Abstract
It is postulated that the specific interactions between cholesterol and lipids in biological membranes are crucial in the formation of complexes leading subsequently to membrane domains (so-called rafts). These interactions are studied in molecular dynamics simulations performed on a dipalmitoylphosphatidylcholine (DPPC)-cholesterol bilayer mixture and a dilauroylphosphatidylcholine (DLPC)-cholesterol bilayer mixture, both having a cholesterol concentration of 40 mol %. Complexation of the simulated phospholipids with cholesterol is observed and visualized, exhibiting 2:1 and 1:1 stoichiometries. The most popular complex is found to be 1:1 in the case of DLPC, whereas the DPPC system carries a larger population of 2:1 complexes. This difference in the observed populations of complexes is shown to be a result of differences in packing geometry and phospholipid conformation due to the differing tail length of the two phosphatidylcholine lipids. Furthermore, aggregation of these complexes appears to form hydrogen-bonded networks in the system containing a mixture of cholesterol and DPPC. The CH⋯O hydrogen bond plays a crucial role in the formation of these complexes as well as the hydrogen bonded aggregates. The aggregation and extension of such a network implies a possible means by which phospholipid:cholesterol domains form.
INTRODUCTION
Eukaryotic cellular life has an undeniable dependence on cholesterol (Miao et al., 2002; Ohvo-Rekilä et al., 2002). It comprises ∼40 mol % of the lipid portion of the eukaryotic plasma membrane and is generally responsible for the modulation of the physico-chemical properties required for viability and cell proliferation (Miao et al., 2002; Ohvo-Rekilä et al., 2002). It is known that cholesterol reduces the passive permeability of membranes, increases membrane mechanical strength, and modulates membrane enzymes (Yeagle, 1993). Among other biological roles, it instigates the formation of membrane “rafts”—domains in which cholesterol, saturated long-chained lipids, and specific proteins are concentrated (Simons and Ikonen, 1997).
Rafts distribute proteins and lipids to organelles and the cell surface, activate immune responses, serve as centers for receptor-mediated signal transduction, and are used by many disease-causing bacteria and viruses as a means to populate host cells (Simons and Ikonen, 1997). The particular contribution cholesterol makes in the formation of rafts is the allowance for maintaining a liquid-ordered, tightly packed membrane domain (Xu and London, 2000). The way in which cholesterol achieves this task, however, is not yet known (Edidin, 2001).
The liquid-ordered phase endemic to membrane rafts consists of very tightly packed sphingolipids, phospholipids (Xu and London, 2000; Simons and Ikonen, 2000), and cholesterol. To understand the physical properties of biological membranes containing cholesterol-induced liquid-ordered phases, studies have been performed on model systems such as monolayers (Keller et al., 2000) or giant unilamellar vesicles containing binary or ternary mixtures of phospholipids and cholesterol (Veatch and Keller, 2002). These studies indicate a possible existence of cholesterol-phospholipid condensed complexes where q molecules of cholesterol, C, and p molecules of phospholipid, P, are considered to react to form a complex CqPp (i.e., 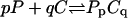 ). It has also been suggested that complex formation is more cooperative when the oligomerization reaction,
). It has also been suggested that complex formation is more cooperative when the oligomerization reaction, 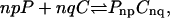 occurs. The existence of this cooperative complexation reaction has been inferred from the observation of two upper miscibility critical points (Keller et al., 2000; McConnell and Radhakrishnan, 2003) in phase diagrams for monolayers containing cholesterol and phosphatidylcholine. It has also been observed in experimental studies that formation of lipid-cholesterol complexes is correlated with the melting temperature of phospholipids. Thus, when cholesterol is mixed with phospholipids having a melting temperature below 296 K, only one critical point in the phase diagram is observed (Keller et al., 2000), meaning that the complexation and oligomerization reaction discussed above does not take place in this case. Recently, a model that explains the thermodynamic behavior of lipid-cholesterol complexes has been developed, which shows consistency with observed phase diagrams (Anderson and McConnell, 2001). Nevertheless, a molecular level description of such complexes still does not exist.
occurs. The existence of this cooperative complexation reaction has been inferred from the observation of two upper miscibility critical points (Keller et al., 2000; McConnell and Radhakrishnan, 2003) in phase diagrams for monolayers containing cholesterol and phosphatidylcholine. It has also been observed in experimental studies that formation of lipid-cholesterol complexes is correlated with the melting temperature of phospholipids. Thus, when cholesterol is mixed with phospholipids having a melting temperature below 296 K, only one critical point in the phase diagram is observed (Keller et al., 2000), meaning that the complexation and oligomerization reaction discussed above does not take place in this case. Recently, a model that explains the thermodynamic behavior of lipid-cholesterol complexes has been developed, which shows consistency with observed phase diagrams (Anderson and McConnell, 2001). Nevertheless, a molecular level description of such complexes still does not exist.
In this work we propose that a hydrogen-bonded network can emerge in bilayers containing a mixture of phospholipids and cholesterol. This network displays a cooperativity whose degree depends on the detailed structure of lipids. It has been assumed that hydrogen bonding between the hydroxyl (OH) group of cholesterol and the headgroup of phospholipid is of considerable importance (McMullen and McElhaney, 1996) in bilayer structure. Indeed, recent computer simulations of bilayers containing cholesterol and phospholipid molecules (Tu et al., 1998; Smondyrev and Berkowitz, 1999a; Pasenkiewicz-Gierula et al., 2000; Róg and Pasenkiewicz-Gierula, 2001; Chiu et al., 2002) confirm the existence of such hydrogen bonds. Hydrogen-bonded water bridges between cholesterol and phospholipids have also been detected (Pasenkiewicz-Gierula et al., 2000). However, the consideration of hydrogen-bonded interactions involving hydrogens from cholesterol and oxygens from lipid provides only for situations where cholesterol is a donor and phospholipid is an acceptor. Although this can explain the existence of 1:1 complexes, the description of larger complexes and their possible cooperative character requires the consideration of hydrogen bonding between cholesterol as an acceptor and phospholipid as a donor. The only route for such an interaction is between the methyl hydrogens of the phospholipid choline group and the hydroxyl oxygen atom of cholesterol.
