Abstract
Cell surface macromolecules such as receptors and ion channels serve as the interface link between the cytoplasm and the extracellular region. Their density, distribution, and clustering are key spatial features influencing effective and proper physical and biochemical cellular responses to many regulatory signals. In this study, the effect of plasma-membrane receptor clustering on local cell mechanics was obtained from maps of interaction forces between antibody-conjugated atomic force microscope tips and a specific receptor, a vascular endothelial growth factor (VEGF) receptor. The technique allows simultaneous measurement of the real-time motion of specific macromolecules and their effect on local rheological properties like elasticity. The clustering was stimulated by online additions of VEGF, or antibody against VEGF receptors. VEGF receptors are found to concentrate toward the cell boundaries and cluster rapidly after the online additions commence. Elasticity of regions under the clusters is found to change remarkably, with order-of-magnitude stiffness reductions and fluidity increases. The local stiffness reductions are nearly proportional to receptor density and, being concentrated near the cell edges, provide a mechanism for cell growth and angiogenesis.
INTRODUCTION
Receptors and ion channels transmit regulatory information from the outside environment to the cell interior and play a crucial role in maintaining metabolic homeostasis and in the sustenance of organisms (Pawson, 1995; Antonova et al., 2001; Horio et al., 1997; Sugiyama et al., 1997; Jefford and Dubreuil, 2000; Rameh and Cantley, 1999; Crouch et al., 2001). The molecular basis of receptor/channel action is being studied extensively using biochemical strategies, but little is known about the spatial localization, density, distribution, and molecular structure involved, although these spatial features are thought to exert localized control over cell function. For example, the clustering of growth factor receptors, e.g., vascular endothelial growth factor receptor (VEGFR) is implicated in endothelial cell growth and migration (Thomas, 1996). To obtain such direct structural information it is crucial to develop and apply techniques for molecular-resolution imaging of cellular specimens in their native hydrated states.
Several new scanning probe techniques are being developed to map membrane macromolecules. Of these, atomic force microscopy (AFM) allows molecular resolution imaging in aqueous media (Lal and John, 1994) and the study of intermolecular interactions using functionalized AFM probes (Zhang et al., 2002; Baumgartner et al., 2000; Yuan et al., 2000; Boland and Ratner, 1995; Dammer et al., 1996; Hinterdorfer et al., 1996). Single-molecule force spectroscopy was previously used to examine cell-cell adhesion properties (Benoit et al., 2000). Although such an approach would be useful for mapping overall interactions between small cells, its relevance to mapping regional distribution of macromolecules in the cell plasma membrane is uncertain. Another approach maps the local ion channel currents, from which the channel density and distribution are derived (Korchev et al., 2000). Application of this approach for mapping membrane receptors without any channel-like activity is limited. However, recently modified commercial AFMs, linked to a confocal microscope, have been successfully used to measure and map adhesion forces between ligands and receptors on the surface of living cells (Horton et al., 2002; Aileen and Moy, 2000).
Here we apply a similar method that uses AFM force-volume mapping (Quist et al., 2000; Rhee et al., 1998) to identify and map regional distribution as well as ligand- or antibody-induced real-time clustering of receptors on the cell surface. The approach allows simultaneous imaging of the resultant changes in micromechanical properties and cytoskeletal reorganization with nanoscale resolution (Quist et al., 2000; Shroff et al., 1995; Parbhu et al., 1999).
As an appropriate physiological sample, we have examined spatial distribution of VEGFR, the receptor for vascular endothelial growth factor (VEGF). VEGF is an important angiogenic factor in human and animal tissues. VEGF acts selectively on vascular endothelial cells to increase their permeability to circulating macromolecules and stimulate endothelial cell growth (Thomas, 1996; Wang et al., 2001). VEGF is a homodimeric glycoprotein that interacts with its receptors in the plasma membrane of vascular endothelial cells. Three cell membrane receptors tyrosine kinases, Flt (also designated as VEGF-R1), Flk-1 (also designated KDR or VEGF-R2), and Flt-4 are involved in endothelial cell growth (Meyer et al., 1999; Neufeld et al., 1999). Two members of this receptor class, Flt and Flk-1, have been shown to represent high affinity receptors for vascular endothelial growth factor. At present, very little is known about the molecular structure, distribution, and clustering of VEGF receptors in endothelial cell plasma membrane. Moreover, the VEGFR clustering-induced short-term and dynamic cellular biophysical changes are not well understood.
We have used AFM tips conjugated with anti-Flk-1 antibody for semiquantitative measurements of binding (or unbinding) forces between anti-Flk-1 antibody and VEGF-R2 (Flk-1). Measurements were made both in vitro as well as in live (in vivo) endothelial cells. Distribution and density of VEGFRs were mapped in the cell plasma membrane using the receptor-antibody interaction and force-volume mapping technique. Antibody and ligand induced real-time clustering and redistribution of VEGFRs. VEGFR clustering altered cell local elastic properties significantly.
MATERIALS AND METHODS
Chemicals and reagents
Silane (3-aminopropylmethyldiethoxysilane) was purchased from Sigma-Aldrich (St. Louis, MO). VEGF was purchased from PeproTech (Rocky Hill, NJ). Flk-1 (a VEGFR2), goat affinity-purified, site-directed, polyclonal anti-Flk-1 antibody, and the blocking peptide (the peptide against which the antibody was raised) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
Bovine aortic endothelial cells (KOM-1 cell line) were cultured on sterile plastic petri dishes as described (Bhatia et al., 2000), in Dulbecco's modified Eagle's medium containing glucose, 10 mM HEPES, 2 mM glutamine, 100 units/ml penicillin, 100 mg/ml streptomycin, and 10% heat-inactivated calf serum (Life Technologies, Rockville, MD) (Yauch et al., 1997). Cells were grown in an incubator maintained at 37°C temperature and 5% CO2.
Immunofluorescence labeling
Donkey anti-rabbit-IgG conjugated with cy-3 was purchased from Jackson Laboratory (Bar Harbor, ME). Cells grown on glass coverslips were fixed with 4% paraformaldehyde for 10 min, and washed with PBS, and PBS containing 3% bovine serum albumin and 1% donkey serum, to minimize any nonspecific binding. Cells were then incubated with antibody against Flk-1 (0.1 μg/ml) in PBS containing 3% BSA and 1% donkey serum for 1 h. After washing, the sample was incubated, for 1 h, with a cy-3 conjugated donkey anti-rabbit antibody (1:400 dilutions) under the same condition as for the primary antibody. Immunofluorescence images were captured with a Bio-Rad MRC 1024 laser confocal microscope (Bio-Rad Laboratories, Hercules, CA) using a 60× Nikon Plan Apo oil-emersion lens with 1.4 NA.
