Abstract
RecBCD is a processive, DNA-based motor enzyme with both helicase and nuclease activities. We used high-resolution optical trapping to study individual RecBCD molecules moving against applied forces up to 8 pN. Fine-scale motion was smooth down to a detection limit of 2 nm, implying a unitary step size below six basepairs (bp). Episodes of constant-velocity motion over hundreds to thousands of basepairs were punctuated by abrupt switches to a different speed or by spontaneous pauses of mean length 3 s. RecBCD occasionally reversed direction, sliding backward along DNA. Backsliding could be halted by reducing the force, after which forward motion sometimes resumed, often after a delay. Elasticity measurements showed that the DNA substrate was partially denatured during backsliding events, but reannealed concomitant with the resumption of forward movement. Our observations show that RecBCD-DNA complexes can exist in multiple, functionally distinct states that persist for many catalytic turnovers: such states may help tune enzyme activity for various biological functions.
INTRODUCTION
The RecBCD enzyme of Escherichia coli plays a central role in diverse aspects of DNA metabolism, including restarting of stalled replication forks, repair of damage via homologous recombination, and the destruction of blunt-ended duplex DNA (Cox et al., 2000; Kowalczykowski et al., 1994; Kuzminov, 1999). RecBCD is one of the most active and processive helicases known: a single molecule can unwind tens of thousands of basepairs (bp) of duplex DNA at rates of 500 bp/s (at 25°C) (Bianco et al., 2001; Roman and Kowalczykowski, 1989). A nuclease activity of RecBCD promotes recombinational repair, but DNA degradation must be limited to maintain genomic integrity (Kowalczykowski, 2000; Kuzminov, 1999).
The diversity of RecBCD functions suggests that the molecule, either alone or complexed with its DNA substrate, might switch among states with different enzymatic properties, each specialized for a particular biochemical activity. One such switch is well established: a fraction of enzymes encountering the χ-sequence (5′-GCTGGTGG-3′) show altered nuclease activity and interactions with the RecA strand exchange protein. Some χ-induced changes persist for thousands of catalytic turnovers (Kowalczykowski, 2000). Consistent with the notion of multiple states are observations of translocation and unwinding in single-molecule studies of RecBCD, which reveal that individual molecules proceed at widely differing yet constant rates, even without χ (Bianco et al., 2001). Those experiments were conducted in the absence of the mechanical forces that may act on molecules during function in vivo. Here, we followed movement of individual RecBCD molecules along duplex DNA in an optical trapping microscope. Applying calibrated loads, we studied RecBCD at significantly improved spatial resolution, and tested whether applied forces promote transitions among alternative states of the enzyme-DNA complex.
MATERIALS AND METHODS
RecBCD-bio and the single-molecule assay
Sample preparation protocols were adapted from those in an earlier single-molecule study based on the biotinated RecBCD derivative RecBCD-bio (Dohoney and Gelles, 2001). A 7138-bp DNA lacking χ-sequences (M13mp18 positions 6518 to 6404; GenBank X02513.1) was prepared by a polymerase chain reaction (PCR; GeneAmp XL PCR kit, Applied Biosystems, Foster City, CA) in which one primer had a 5′-digoxigenin label. Experiments were conducted with this comparatively short DNA and at subsaturating adenosine triphosphate (ATP) concentrations to maximize the spatiotemporal resolution of the measurements. DNA for control experiments without RecBCD was prepared similarly but with one digoxigenin-labeled and one biotin-labeled primer. Anti-digoxigenin antibody (Roche, Applied Sciences, Indianopolis, IN) was coupled to beads (0.46-μm diameter; Interfacial Dynamics, Portland, OR) using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)/sulfo-N-hydroxysuccinimide (NHS) (Pierce, Rockford, IL). Beads were sonicated, then incubated >1 h with DNA (∼3:1 mol ratio; ∼60-pM beads) in wash buffer (WB) (25 mM Tris acetate (pH 7.5), 1 mM Mg(OAc)2, 1 mM NaCl, 1 mM dithiothreitol (DTT), 0.4% Tween-20 (BioRad, Hercules, CA), and 3 mg/ml bovine serum albumin (BSA; concentration cited is that before filtration through a 0.22-μm filter)). Flow chambers with an internal volume of ∼15 μl were constructed from a standard microscope slide, double-stick tape (3M, St. Paul, MN), an ethanolic potassium hydroxide cleaned coverglass (20 × 40 mm), and 5-min epoxy (Devcon, Danvers, MA). Biotinylated bovine serum albumin (Vector, Burlingame, CA) at 10–100 μg/ml in 100 mM Na phosphate buffer (pH 7) was adsorbed to the coverglass by incubation (30 min) at room temperature. After washing with WB, the chamber was incubated with 5 μg/ml streptavidin (Molecular Probes, Eugene, OR) for 30 min. After further washing, RecBCD-bio (Dohoney and Gelles, 2001) at ∼15 nM was introduced and incubated for 2 h. After washing again, bead-DNA complexes at ∼40 pM (beads) were incubated (30–60 min) and then washed with WB. Enzymatic reaction was initiated by flowing in WB supplemented with ATP, 1.1 μM E. coli single-stranded binding protein (SSB) (Promega, Madison, WI), plus an oxygen-scavenging system (6 mg/ml glucose (MP Biomedicals, Irvine, CA), 0.2 mg/ml glucose oxidase (Roche Applied Sciences, Indianapolis, IN), 30 units/ml catalase). At the start of each single-molecule recording, bead-enzyme complexes were pretensioned using the optical trap and selected for enzyme-driven motion. All measurements were done inside a soundproofed, temperature-stabilized cleanroom at 21.1 ± 0.1°C. Time- and population-averaged velocities of forward movement were ∼65% those previously observed at a higher temperature (25°C) (Dohoney and Gelles, 2001).
