Abstract
Paramagnetic contributions to nuclear magnetic spin-lattice relaxation rate constant induced by freely diffusing molecular oxygen measured at hundreds of different protein proton sites provide a direct means for characterizing the exploration of the protein by oxygen. This report focuses on regions of ribonuclease A where the rate constant enhancements are either quite large or quite small. We find that there are several regions of enhanced oxygen affinity for the protein both on the surface and in interior pockets where sufficient free volume permits. Oxygen has weak associative interactions with a number of surface crevices that are generally between secondary structural elements of the protein fold. Several regions near the surface have higher than expected accessibility to oxygen indicating that structural fluctuations in the protein provide intermolecular access. Oxygen penetrates part of the hydrophobic interior, but affinity does not correlate simply with hydrophobicity indices. Oxygen is excluded from regions of high interior packing density and a few surface sites where x-ray diffraction data have indicated the presence of specific hydration with high occupancy.
INTRODUCTION
Intermolecular accessibility is fundamental to understanding molecular recognition and function. Proteins are particularly important because function may be regulated by small molecules. Protein structures derived from both diffraction experiments and nuclear magnetic resonance spectroscopy suggest that some structural flexibility is required for intermolecular access that permits function. For example, myoglobin requires some structural reorganization for oxygen to access the iron atom (Case and Karplus, 1979; Carrero et al., 1995) and ribonuclease does not function below a dynamical transition at 220 K (Rasmssen et al., 1992). Discussions of molecular accessibility are often based on a computational strategy that begins with the high-resolution static or dynamically averaged structural model of the protein (Lee and Richards, 1971). However, these calculations may not include all aspects of the accessibility such as hydration, hydrophobic, van der Waals, or electrostatic interactions that may now be measured by several spectroscopic approaches. We focus here on a nuclear magnetic approach for experimental characterization of intermolecular contacts that provides useful insights to the subtle energetic contributions that govern intermolecular effects.
Experimental methods for characterizing intermolecular accessibility include chemical reactivity, amide-hydrogen exchange kinetics (Woodward and Rosenberg, 1971; Woodward et al., 1982; Kim et al., 1993; Wang et al., 1995; Hitchens and Bryant, 1998; Li and Woodward, 1999; Dixon et al., 2000), fluorescence quenching (Lakowicz and Weber, 1973a,b; Calhoun et al., 1983a,b, 1986; Gratton et al., 1984; Carrero et al., 1995; Mansoor et al., 1999), electron spin relaxation (ESR) (Altenbach et al., 1989a,b; Victor and Cafiso, 1998), as well as the nuclear magnetic resonance (NMR) contact shifts and relaxation (Esposito et al., 1992; Cocco and Lecomte, 1994; Molinari et al., 1997; Prosser et al., 2001). Fluorescence quenching and ESR experiments, although providing both spatial and dynamic information, suffer from the small number of reporter sites. Although site-directed labeling using fluorophores or electron spins may improve the number of reporter sites, these modifications have the potential to shift protein stability and alter the assessibility (Mansoor et al., 1999). The nuclear magnetic relaxation experiment is insensitive, but may utilize one or more reporter spins on each amino acid and thereby, provides spatial as well as dynamic information with minimal chemical or structural perturbation. Moreover, with high-resolution NMR spectroscopy, each nuclear resonance observed in a two-dimensional spectrum may be related to a particular proton site in the protein structure that may be encoded with accessibility information through the effects of intermolecular spin relaxation processes (Niccolai et al., 1982, 1984, 2001a,b; Polnaszek and Bryant, 1984; Zhou et al., 1985; Molinari et al., 1997; Prosser et al., 2000; Teng and Bryant, 2000; Prosser et al., 2001; Hernandez et al., 2002; Pintacuda and Otting, 2002; McNaughton et al., 2003).
An analytical theory for the relaxation of a nuclear spin induced by a freely diffusing paramagnet has been presented by Freed and included the effects of a rapid electron spin relaxation process (Freed, 1978). In the case where the electron spin relaxation time is short compared with the translational correlation time, the efficiency of the nuclear spin relaxation induced by the diffusing paramagnet becomes essentially independent of the translational diffusion constant; that is, the correlation time for the electron-nuclear coupling becomes the electron relaxation time. In this case, the paramagnetic contribution to the nuclear spin relaxation is a direct measure of the effective local concentration of paramagnetic molecule, [S].
We report here nuclear magnetic relaxation rate constant measurements that permit to obtain high-resolution measures of molecular accessibility to different structural regions of a folded protein using oxygen as the small and neutral accessibility probe (Prosser et al., 2000, 2001; Teng and Bryant, 2000; Hernandez et al., 2002; McNaughton et al., 2003). Oxygen is convenient because the electron spin-lattice relaxation time is on the order of 7.5 ps (Teng et al., 2001), which is shorter than most inter- and intramolecular correlation times such as rotation and translation. Therefore, the correlation time for the electron-nuclear dipolar coupling is not dependent on the slower events in the system such as translation of the oxygen or rotation of the protein. The strong distance dependence of oxygen-induced relaxation contribution means that the magnitude is essentially proportional to the contact probability of oxygen for each observed proton, i.e., it provides an excellent measure of molecular accessibility.
This report focuses on the structural characterization of ribonuclease A sites that interact well or poorly with oxygen as measured by the 1H spin-lattice relaxation rate changes. The experimental oxygen accessibility varies from position to position on the protein surface and oxygen penetrates into the loosely packed parts of the protein interior even though the access appears to be sterically prohibited based on the constraints of the static structural model of the protein. In the subsequent paper a quantitative strategy for interpreting relaxation rate changes at each location in terms of local free energy differences will be presented.
MATERIALS AND METHODS
Bovine pancreatic ribonuclease A (RNase A; E.C.3.1.27.5; ∼13.7 mol wt; grade XII-A) was purchased from Sigma Chemical (St. Louis, MO). This protein was purified as follows: 100 mg of RNase A in 3.0 mL deionized water was dialyzed using a Slide-A-Lyzer Dialysis Cassette with ∼10,000 mol wt cutoff (Pierce, Rockford, IL) against 4.0 L deionized water drawn from a Barnstead NANOpure (Barnstead, Dubuque, IA) ultrapure water system that used house-ionized water as the feed. The deionized water was stirred at 4°C to maintain fluid homogeneity, and was refreshed every 6 h for a total of six changes. The pH was adjusted with hydrochloric acid and sodium hydroxide using the protein as the buffer.
After dialysis, the protein was lyophilized and was redissolved in 99.96% D deuterium oxide (D2O; Cambridge Isotope Laboratories, Andover, MA) for the COSY experiments, or was stored at a −20°C refrigerator for later use. Protein concentration was based on the ultraviolet absorption; E (280 nm) = 9473 M−1cm−1 (Pace et al., 1995) for RNase A. An NMR sample for the COSY experiments contained 0.6 mL of ∼3 mM RNase A in D2O.
Although oxygen is a convenient paramagnetic reagent, the solubility is low ∼0.23 mM (Wilhelm et al., 1977) when equilibrated with air at 25°C. To achieve the desired dynamic range in the relaxation measurements, protein samples were equilibrated with 11 atm of O2 using pressure/vacuum valve sample tubes 524-PV purchased from Wilmad Glass (Buena, NJ).
Pressure is applied to the NMR sample tubes by first clearing any dissolved air in the protein solution using freeze-pump-thaw cycles and then by equilibrating the protein solution with 11 atm oxygen (99.997%; BOS Gases, Murray Hill, NJ) or nitrogen (99.9995%; BOS Gases). Eight freeze-pump-thaw cycles were conducted, using dry ice-isopropanol as the cooling agent. Samples were sealed with a gas tight screw that was further secured mechanically with Parafilm.
Computation
Protein proton accessible surface areas are computed based on the ACCESS program (v. 2.2.4) from the Center of Structural Biology at Yale University (http://www.csb.yale.edu/download/download_descrip.html) (Lee and Richards, 1971). The distance of closest approach, b, for the intermolecular electron-nuclear interaction of each protein proton is defined by computing the shortest distance between a proton and the calculated Connolly protein molecular surface defined by a surface probe of 1.4 Å radius (Molecular Simulations, San Diego, CA).
NMR spectroscopy
All NMR data were acquired using a 500-MHz Varian Unity Plus spectrometer (Varian, Palo Alto, CA). Both, T1-weighed gradient double-quantum-filtered COSY (gDQF-COSY) spectra and one-dimensional proton (1H) spectra were acquired for each sample. gDQF-COSY spectra were recorded in the phase-sensitive mode using quadrature detection in the directly detected dimension (D2) and hypercomplex detection in the indirectly detected dimension (D1). The carrier frequency was centered at the HOD resonance frequency that is also used as the internal reference at 4.75 ppm. The RNase A spectra were acquired at 30°C with spectral widths of 5000 Hz in both dimensions. Each acquisition consisted of 16 transients, containing 1024 complex points in D2 and 256 complex points in D1, corresponding to a total acquisition time of ∼3 h for every 1 s of saturation recovery delay.