Atomic force microscopy and preparation of functionalized AFM tips
AFM images and force measurements were recorded with either a commercial multimode AFM (Digital Instruments Nanoscope III, Veeco Instruments, Santa Barbara, CA) or a bioscope AFM with an integrated inverted light microscope (a prototype of Digital Instruments Bioscope) using the NanoScope III software (Version 4.23R2; Digital Instruments) as described (Quist et al., 2000).
Standard commercially available, 200-μm long Si3N4 cantilevers, with integrated tips (Digital Instruments) and a nominal spring constant, k, of 0.06 N/m were used. The approximate nominal spring constant value was verified by measuring the spring constants on five cantilevers from the same batch with the thermal fluctuation method (Hutter and Bechhoefer, 1993) in both air and fluid. Our measurements yielded similar results in air as in a fluid. The measured value was k = 0.063 ± 0.003 N/m, including a 16% correction due to the calibration of the amplitude in static mode. Due to systematic errors, we estimated the uncertainty to be at least 15–20%. The cantilever tips used in our study were conjugated with antibody. The spring constant of silanated and antibody conjugated tips were k = 0.060 ± 0.005 (n = 5). Such correspondence is consistent with previous studies by Cleveland and the Hansma group (Cleveland and Manne, 1993) which indicated that a small increase in the tip mass (as would occur when the antibody is conjugated to the tip) does not change the spring constant significantly. Moreover, whenever feasible, we used the same conjugated tip for experiments involving internal control—for example, in mapping interactions between antibody and receptor alone, adding online inhibitor of their interactions (e.g., blocking peptide). Also, although semiquantitative estimate of interaction forces are described, the main emphasis is on the relative change as a function of specific perturbations.
The cantilever tips were functionalized by silanization with 3-aminopropylmethyl-diethoxysilane by vapor deposition or by immersion in a solution of 2% silane in toluene. A drop of 2 μg/ml of anti-Flk-1 IgG was then placed on the silanized tip for 10 min, to adsorb to the functionalized tip. The antibody-conjugated tips were then washed thoroughly with PBS and water to remove loosely attached antibodies. These tips were used immediately, without being dried, for obtaining the interaction-force curves and the force-volume maps.
Sample preparation and force measurements
In vitro force spectroscopy
The mica surface was silanized and imaged to ensure the uniformity of the silane layer. Flk-1 receptor (10 μg/ml; 10–20 μl vol) was adsorbed on the silanized mica for 10 min at room temperature. Unattached or loosely attached receptors were removed by washing the mica surface thoroughly with water. AFM was used to image the presence and the distribution of Flk-1 receptors in fluid. Once a low concentration of individual VEGF receptors were identified in an AFM image, the tip-receptor interaction force curves were recorded after positioning the tip over a receptor. A series of force curves were recorded in MilliQ water or PBS, in vitro, between the anti-Flk-1-antibody conjugated to a functionalized tip and the Flk-1 receptor adsorbed on mica to measure the specific unbinding forces.
To determine the specificity of the antibody-VEGFR interaction, the blocking peptide (the peptide against which the antibody was raised) was then added online and successive force-curves were measured again after 10 min of incubation with the blocking peptide. As additional control experiments, the force curves were also recorded: 1), with a nonspecific antibody (Mouse IgG) conjugated to the functionalized tip in identical experimental conditions; 2), between the nonfunctionalized tips and the receptors; and 3), between the nonfunctionalized tips and the fresh silanized mica.
In vivo force-volume maps
The same technique used to measure the unbinding force in vitro was used to measure the unbinding forces in vivo, i.e., between the anti-Flk-1 antibody (conjugated to AFM tip) and the VEGF receptors present in the plasma membrane of live, cultured endothelial cells. The force curves were recorded in HEPES-buffered OPTI-MEM reduced serum medium (7.3 pH) in petri dishes on cells 2–3 days after seeding (Bhatia et al., 2000). The usage of this medium was necessary to maintain normal cell morphology and viability (Bhatia et al., 2000). The force mapping between isolated VEGFR and its antibody was conducted in a more defined operating environment—water or PBS. No significant difference in the interaction force between single receptor and antibody was observed in vitro. As the main focus of the whole study is to examine real-time receptor clustering in the cell plasma membrane and this required relative change in overall force field and not the actual single receptor-antibody/ligand adhesion/de-adhesion force, the imaging medium is presumed to play very little (if any) role in our analysis.
Force-volume data were collected by the force-volume map technique as described previously (Quist et al., 2000; Rhee et al., 1998). It allows force curves to be acquired as a function of the lateral position on the specimen surface. One complete force curve was recorded at each position while the AFM tip was raster scanned across the surface of the sample in 64 × 64 measuring points. The force curves were collected with a z-deflection rate of 15–20 nm (ms)−1 and with a maximum cantilever deflection (relative trigger). The total recording time for a complete image was typically 7–8 min. The relative trigger point was typically set to 20 or 40 nm (1.2 or 2.4 nN). A topographic image (64 × 64 pixels) was stored simultaneously in the force-volume map.
The specificity of the antibody-VEGFR interaction in the cell plasma membrane was determined as described above for in vitro study: the blocking peptide was added online and successive force maps were collected at 10-min intervals. As additional control experiments, the same cell was imaged before the addition of VEGF or antibody, with either a regular Si3N4 or a tip functionalized with a nonspecific antibody (Mouse IgG). Real-time receptor clustering was imaged by collecting successive force-volume maps with an anti-Flk-1-antibody-functionalized tip, after the addition of VEGF or anti-Flk-1-antibody in the imaging medium, respectively. All AFM imaging was performed at 25–27°C room temperature and the data were analyzed, as described below, with the previously described protocols developed in our lab.
AFM data analysis
A complete AFM force curve (force-displacement curve) is a plot of the measured forces, on the AFM cantilever probe, as function of the z-piezo-displacement, z, for a complete cycle of the AFM probe approaching and retracting the sample. The measured force is F = kδ, where k is the cantilever spring constant and δ is the cantilever deflection. Force curves were analyzed to investigate: 1), force-induced dissociation of the single molecular bonds; 2), the total de-adhesion between antigen-antibody complexes by evaluating qualitative differences in dissipated energy; and 3), the sample elasticity. The AFM force-volume data was imported and analyzed offline with tools developed within the IGOR Pro 4.00 (Wavemetrics, Lake Oswego, OR) data analysis software.