Such a CH⋯O interaction might come as a surprise, however, the idea of the CH⋯O hydrogen bond is well established (Desiraju, 1991; Gu et al., 1999; Raveendran and Wallen, 2002). This sort of hydrogen bond is weaker and has a less directional character (or is more susceptible to “bending” or nonlinearity) than the typical OH⋯O hydrogen bond. Nonetheless, a recent quantum chemical study of the nature of the CH⋯O interaction has revealed that its strength and directionality qualifies it as a true hydrogen bond (Gu et al., 1999). In addition, the work by Gu et al. showed that the strength of the CH⋯O interaction increases substantially upon the addition of a single electron-withdrawing group to the carbon atom donor. In the case of the choline group of dipalmitoylphosphatidylcholine (DPPC), the –N(CH3)3 substituent could provide for a situation where methyl groups can strongly interact with the hydroxyl group of cholesterol. Furthermore, the CH⋯O interaction dies off much more slowly than the conventional OH⋯O hydrogen bond imparting a larger range of influence to this specific interaction (Gu et al., 1999).
METHODS
To understand the extent of the hydrogen bonding and its network in a system containing cholesterol and zwitterionic phospholipid molecules such as phosphatidylcholine lipids, we performed molecular dynamics (MD) simulations of two bilayer mixtures containing cholesterol. One of these simulated systems was a hydrated DPPC bilayer with cholesterol (referred to as the DPPC + cholesterol system) at a concentration of 40 mol % (Fig. 1). This sort of system was chosen because DPPC is very well characterized in simulated bilayers (Smondyrev and Berkowitz, 1999b; Berger et al., 1997) and has already been studied in bilayer systems containing cholesterol. In addition, results obtained with DPPC should be relevant to membranes having a natural eukaryotic lipid composition. This is so because although in natural membranes, most of the saturated lipids are sphingolipids, DPPC exhibits properties very similar to those of sphingomyelin, which can be the most popular lipid in plasma membranes (Xu and London, 2000). The effect of shortening the hydrocarbon tails of phospholipids on the ability of cholesterol to affect complexation of lipids was investigated in a second simulated system (referred to as the dipalmitoylphosphatidylcholine (DLPC) + cholesterol system) consisting of a hydrated DLPC bilayer with cholesterol (also at 40 mol %).
FIGURE 1.
(Top) Structure of the DPPC and cholesterol molecules and (bottom) a typical snapshot of the DPPC + cholesterol system. Cholesterol is shown as space-filled atoms. DPPC is colored in green. The phosphorus and nitrogen atoms of the DPPC headgroup are shown as yellow and blue spheres, respectively.
Both of our simulations were performed using the GROMACS package (Berendsen et al., 1995; Lindahl et al., 2001). Force-field parameters for phospholipid molecules were based on the work of Berger (Berger et al., 1997) and the cholesterol parameters were those used by Höltje et al. (2001). The LINCS algorithm was used to constrain all bonds in the system (Hess et al., 1997) allowing an integration time step of 4 fs. Periodic boundary conditions were applied in all three dimensions and long-range electrostatics were handled using the SPME algorithm (Essmann et al., 1995). The temperature in the DPPC + cholesterol and DLPC + cholesterol simulations were maintained at 323 K and 279 K, respectively, using the Nose-Hoover scheme with a thermostat oscillatory relaxation period of 0.5 ps. The system was equilibrated in an NPT ensemble using the Parrinello-Rahman pressure coupling scheme (Nose and Klein, 1983; Parrinello and Rahman, 1981) with a barostat time constant of 2.0 ps at a pressure of 1 atm. The temperatures of the DPPC + cholesterol and DLPC + cholesterol systems were chosen because they correspond to the same reduced temperature of ∼0.029 (Mabrey and Sturtevant, 1976). Thus, we were able to compare the structural properties of the lipids in these two systems.
Preparation of the initial configuration of the DPPC + cholesterol system followed the protocol of our previously studied membrane systems (Pandit et al., 2003b). The system contained 120 phospholipids, 80 cholesterol molecules, and 5000 water molecules. Forty cholesterol molecules were placed randomly, along with 60 DPPC molecules in each monolayer of the initial DPPC bilayer configuration. A 35 ns simulation was then performed on this system. The last 20 ns of the trajectory was used for analysis.
The initial configuration of the DLPC + cholesterol system was constructed by taking the initial configuration of the DPPC + cholesterol system and shortening the tails of the DPPC molecules. Thus, any differences observed in the complexation of lipids in the two systems are not due simply to differences in their initial configurations. This is an important consideration, because given the relatively short duration of the simulations compared to the lateral motion of the lipids, observed complexation events will be sensitive to the initial conditions of each simulation. An 18 ns simulation was performed on the DLPC + cholesterol system. The centers of mass of the upper and lower leaflets of the bilayer (the interleaflet distance) were seen to stabilize after 3 ns. The bilayer was then allowed to equilibrate for 5 ns, and the last 10 ns of the trajectory was used for analysis.
RESULTS AND DISCUSSION
Structural properties
We validated the equilibration of our systems by investigating key physical properties. The area per phospholipid molecule and cholesterol in each mixture was determined by following a procedure described recently by Hofsäß et al. (2003). The area per phospholipid was obtained by using the expression
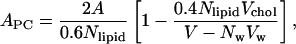 |
where APC is the area per headgroup of phosphatidylcholine lipid (DPPC or DLPC, abbreviated as PC), A is the xy area of the simulation cell, Nlipid is the total number of lipid molecules (i.e., NPC + Nchol = 200), V is the total volume of the simulation cell, Nw is the number of water molecules in each of the systems, Vw is the volume occupied per water molecule (0.0312 nm3), and Vchol is the volume per cholesterol molecule taken to be 0.593 nm3 (Hofsäß et al., 2003). The area per cholesterol molecule was calculated by using the expression
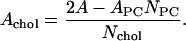 |
The calculated values of the area per phospholipid molecule are ADPPC = 50.3 Å2 and ADLPC = 47.2 Å2. These values clearly demonstrate a condensation effect. The area per cholesterol molecule exhibited an increase with the decrease in PC tail length—from ∼26 Å2 in the DPPC + cholesterol system to ∼31 Å2 in the DLPC + cholesterol system.
Fig. 2 A shows the electron density of the DPPC + cholesterol system. For comparison we have also plotted the electron density of a hydrated pure DPPC bilayer (Pandit et al., 2003b). We see that the thickness of the bilayer is increased by the reasonable extent of 8–9 Å upon the addition of cholesterol. The contributions in electron density due to DPPC + water and cholesterol are also shown separately. Since DPPC and water give a smaller contribution to the total electron density near the center of the bilayer system than in the hydrated pure DPPC bilayer, we can say that much of the electron density in the central portion of the DPPC + cholesterol system is due to cholesterol. A similar trend is seen from Fig. 2 B in the DLPC + cholesterol system.
FIGURE 2.
Electron densities of various components of (A) the DPPC + cholesterol system and (B) the DLPC + cholesterol system. The center of the bilayer corresponds to Z = 0. Electron density for a pure hydrated DPPC system is also shown for comparison in A.