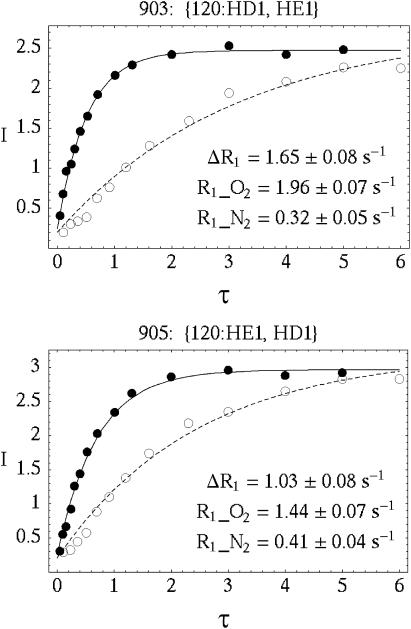
The proton T1-weighed pulse sequence was edited by adding a 1H π/2 pulse (saturation pulse), followed by a saturation recovery delay before a gDQF-COSY pulse sequence (Piantini et al., 1982; Rance et al., 1983). Two additional gradient pulses were added immediately before and after the saturation pulse to spoil any remaining coherences. The delay table was constructed according to the response of the average peak intensity of the protein protons in a one-dimensional inversion recovery experiment for each sample. The spectra were typically measured with a saturation recovery delay list of 0.105, 0.233, 0.357, 0.511, 0.693, 0.911, 1.20, 1.61, 2.30, 3.0, 4.0, 5.0, and 6.0 s for the diamagnetic sample, and 0.05, 0.1, 0.156, 0.244, 0.304, 0.402, 0.528, 0.706, 1.01, 1.31, 2.0, 3.0, 4.0, and 5.0 s for the paramagnetic sample. Examples of the crosspeak intensities as a function of delay times with and without oxygen are shown in Fig. 1.
FIGURE 1.
The saturation recovery response of two representative crosspeaks, (120:HD1, HE1) and (120:HE1, HD1), in the COSY spectra for RNase A measured at pH 3.2 and 30°C. Although 120:HD1 and 120:HE1 are coupled to one another, their respective oxygen induced spin-lattice relaxation rate contributions can be measured separately in the COSY experiment. The spatial resolution of the oxygen accessibility map is limited only by the available spectroscopic resolution.
The time domain data were first filtered with a π/2 phase-shifted sine-bell window function in both dimensions, and then zero filled to 2048 points in D2 and to 1024 points in D1 before Fourier transformations. Absolute values of crosspeak intensities were used to simplify effects of antiphase components. All data were processed using VNMR 6.1B (Varian, Palo Alto, CA) and Felix 2000 NMR software (Molecular Simulations, San Diego, CA). 1H chemical shifts for RNase A were reported at 30°C and pH 3.2 (Robertson et al., 1989), and at 35°C and pH 4.0 (Rico et al., 1989). For our experiments, RNase A accessibility probed by oxygen is measured at 30°C and pH 3.2 and the previous assignments were used (Rico et al., 1989; Robertson et al., 1989).
The peak intensities in a COSY spectrum were measured by integrating the peak magnitude over the peak area. Negative multiplet components of a normal COSY crosspeak are inverted to positive values so that the integration of the peak magnitude over the peak area gives nonzero values of peak intensities. However, peak overlap is not unusual, which may result in composite peak intensities that could interfere with the determination of the relaxation parameters unique to each proton in the protein structure. In such a case, peak areas were defined to include only multiplet components that did not overlap with one another. Spectral peaks where clear isolation was not possible using this strategy were not used in the analysis. Variations in relaxation rates as a result of the choice of peak area were small compared to the standard error from the data-fitting routine.
A total of 463 crosspeaks, including some slowly exchanging amide proton crosspeaks, were unambiguously assigned in the two-dimensional COSY spectra. Because scalar coupling occurs between nuclei that are separated by one to three chemical bonds, a specific proton may be coupled to two distinct protons resulting in two crosspeaks with the same relaxation character in the COSY spectrum. For example, if protons A, B, C are coupled to one another, six crosspeaks at (ωA, ωB), (ωA, ωC), (ωB, ωC), and symmetrically (ωB, ωA), (ωC, ωA), (ωB, ωC) can be observed in the spectrum. However, both crosspeaks at (ωA, ωB) and (ωA, ωC) characterize the relaxation parameters unique to proton A. The resonance frequency in the second dimension does not encode the initial population of proton B or C and merely shows connectivity as a result of scalar coupling.
Ideally, relaxation parameters characterized by crosspeaks at (ωA, ωB) and (ωA, ωC), for example, should give exactly the same ΔR1 for proton A; however, fitting to an exponential is subject to error that is propagated by the necessity of subtracting two relaxation rate constants,
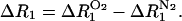 |
(1) |
In the case where the values of ΔR1 for proton A as measured by different crosspeaks at (ωA, ωB) and (ωA, ωC) differed slightly, the crosspeak with greater signal-to-noise, a smaller residual in the fit to a single exponential, or more complete isolation from other crosspeaks was used to characterize the relaxation for proton A. For most of the crosspeaks that carry redundant information, the differences in the relaxation rates are small and are comparable to the standard errors of the fit (∼10%). Some crosspeaks have larger errors because of rapid transverse relaxation or rapid longitudinal relaxation.
In the COSY spectra, 300 out of 463 crosspeaks were uniquely resolved out of a total of 787 protons in RNase A, excluding amide protons. The relaxation parameters for amide protons were not characterized because both the proton relaxation and the hydrogen/deuterium exchange mechanism may affect the peak intensities. Nevertheless, the number of reporter sites within a protein from a COSY spectrum is large; most of the residues provide more than two reporter protons. Some residues, such as 8F, 115Y, and 120F, provide five resolvable reporter protons, and all reporter protons of the same residue render unique relaxation rate constants for measurements of oxygen accessibility at that site.
The apparent proton spin-lattice relaxation rate (R1) characterized by a crosspeak was determined by fitting the peak intensities to a single exponential (Eq. 2) using the nonlinear least-squares fitting routine (Levenberg-Marquardt method) in the Mathematica 3.0 program (Wolfram Research, Champaign, IL).
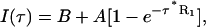 |
(2) |
where I(τ) are the crosspeak intensities measured at the saturation recovery delays, τ. The fitted parameters are: A, the maximum peak intensity at τ = ∞ and R1, the proton spin-lattice relaxation rate constant. The peak intensities in a COSY spectrum were measured by integrating the peak magnitude over the peak area. Negative multiplet components of a normal COSY crosspeak are inverted to positive values so that the integration of the peak magnitude over the peak area gives nonzero values of peak intensities. The quality of the fit is shown by the solid and dashed lines in Fig. 1.
RESULTS AND DISCUSSION
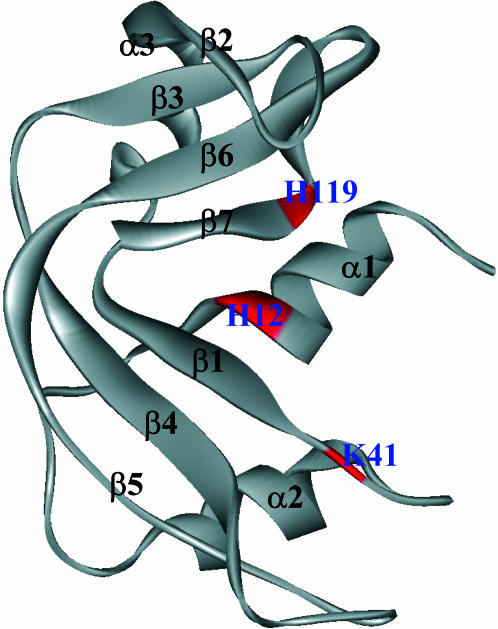
The relaxation rate constants for the 300 well-resolved proton positions span the range from 0.37 s−1 to >6 s−1. The distribution is not sharply peaked but is asymmetric with a tail at high rates. Quantitative discussion of these rate constants will be presented subsequently; we focus this discussion on two classes of proton sites—those with large and those with small oxygen-induced relaxation rate constants. A complete parameter table for each of 300 crosspeaks is included as supplementary material. The primary structural elements of ribonuclease A are summarized in the ribbon diagram of Fig. 2 for reference.