When the antibody-conjugated tip is brought into contact (the approach phase) with the cell surface receptors, the antibody and its specific receptors bind with attractive forces. Upon tip retraction, the antibody-receptor bonds (adhesion) keep them in contact to a certain retract distance (the hysteresis phase). The antibody-receptor bond breaks then and the measured de-adhesion force is equivalent to the antibody-receptor unbinding force. However, the retrace phase of the force curve may contain multiple quantized staircase steps (multiple bond-breaking points), each representing specific molecular interactions or unbinding events. Several different methods were used to evaluate the unbinding forces. The retract force curves were evaluated to find all individual force de-adhesion steps. The first and second derivatives of a curve were used to find all local force minima and maxima and calculate the unbinding forces. To minimize effects from noise, a step was only counted when the second derivative and the step height were above the manually specified threshold values. This automatic evaluation of the unbinding forces from these force curves is nontrivial since the measured rupture forces are small and are the result of multiple unbinding, sequential breaking, and possibly nonspecific molecular interactions, molecular stretching, etc. Sequential unbinding events without any specific effect is recognized in the force curves as the unbinding force drops to zero in between the different ruptures. In the case of multiple specific bindings, for example from the different binding sites of the antibody, parallel breakage, as multiples of the single specific bonds, would be detected. The analysis is complicated if the detected broken interactions arise from a number of different interacting molecules or variable strength of bonds. Then the weakest bonds will break first and the measured forces will not necessarily reflect the unbinding strength. However, at the last unbinding (de-adhesion) step, the measured unbinding force is still accurate and hence eventual single unbinding events can be detected. Therefore, the last step representing the final break point during hysteresis phase of the tip retraction was used to estimate the single-molecule unbinding force (Gad et al., 1997; Benoit et al., 2000).
The last de-adhesion (unbinding) force steps of the antibody-receptor interaction force curves were compiled into histograms or used to plot spatially resolved unbinding force images. Still, it was shown that histograms of all the force steps yielded similar qualitative result as when measuring only the last step. The grouping interval in the histograms was set identical to the sampling interval in the digitally stored AFM force curve data, typically a few picoNewtons. The sampling bin was smaller than the nominal AFM force resolution of 10–15 pN (Cappella and Dietler, 1999). Altogether, since unbinding is a stochastic process in nature, a specific unbinding event should show up as a peak of several bins in the histogram. Since the absolute value of the measured unbinding force depends on the z-position scan rate, we used similar scan rates in all experiments.
The force-volume measurements were also used to visualize spatially resolved maps of the total de-adhesion between antigen-antibody complexes. Generally, such maps, for each data point, show the maximum adhesion force or an area in the force curve plot. Dissipative forces give hysteresis between the trace and retrace force curves; the hysteresis corresponds to the amount of dissipated energy. The relative dissipated energy can be calculated as the area between the trace and retrace force curves (Stark et al., 2001; Marti et al., 1995; Berry et al., 2001). Maps of relative dissipated energy discussed in this article are evaluated by a trapezoidal integration of the area enclosed by the force curves in force-displacement coordinates. As the force curves could contain hydrodynamic drag component, the force values were first subjected to a minor correction before the integration.
Modeling sample elasticity
Sneddon mechanics (Sneddon, 1965) was used to evaluate sample elasticity from the force-volume measurements. Calculations were made for both a spherical indenter and a conical indenter. We used the latter in this study since the so-called Sneddon exponent (VanLandingham et al., 1997) generally indicated a more conical than spherical indentation geometry. In the case of a conical indenter, the piezo position, z, versus cantilever deflection can be expressed as
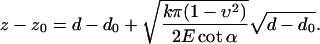 |
(1) |
Here υ is the Poisson's ratio, α is the semivertical angle of the indenter, i.e., π/2 minus half the opening angle (70°) of the cone, and z0, d0 are the contact point and the free deflection value, respectively. We assume υ = 0.5 and use k = 0.06 N/m for the calculations in this article. A nonlinear, two-parameter, numerical curve fit was used to fit the model to the force distance data in a previously used iterative procedure (Almqvist et al., 2001). The contact point was estimated as the intersection between the extrapolated line-fit of the zero-force part of the curve and the curve-fit of the contact part of the curve. Therefore, precautions were taken for any linear change of the force curve in the noncontact region and the d0-value was corrected accordingly. We also reduced the influence of substrate effects due to thinning of the cell. At a certain indentation of the sample, the AFM tip senses the underlying substrate and the force curve is significantly influenced with a steeper slope. For this reason, our automatic task of extracting the Young's (elastic) modulus E makes a curve fit to the model between d0 and the point where the slope of the force curve is 90% of the “hard surface slope” measured on the substrate. In addition, we also used manual inspection of the force curves and of the surface heights.
RESULTS AND DISCUSSION
Unbinding forces
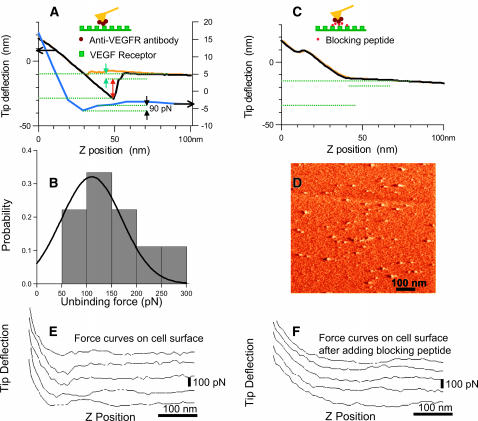
We first examined the specific molecular interaction force between the isolated receptor Flk-1 (VEGF-R2) adsorbed on a silanized mica surface and the antibody (anti-Flk-1 IgG) conjugated to the AFM tip. The functionalized AFM tip was positioned above an individual receptor and a number of force curves (10–50) were measured over a series of experiments to qualitatively measure the unbinding force between receptor-antibody pairs. The upper force curve in Fig. 1 A reveals large unbinding steps (arrows) between the antibody-conjugated tip and the VEGFR adsorbed on to a mica substrate. The lower curve is a more typical curve for the interaction between receptor and antibody. Fig. 1 B shows a histogram of the correspondent manually measured unbinding forces.
