-state properties of the KcsA potassium channel from molecular and Brownian dynamics simulations. Biophys. J. 82:628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado, R., R. L. Rosenberg, and C. Miller. 1980. Ionic selectivity, saturation, and block in a K+-selective channel from sarcoplasmic reticulum. J. Gen. Physiol. 76:425–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry, B., T. W. Allen, S. Kuyucak, and S. H. Chung. 2001. Mechanisms of permeation and selectivity in calcium channels. Biophys. J. 80:195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry, B., S. Kuyucak, and S. H. Chung. 2003. Dielectric self-energy in Poisson-Boltzmann and Poisson-Nernst-Planck models of ion channels. Biophys. J. 84:3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Duan, D., S. Cowley, B. Horowitz, and J. R. Hume. 1999. A serine residue in CLC-3 link phosphorylation-dephosphorylation to chloride channel regulation by cell volume. J. Gen. Physiol. 113:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler, R., E. B. Campbell, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 415:287–294. [DOI] [PubMed] [Google Scholar]
- Dutzler, R., E. B. Campbell, and R. MacKinnon. 2003. Gating the selectivity in ClC chloride channels. Science. 300:108–112. [DOI] [PubMed] [Google Scholar]
- Edwards, S., B. Corry, S. Kuyucak, and S. H. Chung. 2002. Continuum electrostatics fails to describe ion permeation in the gramicidin channel. Biophys. J. 83:1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez, R., B. C. Schroeder, A. Accardi, T. J. Jentsch, and M. Pusch. 2003. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron. 38:47–59. [DOI] [PubMed] [Google Scholar]
- Fahlke, C. 2001. Ion permeation and selectivity in ClC-type chloride channels. 2001. Am. J. Physiol. Renal Physiol. 280:F748–F757. [DOI] [PubMed] [Google Scholar]
- Fahlke, C., R. Rüdel, N. Mitrovic, M. Zhou, and A. L. George, Jr. 1995. An aspartic residue important for voltage-dependent gating of human muscle chloride channels. Neuron. 15:463–472. [DOI] [PubMed] [Google Scholar]
- Friedrich, T., T. Breiderhoff, and T. J. Jentsch. 1999. Mutational analysis demonstrates that ClC-4 and ClC-5 directly mediate plasma membrane currents. J. Biol. Chem. 274:896–902. [DOI] [PubMed] [Google Scholar]
- Frölich, H. 1968. Theory of Dielectrics. Clarendon Press, Oxford, UK.
- Heinz, T. N., W. F. van Gunsteren, and P. H. Hünenberger. 2001. Comparison of four methods to compute the dielectric permittivity of liquids from molecular dynamics simulations. J. Chem. Phys. 115:1125–1136. [Google Scholar]
- Hoyles, M., S. Kuyucak, and S. H. Chung. 1998. Solutions of Poisson's equation in channel-like geometries. Comp. Phys. Comm. 115:45–68. [Google Scholar]
- Jentsch, T. J., K. Steinmeyer, and G. Schwarz. 1990. Primary structure of Torpedo marmorata chloride channel isolated by expression cloning in Xenopus oocytes. Nature. 348:510–514. [DOI] [PubMed] [Google Scholar]
- Jentsch, T. J., T. Friedrich, A. Schriever, and H. Yamada. 1999. The ClC chloride channel family. Pflugers Arch. 437:783–795. [DOI] [PubMed] [Google Scholar]
- Jentsch, T. J., V. Stein, F. Weinreich, and A. A. Zdebik. 2002. Molecular structure and physiological function of chloride channels. Physiol. Rev. 82:503–568. [DOI] [PubMed] [Google Scholar]
- Kawasaki, M., M. Suzuki, S. Uchida, S. Sasaki, and F. Marumo. 1995. Stable and functional expression of the ClC-3 chloride channel in somatic cell lines. Neuron. 14:1285–1291. [DOI] [PubMed] [Google Scholar]
- Kusalik, P. G., M. E. Mandy, and I. M. Swishchev. 1994. The dielectric-constant of polar fluids and the distribution of the total dipole-moment. J. Chem. Phys. 100:7654–7664. [Google Scholar]