Optical trapping instrument and data analysis
Our apparatus was modified from a previous optical trapping instrument (Visscher et al., 1996). A force clamp based upon stage motion was implemented using a three-axis piezoelectric stage (PolytecPI, Karlsruhe, Germany). Laser intensity (and thereby optical trap stiffness) was modulated using acousto-optic deflectors. Trap stiffness was measured by power spectral analysis and confirmed by hydrodynamic drag measurements (Svoboda and Block, 1994a). Data were recorded at 2–10 kHz, median filtered, and decimated to 1 kHz; subsequent smoothing was performed with a boxcar filter. For small stage motions (<50 nm), there was a ∼15-ms latency between commanding stage motion and completing it: this fixed delay was removed during data analysis. The height of the bead over the coverglass surface was set to either 200 nm or 300 nm by monitoring a change in the sum signal for light scattered from the bead onto the quadrant photodetector when the bead contacted the coverslip, then dropping the stage by a precomputed distance. The applied force, F, was calculated from the tether geometry as described previously (Wang et al., 1997), except that a simple inverse formula (modified from Zimm, 1998) was used to find the DNA contour length, L, from F, and elasticity measurements were fit to an analytical approximation of the wormlike chain (Bouchiat et al., 1999). When the DNA is not fully duplex (e.g., during backsliding), the computed values of L are approximate because neither the fractional single-stranded DNA (ssDNA) content nor the number of ssDNA strands (one or two) by which the enzyme is attached is known.
In the optical trapping experiments, we observed occasional discrete displacements of ∼1.4 nm in L records from both experimental and control (no RecBCD; biotinylated-DNA attached directly to the streptavidin coated surface; these instrumental artifacts set an approximate lower limit on the size of enzyme steps detectable in the experiment. In addition, even rarer (once per 170 s on average), abrupt, back-and-forth deflections up to 3 nm were observed in both experimental and control records.
Pauses were detected by an algorithm that fitted the position data (smoothed with a second-order polynomial filter; Savitzky and Golay, 1964) to a series of line segments, took the derivative of these segments, and compared them to a threshold velocity (half the average velocity during a trace). Where found, adjacent pauses (<1 nm) separated by very brief moving segments (<3 s) were combined to form a larger pause. Smoothing bandwidths were dependent upon F (typically, 0.67 Hz for 2 pN; 1.25 Hz for 7 pN). With these parameters, the algorithm reliably discerns (p > 90%) pauses longer than ∼1 s.
Typically, a backsliding event was prefaced by a brief, very high-speed (>500 nm/s) rearward jump in bead position, which we termed a slip (e.g., Fig. 1 c, t = 57 s). Each slip was followed by a longer period of rearward motion at more moderate (but variable) rates, interspersed with pauses, which we termed the backslide. Maximal backsliding velocities were computed from line fits to selected intervals of backslides displaying >36 nm of displacement.
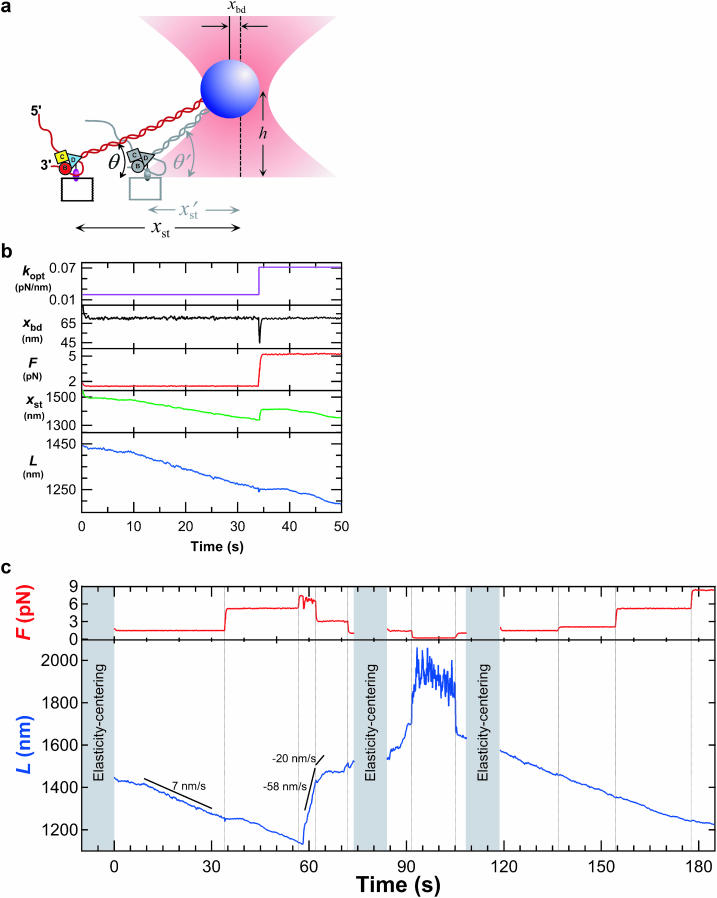
FIGURE 1.