FIGURE 1.
Adhesion (unbinding/binding) forces between VEGF receptor and its antibody. (A) Adhesion force between isolated VEGF receptor (flk-1) adsorbed on the mica substrate and AFM tip conjugated with an antibody (anti-flk-1). The top AFM force curves with deflection scale to the left show the approach (yellow) and retract (black) curve, respectively. The retract curve reveals large multiple de-adhesion steps (arrows) and specific interaction between the antibody (anti-Flk-1) on the tip and the receptors on mica surface. The last unbinding step (green arrow) is attributed to unbinding of a single receptor-antibody pair. The lower blue curve (with deflection scale to the right) shows a more typical interaction. The expected single pair unbinding is ∼100 pN and might correspond to single/double binding. (B) Probability histogram of manually measured unbinding forces from force curves measured as in A. (C) Control; competitive inhibition. The force curve in A between the antibody on the tip and the receptors on mica surface is abolished by online addition of the blocking peptide against which the antibody was made. (D) AFM tapping mode amplitude image of the VEGF receptors sparsely distributed on a silanized mica surface. (E) Adhesion force between VEGF receptor (flk-1) on endothelial cell and AFM tip conjugated with an antibody (anti-flk-1). (F) Force curves on the cell after adding the blocking peptide. The force curves in E indicates larger adhesion force versus no adhesion force in the presence of the blocking peptide (F).
The specificity of the antibody-receptor interaction was determined by complementary sets of control experiments. The unbinding force (Fig. 1 A) was abolished, i.e., the hysteresis in the force curve was eliminated when the blocking peptide (the peptide against which the antibody was raised) was added online (Fig. 1 C). Preliminary data suggest that, by sequential competitive binding, the blocking peptide could reveal multiple receptor-antibody bindings and, as such, could be used to find single binding events even when there are complex interactions. Also, no appreciable unbinding force was observed between the Flk-1 receptors and the regular, non-antibody-conjugated silicon nitride tips (data not shown). Similarly, no hysteresis in the force curve was observed when imaging a freshly cleaved mica surface with non-antibody-conjugated silicon nitride tips under identical fluid conditions.
Generally the unbinding forces varied between 60 and 240 picoNewtons (pN). However, manual inspection yielded frequent occurrence of unbinding forces 60–100 pN, and possibly multiples thereof. For example, unbinding forces of 240 pN could correspond to simultaneous breaking of four 60-pN interactions. The absolute value of the unbinding force is semiquantitative, at best. However, it is apparent that the technique and our approach are sufficient to determine specific receptor-antibody interactions. As described in Materials and Methods, force measurement errors are attributed to the following: 1), a lack of precise determination of the spring constant of the antibody-conjugated AFM tip; 2), a rather large z-position scan rate (overestimated force); 3), neglecting eventual geometric effects of the finite-sized tip radius; and 4), surface forces due to the eventual presence of an electrical double layer.
Previous studies have reported unbinding forces, from in vitro study, of 60 ± 10 pN for biotin/antibiotin (Dammer et al., 1996), 49 ± 10 pN for ferritin/antiferritin, and 40 ± 4 pN for fluorescein/antifluorescein (Ros et al., 1998) single antigen-antibody interaction, and higher multiples for multiple interactions. Our semiquantitative result of a VEGFR-antibody unbinding force of 60 ± 10 pN is consistent with these studies.
The binding/unbinding forces measured in our study are specific to antibody-receptor interactions and not due to unspecific binding processes. An important control for this is the absence of binding events, presumably due to the blocking of the binding sites when the blocking peptide was added in the imaging medium and/or due to a decrease in the total number of binding events when an excess amount of antibody was added in the medium. Moreover, no measurable binding force was observed between the VEGF receptor (Flk-1) adsorbed on mica and a nonspecific antibody mouse IgG onto the tip (unpublished data).
The same technique, as used to measure the unbinding force in vitro (between the isolated VEGF receptor adsorbed on the mica surface and the anti-Flk-1 antibody conjugated to the AFM tip), was used to measure the unbinding forces in vivo, i.e., between the anti-Flk-1-antibody (conjugated to AFM tip) and the VEGF receptors present in the plasma membrane of live cultured endothelial cells. In the in vivo condition, multiple unbinding forces were observed in most of the force curves at each receptor surface (Fig. 1 E), in contrast to predominantly single unbinding steps observed between isolated receptors adsorbed onto mica and the antibody-conjugated tip. In the presence of the blocking peptide (which was added online after the force-curves between the antibody and VEGFRs had been acquired), the interaction between the antibody and VEGF receptor was inhibited, and basically no hysteresis was observed in the force curves (Fig. 1 F), i.e., the unbinding force was eliminated. The force curves in Fig. 1 E indicates a larger adhesion force versus no adhesion force in the presence of the blocking peptide (see also Fig. 1 F).
The binding force measured between the receptors in the cell plasma membrane and the antibody (anti-flk1) anchored to the AFM tip show correspondence to the in vitro experiment. However, on addition of excess anti-Flk-1-antibody in the imaging medium, there was a significant decrease in the total number of binding events but not a complete inhibition. This could be due to the complexity of physicochemical factors and cellular processes involved in the case of receptors present in the cell surface.
The interaction-force histograms obtained using the tip without any antibody and the tip with a nonspecific antibody were similar, suggesting that these tips interact nonspecifically with the cell surface. Various glycoproteins, sugar molecules, adhesion molecules, and other macromolecules present in the cell plasma membrane could contribute to such nonspecific adhesions.
The multiple unbinding steps present in the force measurement curve, when using anti-Flk-1-antibody-conjugated tips, could be due to the presence of two Fab fragments of the antibody. Two Fab fragments could bind to a single receptor producing two separate unbinding (de-adhesion) steps on retraction, or two Fab fragments could bind independently depending on a number of factors like the distribution of receptors on the cell surface and the orientation of the antibody. Moreover, since the cell surface is soft, the contact area between the AFM tip and the cell surface will be large, which further favors the occurrence of multiple molecular bonds and large adhesion. The likelihood of single molecular interactions should be significantly higher in the in vitro experiments, since the density and the distribution of the interacting molecules (VEGFRs, in this case) can be optimized.