FIGURE 2.
The ribbon structure of ribonuclease A labeled with the structural elements. The boxed labels indicate the residues of the active site triad.
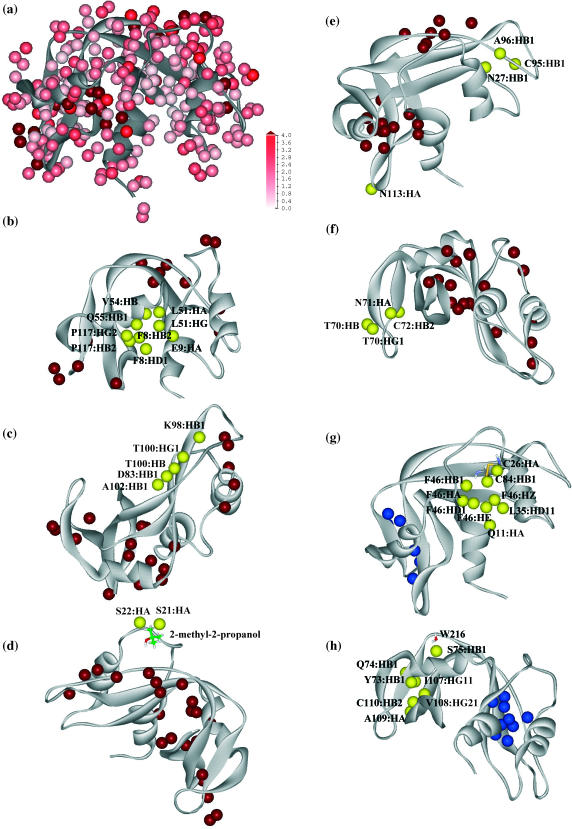
Changes of proton relaxation rates (ΔR1) are mapped onto a color scale and represented as space-filling atoms superimposed onto a ribbon structure of the folded protein and shown in Fig. 3 a. The color ranges from white to pink to red; the color intensity is linearly proportional to the changes of relaxation rates from 0 s−1 to 4 s−1. Any changes of relaxation rates that are larger than 4 s −1 are represented as the deepest red and the relaxation data are summarized in Table 1. The relaxation rate changes are remarkably different at different proton positions. Even among surface protons, ΔR1 differs from the smallest change of 0.47 ± 0.10 s−1 at 75-Ser:HB1 to a large change of 4.84 ± 0.53 s−1 at 51-Lys:HG. Larger relaxation rate changes are also observed but are less accurate. This ensemble of oxygen-induced relaxation rates (ΔR1) reflects the sum of complex energetic contributions that determine the intermolecular exploration of the protein by oxygen.
FIGURE 3.
(a) Color representation of the oxygen-induced proton relaxation rate constant changes (ΔR1). All spectrally resolved protons are represented as space-filling spheres that are superimposed on the ribbon structure of ribonuclease A. The color intensity changes linearly from white to pink to red in proportion to the changes of relaxation rate constants in the range from 0 s−1 to 4 s−1. Relaxation rate constant contributions that are larger than 4 s −1 are colored as dark red. (b–f) Protons whose oxygen-induced relaxation rate constants >4 s −1 are identified in red spheres but are also shown as yellow groups to facilitate discussion in the context of structural features. (b) Phe-8:HD1, Phe-8:HE1, Glu-9:HA, Leu-51:HA, Leu-51:HG, Val-54:HB, Gln-55:HB1, Pro-117:HB2, and Pro-117:HG2 are highlighted in yellow and are in a buried oxygen binding pocket. (c) Asp-83:HB1, Lys-98:HB1, Thr-100:HB, Thr-100:HG1, and Ala-102:HB1 are located at the major hairpin (β4-β5) forming a surface β groove. (d) Ser-21:HA and Ser-22:HA are located near a surface pocket between the helix linker, connecting α1 and α2, and the antiparallel β5. A cocrystallized 2-methyl-2-propanol molecule is found to bind nearby (Wlodawer et al., 1988), which suggests higher hydrophobicity. (e) Asn-27:HB1, Cys-95:HB1, and Ala-96:HB1 are located at the surface cleft between α2 and β5. Asn-113:HA is located right at the sharp turn of the β6-β7 hairpin. (f) Cys-72:HB2, Thr-70:HB, Thr-70:HG1, and Asn-71:HB2 are located at the β2-β3 hairpin region. (g–h) Protons for which the oxygen-induced relaxation rate constants are <0.6 s−1 are shown as blue spheres but are also shown as yellow groups to facilitate discussion in the context of structural features. (g) Gln-11:HA, Cys-26:HA, Leu-35:HD11, Phe-46:HA, Phe-46:HB1, Phe-46:HD1, Phe-46:HE1, Phe-46:HZ, and Cys-84:HB1 are buried in the first structural lobe and are located near the central β-strand. (h) Tyr-73:HB1, Gln-74:HB1, Ile-107:HG11, Val-108:HG21, Ala-109:HA, and Cys-110:HB2 are buried in the second structural lobe and are located at the centers of the β3 and β6 strands. Ser-75:HB1, on the other hand, is located near the protein surface. However, invariant water molecules are found nearby (Sadasivan et al., 1998). The measured and computed parameters for the above high rate and low rate protons are listed in Tables 1 and 2, respectively. The structural model of RNase A (7RSA) is courtesy of the Protein Database (Wlodawer et al., 1988).
TABLE 1.
Large relaxation rate constant summary
| H observed | H crossed | Residue type | ΔR1 (s−1) | Secondary structure | b (Å) | AAREA (Å2) |
|---|---|---|---|---|---|---|
| Buried pockets (Fig. 3 b) | ||||||
| 8:HD1 | HE1 | Phe | 11.8(1.97) | α1 | 2.61 | 0.1 |
| 8:HE1 | HD1 | Phe | 4.56(0.55) | α1 | 4.53 | 0 |
| 9:HA | HB1 | Glu | 4.05(1.4) | α1 | 3.80 | 0 |
| 51:HA | HB1 | Leu | 6.24(0.91) | α3 | 4.32 | 0 |
| 51:HG | HD11 | Leu | 4.84(0.53) | α3 | 2.64 | 0 |
| 54:HB | HA | Val | 20.34(3.22) | α3 | 4.42 | 0 |
| 55:HB1 | HA | Gln | 4.36(0.43) | α3 | 2.60 | 1.4 |
| 117:HB2 | HG2 | Pro | 4.19(0.81) | β7 | 4.59 | 0 |
| 117:HG2 | HB2 | Pro | 5.47(0.99) | β7 | 4.21 | 0 |
| Grooove (Fig. 3 c) | ||||||
| 83:HB1 | HA | Asp | 6.52(1.3) | β4 | 2.60 | 5.1 |
| 98:HB1 | HA | Lys | 4.06(1.39) | β5 | 2.60 | 4.5 |
| 100:HB | HG1 | Thr | 4.2(0.3) | β5 | 2.69 | 0.3 |
| 100:HG1 | HB | Thr | 4.09(0.3) | β5 | 2.60 | 7.9 |
| 102:HB1 | HA | Ala | 7.38(1.35) | β5 | 2.60 | 10.9 |
| Surface pocket between helix linker and β5 (Fig. 3 d) | ||||||
| 21:HA | HB1 | Ser | 4.39(1.45) | T | 2.60 | 11.1 |
| 21:HA | HB1 | Ser | >10 | T | 2.60 | 11.1 |
| Surface cleft between α2 and β5 (Fig. 3 e) | ||||||
| 27:HA | HB1 | Asn | 4.32(0.65) | α2 | 2.71 | 0.4 |
| 95:HA | HB1 | Cys | 8.05(2.56) | β4 | 4.56 | 0 |
| 96:HA | HB1 | Ala | 4.53(0.72) | β4 | 2.63 | 0 |
| β2-β2 hairpin (Fig. 3 f) | ||||||
| 70:HB | HG1 | Thr | 28.84(5.54) | 2.60 | 7.6 | |
| 70:HG1 | HB | Thr | 7.18(2.92) | 2.60 | 18.9 | |
| 71:HA | HB2 | Asn | 65.34(5E3) | β3 | 2.60 | 1.3 |
| 72:HB2 | HB1 | Cys | 4.68(0.73) | β3 | 4.58 | 0 |
| Turn at β6-β7 hairpin (Fig. 3 e) | ||||||
| 113:HA | HB1 | Asn | 4.43(0.82) | T | 3.13 | 0 |
Measured and computed parameters for protons whose oxygen-induced paramagnetic relaxation rate constants are >4.0 s −1 are summarized in Tables 1 and 2. The identities of these protein protons of the relaxation rate measurements are listed in the first column, whereas the scalar coupled protons shown as crosspeaks in the COSY spectra are in the second column. The residue types and the locations of the residues in the secondary protein structure are listed in the third and the fourth columns. The measured oxygen-induced paramagnetic relaxation rate constants and the standard errors in the fit (in parentheses) are listed in the fifth column. The distance of closest approach (b) between these protons and a surface molecular oxygen and the accessibility areas (AAREA) of the protons, computed from a protein structural model (Lee and Richards, 1971; Wlodawer et al., 1988) (7RSA), are listed in the last two columns.