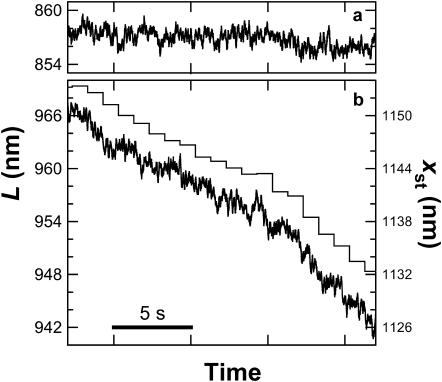
(a) Cartoon illustrating the experimental geometry (not to scale). A RecBCD-bio molecule (B, C, and D) linked to a glass coverslip unwinds duplex DNA (red). The distal end of the DNA is attached to a bead (blue) held at initial height h by the focused beam of an optical trap (pink) centered a lateral distance xst from the enzyme. As the enzyme moves, the stage is automatically repositioned (gray) to maintain the displacement of the bead from the trap center (xbd), and thereby the force. (b) Operation of the force clamp. kopt (purple) is the trap stiffness; xbd (black) is the bead displacement from the trap center; F = kopt × xbd (red) is the applied force; L (blue) is the remaining contour length of the DNA molecule, equivalent to the instantaneous position of RecBCD on the DNA. At t = 33 s, the force set point was raised to 5.5 pN by increasing the laser intensity, transiently deflecting the bead toward the center; the clamp responded by moving the stage until xbd returned to its preset value of 70 nm. (c) Force (red trace) and position (blue trace) records of a RecBCD molecule moving on DNA at 15 μM ATP. Black lines show average velocity over the indicated intervals. Discontinuities in L at t = 91s and 93 s and the increased noise levels in this interval are a consequence of the extremely low (0.2 pN) force setting. Elasticity-centering measurements performed during indicated periods (gray shading) yielded (p, L, and Δ) of (48 nm, 1459 nm, and 1.8 nm), (23 nm, 1596 nm, and 0.3 nm), and (46 nm, 1607 nm, and 4.5 nm), respectively, where Δ represents stage drift since the previous measurement. Data in b are from the first 50 s of this record.
In experiments on a different enzyme using the same instrumentation and analytical procedures (Perkins et al., 2003), a threefold increase in force did not induce detectable pauses; therefore, the observed force induced pauses are not artifacts of the instrumentation.
Bulk DNA unwinding and degradation assays
A 2.6-kbp DNA lacking χ-sequences (5′-TCATAGCT…CGACCACA-3′ from pRW490; Hsieh et al., 1987) was prepared by PCR including [α-32P]-dATP (Amersham). Mixtures (40 μl) of 1.4 nM double-stranded DNA (dsDNA) ends, 0.14 nM active RecBCD-bio, and 1 μM SSB in 25 mM Tris acetate (pH 7.5), 1 mM magnesium acetate, 15 μM ATP, 1 mM dithiothreitol, 1 mM phosphoenolpyruvate, and 4 units/ml pyruvate kinase were quenched at the specified times with 10 μl 0.1 M EDTA, 3% sodium dodecyl sulfate, and 50% glycerol, then extracted with phenol/chloroform/isoamyl alcohol (25:24:1). An aliquot was loaded on a 0.9% agarose 40 mM tris-acetate, 1 mM EDTA, pH 8 electrophoresis gel, and a second aliquot was mixed (1:1) with formamide, denatured 5 min at 96°C and loaded on a 4% polyacrylamide 7-M urea 45 mM tris-borate, 1 mM EDTA, pH 8 gel. Radioactivity was measured using a Storm 840 Phosphor Imager (Amersham Biosciences, Piscataway, NJ). Total radioactivity detected in gel lanes decreased with increasing reaction time, presumably due to disproportionate loss of small DNAs from the gel. Separate experiments (not shown) demonstrated that the addition of excess streptavidin did not affect the rate of DNA cleavage by RecBCD-bio. The extent to which full-length ssDNA strands produced by RecBCD-bio were degraded before electrophoresis was measured in otherwise identical control samples with unlabeled dsDNA that were supplemented with equimolar uniformly 32P-labeled ssDNA. At the longest (60 min) reaction time, ∼40% of the added ssDNA was degraded in these experiments.
RESULTS AND DISCUSSION
To facilitate the application of force in vitro, we used RecBCD-bio, a fully functional derivative that is biotinylated on a carboxy-terminal extension of the RecD subunit (Dohoney and Gelles, 2001). RecBCD-bio molecules were attached to a streptavidin-coated coverglass via their biotin moiety. In the presence of ATP, enzyme molecules bind to and translocate along the experimental substrate, a 7.1-kbp double-stranded DNA. The distal end of this DNA was attached to a 0.46-μm diameter polystyrene bead, which was captured and held in an optical trap (Fig. 1 a). Our trapping instrument incorporates a force clamp implemented through the servo motion of a piezoelectric stage (T. T. Perkins and S. M. Block, unpublished results) that maintains the DNA tether at a preset tension F (Fig. 1 b). Such a feedback arrangement enables real-time monitoring of enzyme position along the DNA with improved spatiotemporal precision (0.8-nm root mean-square positional noise at 13 Hz over a 10-s interval on an 852-nm long tether with F = 7.4 pN). Before each recording, a 2D elasticity-centering procedure was conducted using the instrument in a position-clamp mode. This procedure determines with nanometer-level accuracy the spatial coordinates where the DNA tether is attached to the coverglass. Additionally, the routine returns the contour length, L, and the bending elasticity of the DNA, expressed as a persistence length, p. Measured values of p were used to verify that each bead selected for study was linked to the coverglass through only one dsDNA molecule (Bustamante et al., 1994; Marko and Siggia, 1995; Wang et al., 1997).