Redistribution and clustering of plasma membrane receptors
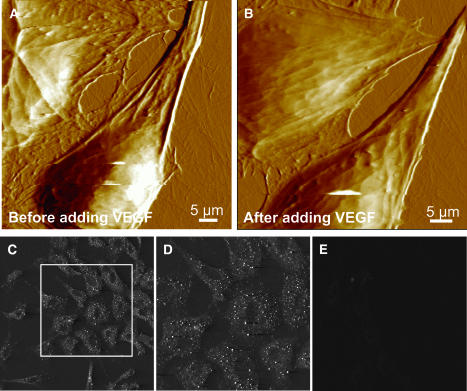
With standard contact mode AFM imaging, the endothelial cells could be imaged without any imaging-induced structural artifacts for an extended period of time as described previously (Bhatia et al., 2000). Fig. 2, A and B, shows examples of real-time AFM imaging of endothelial cells. Online addition of 25 nM VEGF, an important angiogenic factor in human and animal tissues, induced cytoskeletal reorganization and cellular growth. However, as expected, within a short period (2 h) of imaging, the cell growth/reorganization was very small, and was normally not observable under light microscopy. AFM images show a clear difference in the image contrast reflective of differing viscoelasticity; cells after VEGF incubation show fewer cytoskeletal details (Fig. 2 B, top cell) and a changing cell boundary. Previous studies have reported considerably pronounced cell growth after a longer duration (8–10 h or longer) of VEGF incubation. In the present study, the cell elastic property changed considerably (see below) and the endothelial cells became softer after the addition of VEGF or anti-VEGFR-antibody in the imaging medium. A qualitative change in the cell softness is reflected as a loss of ultrastructural details (Fig. 2, A–B).
FIGURE 2.
AFM images of endothelial cells showing VEGF induced cytoskeletal reorganization, (A) before adding VEGF and (B) 2 h after adding VEGF (25 nM). Cytoskeletal reorganization as well as a change in the elasticity is observed. Cell softness is reflected in a loss of fine ultrastructural details. (C–D) Immunofluorescence labeling of Flk-1 receptors in the plasma membrane. Endothelial cells show immunolabeling with a polyclonal anti-Flk-1 antibody followed by cy-3 conjugated secondary antibody. D shows a zoomed image of a portion of C. Receptors are distributed throughout the cell surface with a higher density along the cell periphery. (E) Endothelial cells show no immunolabeling with a nonspecific antibody followed by cy-3 conjugated secondary antibody.
The presence of VEGF receptors in the endothelial cell plasma membrane is evident from immunofluorescence labeling of these receptors with anti-Flk-1-antibody and fluorescently labeled with cy3 conjugated secondary antibody (Fig. 2, C and D). Fig. 2, C and D, show single slices in confocal images of immunofluorescence labeling and show a lack of label above the cell nucleus. However, the integrated confocal image of whole-cell immunolabeling indicates that VEGF receptors are distributed throughout the cell surface but at a slightly higher density along the cell periphery (data not shown). The punctuated immunolabeling pattern with a higher density along the cellular periphery (Fig. 2, C and D) is consistent with the presence of punctuate regions of different elasticity measured in our study, as discussed later.
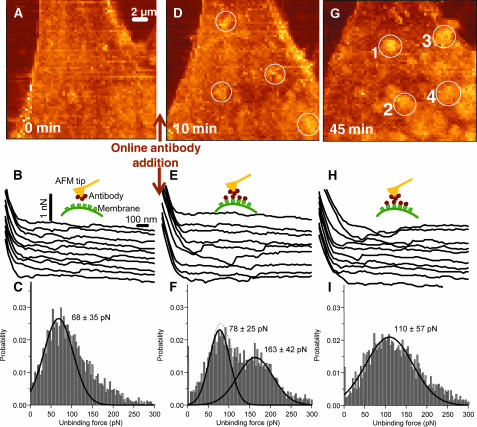
Receptor clustering and distribution were examined using force-volume mapping as described earlier and not by the method used for the force measurements shown in Fig. 1. The force maps were first recorded with a regular Si3N4 tip (which serves as a control), then with a tip conjugated with anti-Flk-1 antibody, and then again after the excess antibody was added in the solution. Fig. 3, A–C, shows the result with the conjugated tip before addition of the antibodies. The unbinding forces in the in vivo whole-cell experiments varied between 30 and 400 pN. There was a more complex set of unbinding forces than in the in vitro experiments. Significantly, there were multiple interactions between individual receptors with the antibody present on the tip and the receptors on the cell surface (Fig. 3 B). The final force step, after which the receptor and antibody completely separated, was analyzed to estimate the single antibody-receptor unbinding forces. Fig. 3 C shows one example of the data compiled in such histogram. Typically, >3000 steps were used for the plot. The histogram of the last unbinding forces, shown in Fig. 3 C, has the predominant unbinding force ∼60–70 pN. This is similar to the unbinding force observed in vitro. Maps of the spatial distribution of the last unbinding force between the antibody functionalized AFM tip and the cell membrane did not reveal any characteristic features in distribution of plasma membrane receptors. This is consistent with the uniformly distributed receptors found with immunofluorescence labeling (Fig. 2). The distribution of unbinding forces (data not shown), when the tip conjugated with the nonspecific antibody (another control) was used on the same cell, did not reveal this unbinding force. Hence, the result from the in vivo experiments semiquantitatively agrees with the characteristic unbinding force measured between the antibody-functionalized AFM tip and the isolated VEGF receptors in vitro. The correspondence between unbinding forces measured in vitro and in vivo, suggests that the force-volume map can be used to map the heterogeneous distribution of cell surface macromolecules (channels and receptors) using functionalized AFM tips.
FIGURE 3.
Force maps on endothelial cells in real-time. (A–C) Specific interaction probed with a Si3N4 tip functionalized with anti-Flk-1. (A) Force map. (B) Force curves taken at various points on the cell from the map shown in A. The curves are offset with respect to zero force. (C) Probability histogram of the unbinding forces of the force curves from the force map in A. The histogram is fitted with a Gaussian and the corresponding maxima and σ is indicated in the figure. The dominant unbinding force ∼60–70 pN suggests breakage of single receptor-antibody bonds. (D–F) Competitive inhibition probed with the anti-Flk-1 functionalized Si3N4 tip, 10 min after adding antibody in the recording medium. The panels correspond to A–C. The characteristic unbinding force is suppressed and the measured forces are shifted toward higher values. (G–I) Corresponds to A–C, but at 45 min after adding antibody in the recording medium. The micrometer-sized brighter spots in G are identified as receptor clusters. A few of the clusters are marked by numbers 1–4.