Twenty-one proton sites, shown as dark red spheres in Fig. 3 a, experienced oxygen-induced relaxation rate enhancements larger than 4 s−1, which is more than twice as large as the rate predicted for the experimental conditions by the Freed force-free model (Hwang and Freed, 1975; Freed, 1978). This force-free model is suitable for hard-sphere collisions between freely diffusing spins and predicts an oxygen-induced rate enhancement 2.7 s−1 if the distance of intermoment closest approach is 2.6 Å and the effective correlation time is 7.5 ps. This value corresponds to the case where the paramagnet and proton have uniform distances of closest approach from all sides. For a proton at a surface position, however, there is an excluded region of approach because of steric hindrance from the protein. This geometric factor may reduce the relaxation rate to approximately half. Thus, we do not expect to observe oxygen-induced relaxation rates that are even as large as 2.7 s−1 for the limit of free diffusion. Note, this conclusion is independent of the translational mobility at the surface because the oxygen T1e is so short. Therefore, the regions with large paramagnetic relaxation rates correspond to regions with favorable energies for the oxygen-protein interaction. That is, the oxygen–protein interaction is not describable in terms of a hard-sphere model.
Large oxygen-induced relaxation
It is useful to compute the effects of a protein-bound oxygen molecule at some site on the protein. If we assume that oxygen binds to a site, the correlation time for the electron-proton dipolar coupling is the oxygen T1e or 7.5 ps not the rotational correlation time for the protein. Nevertheless, because we presume for this limiting case that the oxygen occupancy is unity, i.e., it is bound all the time, the Solomon-Bloembergen equations (Bloembergen et al., 1948; Solomon, 1955) predict an oxygen-induced proton relaxation rate constant of ∼6.2 × 103 s−1 for a proton in van der Waals contact with the oxygen. Thus, for a weak binding probability of as small as 0.1%, the oxygen-induced rate constant enhancement would be 6.2 s−1. The strength of the oxygen-proton dipolar coupling is large enough that even weak oxygen-protein interactions provide readily observable relaxation rate changes that then provide an excellent tool for characterizing weak intermolecular interactions. There are several regions in the protein with paramagnetic relaxation rate increments in excess of 4.0 s−1, which we discuss in terms of specific but still clearly weak binding interactions.
Buried pocket
Oxygen-induced relaxation rate constants >4.0 s−1 are observed for Phe-8:HD1, Phe-8:HE1, Glu-9:HA, Leu-51:HA, Leu-51:HG, Val-54:HB, Gln-55:HB1, Pro-117:HB2, and Pro-117:HG2. These protons are colored yellow in Fig. 3 b; all other protons with large oxygen-induced relaxation rates are located and colored in red in this same figure. RNase A has a major, a minor, and a small hydrophobic core (Scheraga et al., 2001). The yellow group forms a cluster located between helix 1, helix 3, and the C-terminal β-strand of the RNase A structure and is part of the major hydrophobic core. The major hydrophobic core is composed primarily of residues from the C-terminal β-hairpin (Ile-106, Val-108, Cys-110, Pro-114, Tyr-115, Val-116, Pro-117, Val-118, Phe-120), as well as the third α-helix (Leu-51, Val-54, Val-57, Cys-58), and the first α-helix (Ala-4, Ala-5, Phe-8, Met-13) (Scheraga et al., 2001). Except for Pro-8:HD1, Leu-51:HG, and Gln-66:HB1, all the other protons in this cluster are buried with depths averaging ∼4.3 ± 0.3 Å, which is indicative of a buried oxygen binding pocket.
Although local unfolding and protein structural fluctuations could allow close interactions between the oxygen and these buried protons, amide proton hydrogen/deuterium exchange data show that the entire β-sheet (with exception of bulges) together with helix 3 are the most stable segment of RNase A (Santoro et al., 1993; Wang et al., 1995). Helix 3 lies on top of a four-stranded β-sheet (β2, β3, β6, β7) with residues Val-57 and Cys-58 in close contact with Tyr-73 and Ser-75 and Ile-106 and Val-108 creating a hydrophobic core of exceptional stability against unfolding. Thus, it is unlikely that these interior protons are strongly coupled to surface oxygen as a result of local unfolding. In addition, unfolding could expose the protein interior and cause a strong correlation in the observed oxygen-induced relaxation with neighboring protons, but this correlation is not observed.
An early computational study by Lee and Richards reported three cavities in ribonulease A large enough to accommodate one water molecule (Lee and Richards, 1971). Long-lived water molecules have not been found experimentally (Halle and Denisov, 1995; Bryant, 1996). Although the oxygen binding region identified in these experiments is displaced somewhat from that suggested by Lee and Richards, this and subsequent studies suggest that the internal packing has sufficient voids that small rearrangements to permit access and accommodate a guest molecule may be made without a large energetic cost. This finding is consistent with other studies where low molecular weight hydrocarbons may fill protein cavities (Feher et al., 1996; Otting et al., 1997).
β-Groove
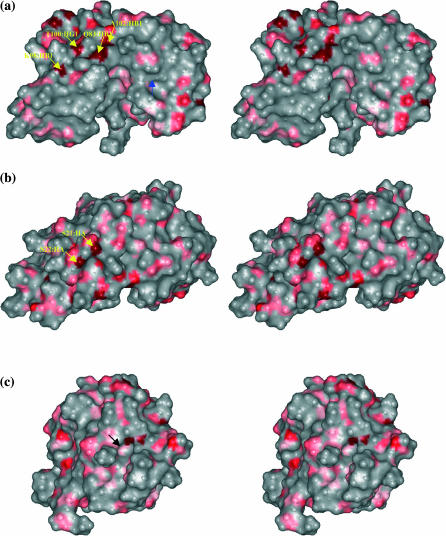
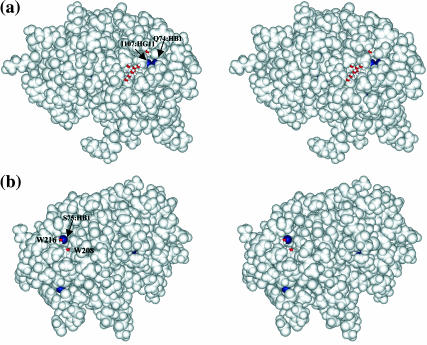
In the major β-hairpin region (β4-β5), protons at Asp-83:HB1, Lys-98:HB1, Thr-100:HB, Thr-100:HG1, and Ala-102:HB1 are observed with large paramagnetic relaxation rates and highlighted in yellow in Fig. 3 c. Unlike the interior pocket above, these protons are located near (Thr-100:HB) and are at the protein surface (Asp-83:HB1, Lys-98:HB1, Thr-100:HG1, Ala-102:HB1). Fig. 4 a shows a stereo pair of the molecular contour surface of ribonuclease A with protons having large relaxation rates colored in red. Asp-83:HB1, Lys-98:HB1, Thr-100:HB, Thr-100:HG1, and Ala-102:HB1 are located at a rather deep surface groove between the major hairpin (β4-β5). The buried side of this major hairpin (Cys-84, Tyr-97) forms the minor hydrophobic core with the second α-helix (Tyr-25, Cys-26, Met-29, Met-30) and the adjoining residues (Leu-35, Phe-46), which is not accessible to the solvent molecules based on calculations using the Lee and Richards methods (Lee and Richards, 1971). This β-surface groove, located on top of the minor hydrophobic core, is connected directly to the major catalytic cleft between the lobes of RNase A shown in Fig. 4 a. Nearby residues Asp-83, Asp-85, and Lys-98 are found to have alternative conformations in the x-ray model (Wlodawer et al., 1988), which indicates a low packing density or low activation energy for conformational reorientations. In addition, the hydrogen-bond opportunities offered at the side chains of Asp-83, Lys-98, and Thr-100 may further contribute to the energetic heterogeneity at the protein surface sensed by oxygen.
FIGURE 4.