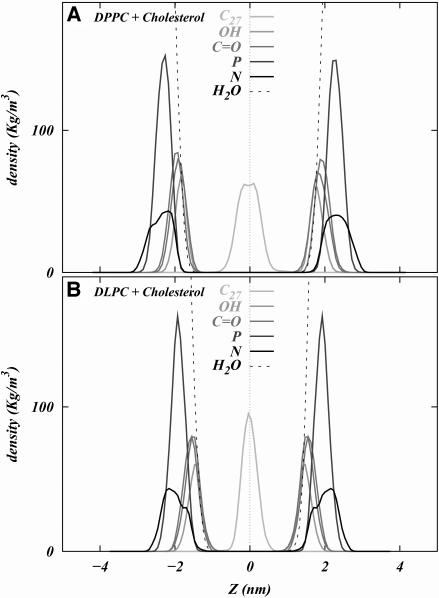
The z density profiles of various atoms in both systems are shown in Fig. 3, A and B. In both systems, the –OH group of cholesterol is hydrated and its location roughly coincides with the location of the carbonyl oxygens of DPPC. Another striking feature is the relative peak positions of the phosphorus and nitrogen atoms of the phospholipid headgroups. It is seen that in the case of the DPPC + cholesterol system, the peaks of phosphorus and nitrogen densities nearly coincide, whereas in the DLPC + cholesterol system, the peak of the nitrogen density rests slightly outside the peak of the phosphorus density. Thus we may expect that the vector joining the phosphorus and nitrogen atoms of the phospholipid headgroups should point more outwardly in the case of DLPC than of DPPC. It is also seen that the “tail” carbon atom of cholesterol, C27, shows more overlap across the bilayer leaflets in the DLPC + cholesterol system than in the DPPC + cholesterol system. This can be expected, given the smaller thickness of the DLPC + cholesterol bilayer.
FIGURE 3.
Density profiles of various components of the (A) DPPC + cholesterol system and (B) DLPC + cholesterol system as a function of Z.
The increase in thickness and decrease in ADPPC (with respect to the pure DPPC bilayer) in the DPPC + cholesterol system is concurrent with the expected increase in the deuterium order parameters for the hydrocarbon tails of DPPC (Fig. 4). The chain order of the lipids in the DPPC + cholesterol mixture is nearly twice that of pure DPPC bilayer, validating that the simulated bilayer is in the liquid-ordered phase. Indeed, this phase is consistent with the phase point corresponding to the simulated temperature and pressure conditions (Thewalt and Bloom, 1992; Scott, 1993). The order parameters for the first few carbons of either chain of DPPC coincide with those for DLPC in the DLPC + cholesterol system. However, the last few carbons of DLPC exhibit substantially lower order parameter values. This DLPC tail disorder is a result of the change in molecular packing due to the overall “tilt” of the cholesterol molecules. The difference in tilt is exhibited in the distributions of Fig. 5. The tilt was defined as the angle between the vector joining the C21 and the C5 (see Fig. 1) atoms of a cholesterol molecule and the outwardly directed bilayer normal. The average tilt angle of cholesterol for the DLPC + cholesterol system is 17.1°, and for the DPPC + cholesterol system it is 14.7°.
FIGURE 4.
Sn-1 (A) and Sn-2 (B) deuterium order parameters for the tails of DPPC lipid in the DPPC + cholesterol system and in a pure hydrated bilayer, and of DLPC lipid in the DLPC + cholesterol system.
FIGURE 5.
Distribution of the cholesterol tilt angle in both simulated systems.
Hydrogen bonding among phospholipids, water, and cholesterol
We now turn our attention to the specific interactions occurring between phospholipids and cholesterol. Given the species of molecules in our systems (PC, cholesterol, and water), it is possible to distinguish three possible distinct modes of binding: i), a direct OH⋯O hydrogen bond between a cholesterol hydroxyl group (donor) and any oxygen (acceptor) in a PC molecule (denoted by OHO); ii), a water bridge with the bridging water acting as a donor/acceptor to a cholesterol hydroxyl group and as an acceptor/donor to a PC molecule (denoted by WB); and iii), a CH⋯O hydrogen bond between the CH3 group in the choline of a PC molecule and an oxygen in a cholesterol hydroxyl group (denoted by CHO). Hence, in our analysis of PC-cholesterol interactions, we will consider a PC molecule to be connected to cholesterol if there exists one or more binding modes of the above three types (i–iii) between them. Similar interactions between PC and cholesterol have been observed in recent simulation studies with the exception that the interaction described in mode iii, above, is usually referred to as a “charge pair” interaction (Pasenkiewicz-Gierula et al., 2000).
The investigation of these three possible modes of binding requires the establishment of criteria for determining the existence of hydrogen bonds involving the relevant functional groups of the lipid molecules and water in our systems. Generally speaking, the objective definition of a hydrogen bond can be slightly ambiguous in classical treatments of liquids employing an empirical potential. Many studies have explored this problem for systems containing water and other molecules (see, for example, Luzar and Chandler, 1993; Ferrario et al., 1990; Xu and Berne, 2001; Jedlovsky and Turi, 1997). When treating surfactant molecules in water as in our case, some studies place geometric criterion on the H⋯O distance and the angle, θ(H-O⋯O), in discerning the existence of an OH⋯O hydrogen bond (Bruce et al., 2002; Pandit et al., 2003a) or the C⋯O distance and θ(N-C⋯O) for a CH⋯O hydrogen bond (Pandit et al., 2003a). Such geometric criteria are intuitively appealing and their utilization is convenient in the analysis of such systems. On the other hand, although a distance (H⋯O or C⋯O) criterion can be rationalized by observing the position of the first minimum in the corresponding pair-correlation function, the angular criterion would seem to lack rigor (Pal et al., 2003). This caveat can be avoided by the use of an energetic cutoff criterion for identifying a hydrogen bond.
