Abstract
Computational methods that predict three-dimensional structures from amino acid sequences have become increasingly accurate and have provided insights into structure-function relationships for proteins in the absence of structural data. However, the accuracy of computational structural models requires experimental approaches for validation. Here we report direct testing of the predictions of a previously reported structural model of the C-terminus of the human heart Na+ channel. We focused on understanding the structural basis for the unique effects of an inherited C-terminal mutation (Y1795C), associated with long QT syndrome variant 3 (LQT-3), that has pronounced effects on Na+ channel inactivation. Here we provide evidence that this mutation, in which a cysteine replaces a tyrosine at position 1795 (Y1795C), enables the formation of disulfide bonds with a partner cysteine in the channel. Using the predictions of the model, we identify the cysteine and show that three-dimensional information contained in the sequence for the channel protein is necessary to understand the structural basis for some of the effects of the mutation. The experimental evidence supports the accuracy of the predicted structural model of the human heart Na+ channel C-terminal domain and provides insight into a structural basis for some of the mutation-induced altered channel function underlying the disease phenotype.
INTRODUCTION
Disease-linked inherited mutations of ion channel proteins are so prevalent that the disorders caused by them, including epilepsy, febrile seizures, Dent's disease, and cardiac arrhythmias, are now referred to as channelopathies (Marban, 2002; Jentsch, 2000; Jentsch et al., 2000; Schwake et al., 2001; Lossin et al., 2002; Jurkat-Rott and Lehmann-Horn, 2001; Kullmann, 2002). Expression of ion channels in heterologous systems allows for investigation of inherited ion channel defects at the single protein and cellular level to directly identify the disease-associated alteration in ion channel function (Clancy and Kass, 2002). However, despite a wealth of experimental evidence documenting the functional consequences of the disease-linked mutations in ion channels, understanding the physical mechanisms affected by the mutations at the structural level has been lacking. This is due in large part to the difficulty in obtaining structural information with high-resolution experimental techniques such as x-ray crystallography and multidimensional NMR methods in integral membrane proteins. To date, structural information is available for only a limited number of ion channel proteins, despite the fact that ion channels regulate key physiological cellular and subcellular functions in health and disease (Miller, 2000a,b).
Computational methods that predict three-dimensional structures from amino acid sequences have become increasingly accurate and have provided insights into structure-function relationships for proteins without structural information (Yang and Honig, 2000; Yang, 2002; Yang and Wang, 2002). Although the information required to generate a protein structure is expected to be embedded in its amino acid sequence, current computational methodologies that unravel how three-dimensional information is mapped onto a linear sequence predict low-resolution structures at best when close homologous structural templates from protein structural databases are absent. The accuracy of computational structural models requires experimental approaches for validation.
Here we report direct testing of the predictions of a structural model reported by us of the C-terminal domain of the human heart Na+ channel (Cormier et al., 2002). We focused on understanding the structural basis for the unique effects of an inherited C-terminal mutation (Y1795C (YC)) that is associated with variant 3 of the long QT syndrome (LQT-3) and has pronounced effects on the entry of Na+ channels into a nonconducting inactivated state (Rivolta et al., 2002; Clancy et al., 2002). The model structure of the C-terminal domain was based on a remote relationship of the C-terminal domain and calmodulin structure: the C-terminal domain was predicted to have a calmodulin-like fold with one pair of EF-hands packed against each other as in calmodulin. In this article, we provide evidence that the naturally occurring mutation, in which a cysteine replaces a tyrosine at position 1795 (Y1795C), enables the formation of disulfide bonds with a partner cysteine in the channel. Using the predictions of the model, we identify the cysteine and show that three-dimensional information contained in the sequence for the channel protein is necessary to understand the structural basis of the effects of the mutation. The experimental evidence supports the accuracy of the predicted structural model of the C-terminal domain in the human heart Na+ channel and provides insight into a structural basis for the mutation-induced altered channel function underlying the disease phenotype.
METHODS
Molecular biology
The C-terminal mutations of SCN5A were engineered into wild-type (WT) cDNA cloned in pcDNA3.1 (Invitrogen, Carlsbad, CA) by overlap extension using mutation-specific primers and Quick Change Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA). The presence of the mutation was confirmed by sequence analysis (Rivolta et al., 2001). All constructs were transiently expressed in HEK293 cells. Transient transfection was carried out using lipofectamine (Invitrogen) according to the protocol suggested by the manufacturer. Cells were transfected with equal amounts of the α-subunit and hβ1-subunit (SCNB1), subcloned individually into the pcDNA3.1 (Invitrogen) vector. In addition, the same amount of CD8 cDNA was cotransfected as a reporter gene (EBo-pCD vector, American Type Culture Collection, total cDNA 2.5 μg). CD8 positive cells were patch clamped 48 h after transfection. Expression of CD8 did not alter the channel properties, compared with those of stably expressed channel (data not shown). Control experiments in which CD8 and hβ1 were transfected together using a bicistronic vector (pIRES, Clontech, Palo Alto, CA) were used to confirm coexpression of hβ1 and α-subunits as described above. Expressed channel biophysical properties were independent of the above conditions (data not shown).