In an example recording (Fig. 1 c), a single RecBCD molecule spent long intervals (9–33 s; 119–154 s) moving unidirectionally at nearly constant speed (∼7 nm/s, corresponding to 21 bp/s using a conversion factor of 0.338 nm/bp for dsDNA; Wang et al., 1997). (By convention, we define movements in the direction of DNA unwinding to have positive velocities.) Other molecules moved at different velocities. The heterogeneity is consistent with the range of velocities found in previous single-molecule experiments with the enzyme free in solution (Bianco et al., 2001) and is therefore not likely to be due to surface immobilization. At selected times during our recordings, the force was raised stepwise by increasing the laser trap power. When such increases were sufficiently large, they induced transient pauses in enzyme progress (Fig. 1 c, 33–39 s): in 31 of 32 cases after an increase from an average force of 1.7 pN to 6.3 pN, a pause followed of average duration 7.8 ± 1.1 s (mean ± SE), after which forward motion resumed.
We also observed frequent pausing not associated with changes in applied force. A comparison of pauses observed in 3–4 μM ATP at low force (F = 1.5–2.2 pN; N = 487) and high force (F = 6.3–7.6 pN; N = 203) revealed no changes in the mean frequency (0.14 ± 0.01 (SE) and 0.14 ± 0.01 s−1, respectively) or duration (3.1 ± 0.1 and 3.0 ± 0.2 s) of these events. Also, the frequencies of pauses at 3–4 and 15 μM ATP (at F = 1.5–3.3 pN; N = 487 and 34) were indistinguishable (0.14 ± 0.01 and 0.12 ± 0.01 s−1, respectively). These findings are consistent with the entry into pauses arising from a spontaneous process not driven by ATP hydrolysis and not involving significant movement of the enzyme relative to the DNA. Previous single-molecule RecBCD studies did not report spontaneous pausing (Bianco et al., 2001; Dohoney and Gelles, 2001; Spies et al., 2003), possibly because these lacked sufficient spatiotemporal resolution or because pause duration is reduced at higher ATP concentrations. Pauses also have been reported with a different helicase (Ha et al., 2002).
It is possible that pausing might be caused by transient inactivation of the enzyme due to interaction with the surface. However, velocities averaged over the molecular populations in the previous single-molecule studies of both wild-type RecBCD and RecBCD-bio (Bianco et al., 2001; Dohoney and Gelles, 2001; Spies et al., 2003) agree, within experimental uncertainty, with the rates of unwinding for RecBCD measured in macroscopic (bulk) solution experiments. Thus, neither biotinylation nor surface immobilization of the enzyme induces a change in the population-averaged velocity. Although we cannot rigorously exclude the possibility that enzyme interactions with the surface trigger minor, transient changes in velocity (of sufficiently short duration, or of compensating magnitudes, so as not to significantly alter the population average), we consider such changes unlikely. In addition, control experiments in which streptavidin was omitted demonstrate that essentially all enzyme molecules in our experiment are attached through the same, specific biotin-streptavidin linkage; thus, activity changes caused by the formation of stable nonspecific attachments between enzyme and surface are excluded.
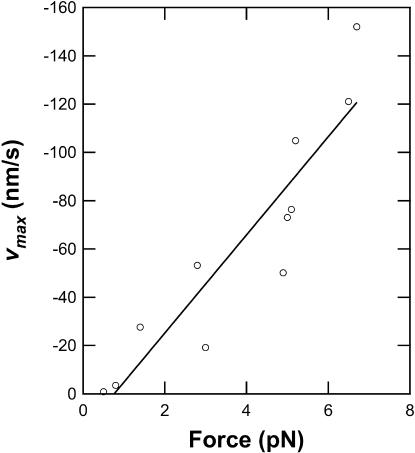
The forward motion of RecBCD was occasionally interrupted by episodes of a qualitatively different behavior, where the enzyme abruptly reversed direction under constant load, resulting in rapid, rearward movements over variable distances up to 900 nm (Fig. 1 c, 58 s). Such movements were never seen in control samples where the DNA was linked directly to the coverglass without the RecBCD enzyme. During these backsliding events, the enzyme-DNA complex continued to support the external load, indicating that RecBCD remained bound to at least one DNA strand. The dramatic reverse movements were unexpected and have not been reported in previous studies of RecBCD, all of which were conducted under negligible load. (In single-molecule studies of other nucleic acid-based motors, large reverse movements have been reported only for T7 DNA polymerase (Wuite et al., 2000), which combines a forward polymerase activity with a reverse exonuclease activity.) Typically, the backslides exhibited an uneven, jerky characteristic, and were occasionally punctuated by pauses (Fig. 1 c, 65–70 s and 86–88 s). Rearward speeds varied considerably, both within and among individual records, but peak velocities were roughly proportional to load (Fig. 2). This is consistent with the hypothesis that the energy for the unidirectional backsliding comes from the applied force (i.e., from the optical trap), not from the catalysis of a chemical reaction (e.g., ATP hydrolysis) by RecBCD. The lack of detectable backsliding in earlier single-molecule studies of RecBCD most likely was caused by low rates of entry into the backsliding state and/or undetectably slow backsliding velocities at the near-zero applied forces used previously.