The energetic approach was first used for the determination of hydrogen bonds between water molecules by Stillinger and Rahman (1974) and has since been extended for the determination of CH⋯O bonds by Jedlovsky and Turi (1997). It was also used by Pal et al. (2003) in the analysis of OH⋯O bonding between water and surfactant molecules. Such an energy cutoff can be defined by first determining the distribution of donor-acceptor interaction energies in the system without making any prior assumptions about the geometry or energy of a hydrogen-bonded interaction. The hydrogen bond-donating groups for PC and cholesterol are marked in Fig. 1. Each PC molecule was divided such that it contained three “acceptor” regions (phosphate, Sn1 carbonyl, and Sn2 carbonyl) and one possible “donor” moiety (choline). The cholesterol has a hydroxyl group that may act as either a donor or acceptor. A water molecule, in its entirety, may also act as a donor or acceptor. The energy distributions for all of the possible direct hydrogen-bonded interactions between PC and cholesterol are shown in Fig. 6. Interactions involving a hydrogen bond between water and either PC or cholesterol are characterized by the energy distribution plots in Fig. 7. The calculated pair energy for each distribution generally involved neutral groups of atoms within the molecules. As shown in Fig. 1, the PC phosphate group (PHO) consists of PO4 plus three united CH2 carbon atoms whereas the Sn1 and Sn2 carbonyl groups (1CO and 2CO) simply involve one carbon and one oxygen atom. The putative donor of PC, N(CH3)3, is named NC3, and the entire headgroup is named HG. The hydrogen-bonding group on cholesterol used in our calculation of pair energy distributions consists of the CH united atom, labeled C5, and OH (C5−OH in Fig. 7). When calculating pair interaction energies involving water, the entire water molecule was used.
FIGURE 6.
Distribution of pair energies for the groups of PC and cholesterol that are capable of forming direct hydrogen bonds.
FIGURE 7.
Distribution of pair energies for water and (A) the PC headgroup and (B) the cholesterol headgroup.
Each distribution in Figs. 6 and 7 exhibits the characteristic large peak near zero energy (data not shown), which indicates (as expected) that the majority of the pair energies correspond to cases where the two interacting groups are far away. Additional distinct extrema in the negative regime of energy signify vicinal pairs that are hydrogen bonded. We see from Fig. 6 that it is possible for any of the designated groups on a PC molecule to form a hydrogen bond with the C5–OH of cholesterol, and that, indeed, a water molecule may hydrogen bond with either PC or cholesterol (or both—Fig. 7). For each distribution, we may take the first minimum in the negative domain of energy closest to the zero energy peak to define an energy cutoff for a hydrogen bond between the corresponding pair. That is, any particular pair whose energy falls below its cutoff can be considered hydrogen bonded. The energy criterion for each type of hydrogen bond in our systems is summarized in Table 1.
TABLE 1.
Energy cutoff criterion for each pair interaction
| Group 1 | Group 2 | Energy cutoff (kcal/mol) |
|---|---|---|
| NC3 | C5–OH | −2.8 |
| 2CO | C5–OH | −4.0 |
| 1CO | C5–OH | −3.5 |
| PHO | C5–OH | −8.0 |
| HG | H2O | −6.2 |
| C5–OH | H2O | −2.4 |
Comparing the energy distributions in Fig. 6, A and B, it is seen that the CH⋯O hydrogen bond between NC3 and C5–OH (with a peak at ∼−4.5 kcal/mol) is roughly half as strong as the OH⋯O hydrogen bond between 2CO and C5–OH (with a peak at ∼−9.5 kcal/mol). This observation is consistent with the current understanding of the strength of the CH⋯O hydrogen bond (Jeffrey, 1997). The OH⋯O hydrogen bond between 2CO and C5−OH in cholesterol is similar in energy to the water hydrogen bond with the headgroup as depicted in Fig. 7 A. However, the peak in the distribution of energies below the cutoff is much sharper (Fig. 6 B). This indicates that the interaction between the Sn2 carbonyl and the hydroxyl of cholesterol is limited by the restricted motion of these two groups as compared to water interacting with HG. In addition, the energy cutoff of the HG interaction with water is very similar to that established for cesium perfluorooctanoate surfactant with water (−6.5 kcal/mol) in the work of Pal et al. (2003). The energy of the 1CO hydrogen bond with C5–OH is also an OH⋯O hydrogen bond, but is slightly weaker (∼−6.0 kcal/mol) than that of 2CO and C5−OH. The strongest hydrogen bond of all is that of PHO with C5–OH as seen in Fig. 6 D. This interaction is ∼−18.5 kcal/mol, but the distribution shows that this bond is more rare than the others between PC and cholesterol. The rarity of this interaction makes sense, because the phosphate portion of the headgroup lies substantially far away from the hydroxyl of cholesterol (see the distributions in Fig. 3). Also, the energy has such a large negative value, because the cholesterol hydroxyl interaction with the phosphate group involves several electronegative phosphate oxygens. This interaction would naturally lead to a larger negative value in energy when compared to its interaction with a singular carbonyl oxygen of PC (as in Fig. 6, B and C).
Of particular interest is the distribution in Fig. 7 B for pair energies between the hydroxyl of cholesterol and a molecule of water. This distribution shows two maxima and minima (excluding the peak at zero energy), indicating that this pair participates in two types of hydrogen bonded interactions—one where C5–OH usually serves as a donor to a water molecule, and one where it usually serves as an acceptor. In the case of water and PC, there is only one minimum, corresponding to the situation where water is a donor to any PC oxygen. In our analysis of complexation of lipids via water bridging, we do not distinguish between the two types of hydrogen-bonded interactions between cholesterol and water, placing them both in the general binding mode category, WB.
The distributions shown in Figs. 6 and 7 provide a solid basis for establishing the existence of hydrogen-bonded interactions between pairs of molecules and allow us to evade the ambiguity that might be caused by adopting geometric criterion. This is particularly true in establishing the existence of the CHO binding mode. Even though the hydrogens of the N(CH3)3 moiety of PC are represented implicitly by positive partial charges on the united carbon atoms of the NC3 group, the absence of explicit hydrogens presents some difficulty in establishing a reasonable geometric criterion for the CH⋯O hydrogen bond. However, when utilizing the energetic cutoff given in Table 1, we see that this bond has the expected geometric tendencies. Fig. 8 A shows the distribution of distances between the hydrogen-bonded united methyl group from NC3 of DPPC and the hydroxyl oxygen of cholesterol for the pair interactions meeting the CH⋯O hydrogen bond energetic criterion. The most probable C⋯O distance from this distribution is 3.3 ± 0.3 Å. Recent quantum chemical calculations for this hydrogen bond have shown that this interaction is optimal at ∼3.4 Å (Gu et al., 1999), falling well within the range that we observe in our simulation. Fig. 8 B shows the distribution of N-C⋯O angles for (NC3)-(C5–OH) pairs meeting the energetic CH⋯O hydrogen-bonding criterion. This distribution is seen to be quite broad, with a peak at ∼104°. A perfectly linear CH⋯O bond would require this angle to be 109.5°. Thus, the distribution in Fig. 8 B shows that this interaction, indeed, has the appropriate directionality for a CH⋯O hydrogen bond.
FIGURE 8.
(A) Distribution of distances between the united methyl group of choline and the hydroxyl oxygen of cholesterol satisfying the energetic criterion of the CH⋯O hydrogen bond. (B) Distribution of the N-C⋯O angles for the pairs satisfying the energetic criterion of the CH⋯O hydrogen bond.