Electrophysiology
Membrane currents were measured using whole cell and single channel patch-clamp procedures with Axopatch 200B amplifiers and Pclamp 8 (Axon Instruments, Foster City, CA). All measurements were obtained at room temperature (22°C). Single channel recordings employed the following bath solution (mM): 140 KCl, 5 HEPES, 1 MgCl2, and pH adjusted to 7.4. Whole cell procedures have been published previously (Abriel et al., 2001). For single channel experiments, pipettes were coated with Sylgard (Dow Chemical, Midland, MI) to decrease noise and capacitance of the glass. Electrode resistance was typically 5–7 MΩ when filled with internal solution (110 mM NaCl, 10 mM HEPES, pH adjusted to 7.4). In single channel experiments, after establishing the cell-attached configuration (seal resistance > 10 GΩ), the membrane was held at a holding potential of −120 mV. Test pulses (−30 mV, 100 ms) were applied every 0.5 s. Single channel currents were filtered by a low pass filter in the clamp amplifier with a cutoff frequency of 5 kHz and digitized for storage on computer at a sampling frequency of 20 kHz.
DTT (Dithiothreitol; Sigma, St. Louis, MO) was dissolved in DMSO at a stock concentration of 1 M. Stock solutions were kept no longer than 1 week. DTT stock solution was diluted into extracellular (bath) solutions to a final concentration of 10 mM immediately before experimental use. Because DTT is membrane permeable and can be used to modify candidate intracellular sites by external application (Nassir et al., 2003), cells were first incubated in DTT-containing solutions for at least 30 min at room temperature before patch-clamp experiments. DTT (10 mM) was maintained in external solutions during each experiment, and KCl (3 M) was used as a reference electrode to compensate for junction potentials caused by 10 mM DTT. Alternatively, in specified experiments, DTT (50 mM) was added to the external solution and then applied by perfusion to patched cells. Effects on membrane currents reached steady state and were recorded in the presence of DTT after 10-min exposure periods. These effects were reversible upon the return to DTT-free external solutions (within 10 min). The effects recorded on the decay of Na+ channel currents were the same for each method of DTT application.
Data analysis
Capacitative and leak currents were eliminated by digital subtraction of averaged null sweeps. Idealization of single channel currents and the measurement of open time were carried out with the program SKM (QUB suit, U. Buffalo; Premkumar et al., 1997; Qin et al., 1996). Further analysis was carried out using Excel (Microsoft, Seattle, WA) and Origin (Microcal Software, Northampton, MA). Data are represented as mean ± SE. Dannet's t-test was used to compare statistical significance among four groups; p < 0.05 was considered statistically significant.
Model
Details of the model used to predict the C-terminal domain of the Na+ channel have been published previously by us (Cormier et al., 2002).
RESULTS
The inherited mutation Y1795C slows the onset of inactivation
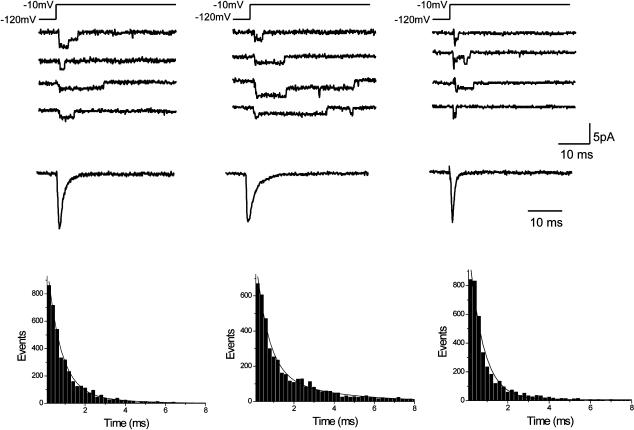
Open state Na+ channel inactivation, due to rapid block of the inner mouth of the channel pore by the (α-subunit) cytoplasmic linker between domains III and IV of the channel that occurs within milliseconds of membrane depolarization (Stuhmer et al., 1989; West et al., 1992; Vassilev et al., 1988, 1989), is evident as brief channel openings followed by quiescent periods in single channel recordings and in the decay of peak inward whole cell or macroscopic averaged currents. We previously reported that the inherited mutations Y1795C (associated with long QT variant 3) and Y1795H (associated with Brugada Syndrome) had opposite effects on both single channel gating kinetics and the time course of open state inactivation measured in whole cell recordings (Liu et al., 2002). Here we varied systematically the amino acid substitution for Tyr1795 to determine whether a physical chemical property such as charge or molecular size could be causally linked to the altered rate of open state inactivation. Fig. 1 illustrates the experiments by showing the effects of replacement of Tyr1795 by Ser (Y1795S (YS)). This mutation does not significantly alter channel mean open time (MOT) or open state inactivation kinetics (right traces and histogram, Fig. 1), in contrast to the effects of the Y1795C mutation and the previously reported effects of the Y1795H mutation. What properties of the Cys in this position underlie the apparently distinct slowing of inactivation and prolongation of channel mean open time?
FIGURE 1.