FIGURE 2.
Peak backsliding velocities are roughly proportional to the applied force. Peak velocity vmax was computed over a minimal distance of 35 nm for each of the 11 backsliding events detected at 15 μM ATP. Line is the unweighted linear fit to the data.
Two observations indicate that backsliding is not simply a load-induced reversal of the RecBCD helicase activity. First, elasticity-centering measurements made during backslides consistently yield persistence lengths significantly lower (e.g., 23 nm at 74–84 s in Fig. 1 c) than the ∼45-nm initial values characteristic of dsDNA (Hagerman, 1988; Wang et al., 1997). This indicates that a nonduplex DNA structure is formed by backsliding RecBCD. This structure most likely consists of ssDNA, possibly complexed with single-stranded binding protein, in the region between the enzyme and the bead. Mere reversal of the helicase reaction would, by definition, regenerate dsDNA with unaltered persistence length. Second, forward-moving molecules routinely proceed against retarding forces of ∼2 pN for up to minutes at a time, whereas comparable loads applied to backsliding complexes generally lead to continued slippage. This observation implies that the backsliding complex is in a different structural and functional state from that present during forward motion.
The results of the elasticity-centering measurements show that backslides are not artifactual signals arising from abrupt, unintended changes in the trapping geometry. Between successive measurements, the tether position typically moved less than 10 nm (1.8 nm, 0.3 nm, and 4.5 nm in Fig. 1 c), a distance sufficiently small as to rule out any significant lateral motion of the enzyme on the coverglass surface or the unzippering of DNA nonspecifically attached to this surface. The observed decrease in persistence length during backslides, along with the comparatively large displacements involved, also exclude a scenario where length changes arise from the sudden release of dsDNA stuck to the bead.
We lowered the applied force during backslides to see if the enzyme molecule would resume forward motion. When the force was reduced below 0.5 pN, rearward movement ceased after a variable delay ranging from 0 s to 21 s (91–93 s in Fig. 1 c). In 36 of 46 slides, forward motion subsequently resumed, often only after a second, variable delay (93–98 s in Fig. 1 c). Resumption of forward motion demonstrates that backsliding is not accompanied by any irreversible damage to the enzyme or to the DNA.
To determine whether RecBCD moves forward along denatured (unwound) DNA after recovering from a backslide, we performed elasticity-centering measurements subsequent to the resumption of forward motion by the enzyme. These measurements indicated a complete reannealing to duplex DNA in the region between the bead and the enzyme (for example, p = 46 nm at 108–119 s; Fig. 1 c), even when performed while the enzyme was still positioned in a region of the DNA that had been unwound before backsliding. Forward motion resumed and the persistence length recovered to its full duplex value (∼45 nm) on tethers where p had dropped as low as 9 nm during backsliding.
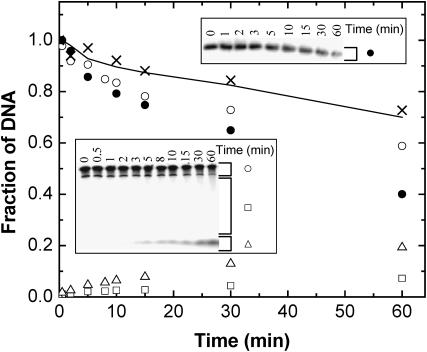
Reannealing of the DNA tether and the resumption of forward progress by RecBCD molecules after sliding backward (by hundreds to thousands of nucleotides) implies that neither strand is nucleolytically degraded within the reannealed portion. How can this be reconciled with previous studies suggesting that a nuclease site in RecB degrades one DNA strand (or, at least, the end of one strand) to short (i.e., acid-soluble) oligonucleotides as RecBCD unwinds dsDNA (Dixon and Kowalczykowski, 1993, and references therein; Yu et al., 1998)? To check whether RecBCD-bio retains this nuclease activity of wild-type RecBCD, we reacted samples of uniformly 32P-labeled DNA with RecBCD-bio in solution. The degradation accompanying DNA unwinding was measured (Fig. 3) by comparing the loss of full-length dsDNA in native gels, which reports unwinding, with loss of full-length ssDNA in denaturing gels, which reports nucleolytic degradation. The data demonstrate that dsDNA unwinding by RecBCD-bio is accompanied by cleavage of half of the strands, just as reported for wild-type RecBCD (Dixon and Kowalczykowski, 1993). The degraded DNA is mostly reduced to small fragments <15 nt (Fig. 3, inset, and data not shown), consistent with models (Kowalczykowski, 2000) where the entire strand, not merely its upstream end, is degraded after unwinding. It is conceivable that such degradation might be suppressed by the application of force in the single-molecule experiments. However, a straightforward way to reconcile the complete degradation of one strand with the observed reannealing after backsliding is to hypothesize that there is a considerable lag between the unwinding of a DNA segment and its subsequent degradation, caused by the accumulation of a large ssDNA loop (Kowalczykowski, 2000; Taylor and Smith, 1980) between nuclease and helicase domains of the enzyme.