Complexation of cholesterol with phospholipids
With the establishment of hydrogen-bonding criteria, we are able to obtain the distribution of PC molecules connected to cholesterol in our simulations. This is shown in the histograms depicted in Fig. 9, A and B. As we can see, in both simulated systems, PC:cholesterol complexes prefer to occur in stoichiometric ratios of 1:1 and 2:1. In the DLPC + cholesterol system, there are more cholesterol molecules having no complexation with PC than in the DPPC + cholesterol system. In addition, whereas cholesterol complexation in the DPPC + cholesterol system prefers a 2:1 stoichiometry, it prefers a 1:1 stoichiometry in the DLPC + cholesterol system. Thus, there is a clear preference for smaller-sized complexes in the DLPC + cholesterol system. As we will argue below, this preference may be due to the larger average orientation of cholesterol (tilt) in the DLPC bilayer.
FIGURE 9.
Distribution of stoichiometries of PC binding to cholesterol in (A) the DPPC + cholesterol system and (B) the DLPC + cholesterol system.
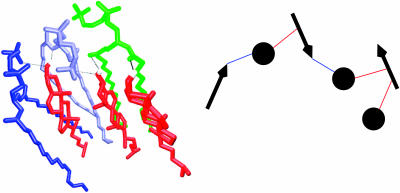
Despite the systems' unique preferences for complex stoichiometries, the distributions of types of 2:1 and 1:1 complexes within each system are seen to be very similar. Further analysis of 1:1 complexes in both systems shows that a majority of these complexes favor an OHO binding mode (∼57% for DPPC + cholesterol and ∼54% for DLPC + cholesterol); ∼28% of the 1:1 complexes favor a CHO binding mode in the case of DPPC + cholesterol and ∼26% favor this mode with DLPC + cholesterol (Fig. 10). Upon analyzing complexes occurring at a 2:1 ratio, it is seen that cholesterol predominantly engages in the CHO and OHO binding modes with PC simultaneously (Fig. 11). Thus, the most preferred 2:1 complexation is through interlipid direct hydrogen bonding in both simulated systems. There is a smaller, yet significant number of complexes involving a water bridge. However, the most preferred 2:1 complexes involving a water bridge always incorporate an OH⋯O or CH⋯O direct bond. Thus, direct OH⋯O and CH⋯O bonding plays a nearly equivalent and most crucial role in the formation of 2:1 complexes. Fig. 12 shows some examples of complexes employing such CHO and OHO binding modes.
FIGURE 10.
Distribution of binding modes in the observed 1:1 complexes of PC:cholesterol in (A) the DPPC + cholesterol system and (B) the DLPC + cholesterol system. The drawing above each bar in the histogram is a schematic representation of the particular binding mode. Water bridges and all direct bonds are shown in red. Cholesterol is represented by a rigid box.
FIGURE 11.
Distribution of binding modes in the observed 2:1 complexes of PC:cholesterol in (A) the DPPC + cholesterol system and (B) the DLPC + cholesterol system. The schematic drawings are similar to those in Fig. 10.
FIGURE 12.
Snapshots of the most popular 2:1 complex of DPPC:cholesterol.
We studied the distribution of the angle made by the vector joining the phosphorus and nitrogen of the PC headgroup 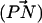 and the outwardly directed bilayer normal. Fig. 13 shows this distribution for both simulated systems. It is seen that in the DPPC + cholesterol system, it is favorable for
and the outwardly directed bilayer normal. Fig. 13 shows this distribution for both simulated systems. It is seen that in the DPPC + cholesterol system, it is favorable for  to direct itself more nearly parallel to the bilayer plane (and slightly inwardly with respect to the bilayer normal) compared to either the DLPC + cholesterol or the pure hydrated DPPC bilayer systems. The more inwardly directed DPPC headgroup of the DPPC + cholesterol system helps to enhance the CHO binding mode, because it brings the choline methyl groups close to the hydroxyl oxygen of cholesterol.
to direct itself more nearly parallel to the bilayer plane (and slightly inwardly with respect to the bilayer normal) compared to either the DLPC + cholesterol or the pure hydrated DPPC bilayer systems. The more inwardly directed DPPC headgroup of the DPPC + cholesterol system helps to enhance the CHO binding mode, because it brings the choline methyl groups close to the hydroxyl oxygen of cholesterol.
FIGURE 13.
Distribution of angles between  and the bilayer normal for all systems.
and the bilayer normal for all systems.
To better understand why the complex formation has a cooperative character, we also consider complexes containing one DPPC molecule and two cholesterol molecules (1:2 complexes) in Fig. 14. Upon observing the populations of each possible combination of direct binding (CHO and OHO) modes occurring in 1:2 complexes, it is seen that the most significant contribution comes from situations where DPPC is bound to one cholesterol via a CHO mode, and to another cholesterol via an Sn2 carbonyl OH⋯O (an OHO mode—see Fig. 14). It is also seen that there is a significant fraction of DPPC molecules that are bound to two cholesterol molecules via two CHO modes. With these observations in mind, one can begin to put together a picture of how cooperative networks might form. The observed behavior in our systems suggests that the formation of a complex between a cholesterol molecule and a phospholipid can give rise to a conformational change in the PC molecule's headgroup that leads to the establishment of a CH⋯O hydrogen bond between that PC and another cholesterol. Given the statistics shown in Fig. 14 (which tell us that 1:2 DPPC:cholesterol complexes are mostly composed of both a CHO and OHO mode), and the statistics shown in Fig. 11 (which tell us that 2:1 DPPC:cholesterol complexes are mostly composed of both a CHO and OHO mode), we can conclude that the alternation of 2:1 complexes and 1:2 complexes can lead to self-propagating networks of PC and cholesterol. Fig. 15 shows a simulation snapshot of a self-propagated network of complexes. The schematic drawing in Fig. 15 illustrates the alternating 1:2 and 2:1 complexes.
FIGURE 14.
Distribution of binding modes in the observed 1:2 complexes of PC:cholesterol involving direct bonds. On the abscissa, each bar is labeled with the two involved modes and each mode's specific binding location. For the CHO mode, it is understood that the binding is between NC3 and C5–OH. Other modes are labeled explicitly.
FIGURE 15.
Network of DPPC:cholesterol complexes linked through alternating OH⋯O and CH⋯O hydrogen bonds. The schematic drawing represents the pictured network. The thick arrows represent the DPPC molecules (arrowhead indicates the choline group). The filled circles represent cholesterol. Direct OH⋯O hydrogen bonds are represented by red lines and direct CH⋯O hydrogen bonds are represented by blue lines. Note that the networks exhibit the tendency to form a linear chain.