The interaction force was further measured in the presence of excess antibody in the imaging media. The evaluation of these data revealed the appearance of micrometer-sized spots (clusters) on the cell surface in the excess antibody media. Representative force-volume images from the Nanoscope software are shown in Fig. 3, A, D, and G. These images, in each point, show the tip-sample force at a specified z-piezo position. There was continuous reorganization of these clusters and the cluster size increased with time as evident in the force maps (Fig. 3, A, D, and G), the maps of dissipated energy (not shown), and in the elasticity maps (Fig. 4 in the next subsection). Maximum clustering was observed 45 min after online addition of antibody, and a few of these clusters are marked with circles and numbered 1–4 in Fig. 3 G. The clusters appeared to be concentrating more toward the cellular boundaries, consistent with the mitogenic activity of VEGFR and the cell growth pattern. Similar clustering of VEGFRs was also observed after adding VEGF online, although the VEGF-induced clustering was less pronounced (data not shown).
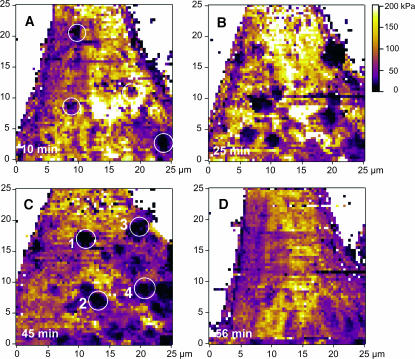
FIGURE 4.
Elasticity maps of the evaluated Young's modulus on endothelial cells in real-time, showing clustering of VEGF receptors on the cell surface. The images are color-coded according to the color bar, from 0 kPa (dark) to 200 kPa (bright yellow). The images show the elasticity at different time points after adding anti-flk-1 antibody in the imaging solution: (A) 10 min after addition; (B) 25 min after addition; (C) 45 min after addition; and (D) 56 min after addition. A few regions with lower elasticity are marked with numbers 1–4 in C. These are the same regions showing receptor clusters in Fig. 3 G (marked as 1–4). The regions underlying the receptor clusters appeared as less stiff.
As before, the individual unbinding forces between the antibody and VEGFRs were analyzed from the single force curves within a force map (Fig. 3, B, E, and H). The unbinding mostly occurred in several steps for both the sequential-type unbindings and the more complex multiple unbindings. For example, in the force curves of the force map shown in Fig. 3 A, 43% of the force curves have two or more unbinding points and 66% of these curves had increasing unbinding steps outwards from the surface.
In the presence of excess antibody, the number of measured characteristic antibody-receptor unbinding events (at 60–70 pN) were reduced (compare Fig. 3 C with F and I), possibly due to competing inhibition of interactions of the AFM tip-attached antibody and the plasma membrane receptors. Hence, we could not identify the spatial distribution of clusters simply from maps of the final unbinding step. However, the unbinding forces did not disappear (Fig. 3, E and H). Instead, as seen in Fig. 3 F, additional unbinding forces appear at a higher magnitude (170–190 pN) and the force distribution widens out in time (Fig. 3 I). This could be due to the complexity of other factors and cellular processes not expected in the in vitro study. Even in the histogram for the force curves obtained when a regular Si3N4 tip (without any antibody and serving as a control) was used on the same cell, some adhesion between the tip and the cell surface was observed (data not shown). However, the magnitude of the unbinding forces was then more uniformly distributed in the range of 30–95 pN. The images of calculated total de-adhesion (maps not shown), i.e., the dissipated energy calculated as the area between the trace and retrace force curves, reveal the same qualitative features as the force-volume images. Large total interactions (unbinding) were detected on cluster-like regions, whereas these large de-adhesions were not detected at noncluster regions.
The nature of the experimental paradigm suggests that the excess of the antibody in the imaging medium should compete with the antibody conjugated to the AFM tip for the same binding sites on the VEGFRs. Such condition should give rise to reduced adhesion forces, although individual antibody-receptor binding would still be present. Consistent with such possibility, the clusters diminished significantly 60 min after the addition of IgG as was seen in the maps of dissipated energy (and is further demonstrated in the next subsection).
The clustering and activation of growth factor receptors in general has been reported to precede the formation of receptor dimmers and the subsequent receptor autotyrosin phosphorylation (Jefford and Dubreuil, 2000). Such receptor clusterings could be mediated by appropriate ligands (the ligand-induced clustering) (Kirsch and Betz, 1998) as well as by antibodies (Yauch et al., 1997; Gao et al., 2000). In our study, ligand-induced clustering (after online addition of VEGF) was less pronounced. This is consistent with the results from several other studies and suggests that VEGF binding to endothelial cell receptors induces receptor dimerization, i.e., for each VEGF, two receptors will be clustered. On the other hand, each antibody could bind to several receptors and hence the receptor size would be considerably larger and the clustering would be faster as well. This results in subsequent binding and phosphorylation of the downstream mediators, and also leads to increased cellular calcium (Meyer et al., 1999) and the reorganization of the actin and microtubule cytoskeleton necessary for the cell growth and migration. During such reorganization, the cellular elastic properties would change as well. We examined such changes in real-time and simultaneously, as discussed in the next subsection.
Elasticity maps and underlying cytoskeletal elements
The elasticity maps obtained at different times after online addition of anti-Flk-1-antibody are shown in Fig. 4. Initially, the number and total area of low elasticity spots, i.e., clusters, increased and after 45 min almost 20% of the total cell surface was composed of clusters. The difference in elasticity between the individual clusters and the surrounding cytoskeleton significantly increased during the first 25 min after the addition of anti-VEGFR-antibody in the imaging medium (Fig. 4 B). The elasticity maps derived from the force-maps showed significant changes in the local elasticity. Interestingly, the regions underlying the receptor clusters appeared less stiff (compare Fig. 3 G with Fig. 4 C, positions 1–4). The Young's modulus changes were unexpectedly large. The modulus was between 50 and 120 kPa on the outer cell regions and at the leading edge, and ∼3–6 kPa around the nucleus and under the receptor clusters.