Relaxed-eyed steric pairs of the computed molecular surface of ribonuclease A sensed by a surface probe of radius 1.4 Å (the shorter axis of molecular oxygen) in various orientations. Those protein protons for which the oxygen-induced relaxation rate constant is >4 s−1 are colored red. (a) Asp-83:HB1, Lys-98:HB1, Thr-100:HG1, and Ala-102:HB1, indicated by black arrows from left to right, are located at a rather deep surface groove between the major hairpin (β4-β5) on top of the minor hydrophobic core that connects directly to the major catalytic cleft. Blue arrow points to another surface pocket where Cys-72:HB2 is located and has large oxygen association. (b) Ser-22:HA and Ser-21:HA, indicated by black arrows from left to right, are located at a surface pocket in which a cosolute molecule, 2-methyl-2-propanol cosolvent molecule is found to bind in the x-ray structure model (Wlodawer et al., 1988).
Surface pocket between helix linker and β5
Large paramagnetic relaxation rates are observed at Ser-21:HA and Ser-22:HA. Ser-21, Ser-22, and Ser-23 border a surface pocket, which is between the linker regions connecting α1 and α2, and the antiparallel β-strand (β4). The molecular surface contour is shown in Fig. 4 b; the arrows point to the protons at Ser-21:HA and Ser-22:HA in red. Unlike the previously discussed buried binding pocket between α1 and α3 (Fig. 3 b), this binding pocket is well exposed to the solvent. In the x-ray structure (Wlodawer et al., 1988), a 2-methyl-2-propanol cosolvent molecule binds here, which suggests higher local hydrophobicity in this region. In Fig. 3 d, Ser-21:HA and Ser-22:HA are highlighted in yellow on the ribbon representation of RNase A with all the protons with large oxygen-induced relaxation rates marked as red spheres. The cocrystallized 2-methyl-2-propanol molecule is also shown in the structure.
Besides Ser-21:HA and Ser-22:HA, protons with large oxygen-induced relaxation rates are observed throughout this surface binding site, especially at Tyr-25:HA (ΔR1 = 3.56 ± 0.48 s−1) and at Tyr-25HB (ΔR1 = 2.49 ± 0.48 s−1), both of which are located at the bottom of this surface pocket. Note in Table 1, the very large oxygen-induced relaxation rate at Ser-22:HA has a large error because the vector of delay times does not sample the relaxation well for rates over 5 s−1; however, both the longitudinal and transverse relaxation rates are large and the effects of paramagnetic rate enhancement are significant.
Surface cleft between α2 and β5
Three protons (Asn-27:HB1, Cys-95:HB1, Ala-96:HB1) situated in the surface cleft between α2 and the antiparallel β-strand (β5) have large oxygen-induced relaxation rates and are highlighted in yellow and superimposed on the ribbon structure of RNase A in Fig. 3 e. In a van der Waals space-filling model of the protein, Asn-27:HB1 and Ala-96:HB1 are partially buried in the protein structure and are located near the mouth of the inaccessibly minor hydrophobic core. Shown in the molecular contour surface, two of the protons (Asn-27:HB1 and Ala-96:HB1) are located at a surface pocket (indicated by an arrow in Fig. 4 c), and are surrounded by the polar or charged side chains of Asn-27, Lys-31, and Asn-34. Although the static accessibility calculations suggest little (0.4 Å2 for Asn-27:HB1) or no (Cys-95:HB1, Ala-96:HB1) solvent accessible area at these proton sites, surface side-chain fluctuations, especially in Lys-31, or perhaps internal protein motion between α2 and β4 may provide oxygen access. The structural topology, in addition to favorable interactions between the oxygen and the minor hydrophobic core residues at Met-30 and Tyr-79, may increase the oxygen affinity and the induced proton relaxation for Asn-27:HB1, Cys-95:HB1, Ala-96:HB1.
β2-β3 hairpin
Additional protons (Thr-70:HB, Thr-70:HG1, Asn-71:HB2, Cys-72:HB2) with large oxygen-induced relaxation rates are found in the β2-β3 hairpin region and colored yellow in Fig. 3 f. Except for proton Cys-72:HB2, all the other protons (Thr-70:HB, Thr-70:HG1, and Asn-71:HB2) are located at the edge of the third hydrophobic core, which is comprised primarily of the 62-72 hairpin (Val-63, Cys-65, Cys-72) and the C-terminal hairpin (Ile-107, Ala-122, Val-124). The Thr-70:HB, Thr-70:HG1, and Asn-71:HB2 protons have large computed accessibilities, which are summarized in Table 1 . Additional hydrogen-bond opportunities at the polar Thr-70 and the Asn-71 side chains may further contribute to the energetic heterogeneity at the protein molecular surface as measured by oxygen accessibility.
Cys-72:HB2, on the other hand, is located in the third hydrophobic core, a buried position at the bottom of a surface pocket indicated by the blue arrow in the molecular surface in Fig. 4 a. Unlike the surface pocket near Asn-27:HB1 and Ala-96:HA, which is surrounded by polar or charged residues, the surface pocket at Cys-72:HB2 is mostly surrounded by hydrophobic residues including Ile-107, Val-63, and Val-64.
Turn at β6-β7 hairpin
The Asn-113:HA relaxation rate is large although this site is somewhat isolated from all the other proton sites with large oxygen-induced rates (Fig. 3 e). The computed surface accessible area shows that the Asn-113:HA proton is inaccessible to solvent molecules. However, the unusually large B-factor (22.94 Å2) (Wlodawer et al., 1988) at this proton site suggests structural flexibility that may permit close oxygen contact. Structurally, Asn-113:HA is situated before the cis-peptide bond between residues Asn-113 and Pro-114, and is located at the sharp turn of the β6-β7 hairpin. This turn at the β6-β7 hairpin appears to provide a natural hook that may create a binding site for oxygen, which is reflected in the high oxygen-induced proton relaxation rate.
The high paramagnetic relaxation rate constants found for a number of interior proton sites require that the protein structure move to permit oxygen access. Molecular dynamics simulations of ribonuclease A have suggested that there are rapid breathing motions involving relative motions of the two β-sheet regions as well as three helices (Merlino et al. 2002, 2003). Some of the simulated displacements are reported to be as large as 9 Å. These simulations are consistent with this finding that the interior protein protons in the hinge region marked in red in Fig. 2, are accessible to oxygen. The simulations also suggest flexibility in the disulfide loop region (65-72) where we see significant oxygen accessibility to the apparently buried cystine protons. In comparing these results, it is important to point out that the NMR relaxation measurement of accessibility is a time and ensemble average where the averaging period is of the order of the relaxation time or ∼100 ms. Because the measurement is strongly sensitive to distance, the modification of accessibility to one region of the protein may change the relaxation rates in other or nonflexible regions because the effective distance of closest approach of the interacting electron and observed nuclear magnetic moments may be significantly modified.
Low rates
Small oxygen-induced relaxation rates are defined as ΔR1 < 0.6 s−1 and are summarized in Table 2. Mapping protons that have paramagnetic relaxation rates ΔR1 < 0.6 s−1 onto a ribbon representation, we find that except for 75:HB1, which is on the surface, these protons are located deeply inside the two lobes of the RNase A structure, on either side of the major catalytic cleft. All other protons are buried with an average distance to the accessible surface ∼6.05 ± 1.19 Å. At this depth, much smaller paramagnetic relaxation rates are expected because of the large distance of closest approach between the electron and nuclear moment detected. We note that a low rate may result if any solvent or cosolute prevents free access of the diffusing oxygen to the site. The obstacles may include solvent, counterions, and the protein itself. The protein concentration used in these experiments is sufficiently large that some aggregation is possible if not likely (Liu et al., 1998, 2002; Gotte et al., 1999). Exclusion of oxygen or other freely diffusing paramagnet provides a simple method for defining the protein-protein interface; however, in this case, the low rates are very largely restricted to the interior of the protein as noted. The surface positions with particularly low oxygen induce relaxation rates that do not correlate with the reported interface regions of the dimers or trimers, which suggests that the aggregation does not dominate the observations under the conditions used here or that the protein-protein interfaces are accessible to oxygen over the long timescale of the experiment.
TABLE 2.