As we can see, the orientational distribution of  could conceivably play a crucial role in the establishment of hydrogen-bonded networks in bilayers containing a mixture of cholesterol and phospholipid molecules. This orientational distribution is different for each simulated system. Therefore, since the
could conceivably play a crucial role in the establishment of hydrogen-bonded networks in bilayers containing a mixture of cholesterol and phospholipid molecules. This orientational distribution is different for each simulated system. Therefore, since the 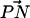 orientation is linked to the formation of CH⋯O hydrogen bonds, we can expect that the pattern of the hydrogen-bonded network will also be different. What is the reason for this difference? The lipid possessing the longer hydrocarbon tail, DPPC, demonstrated a preference for forming larger complexes (in particular, with a 2:1 PC:cholesterol stoichiometry). On the other hand, the shorter-tailed lipid, DLPC, exhibited a preference for the smaller 1:1 complexes. We have alluded that the tilt of cholesterol intrinsic to the bilayer thickness is the main cause of the difference in the unique stoichiometric preferences in DPPC and DLPC complexation. The most significant contribution to complexation involves direct CH⋯O or OH⋯O hydrogen bonding between lipids (see Figs. 11 and 14). Thus, the greater average tilt of cholesterol in the DLPC + cholesterol system shown in Fig. 5 can give rise to a situation where the “head” of cholesterol (containing the hydroxyl) might be near either a donating or accepting group of one DLPC molecule, but further away from the donating or accepting group of another DLPC molecule. In the case of DPPC + cholesterol, the more upright orientation of cholesterol might give rise to a situation where the hydroxyl group of cholesterol can easily access the donating/accepting groups of two DPPC molecules. Essentially, a larger tilt in cholesterol leads to a larger “effective” surface area for this molecule in the bilayer. Given the previously described putative mechanism for the propagation of the PC-cholesterol network, the enhanced tilt of cholesterol in the DLPC + cholesterol system would likely lead to the allowance of only 1:1 complexes and would stop the network's propagation.
orientation is linked to the formation of CH⋯O hydrogen bonds, we can expect that the pattern of the hydrogen-bonded network will also be different. What is the reason for this difference? The lipid possessing the longer hydrocarbon tail, DPPC, demonstrated a preference for forming larger complexes (in particular, with a 2:1 PC:cholesterol stoichiometry). On the other hand, the shorter-tailed lipid, DLPC, exhibited a preference for the smaller 1:1 complexes. We have alluded that the tilt of cholesterol intrinsic to the bilayer thickness is the main cause of the difference in the unique stoichiometric preferences in DPPC and DLPC complexation. The most significant contribution to complexation involves direct CH⋯O or OH⋯O hydrogen bonding between lipids (see Figs. 11 and 14). Thus, the greater average tilt of cholesterol in the DLPC + cholesterol system shown in Fig. 5 can give rise to a situation where the “head” of cholesterol (containing the hydroxyl) might be near either a donating or accepting group of one DLPC molecule, but further away from the donating or accepting group of another DLPC molecule. In the case of DPPC + cholesterol, the more upright orientation of cholesterol might give rise to a situation where the hydroxyl group of cholesterol can easily access the donating/accepting groups of two DPPC molecules. Essentially, a larger tilt in cholesterol leads to a larger “effective” surface area for this molecule in the bilayer. Given the previously described putative mechanism for the propagation of the PC-cholesterol network, the enhanced tilt of cholesterol in the DLPC + cholesterol system would likely lead to the allowance of only 1:1 complexes and would stop the network's propagation.
SUMMARY
We observe in our simulation complexation of cholesterol with PC. Much of the complexation that we observe has a dependence upon the CH⋯O hydrogen bond—a subject that has of late been discussed quite intensely (Desiraju, 1991; Gu et al., 1999; Raveendran and Wallen, 2002). It is normally perceived that a hydrogen bond results upon the approach of a donor molecule to an acceptor molecule. This approach yields the interaction D-H⋯A, where the donor and acceptor, D and A respectively, are thought to be very electronegative atoms such as oxygen or nitrogen. Although carbon is not as extremely electronegative as oxygen or nitrogen, the CH⋯O hydrogen bond has been implicated in many biological systems such as nucleic acids (Auffinger and Westhof, 1996; Berger et al., 1996; Egli and Gessner, 1995), proteins (Bella and Berman, 1996; Derewenda et al., 1995; Musah et al., 1997), and carbohydrates (Steiner and Saenger, 1992; Steiner and Saenger, 1993). This interaction does not seem to arise simply due to geometrical constraints imposed by other contacts, but contributes to the overall stabilization of these macromolecules and their complexes (Wahl and Sundaralingam, 1997). Given its ubiquity, it might not be surprising to find the CH⋯O interaction among the fourth genre of macromolecules—lipids within a bilayer.
Our study shows that a larger capacity for complexation is concurrent with a larger angle made by 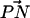 with the outward bilayer normal. This coupling between the headgroup orientation and the capacity to participate in a CHO binding mode arises because PC-cholesterol binding requires that the –CH from choline of one PC molecule should be placed in a position to donate a proton to an acceptor oxygen atom of a cholesterol molecule. The MD studies of Tu et al. (1998) and of Pasenkiewicz-Gierula et al. (2000) also revealed that the strong N-CH3⋯OH interaction was coupled with an inward orientation of the headgroup, although the studies referred to the interaction as a “charge pair” interaction (Pasenkiewicz-Gierula et al., 2000; Tu et al., 1998). In addition, recent studies have shown that in bilayer systems containing DPPC along with other “impurities”, such as salt or dipalmitoylphosphatidylserine (DPPS) plus counterions and salt, the angular distribution of
with the outward bilayer normal. This coupling between the headgroup orientation and the capacity to participate in a CHO binding mode arises because PC-cholesterol binding requires that the –CH from choline of one PC molecule should be placed in a position to donate a proton to an acceptor oxygen atom of a cholesterol molecule. The MD studies of Tu et al. (1998) and of Pasenkiewicz-Gierula et al. (2000) also revealed that the strong N-CH3⋯OH interaction was coupled with an inward orientation of the headgroup, although the studies referred to the interaction as a “charge pair” interaction (Pasenkiewicz-Gierula et al., 2000; Tu et al., 1998). In addition, recent studies have shown that in bilayer systems containing DPPC along with other “impurities”, such as salt or dipalmitoylphosphatidylserine (DPPS) plus counterions and salt, the angular distribution of 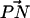 with the outward bilayer normal is distinctly affected. The change in PC headgroup conformation is seen to give rise not only to a larger propensity for DPPS-DPPC complexation in mixed bilayers, but also DPPC-DPPC complexation in pure bilayers where salt is present in the surrounding aqueous baths (Pandit et al., 2003a). The parallel behavior to our DPPC + cholesterol system suggests that in cases where species such as cations or cholesterol interact strongly with the carbonyl oxygens of DPPC, the changes that occur in the headgroup give rise to a greater interlipid binding propensity. Such a behavior appears to be peculiar to PC. One might speculate that it aids in the propagation of cholesterol-DPPC complexes into networks (like in Fig. 15). The tilt of cholesterol in the DLPC + cholesterol system forced by the shorter DLPC molecules nullifies the effect of the change in the headgroup upon cholesterol binding via an OHO mode, allowing only 1:1 complexes and stopping the propagation of the network.