The decreased elasticity on the clusters causes the AFM tip to indent the surface and form a large set of bonds, increasing the total adhesion upon removal. However, it is difficult to directly correlate the binding force and the change in the elasticity. The elasticity distribution depends on the interaction between the receptors and the cytoskeletal filaments beneath the cellular surface. The localized reduction in cell stiffness is consistent with a signal transduction mechanism wherein a localized clustering of VEGF receptors would induce reorganization of the underlying cytoskeletal network (as required for the cellular growth and migration). In addition, the VEGF-induced elevation of cellular calcium (Meyer et al., 1999) will also alter cell elasticity (Quist et al., 2000). Significantly, the change in stiffness was observed within minutes of real-time receptor clustering and was reciprocal to the cluster size, i.e., reciprocal to the receptor density. Thus increased clustering resulted in decreased stiffness. Such real-time mapping of the receptor clustering and of the resultant rapid changes in the cellular biophysical properties is unique to the technique described in this study.
Previous studies have used a longer duration, usually 8–10 h, of ligand- or antibody-induced receptor clustering, to examine changes in the resulting cellular biophysical properties. Such a paradigm suggests that the receptor-clustering induced signal transduction process underlying cellular growth and migration is relatively slow. Alternately, this could reflect a lack of high temporal and spatial resolution techniques for simultaneously mapping receptor clustering as well as the resultant change in the cellular biophysical properties. Moreover, the change in the elastic properties was transient in our study, suggesting a wavelike change in the local cytoskeletal network as the cell undergoes progressive growth and development.
Implications of results
The VEGF receptor clustering, illustrated in this work, is an example of the dynamic receptor clustering process that mediates various physiological cellular activities. For example, clustering of neurotransmitter receptors at presynaptic regions provides the anatomical basis for receiving an array of synaptic inputs necessary for information processing in the vertebrate brain (Antonova et al., 2001; Horio et al., 1997; Sugiyama et al., 1997; Kirsch and Betz, 1998). Receptor clustering is required for polarized assembly of ankyrins (Jefford and Dubreuil, 2000). Receptor clustering and cytoskeletal association is required for receptor-mediated adhesion to extracellular matrix and for the differential clustering of CD4, and T-cell receptors are required for ligand recognition (Krummel et al., 2000), etc. Measurements of such real-time receptor clustering and of the resultant rapid and order-of-magnitude changes in cellular elastic properties have not been reported previously. The technique described in this study is uniquely suited for such investigations, i.e., to map the distribution and clustering of various biological macromolecules on the cell surface as well as their resulting effects on cellular physical properties.
The remarkably large decreases in local cell stiffness induced by the clustering of VEGF receptors provide a mechanism for cell growth. The rapidity with which these effects can be induced by the appropriate stimuli could have interesting implications, such as enhancing the prospects for rapid angiogenesis and accelerated vascularization, which in turn could help in the recovery of injured vascular tissue (Gill et al., 2001).
Acknowledgments
We acknowledge technical help in the AFM studies from Drs. Arjan Quist, Hai Lin, and Ashok Parbhu. We also acknowledge helpful suggestions of the reviewers.
This work was supported by grants from the National Science Foundation (to R.L., S.B.), the National Institutes of Health (to R.L.), the Philip Morris External Grant Program (to R.L.), the Santa Barbara Cottage Hospital Research Grant Program (to R.L., G.P.), and the Swedish Research Council's contract 2001-2585 (to N.A.).
N. Almqvist and R. Bhatia contributed equally to this work.
References
- Aileen, C., and T. M. Moy. 2000. Cross-linking of cell surface receptors enhances cooperativity of molecular adhesion. Biophys. J. 78:2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almqvist, N., Y. del Amo, B. L. Smith, N. H. Thomson, Å. Bartholdson, M. Brzenski, R. Lal, and P. K. Hansma. 2001. Micromechanical and structural properties of a pennate diatom investigated by atomic force microscopy. J. Microsc. 202:518–532. [DOI] [PubMed] [Google Scholar]
- Antonova, I., O. Arancio, A. C. Trillat, H.-G. Wang, L. Zablow, H. Udo, E. R. Kandel, and R. D. Hawkins. 2001. Rapid increase in clusters of presynaptic proteins at onset of long-lasting potentiation. Science. 294:1547–1550. [DOI] [PubMed] [Google Scholar]
- Baumgartner, W., P. Hinterdorfer, W. Ness, A. Raab, D. Vestweben, H. Schindler, and D. Drenckhahn. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 97:4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit, M., D. Gabriel, G. Gerish, and H. E. Gaub. 2000. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2:313–317. [DOI] [PubMed] [Google Scholar]
- Berry, M., T. J. McMaster, A. P. Corfield, and M. J. Miles. 2001. Exploring the molecular adhesion of ocular mucins. Biomacromolecules. 2:498–503. [DOI] [PubMed] [Google Scholar]
- Bhatia, R., H. Lin, and R. Lal. 2000. Fresh and globular amyloid-β protein (1–42) induces rapid cellular degeneration: evidence for AβP channel-mediated cellular toxicity. FASEB J. 14:1233–1243. [DOI] [PubMed] [Google Scholar]
- Boland, T., and B. D. Ratner. 1995. Direct measurement of hydrogen bonding in DNA nucleotide bases by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 92:5297–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappella, B., and G. Dietler. 1999. Force-distance curves by atomic force microscopy. Surf. Sci. Rep. 34:1–104. [Google Scholar]
- Cleveland, J., and S. Manne. 1993. A nondestructive method for determining the spring constant of cantilevers for scanning force microscopy. Rev. Sci. Instrum. 64:403–405. [Google Scholar]