FIGURE 3.
RecBCD-bio degrades one DNA strand into small fragments during unwinding. RecBCD degradation of uniformly 32P-labeled DNA lacking χ-sites was detected by denaturing gel electrophoresis (lower inset). At increasing reaction times, concentration of full-length DNA strands (○) decreases, whereas intermediate (□) and short (▵) DNA fragment concentrations increase. Concentrations are calculated by integrating the bracketed areas of the gel lanes (inset) and normalizing to the full-length DNA band at zero time. Analysis of identical samples on a nondenaturing gel (upper inset) reveals concomitant loss of full-length dsDNA (•). The concentration of full-length DNA strands is predicted (——) to decrease at half the rate of full-length dsDNA loss if degradation of one of the two strands is simultaneous with unwinding. This prediction agrees well with the measured full-length DNA concentrations corrected (see Materials and Methods) for postunwinding degradation of full-length ssDNA (×).
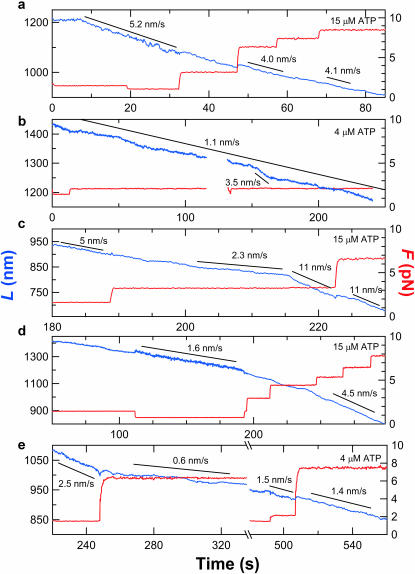
In addition to backsliding, further evidence for kinetically distinct, persistent states of the enzyme-DNA complex comes from high-resolution measurements of RecBCD forward velocity. This velocity varied not only from molecule to molecule, but also within records of individual molecules (Fig. 4). Some molecules proceeded at nearly uniform rates for long intervals (Figs. 1 c, 8–33 s, and 4 a); others moved with fluctuating speeds (Fig. 4 b). Although some fluctuations arise simply from the stochastic nature of motor enzyme stepping (Svoboda et al., 1994), a minimal stepping model does not account for the abrupt and persistent changes in speed lasting from seconds to minutes that often were observed (Fig. 4, c and d). Based on comparisons of records from different molecules, we found no clear correlations between the positions of these rate transitions and particular DNA sequences, even though the instrumentation is capable of resolving such sequence dependences of enzyme movement (Perkins et al., 2003). These observations are inconsistent with proposed mechanisms that assume a uniform population of RecBCD-DNA complexes with invariant unwinding rate constants. Substantial fluctuations in velocity were not seen in previous studies (Bianco et al., 2001; Dohoney and Gelles, 2001; Spies et al., 2003), possibly due to the limited spatiotemporal resolution of earlier work or to the differences in experimental conditions such as applied force or ATP concentration. As previously discussed, surface immobilization is specific and does not alter the population velocity distribution; therefore, we regard as unlikely the hypothesis that the velocity fluctuations resulted from the time-dependent alterations in enzyme interaction with the surface.
FIGURE 4.
Contour length records at various loads and ATP concentrations reveal diverse enzyme behaviors. (a) A trace displaying relatively uniform motion (blue) irrespective of the external load (red) up to 8 pN. Line fits over indicated intervals (black lines) show average velocities. The minor discontinuity in L at 47 s as F was doubled to 6.3 pN corresponds to a 1% error in determining L. (b) Record showing spontaneous variations in enzyme speed. (c–e) Examples of records with abrupt velocity changes. Some changes are spontaneous; others are concomitant with changes in applied force. A backslide occurred during the interruption in e.
Measuring how velocity varies with external force can provide insight into the mechanochemistry of motor enzymes (Finer et al., 1994; Leibler and Huse, 1993; Svoboda and Block, 1994b; Wang et al., 1998). In contrast to RNA polymerase molecules, which display consistent changes in velocity with increasing load (Wang et al., 1998), the responses of RecBCD molecules were high ly variable. For some molecules, forward speeds were unaffected by moderate increases in force (Figs. 1 c and 4 a); we were not able to reliably test the effects of further increases because substantially higher forces induced backsliding. For other molecules, a moderate force increment was associated with a decrease in movement velocity. In several cases, the same molecule responded differently to an increase in load at different times (Fig. 4 e), consistent with a change in the functional state of the enzyme-DNA complex. The independence of velocity with load during some periods implies that the rate-limiting step in the enzymatic cycle during these periods does not entail significant (>1 bp) motion along the DNA (Schnitzer and Block, 1997; Wang et al., 1998). This is expected, for example, in a simple reaction mechanism in which ATP binding—not a translocation step—limits the turnover rate at the subsaturating ATP concentrations and moderate forces used here. The sensitivity to applied force observed at other times implies a significant change in the rate constants, or perhaps an entirely different reaction pathway, from that in the force-insensitive periods.