with the outward bilayer normal is distinctly affected. The change in PC headgroup conformation is seen to give rise not only to a larger propensity for DPPS-DPPC complexation in mixed bilayers, but also DPPC-DPPC complexation in pure bilayers where salt is present in the surrounding aqueous baths (Pandit et al., 2003a). The parallel behavior to our DPPC + cholesterol system suggests that in cases where species such as cations or cholesterol interact strongly with the carbonyl oxygens of DPPC, the changes that occur in the headgroup give rise to a greater interlipid binding propensity. Such a behavior appears to be peculiar to PC. One might speculate that it aids in the propagation of cholesterol-DPPC complexes into networks (like in Fig. 15). The tilt of cholesterol in the DLPC + cholesterol system forced by the shorter DLPC molecules nullifies the effect of the change in the headgroup upon cholesterol binding via an OHO mode, allowing only 1:1 complexes and stopping the propagation of the network.
Complexation and aggregation events such as those we observe and describe may shed light on the formation process of raft-like domains. Experimental studies aim to understand the nature of the formation of cholesterol-rich membrane domains. Molecular dynamics simulation offers a means by which one might probe the molecular details of such domains; however, direct observation of their formation using MD techniques is intractable today. Nonetheless, if indeed the complexation of cholesterol with phospholipid precedes the formation of domains as suggested by our simulation study, then we can project a possible way by which the complexes may cooperatively form aggregates. Such an implied cooperative enhancement of complex aggregation is in support of the notion of lipid complex oligomerization (Radhakrishnan and McConnell, 1999; Radhakrishnan et al., 2000). The structural changes evoked by DPPC-cholesterol complexes can lead to yet larger networks of complexes (as in Fig. 15) such that the hydrogen-bonded network of lipids in the mixed bilayer is self propagating. This conclusion agrees with the findings obtained from experiments performed on cholesterol-phospholipid monolayers (Keller et al., 2000).
Finally, we would like to suggest that the presence of the CH⋯O hydrogen bond's role in the above-proposed cooperative network might be investigated using infrared spectroscopy. Since the C-H stretch frequency can be expected to be blue-shifted upon the formation of such hydrogen bonds (Gu et al., 1999), it may be possible that changes in the infrared spectrum might be characterized as a function of the cholesterol concentration of a bilayer.
Acknowledgments
Computational support from the North Carolina Supercomputing Center is gratefully acknowledged. Author S.A.P. thanks Gauri Pradhan for discussions.
This work was supported by the National Science Foundation under grant MCB0315502 and also in part by the Program in Molecular and Cellular Biophysics at the University of North Carolina at Chapel Hill under United States Public Health Service training grant T32 GM08570.
Sagar A. Pandit's present address is Dept. of Biological, Chemical, and Physical Sciences, Illinois Institute of Technology, 3101 S. Dearborn, Chicago, IL 60616.
References
- Anderson, T. G., and H. M. McConnell. 2001. Condensed complexes and the calorimetry of cholesterol-phospholipid bilayers. Biophys. J. 81:2774–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger, P., and E. Westhof. 1996. H-bond stability in the tRNA anticodon hairpin: 3 ns of multiple molecular dynamics simulations. Biophys. J. 71:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella, J., and H. M. Berman. 1996. Crystallographic evidence for Cα-O⋯O=C hydrogen bonds in a collagen triple helix. J. Mol. Biol. 264:734–742. [DOI] [PubMed] [Google Scholar]
- Berendsen, H., D. van der Spoel, and R. van Drunen. 1995. Gromacs: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91:43–56. [Google Scholar]
- Berger, I., M. Egli, and A. Rich. 1996. Inter-strand C-H⋯O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. USA. 93:12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, O., O. Edholm, and F. Jahnig. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, C., S. Senapati, M. Berkowitz, L. Perera, and M. Forbes.2002. Molecular dynamics simulations of sodium dodecyl sulfate micelle in water: the behavior of water. J. Phys. Chem. B. 106:10902–10907. [Google Scholar]
- Chiu, S. W., E. Jakobsson, R. J. Mashl, and H. L. Scott. 2002. Cholesterol-induced modifications in lipid bilayers: a simulation study. Biophys. J. 83:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda, Z. S., L. Lee, and U. Derewenda. 1995. The occurrence of C-H⋯O hydrogen bonds in proteins. J. Mol. Biol. 252:248–262. [DOI] [PubMed] [Google Scholar]
- Desiraju, G. R. 1991. The C-H⋯O hydrogen bond in crystals: what is it? Acc. Chem. Res. 24:290–296. [Google Scholar]
- Edidin, M. 2001. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 11:492–496. [DOI] [PubMed] [Google Scholar]
- Egli, M., and R. V. Gessner. 1995. Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proc. Natl. Acad. Sci. USA. 92:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- Ferrario, M., M. Haughney, and I. McDonald. 1990. Molecular dynamics simulation of aqueous mixtures: methanol, acetone, and ammonia. J. Chem. Phys. 93:5156–5166. [Google Scholar]
- Gu, Y., T. Kar, and S. Scheiner. 1999. Fundamental properties of the CH⋯O interaction: is it a true hydrogen bond? J. Am. Chem. Soc. 121:9411–9422. [Google Scholar]
- Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaige. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- Hofsäß, C., E. Lindahl, and O. Edholm. 2003. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 84:2192–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje, M., T. Förster, B. Brandt, T. Engels, W. von Rybinski, and H.-D. Höltje. 2001. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 1511:156–167. [DOI] [PubMed] [Google Scholar]
- Jedlovsky, P., and L. Turi. 1997. Role of the C-H⋯O hydrogen bonds in liquids: a Monte Carlo simulation study of liquid formic acid using a newly developed pair-potential. J. Phys. Chem. B. 101:5429–5436. [Google Scholar]
- Jeffrey, G. A. 1997. An Introduction to Hydrogen Bonding. Oxford University Press, New York.