Small relaxation rate constant summary
| H observed | H crossed | Residue type | ΔR1 (s−1) | Secondary structure | b (Å) | AAREA (Å2) |
|---|---|---|---|---|---|---|
| Central β-strand (Fig. 3 g) | ||||||
| 11:HA | HB1 | Gln | 0.59(0.18) | α1 | 6.02 | 0 |
| 26:HA | HB1 | Cys | 0.57(0.15) | α2 | 6.34 | 0 |
| 35:HD11 | HG | Leu | 0.24(0.17) | 5.47 | 0 | |
| 46:HA | HB1 | Phe | 0.37(0.33) | β1 | 6.87 | 0 |
| 46:HB1 | HA | Phe | 0.44(0.16) | β1 | 7.85 | 0 |
| 46:HD1 | HE1 | Phe | 0.59(0.1) | β1 | 6.20 | 0 |
| 46:HE1 | HZ | Phe | 0.54(0.1) | β1 | 6.31 | 0 |
| 46:HZ | HE1 | Phe | 0.46(0.09) | β1 | 7.26 | 0 |
| 84:HB1 | HA | Cys | 0.45(0.13) | β4 | 5.98 | 0 |
| β3 and β6 (Fig. 3 h) | ||||||
| 73:HB1 | HB2 | Tyr | 0.45(0.18) | β3 | 6.58 | 0 |
| 74:HB1 | HA | Gln | 0.53(0.32) | β3 | 3.67 | 0 |
| 107:HG11 | HB | Ile | 0.59(0.13) | β6 | 4.15 | 0 |
| 108:HG21 | HB | Val | 0.54(0.17) | β6 | 7.73 | 0 |
| 109:HA | HB1 | Ala | 0.59(0.11) | β6 | 4.95 | 0 |
| 110:HB2 | HB1 | Cys | 0.55(0.14) | β6 | 5.39 | 0 |
| Ser-75: HB1 (Fig. 3 h) | ||||||
| 75:HB1 | HA | Ser | 0.47(0.1) | 2.61 | 0 | |
Central β-strand
Fig. 3 g shows the nine protons (Gln-11:HA, Cys-26:HA, Leu-35:HD11, Phe-46:HA, Phe-46:HB1, Phe-46:HD1, Phe-46:HE1, Phe-46:HZ, Cys-84:HB1) located near the central β-strand that have small paramagnetic relaxation contributions. These protons are buried and surrounded by α1, α2, and the antiparallel β-strands (β1 and β4), which constitute the first structural lobe of RNase A. Besides Gln-11:HA, which is deeply buried, protons of Cys-26, Len-35, Phe-46, and Cys-84 are part of the minor hydrophobic core associated with the interior Cys-26–Cys-84 disulfide bond (indicated in the yellow molecular bond in Fig. 3 g). None of the protons measured with high rates are found in this minor hydrophobic core.
β3 and β6
Six protons (Tyr-73:HB1, Gln-74:HB1, Ile-107:HG11, Val-108:HG21, Ala-109:HA, Cys-110:HB2), which are buried near the center (β3, β6) of the four-stranded (β2-β3, β6-β7) antiparallel β-sheet, have low oxygen-induced relaxation rates. Tyr-73:HB1 and Ile-107:HG11 are part of the third hydrophobic core where Gln-74:HB1 is structurally connected to Val-108:HG21 and Cys-110:HB2, which are in the middle of β4 that forms an interior side of the major hydrophobic core. Ala-109:HA on the other hand, is between β3 and β6 and is adjacent to both the major and the third hydrophobic cores. The protons in the first lobe are more deeply buried than the protons in this second lobe as measured by their average distance of closest approach, <b>, to a surface molecular oxygen computed from the crystal structure, i.e., <b> = 6.48 ± 0.73 Å vs. <b> = 5.41 ± 1.52 Å. Correspondingly, both the major hydrophobic and the third hydrophobic core have large solvent-exposed areas indicated by the static surface accessibility calculations, whereas the minor hydrophobic core comprising the interior of the first lobe is largely inaccessible.
Gln-74:HB1 and Ile-107:HG11 are located at a surface crevice surrounded by hydrophobic residues. An extensive interpenetrating water network is reported in the region (Savage and Wlodawer, 1986; Wlodawer et al., 1988) shown in Fig. 5 a. If water molecules consistently occupy surface sites, the minimal intermoment distances between the surface 1H and the oxygen magnetic moment increase by ∼3 Å, which will decrease the oxygen-induced spin relaxation rate constant by 90%. Thus, the small oxygen induced relaxation rates at Gln-74:HB1 and Ile-107:HG11 are consistent with the steric exclusion by water molecules as suggested by the x-ray data.
FIGURE 5.
Relaxed-eyed steric pairs of the van der Waals surface of ribonuclease A. Those protein protons for which the oxygen-induced relaxation rate constant is <0.6 s−1 are colored blue. (a) Gln-74:HB1 and Ile-107:HG11 (arrows) are located at a surface crevice surrounded by hydrophobic residues. An interpenetrating water network is reported in the neighborhood (Savage and Wlodawer, 1986; Wlodawer et al., 1988). (b) Ser-75:HB1 is the only surface proton with low oxygen-induced relaxation rate. Two of the invariant water molecules (W216 and W208) are found nearby (Sadasivan et al., 1998), which is consistent with the steric exclusion of oxygen caused by hydration effects.
Ser-75:HB1
Ser-75:HB1 is remarkable in that it is a surface proton that has a low paramagnetic relaxation rate contribution, ΔR1 = 0.47 ± 0.1 s−1. This proton is located in a surface cleft created by the α3 and the linker region connecting β3 and β4. Comparing nine x-ray structures of RNase A, Sadasivan et al. (1998) found 14 invariant water molecules. Two of these invariant water molecules are in the vicinity of Ser-75:HB1. Wat-216, which forms hydrogen bonds with Gln-60:OE1, Tyr-76:N, and Ser-77:N, is only 2.55 Å from Ser-75:HB1, and Wat-208, which is 4.75 Å from Ser-75:HB1, makes hydrogen bonds with Asp-53:O and Gln-60:NE2. The locations for Ser-75:HB1 as well as for Wat-216 and Wat-208 (not drawn to scale) are shown in Fig. 5 b. Although multiple side-chain conformations are found for residue Ser-77 near Ser-75:HB1 (Wlodawer et al., 1988), the χ1 rotation is not sufficient to bury Ser-75:HB1. We cannot exclude the possibility that some cooperative protein structural fluctuations may reduce or increase the solvent exposure of Ser-75:HB1; however, the fact that these water molecules are conserved in many x-ray structural models suggests local energy minima near Ser-75:HB1 for the water-protein interactions. Although we cannot rule out the effects of protein aggregation, the small paramagnetic relaxation rate observed at Ser-75:HB1 is consistent with steric exclusion of oxygen caused by these hydration effects.
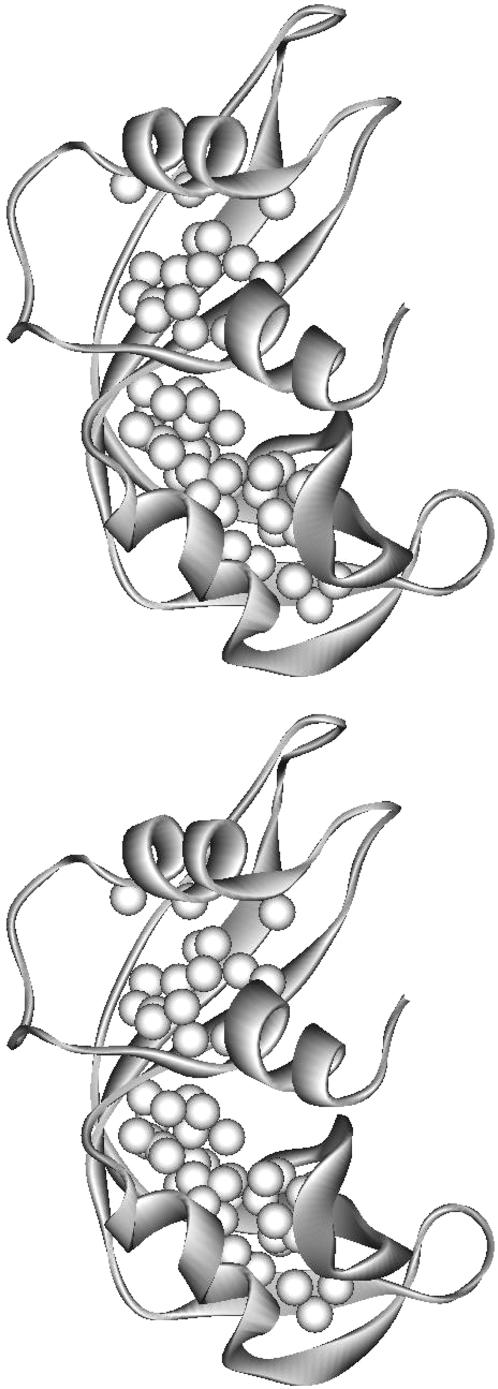
The interior protons at depths of >6 Å from the intermolecular accessible surface should have very low paramagnetic relaxation rates because of the large intermoment distances between these proton sites and the exterior oxygen. Fig. 6 shows a ribbon representation of RNase A with all the interior protons measured at depths >6 Å indicated as white spheres. These interior protons are not located only in the bilobe centers where protons have low oxygen-induced relaxation rates (Fig. 3, g and h). The protons in the space surrounded by α2, the end of central β-strand, the beginning of the β4, and the C-terminal end of the β5-β6 hairpin are also as deeply buried. However, moderate paramagnetic relaxation rates are measured for these interior protons, which implies modest dipolar coupling between the oxygen and the interior protons in this hinge region. These measurements indicate that the oxygen-induced relaxation rates are not a simple function of the proton depth calculated from the x-ray protein structural model. Protein structural fluctuations permitting transient penetration in this region are consistent with the molecular dynamics simulations, which indicate that breathing motions open the active site region (Merlino et al., 2002). As mentioned earlier, proton-proton cross relaxation or spin-diffusion may enter the observations and is increasingly important for the smallest paramagnetic relaxation rate enhancements.