- Li, S. C., M. Hoyles, S. Kuyucak, and S. H. Chung. 1998. Brownian dynamics study of ion transport in the vestibule of membrane channels. Biophys. J. 74:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, C., M. Pusch, and T. J. Jentsch. 1996. Heteromultimeric CLC chloride channels with novel properties. Proc. Natl. Acad. Sci. USA. 93:13362–13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., M. Pusch, and T. J. Jentsch. 1996. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature. 383:340–343. [DOI] [PubMed] [Google Scholar]
- Maduke, M., C. Miller, and J. A. Mindell. 2000. A decade of ClC chloride channels: structure, mechanism, and many unsettled questions. Annu. Rev. Biophys. Biomol. Struct. 29:411–438. [DOI] [PubMed] [Google Scholar]
- Middleton, R. E., D. J. Pheasant, and C. Miller. 1996. Homodimeric architecture of a ClC-type chloride channel. Nature. 383:337–340. [DOI] [PubMed] [Google Scholar]
- Miller, C. 1982. Open-state substructure of single chloride channels from Torpedo electroplax. Phil. Trans. Roy. Soc. Lond. B299:401–411. [DOI] [PubMed] [Google Scholar]
- Moy, G., B. Corry, S. Kuyucak, and S. H. Chung. 2000. Tests of continuum theories as models of ion channels. I. Poisson-Boltzmann theory versus Brownian dynamics. Biophys. J. 78:2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitera, J. W., M. Falta, and W. F. van Gunsteren. 2001. Dielectric properties of proteins from simulation: the effects of solvent, ligands, pH, and temperature. Biophys. J. 80:2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press, W. H., B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling. 1989. Numerical Recipes. Cambridge University Press, Cambridge, UK.
- Pusch, M., U. Ludewig, A. Rehfeldt, and T. J. Jentsch. 1995. Gating of the voltage-dependent chloride channel ClC-0 by the permeant anions. Nature. 373:527–531. [DOI] [PubMed] [Google Scholar]
- Rychkov, G. Y., M. Pusch, M. L. Roberts, and A. H. Bretag. 2001. Interaction of hydrophobic anions with the rat skeletal muscle chloride channel ClC-1: effects on permeation and gating. J. Physiol. 530:379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviane, C., F. Conti, and M. Pusch. 1999. The muscle chloride channel ClC-1 has a double-barreled appearance that is differentially affected in dominant recessive myotonia. J. Gen. Physiol. 113:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz, C. N., and A. Warshel. 2001. What are the dielectric “constants” of proteins and how to validate electrostatic models. Proteins. 44:400–417. [DOI] [PubMed] [Google Scholar]
- Simonson, T., and C. L. Brooks, Iii.1996. Charge screening and the dielectric constant of proteins: insights from molecular dynamics. J. Am. Chem. Soc. 118:8452–8458. [Google Scholar]
- Smith, P. E., R. M. Brunne, A. E. Mark, and W. F. van Gunsteren. 1993. Dielectric properties of trypsin inhibitor and lysozyme calculated from molecular dynamics simulations. J. Phys. Chem. 97:2009–2014. [Google Scholar]
- Steinmeyer, K., B. Schwappach, M. Bens, A. Vandewalle, and T. J. Jentsch. 1995. Cloning and functional expression of rat CLC-5, a chloride channel related to kidney disease. J. Biol. Chem. 270:31172–31177. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. J. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren, W. F., and H. J. C. Berendsen. 1982. Algorithms for Brownian dynamics. Mol. Phys. 45:637–647. [Google Scholar]
- Vonoye, C. G., and A. L. George, Jr. 2002. Functional characterization of recombinant human ClC-4 chloride channels in cultured mammalian cells. J. Physiol. 539:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, F., and T. J. Jentsch. 2001. Pores formed by single subunits in mixed dimers of different CLC chloride channels. J. Biol. Chem. 276:2347–2353. [DOI] [PubMed] [Google Scholar]
- White, M. M., and C. Miller. 1981. Probes of the conduction process of a voltage-gated Cl− channel. J. Gen. Physiol. 78:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]