- Keller, S. L., A. Radhakrishnan, and H. M. McConnell. 2000. Saturated phospholipids with high melting temperatures form complexes with cholesterol in monolayers. J. Phys. Chem. B. 104:7522–7527. [Google Scholar]
- Lindahl, E., B. Hess, and D. van der Spoel. 2001. Gromacs 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- Luzar, A., and D. Chandler. 1993. Structure and hydrogen bond dynamics of water-dimethyl sulfoxide mixtures by computer simulation. J. Clin. Pathol. 98:8160–8173. [Google Scholar]
- Mabrey, S., and J. M. Sturtevant. 1976. Investigation of phase transitions of lipids and lipid mixtures by high sensitivity differential scanning calorimetry. Proc. Natl. Acad. Sci. USA. 73:3862–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, H. M., and A. Radhakrishnan. 2003. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta. 1610:159–173. [DOI] [PubMed] [Google Scholar]
- McMullen, T. P. W., and R. N. McElhaney. 1996. Physical studies of cholesterol-phospholipid interactions. Curr. Opin. Coll. Interf. Sci. 1:83–90. [Google Scholar]
- Miao, L., M. Nielsen, J. Thewalt, J. H. Ipsen, M. Bloom, M. J. Zuckermann, and O. G. Mouritsen. 2002. From lanosterol to cholesterol: structural evolution and differential effects on lipid bilayers. Biophys. J. 82:1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah, R. A., G. M. Jensen, R. J. Rosenfeld, D. E. McRee, D. B. Goodin, and S. W. Bunte. 1997. Variation in strength of an unconventional C-H to O hydrogen bond in an engineered protein cavity. J. Am. Chem. Soc. 119:9083–9084. [Google Scholar]
- Nose, S., and M. L. Klein. 1983. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50:1055–1076. [Google Scholar]
- Ohvo-Rekilä, H., B. Ramstedt, P. Leppimäki, and J. P. Slotte. 2002. Cholesterol interactions with phospholipids in membranes. Prog. Lipid Res. 41:66–97. [DOI] [PubMed] [Google Scholar]
- Pal, S., S. Balasubramanian, and B. Bagchi. 2003. Identity, energy, and environment of interfacial water molecules in a micellar solution. J. Phys. Chem. B. 107:5194–5202. [Google Scholar]
- Pandit, S. A., D. Bostick, and M. L. Berkowitz. 2003a. Mixed bilayer containing dipalmitoylphosphatidylcholine and dipalmitoylphosphatidylserine:lipid complexation, ion binding, and electrostatics. Biophys. J. 85:3120–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, S. A., D. Bostick, and M. L. Berkowitz. 2003b. Molecular dynamics simulation of a dipalmitoylphosphatidylcholine bilayer with NaCl. Biophys. J. 86:3743–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., and A. Rahman. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52:7182–7190. [Google Scholar]
- Pasenkiewicz-Gierula, M., T. Róg, K. Kitamura, and A. Kusumi. 2000. Cholesterol effects on the phosphatidylcholine bilayer polar region: a molecular simulation study. Biophys. J. 78:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan, A., T. G. Anderson, and H. M. McConnell. 2000. Condensed complexes, rafts, and the chemical activity of cholesterol in membranes. Proc. Natl. Acad. Sci. USA. 97:12422–12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan, A., and H. M. McConnell. 1999. Cholesterol-phospholipid complexes in membranes. J. Am. Chem. Soc. 121:486–487. [Google Scholar]
- Raveendran, P., and S. L. Wallen. 2002. Cooperative C-H⋯O hydrogen bonding in CO2-Lewis base complexes: implications for solvation in supercritical CO2. J. Am. Chem. Soc. 124:12590–12599. [DOI] [PubMed] [Google Scholar]
- Róg, T., and M. Pasenkiewicz-Gierula. 2001. Cholesterol effects on the phosphatidylcholine bilayer nonpolar region: a molecular simulation study. Biophys. J. 81:2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, H. L. 1993. Lipid-cholesterol phase diagrams: theoretical and numerical aspects. In Cholesterol in Model Membranes. L. Finegold, editor. CRC Press, Boca Raton, FL. 197–222.
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science. 290:1721–1726. [DOI] [PubMed] [Google Scholar]
- Smondyrev, A. M., and M. L. Berkowitz. 1999a. Structure of dipalmitoylphosphatidylcholine/cholesterol bilayer at low and high cholesterol concentrations: molecular dynamics simulation. Biophys. J. 77:2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smondyrev, A. M., and M. L. Berkowitz. 1999b. United atom force field for phospholipid membranes: constant pressure molecular dynamics simulation of dipalmitoylphosphatidicholine/water system. J. Comput. Chem. 50:531–545. [Google Scholar]
- Steiner, T., and W. Saenger. 1992. Geometry of C-H⋯O hydrogen bonds in carbohydrate crystal structures. Analysis of neutron diffraction data. J. Am. Chem. Soc. 114:10146–10154. [Google Scholar]
- Steiner, T., and W. Saenger. 1993. Role of C-H⋯O hydrogen bonds in the coordination of water molecules. Analysis of neutron diffraction data. J. Am. Chem. Soc. 115:4540–4547. [Google Scholar]
- Stillinger, F., and A. Rahman. 1974. Improved simulation of liquid water by molecular dynamics. J. Chem. Phys. 60:1545–1557. [Google Scholar]
- Thewalt, J. L., and M. Bloom. 1992. Phophatidylcholine: cholesterol phase diagram. Biophys. J. 63:1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, K., M. L. Klein, and D. J. Tobias. 1998. Constant-pressure molecular dynamics investigation of cholesterol effects in a dipalmitoylphosphatidylcholine bilayer. Biophys. J. 75:2147–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch, S. L., and S. L. Keller. 2002. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 89:268101–1. [DOI] [PubMed] [Google Scholar]
- Wahl, M. C., and M. Sundaralingam. 1997. C-H⋯O hydrogen bonding in biology. Trends Biochem. Sci. 22:97–101. [DOI] [PubMed] [Google Scholar]
- Xu, H., and B. Berne. 2001. Hydrogen-bond kinetics in the solvation shell of a polypeptide. J. Phys. Chem. B. 105:11929–11932. [Google Scholar]
- Xu, X., and E. London. 2000. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 39:843–849. [DOI] [PubMed] [Google Scholar]
- Yeagle, P. L. 1993. The biophysics and cell biology of cholesterol: an hypothesis for the essential role of cholesterol in mammalian cells. In Cholesterol in Membrane Models. L. Finegold, editor. CRC Press, Boca Raton, FL. 1–12.