FIGURE 6.
Interior protons relaxation rate differences were measured at depths >6 Å and are indicated as white spheres in the ribbon representation of RNase A. Comparison with Fig. 3, g and h, shows that oxygen-induced relaxation efficiency is not simply correlated with the depth of an interior proton.
CONCLUSION
In conclusion, we find that oxygen has weak associative interactions at many surface crevices. A surface groove between the major hairpin, a surface cleft between α2 and the loop region connecting β4-β5 hairpin, a surface pocket near the loop region connecting α1 and α2, and a surface pocket at the exposed surface of third hydrophobic core, are observed with significantly enhanced oxygen contact. These surface crevices are generally located between secondary structures. Although not all protons with large paramagnetic relaxation contributions have large accessible areas for oxygen contact as suggested by calculations based on the x-ray structural model, protein structural fluctuations apparently permit oxygen access. Polar residues are often found near protons with high paramagnetic relaxation rates.
Oxygen is also found to penetrate part of the major hydrophobic core interior. However, oxygen associations do not correlate simply with hydrophobicity indices. We find both high and low oxygen-induced relaxation rates at the hydrophobic cores of the protein. The high oxygen-induced relaxation rates found in the hydrophobic interior pocket are correlated with the presence of nearby cavities. The presence of the cavities suggests that sufficient void space is provided for protein structural fluctuations to allow oxygen penetration. When the protein is densely packed as in the inner hydrophobic centers of the two structural lobes, weak paramagnetic contributions to the proton relaxation rates are found. We note that the oxygen induced relaxation efficiency is not simply correlated with the depth of a buried proton as computed from x-ray data. The absence of small paramagnetic relaxation rates at the interior of the hinge region indicates that there is reasonably efficient oxygen access to the hinge area.
The ineffective oxygen-protein interaction at a surface proton site near Ser-75:HB1 is consistent with x-ray data that indicates the presence of persistent water molecule occupancy that reduces oxygen-protein intermolecular contact. However, large oxygen-induced relaxations are observed at T70:HG1 and T70:HB where the hydroxyl groups of the Thr residue provide hydrogen-bond opportunities. These observations indicate that the subtle differences in the enthalpic and entropic factors governing the local solvation of protein residues may modulate the intermolecular accessibility.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
For helpful discussions, we thank David Cafiso, Rodney Biltonen, Jack Freed, Alfred Redfield, Herald Scheraga, Irwin Kuntz, John Buschweller, David LeMaster, and Griselda Hernandez.
The authors gratefully acknowledge financial support from the University of Virginia and the National Institutes of Health (5R01-GM34541).
References
- Altenbach, C., S. L. Flitsch, H. G. Khorana, and W. L. Hubbell. 1989a. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 28:7806–7812. [DOI] [PubMed] [Google Scholar]
- Altenbach, C., W. Froncisz, J. S. Hyde, and W. L. Hubbell. 1989b. Conformation of spin-labeled melittin at membrane surfaces investigated by pulse saturation recovery and continuous wave power saturation electron paramagnetic resonance. Biophys. J. 56:1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloembergen, N., E. M. Purcell, and R. V. Pound. 1948. Relaxation effects in nuclear magnetic resonance absorption. Physical Review. 73:679–712. [Google Scholar]
- Bryant, R. G. 1996. The dynamics of water-protein interactions. Annu. Rev. Biophys. Biomol. Struct. 25:29–53. [DOI] [PubMed] [Google Scholar]
- Calhoun, D. B., J. M. Vanderkooi, and S. W. Englander. 1983a. Penetration of small molecules into proteins studied by quenching of phosphorescence and fluorescence. Biochemistry. 22:1533–1539. [DOI] [PubMed] [Google Scholar]
- Calhoun, D. B., J. M. Vanderkooi, G. R. Holtom, and S. W. Englander. 1986. Protein fluorescence quenching by small molecules: protein penetration versus solvent exposure. Proteins. 1:109–115. [DOI] [PubMed] [Google Scholar]
- Calhoun, D. B., J. M. Vanderkooi, G. V. Woodrow III, and S. W. Englander. 1983b. Penetration of dioxygen into proteins studied by quenching of phosphorescence and fluorescence. Biochemistry. 22:1526–1532. [DOI] [PubMed] [Google Scholar]
- Carrero, J., D. M. Jameson, and E. Gratton. 1995. Oxygen penetration and diffusion into myoglobin revealed by quenching of zincprotoporphyrin IX fluorescence. Biophys. Chem. 54:143–154. [DOI] [PubMed] [Google Scholar]
- Case, D. A., and M. Karplus. 1979. Dynamics of ligand binding to heme proteins. J. Mol. Biol. 132:343–368. [DOI] [PubMed] [Google Scholar]
- Cocco, M. J., and J. T. Lecomte. 1994. The native state of apomyoglobin described by proton NMR spectroscopy: interaction with the paramagnetic probe hytempo and the fluorescent dye ans. Protein Sci. 3:267–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M. E., T. K. Hitchens, and R. G. Bryant. 2000. Comparisons of pressure and temperature activation parameters for amide hydrogen exchange in t4 lysozyme. Biochemistry. 39:248–254. [DOI] [PubMed] [Google Scholar]
- Esposito, G., A. M. Lesk, H. Molinari, A. Motta, N. Niccolai, and A. Pastore. 1992. Probing protein structure by solvent perturbation of nuclear magnetic resonance spectra. Nuclear magnetic resonance spectral editing and topological mapping in proteins by paramagnetic relaxation filtering. J. Mol. Biol. 224:659–670. [DOI] [PubMed] [Google Scholar]
- Feher, V. A., E. P. Baldwin, and F. W. Dahlquist. 1996. Access of ligands to cavities within the core of a protein is rapid. Nat. Struct. Biol. 3:516–521. [DOI] [PubMed] [Google Scholar]
- Freed, J. H. 1978. Dynamic effects of pair correlation functions on spin relaxation by translational diffusion in liquids. II. Finite jumps and independent t1 processes. J. Chem. Phys. 68:4034–4037. [Google Scholar]
- Gotte, G., M. Bertoldi, and M. Libonati. 1999. Structural versatility of bovine ribonuclease A. Distinct conformers of trimeric and tetrameric aggregates of the enzyme. Eur. J. Biochem. 265:680–687. [DOI] [PubMed] [Google Scholar]
- Gratton, E., D. M. Jameson, G. Weber, and B. Alpert. 1984. A model of dynamic quenching of fluorescence in globular proteins. Biophys. J. 45:789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle, B., and V. P. Denisov. 1995. A new view of water dynamics in immobilized proteins. Biophys. J. 69:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., C. L. Teng, R. G. Bryant, and D. M. LeMaster. 2002. O2 penetration and proton burial depth in proteins: applicability to fold family recognition. J. Am. Chem. Soc. 124:4463–4472. [DOI] [PubMed] [Google Scholar]
- Hitchens, T. K., and R. G. Bryant. 1998. Pressure dependence of amide hydrogen-deuterium exchange rates for individual sites in t4 lysozyme. Biochemistry. 37:5878–5887. [DOI] [PubMed] [Google Scholar]
- Hwang, L., and J. H. Freed. 1975. Dynamic effects of pair correlation functions on spin relaxation by translational diffusion in liquids. J. Chem. Phys. 63:4017–4025. [Google Scholar]
- Kim, K. S., J. A. Fuchs, and C. K. Woodward. 1993. Hydrogen exchange identifies native-state motional domains important in protein folding. Biochemistry. 32:9600–9608. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J. R., and G. Weber. 1973a. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 12:4161–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz, J. R., and G. Weber. 1973b. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 12:4171–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379–400. [DOI] [PubMed] [Google Scholar]