Understanding the molecular mechanism of a motor enzyme requires determining the size of the elementary step by which it moves. Our apparatus was optimized so that steps as small as 2 nm (∼6 bp) could be detected (at a 1-Hz rate on 600–900-nm DNA segments under loads of 6–8 pN; Fig. 5 a). None of our records revealed repeated steps, and most traces were smooth down to the 2-nm experimental limit (Fig. 5 b). Because RecBCD-bio was anchored to the coverglass surface through the RecD subunit, we conclude that RecD does not advance relative to DNA in uniform steps greater than 2 nm (6 bp). This is consistent with RecBCD presteady-state unwinding kinetics experiments showing ∼4 bp unwound per rate-limiting step (Lucius et al., 2002). A much larger step of ∼23 bp (7.8 nm) has been inferred indirectly from gap-traversal efficiencies (Bianco and Kowalczykowski, 2000). However, those experiments were performed on a RecBC preparation lacking the RecD subunit. Since RecD is thought to be the helicase responsible for dsDNA unwinding by the intact trimer (Dillingham et al., 2003), one would not expect the step size for an enzyme missing this subunit to necessarily agree with results reported here and elsewhere for the RecBCD holoenzyme.
FIGURE 5.
Fine-scale motion. (a) Control trace of a bead-DNA complex attached directly to the coverglass without RecBCD illustrates the low drift and noise of the system. Filter bandwidth, 13 Hz; force, 7.5 pN. (b) RecBCD motion at 4 μM ATP (black). There is no evidence for uniform steps > 2 nm, and even isolated abrupt displacements > 2 nm are infrequent. Filter bandwidth, 13 Hz; force, 7.4 pN. The microscope stage position (gray) was adjusted every 1 s by the feedback system to maintain constant force. The stage motions are not detected as either pauses or abrupt changes in contour length, demonstrating the validity of the derived contour length measurement.
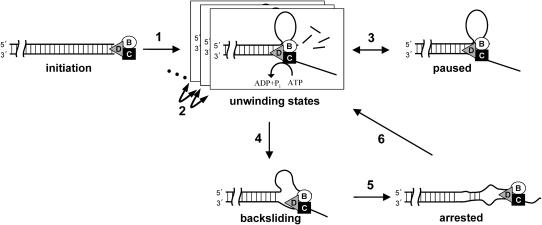
Taken together, the single-molecule data imply that RecBCD-DNA complexes interconvert between persistent states with distinctly different physical and catalytic properties (Fig. 6), even in the absence of χ-sequences. After binding to a blunt DNA end to form an initiation complex (Lucius et al., 2002), the complex isomerizes (Fig. 6; step 1) to one of several possible unwinding states. During unwinding, ATP hydrolysis catalyzed by RecD drives movement of the trimeric enzyme along duplex DNA. Degradation of the top strand by the lagging RecB nuclease and translocase domains is slower than unwinding by the leading RecD helicase domain, resulting in the production of a growing DNA loop (Taylor and Smith, 1980, 2003), as proposed (Dillingham et al., 2003; Kowalczykowski, 2000; Taylor and Smith, 2003). In this model, uniform stepwise movement of RecD is responsible for advancing the trimeric enzyme along dsDNA. We find that such steps are smaller than 6 bp. In contrast, RecB does not unwind, but simply moves along ssDNA at the trailing edge of the loop. This movement may occur in larger steps (Bianco and Kowalczykowski, 2000). Our data demonstrate the presence of different unwinding states of the RecBCD-DNA complex, each with its own characteristic unwinding rate. Interconversions among states (steps 2) occur spontaneously but can also be induced by changes in applied load. Moving complexes can reversibly isomerize (step 3) to one or more inactive paused configurations. Application of moderate opposing forces (∼4–8 pN) much smaller than those required to stall other DNA motors (Smith et al., 2001; Wang et al., 1998) occasionally triggers the conversion to a backsliding state (step 4) where the enzyme can rapidly move rearward. One possible scenario is that in this state the enzyme slides along the lower strand while remaining bound to a fixed position on the top strand. The accumulation of significant amounts of nonduplexed DNA during backsliding suggests that reannealing occurs slower than reported with naked DNA (Bockelmann et al., 2002), perhaps due to the sequestration of ssDNA by SSB in our experiments. Reducing the opposing force arrests backward motion (step 5). DNA in the loop, which has not yet been degraded by RecB, can then reanneal, allowing unwinding to resume (step 6).
FIGURE 6.
Scheme of state switching inferred from single-molecule experiments, together with hypothesized structures of the RecBCD-DNA complex in the different states (see text). The enzyme moves leftward relative to the DNA during unwinding and rightward during backsliding.
DNA tangles and proteins bound to DNA present mechanical obstacles that must be overcome to allow the cellular functions of RecBCD in DNA replication, repair, and degradation. We speculate that the ability of the enzyme to backslide and restart improves function by allowing RecBCD to make repeated attempts at such mechanical blockages. The existence of paused, backsliding, arrested, multiple unwinding, and χ-modified unwinding states demonstrates that the RecBCD-DNA complex exhibits remarkable functional polymorphism, the biological significance of which is largely unexplored.
Acknowledgments
We thank E. Abbondanzieri, J. Haber, K. Neuman, and J. Shaevitz for helpful discussions.