- Crouch, M. F., D. A. Davy, F. S. Willard, and L. A. Berven. 2001. Activation of endogenous thrombin receptors causes clustering and sensitization of epidermal growth factor receptors of Swiss 3T3 cells without transactivation. J. Cell Biol. 152:263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer, U., M. Hegner, D. Anselmetti, P. Wagner, M. Dreier, W. Huber, and H. J. Guntherodt. 1996. Specific antigen/antibody interactions measured by force microscopy. Biophys. J. 70:2437–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad, M., A. Itoh, and A. Ikai. 1997. Mapping cell wall polysaccharides of living microbial cells using atomic force microscopy. Cell Biol. Int. 21:697–706. [DOI] [PubMed] [Google Scholar]
- Gao, B., C. Blumenstock, F. A. Minnear, and T. M. Saba. 2000. Increased recycling of integrins by lung endothelial cells in response to tumor necrosis factor. J. Cell Sci. 113:247–257. [DOI] [PubMed] [Google Scholar]
- Gill, M., S. Diaz, K. Hattori, M. L. Rivera, D. Hicklin, L. Witte, L. Girardi, R. Yurt, H. Himel, and S. Rafii. 2001. Vascular trauma induces rapid but transient mobilization of VEGFR2+AC133+1 endothelial precursor cells. Circ. Res. 88:167–174. [DOI] [PubMed] [Google Scholar]
- Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, Y., H. Hibino, A. Inanobe, M. Yamada, M. Ishii, Y. Tada, E. Satoh, Y. Hata, Y. Takai, and Y. Kurachi. 1997. Clustering and enhanced activity of an inwardly rectifying potassium channel, Kir4.1, by an anchoring protein. J. Biol. Chem. 272:12885–12888. [DOI] [PubMed] [Google Scholar]
- Horton, M., G. Charras, and P. Lehenkari. 2002. Analysis of ligand-receptor interactions in cells by atomic force microscopy. J. Recept. Signal Transd. 22:169–190. [DOI] [PubMed] [Google Scholar]
- Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic force microscope tips. Rev. Sci. Instr. 64:1868–1873. [Google Scholar]
- Jefford, G., and R. Dubreuil. 2000. Receptor clustering drives polarized assembly of ankyrin. J. Biol. Chem. 275:27726–27732. [DOI] [PubMed] [Google Scholar]
- Kirsch, J., and H. Betz. 1998. Glycine-receptor activation for receptor clustering in spinal neurons. Nature. 392:717–720. [DOI] [PubMed] [Google Scholar]
- Korchev, Y. E., Y. A. Negulyaev, C. R. W. Edwards, I. Vodyanoy, and M. J. Lab. 2000. Functional localization of single active ion channels on the surface of a living cell. Nat. Cell Biol. 2:616–619. [DOI] [PubMed] [Google Scholar]
- Krummel, M. F., M. D. Sjaastad, C. Wülfing, and M. M. Davis. 2000. Differential clustering of CD4 and CD3 zeta during T-cell recognition. Science. 289:1349–1352. [DOI] [PubMed] [Google Scholar]
- Lal, R., and S. A. John. 1994. Biological applications of atomic force microscopy. Am. J. Physiol. 266:C1–C21. [DOI] [PubMed] [Google Scholar]
- Marti, A., G. Hähner, and N. D. Spencer. 1995. Sensitivity of frictional forces to pH on a nanometer scale: a lateral force microscopy study. Langmuir. 11:4632–4635. [Google Scholar]
- Meyer, M., M. Clauss, A. L. Wienhues, J. Waltenberger, H. G. Augustin, M. Ziche, C. Lanz, M. Buttner, H.-J. Rziha, and C. Dahio. 1999. A novel vascular endothelial growth factor encoded by Orf virus, VEGF-E, mediates angiogenesis via signaling through VEGFR-2 (KDR) but not VEGFR-1 (Flt-1) receptor tyrosine kinases. EMBO J. 18:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, G., T. Cohen, S. Gengrinovitch, and Z. Poltorak. 1999. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 13:9–22. [PubMed] [Google Scholar]
- Parbhu, A., W. Bryson, and R. Lal. 1999. Disulfide bonds in the outer layer of keratin fibers confer higher mechanical rigidity: correlative nano-indentation and elasticity measurement with an AFM. Biochemistry. 38:11755–11761. [DOI] [PubMed] [Google Scholar]
- Pawson, T. 1995. Protein modules and signaling networks. Nature. 373:573–580. [DOI] [PubMed] [Google Scholar]
- Quist, A. P., S. K. Rhee, H. Lin, and R. Lal. 2000. Physiological role of gap-junctional hemichannels: extracellular calcium-dependent isoosmotic volume regulation. J. Cell Biol. 148:1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh, L. E., and L. C. Cantley. 1999. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 274:8347–8350. [DOI] [PubMed] [Google Scholar]
- Rhee, S. K., A. P. Quist, and R. Lal. 1998. Amyloid-β protein-(1–42) forms calcium-permeable Zn2+-sensitive channel. J. Biol. Chem. 273:13379–13382. [DOI] [PubMed] [Google Scholar]
- Ros, R., F. Schwesinger, D. Anselmetti, M. Kubon, R. Schäfer, A. Plückthun, and L. Tiefenauer. 1998. Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. USA. 95:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff, S. G., D. R. Saner, and R. Lal. 1995. Dynamic micromechanical properties of cultured rat atrial myocytes measured by atomic force microscopy. Am. J. Physiol. 38:C286–C292. [DOI] [PubMed] [Google Scholar]
- Sneddon, I. N. 1965. The relation between load and penetration in the axisymmetric Boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 3:47–57. [Google Scholar]
- Stark, M., C. Möller, D. J. Müller, and R. Guckenberger. 2001. From images to interactions: high-resolution phase imaging in tapping-mode atomic force microscopy. Biophys. J. 80:3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, J. E., D. J. Glass, G. D. Yancopoulos, and Z. W. Hall. 1997. Laminin-induced acetylcholine receptor clustering: an alternative pathway. J. Cell Biol. 139:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, K. A. 1996. Vascular endothelial growth factor, a potent and selective angiogenic agent. J. Biol. Chem. 271:603–606. [DOI] [PubMed] [Google Scholar]
- VanLandingham, M. R., S. H. McKnight, G. R. Palmese, X. Huang, T. A. Bogetti, R. F. Eduljee, and J. W. Gillespie, Jr. 1997. Nanoscale indentation of polymer systems using the atomic force microscope. J. Adhes. 64:31–59. [Google Scholar]
- Wang, W., W. L. Dentler, and R. T. Borchardt. 2001. VEGF increases BMEC monolayer permeability by affecting occluding expression and tight junction permeability. Am. J. Physiol. Heart Circ. Physiol. 280:H434–H440. [DOI] [PubMed] [Google Scholar]
- Yauch, R. L., D. P. Felsenfeld, S. K. Kraeft, L. B. Chen, M. P. Sheetz, and M. E. Hemler. 1997. Mutational evidence for control of cell adhesion through integrin diffusion/clustering, independent of ligand binding. J. Exp. Med. 186:1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C. B., A. Chen, P. Kolb, and V. T. Moy. 2000. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 39:10219–10223. [DOI] [PubMed] [Google Scholar]
- Zhang, X. H., E. Wojcikiewicz, and V. T. Moy. 2002. Force spectroscopy of the leukocyte function-associated antigen-1 intercellular adhesion molecule-1 interaction. Biophys. J. 83:2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]