- Li, R., and C. Woodward. 1999. The hydrogen exchange core and protein folding. Protein Sci. 8:1571–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., G. Gotte, M. Libonati, and D. Eisenberg. 2002. Structures of the two 3d domain swapped rnase a trimers. Protein Sci. 11:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., P. J. Hart, M. P. Schlunegger, and D. Eisenberg. 1998. The crystal structure of a 3d domain-swapped dimer of rnase a at a 2.1 a resolutioin. Proc. Natl. Acad. Sci. USA. 95:3437–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor, S. E., H. S. McHaourab, and D. L. Farrens. 1999. Determination of protein secondary structure and solvent accessibility using site-directed fluorescence labeling. Studies of t4 lysozyme using the fluorescent probe monobromobimane. Biochemistry. 38:16383–16393. [DOI] [PubMed] [Google Scholar]
- McNaughton, L., G. Hernandez, and D. M. LeMaster. 2003. Equilibrium o2 distribution in zn2+-protoporphyrin ix deoxymyoglobin mimic: application to oxygen migration pathway analysis. J. Am. Chem. Soc. 125:3813–3820. [DOI] [PubMed] [Google Scholar]
- Merlino, A., L. Vitagliano, M. A. Ceruso, A. Di Nola, and L. Mazzarella. 2002. Global and local motions in ribonuclease A: a molecular dynamics study. Biopolymers. 65:274–283. [DOI] [PubMed] [Google Scholar]
- Merlino, A., L. Vitagliano, M. A. Ceruso, and L. Mazzarella. 2003. Subtle functional collective motions in pancreatic-like ribonucleases: from ribonuclease A to angiogenin. Proteins. 53:101–110. [DOI] [PubMed] [Google Scholar]
- Molinari, H., G. Esposito, L. Ragona, M. Pegna, N. Niccolai, R. M. Brunne, A. M. Lesk, and L. Zetta. 1997. Probing protein structure by solvent perturbation of NMR spectra: the surface accessibility of bovine pancreatic trypsin inhibitor. Biophys. J. 73:382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccolai, N., A. Ciutti, O. Spiga, M. Scarselli, A. Bernini, L. Bracci, D. Di Maro, C. Dalvit, H. Molinari, G. Esposito, and P. A. Temussi. 2001a. NMR studies of protein surface accessibility. J. Biol. Chem. 276:42455–42461. [DOI] [PubMed] [Google Scholar]
- Niccolai, N., G. Valensin, C. Rossi, and W. Gibbons. 1982. The stereochemistry and dynamics of natural products and biopolymers from proton relaxation spectroscopy: spin label delineation of inner and outer protons of gramicidin s including hydrogen bonds. J. Amer. Chem. Soc. 104:1534–1537. [Google Scholar]
- Niccolai, N., C. Rossi, G. Valensin, P. Mascagni, and W. A. Gibbons. 1984. An investigation of the mechanisms of nitroxide-induced proton relaxation enhancements in biopolymers. J. Phys. Chem. 88:5689–5692. [Google Scholar]
- Niccolai, N., R. Spadaccini, M. Scarselli, A. Bernini, O. Crescenzi, O. Spiga, A. Ciutti, D. Di Maro, L. Bracci, C. Dalvit, and P. A. Temussi. 2001b. Probing the surface of a sweet protein: NMR study of MNEI with a paramagnetic probe. Protein Sci. 10:1498–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting, G., E. Liepinsh, B. Halle, and U. Frey. 1997. NMR identification of hydrophobic cavities with low water occupancies in protein structures using small gas molecules. Nat. Struct. Biol. 4:396–404. [DOI] [PubMed] [Google Scholar]
- Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantini, U., O. W. Sorensen, and R. R. Ernst. 1982. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104:6800–6801. [Google Scholar]
- Pintacuda, G., and G. Otting. 2002. Identification of protein surfaces by NMR measurements of a paramagnetic gd(III) chelate. J. Am. Chem. Soc. 124:372–373. [DOI] [PubMed] [Google Scholar]
- Polnaszek, C. F., and R. G. Bryant. 1984. Nitroxide radical induced solvent proton relaxation: measurement of localized translational diffusion. J. Chem. Phys. 81:4038–4045. [Google Scholar]
- Prosser, R. S., P. A. Luchette, and P. W. Westerman. 2000. Using O2 to probe membrane immersion depth by 19F NMR. Proc. Natl. Acad. Sci. USA. 97:9967–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, R. S., P. A. Luchette, P. W. Westerman, A. Rozek, and R. E. Hancock. 2001. Determination of membrane immersion depth with O(2): a high-pressure (19)F NMR study. Biophys. J. 80:1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance, M., O. W. Sorensen, G. Bodenhausen, G. Wagner, R. R. Ernst, and K. Wuthrich. 1983. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117:479–485. [DOI] [PubMed] [Google Scholar]
- Rasmssen, B. F., A. M. Stock, D. Ringe, and G. A. Petsko. 1992. Crystalline ribonuclease A loses function below the dynamical transition at 220 k. Nature. 357:423–424. [DOI] [PubMed] [Google Scholar]
- Rico, M., M. Bruix, J. Santoro, C. Gonzalez, J. L. Neira, J. L. Nieto, and J. Herranz. 1989. Sequential 1H-NMR assignment and solution structure of bovine pancreatic ribonuclease A. Eur. J. Biochem. 183:623–638. [DOI] [PubMed] [Google Scholar]
- Robertson, A. D., E. O. Purisima, M. A. Eastman, and H. A. Scheraga. 1989. Proton NMR assignments and regular backbone structure of bovine pancreatic ribonuclease A in aqueous solution. Biochemistry. 28:5930–5938. [DOI] [PubMed] [Google Scholar]
- Sadasivan, C., H. G. Nagendra, and M. Vijayan. 1998. Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant. Acta Crystallogr. D Biol. Crystallogr. 54:1343–1352. [DOI] [PubMed] [Google Scholar]
- Santoro, J., C. Gonzalez, M. Bruix, J. L. Neira, J. L. Nieto, J. Herranz, and M. Rico. 1993. High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 229:722–734. [DOI] [PubMed] [Google Scholar]
- Savage, H., and A. Wlodawer. 1986. Determination of water structure around biomolecules using x-ray and neutron diffraction methods. Methods Enzymol. 127:162–183. [DOI] [PubMed] [Google Scholar]
- Scheraga, H. A., W. J. Wedemeyer, and E. Welker. 2001. Bovine pancreatic ribonuclease A: oxidative and conformational folding studies. Methods Enzymol. 341:189–221. [DOI] [PubMed] [Google Scholar]
- Solomon, I. 1955. Relaxation processes in a system of two spins. Physcial Review. 99:559–565. [Google Scholar]
- Teng, C. L., and R. G. Bryant. 2000. Experimental measurement of nonuniform dioxygen accessibility to ribonuclease A surface and interior. J. Am. Chem. Soc. 122:2667–2668. [Google Scholar]
- Teng, C. L., H. Hong, S. Kiihne, and R. G. Bryant. 2001. Molecular oxygen spin-lattice relaxation in solutions measured by proton magnetic relaxation dispersion. J. Magn. Reson. 148:31–34. [DOI] [PubMed] [Google Scholar]
- Victor, K., and D. S. Cafiso. 1998. Structure and position of the n-terminal membrane-binding domain of pp60src at the membrane interface. Biochemistry. 37:3402–3410. [DOI] [PubMed] [Google Scholar]
- Wang, A., A. D. Robertson, and D. W. Bolen. 1995. Effects of a naturally occurring compatible osmolyte on the internal dynamics of ribonuclease A. Biochemistry. 34:15096–15104. [DOI] [PubMed] [Google Scholar]
- Wilhelm, E., R. Battino, and R. J. Wilcock. 1977. Low-pressure solubility of gasses in liquid water. Chem. Rev. 77:219–262. [Google Scholar]
- Wlodawer, A., L. A. Svensson, L. Sjolin, and G. L. Gilliland. 1988. Structure of phosphate-free ribonuclease A refined at 1.26 A. Biochemistry. 27:2705–2717. [DOI] [PubMed] [Google Scholar]
- Woodward, C., I. Simon, and E. Tuchsen. 1982. Hydrogen exchange and the dynamic structure of proteins. Mol. Cell. Biochem. 48:135–160. [DOI] [PubMed] [Google Scholar]
- Woodward, C. K., and A. Rosenberg. 1971. Studies of hydrogen exchange in proteins. V. The correlation of ribonuclease exchange kinetics with the temperature-induced transition. J. Biol. Chem. 246:4105–4113. [PubMed] [Google Scholar]
- Zhou, N., P. Mascagni, W. Gibbons, N. Niccolai, C. Rossi, and H. Wyssbrod. 1985. Confirmation of the solution structure of tyrocidine a using perturbation of proton relaxation rates by nitroxide spin labels. J. Chem. Soc. Perkin Trans. 2:581–587. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.