Supported by a Burroughs-Wellcome Fund Career Award in the Biomedical Sciences and National Institute of Standards and Technology (T.T.P.), a Damon Runyon-Walter Winchell Cancer Research Fund Fellowship (H.W.L.), and grants from National Institute of General Medical Sciences (J.G. and S.M.B.).
Thomas T. Perkins' present address is JILA, National Institute of Standards and Technology and University of Colorado, 440 UCB, Boulder, CO 80309-0440.
References
- Bianco, P. R., L. R. Brewer, M. Corzett, R. Balhorn, Y. Yeh, S. C. Kowalczykowski, and R. J. Baskin. 2001. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 409:374–378. [DOI] [PubMed] [Google Scholar]
- Bianco, P. R., and S. C. Kowalczykowski. 2000. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 405:368–372. [DOI] [PubMed] [Google Scholar]
- Bockelmann, U., P. Thomen, B. Essevaz-Roulet, V. Viasnoff, and F. Heslot. 2002. Unzipping DNA with optical tweezers: high sequence sensitivity and force flips. Biophys. J. 82:1537–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchiat, C., M. D. Wang, J. Allemand, T. Strick, S. M. Block, and V. Croquette. 1999. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 76:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C., J. F. Marko, E. D. Siggia, and S. B. Smith. 1994. Entropic elasticity of λ-DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature. 404:37–41. [DOI] [PubMed] [Google Scholar]
- Dillingham, M. S., M. Spies, and S. C. Kowalczykowski. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature. 423:893–897. [DOI] [PubMed] [Google Scholar]
- Dixon, D. A., and S. C. Kowalczykowski. 1993. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 73:87–96. [DOI] [PubMed] [Google Scholar]
- Dohoney, K. M., and J. Gelles. 2001. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 409:370–374. [DOI] [PubMed] [Google Scholar]
- Finer, J. T., R. M. Simmons, and J. A. Spudich. 1994. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 368:113–119. [DOI] [PubMed] [Google Scholar]
- Ha, T., I. Rasnik, W. Cheng, H. P. Babcock, G. H. Gauss, T. M. Lohman, and S. Chu. 2002. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 419:638–641. [DOI] [PubMed] [Google Scholar]
- Hagerman, P. J. 1988. Flexibility of DNA. Ann. Rev. Biophys. Biophys. Chem. 17:265–286. [DOI] [PubMed] [Google Scholar]
- Hsieh, W. T., P. A. Whitson, K. S. Matthews, and R. D. Wells. 1987. Influence of sequence and distance between two operators on interaction with the lac repressor. J. Biol. Chem. 262:14583–14591. [PubMed] [Google Scholar]
- Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156–165. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibler, S., and D. A. Huse. 1993. Porters versus rowers: a unified stochastic model of motor proteins. J. Cell Biol. 121:1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucius, A. L., A. Vindigni, R. Gregorian, J. A. Ali, A. F. Taylor, G. R. Smith, and T. M. Lohman. 2002. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 324:409–428. [DOI] [PubMed] [Google Scholar]
- Marko, J. F., and E. D. Siggia. 1995. Stretching of DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- Perkins, T. T., R. V. Dalal, P. G. Mitsis, and S. M. Block. 2003. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 301:1914–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 28:2863–2873. [DOI] [PubMed] [Google Scholar]
- Savitzky, A., and M. J. E. Golay. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36:1627–1639. [DOI] [PubMed] [Google Scholar]
- Schnitzer, M. J., and S. M. Block. 1997. Kinesin hydrolyses one ATP per 8-nm step. Nature. 388:386–390. [DOI] [PubMed] [Google Scholar]
- Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 413:748–752. [DOI] [PubMed] [Google Scholar]
- Spies, M., P. R. Bianco, M. S. Dillingham, N. Handa, R. J. Baskin, and S. C. Kowalczykowski. 2003. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 114:647–654. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., and S. M. Block. 1994a. Biological applications of optical forces. Ann. Rev. Biophys. Biophys. Chem. 23:247–285. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., and S. M. Block. 1994b. Force and velocity measured for single kinesin molecules. Cell. 77:773–784. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., P. P. Mitra, and S. M. Block. 1994. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc. Natl. Acad. Sci. USA. 91:11782–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A., and G. R. Smith. 1980. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 22:447–457. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 423:889–893. [DOI] [PubMed] [Google Scholar]
- Visscher, K., S. P. Gross, and S. M. Block. 1996. Construction of multiple-beam optical traps with nanometer-resolution. IEEE J. Sel. Top. Quant. Electr. 2:1066–1076. [Google Scholar]
- Wang, M. D., M. J. Schnitzer, H. Yin, R. Landick, J. Gelles, and S. M. Block. 1998. Force and velocity measured for single molecules of RNA polymerase. Science. 282:902–907. [DOI] [PubMed] [Google Scholar]
- Wang, M. E., H. Yin, R. Landick, J. Gelles, and S. M. Block. 1997. Stretching DNA with optical tweezers. Biophys. J. 72:1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuite, G. J., S. B. Smith, M. Young, D. Keller, and C. Bustamante. 2000. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 404:103–106. [DOI] [PubMed] [Google Scholar]
- Yu, M., J. Souaya, and D. A. Julin. 1998. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA. 95:981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimm, B. H. 1998. Extension in flow of a DNA molecule tethered at one end. Macromolecules. 31:6089–6098. [Google Scholar]