Inherited mutation Y1795C alters single channel properties of wild-type cardiac sodium channels. Cell-attached patch recordings are shown for wild-type (left), Y1795C (middle), and Y1795S (right) channels. Recordings were obtained in response to test pulses (−10 mV, 100 ms) applied at 2 Hz from −120 mV. Current recordings of individual and consecutive sweeps are shown to emphasize the effects of inherited mutations on channel opening kinetics. Ensemble currents (constructed by averaging 500 consecutive sweeps) are shown for each construct below the individual sweeps. Open time histograms were generated for each construct using 200-μs bins of all events recorded from 500 to 1000 sweeps. Patches used included three or fewer channels. MOT was estimated by the best-fit double exponential functions to each histogram. The fitted curves are illustrated as dashed lines in each panel. The resulting MOT estimates based on the extracted time constants are 0.76 ± 0.05 ms (WT, n = 4); 1.09 ± 0.8 ms (YC, n = 5); and 0.72 ± 0.04 ms (YS, n = 3); p = 0.01 for YC versus WT, YS versus WT (NS).
To explore this, we focused on the kinetics of open state inactivation measured under whole cell recording conditions and over a broad range of voltages. Na+ channel open state inactivation kinetics are voltage-dependent: inactivation of wild-type channels becomes faster with increasing depolarization. This voltage dependence of WT channels, which is most prominent at voltages negative to −10 mV, is evident in the data summarized in Fig. 2 A, which plots the time for peak inward current to decay by a factor of two as a function of test voltage. The Y1795C (♦) mutation slows open state inactivation over the range of voltages tested. Importantly, the slowing (increase in mean open time) is most apparent at voltages positive to −10 mV, where there is little change in the time course of inactivation with further depolarization, i.e., where there is a weak voltage dependence to inactivation kinetics. The effect of this mutation on the kinetics of the onset of inactivation appears to be linked to the presence of the cysteine thiol group, because replacement of Tyr1795 by either alanine or serine, amino acids with similar structures but lacking sulfur atoms, fails to slow inactivation kinetics (Fig. 2, A and B), and none of the mutations affected the voltage dependence of activation (Table 1). We found similar results when we replaced Tyr1795 by Arg, Glu, and His (see Table 1) and, importantly, found no effect on gating kinetics when the tyrosine was replaced by phenylalanine. As summarized in Table 1, the time to half decay of peak whole cell current varies with the following order for residues substituted at codon 1795: C ≫ A > Y (WT) = F > S > H > E = R. Alteration in inactivation gating does not vary systematically with either charge or size of substituted residues at position 1795, suggesting that mutation of Tyr1795 disrupts a structural motif of the C-terminal tail, which, in turn, plays a key role in controlling channel gating. Furthermore, because of the unique effects of the cysteine substitution, our results raise the possibility that the slowing of inactivation by the Y1795C mutation may be uniquely related to the presence of a Cys residue at this location. This suggests that disulfide bonds may form in the channel C-terminus when Tyr1795 is replaced by Cys in the disease-associated Y1795C mutation. Further, the data suggest that putative disulfide bond formation may impede transitions from the open to inactivated states (slow the onset of inactivation and prolongs channel mean open time).
FIGURE 2.
Distinct properties of Y1795C channels: effects on the kinetics of the onset of inactivation. Residue Y1795 (WT, Y, ○) was mutated to Cys (Y1795C, C, ♦), Ala (Y1795A, A, □), Phen (Y1795F, F, •), and Ser (Y1795S, S, ▪). (A) The plot shows the time for peak inward current elicited by the test voltages to decay to 50% of peak (t1/2) plotted versus test pulse voltage for each construct. Data shown are mean ± SE: n = 8 (WT); n = 7 (C); n = 6 (S); n = 8 (A). (B) Summary plot of time to half decay (t1/2) measured at +10 mV versus channel construct. Experimental numbers are as in A. *p < 0.01 versus WT; **NS versus WT.
TABLE 1.
Whole cell properties of Y1795 mutations
| Residue at 1795 | Activation (V1/2) (mV) | t1/2 (+10 mV) (ms) |
|---|---|---|
| Y (WT) | −23.0 ± 1.5 | 0.53 ± 0.03 |
| C | −24.6 ± 1.8NS | 0.90 ± 0.05*,# |
| A | −19.2 ± 1.5NS | 0.58 ± 0.02NS,# |
| S | −19.8 ± 1.1NS | 0.47 ± 0.01NS,# |
| H | −20.1 ± 2.1NS | 0.42 ± 0.03**,# |
| E | −21.3 ± 1.8NS | 0.32 ± 0.03*,# |
| R | −20.9 ± 0.5NS | 0.29 ± 0.04*,# |
| F | −22.9 ± 0.25NS | 0.52 ± 0.01NS,# |
The residue at position 1795 (i.e., C = C1795) defines the notation used in the table. Onset of inactivation kinetics is summarized as t1/2 (time of half to peak in milliseconds) measured at +10 mV. Each value is mean ± SE from six to eight experiments.
*p < 0.01 versus WT; **p < 0.05 versus WT; NS, not significant versus WT; #p < 0.01 versus C1795.
Disulfide bond formation underlies the effects of the Y1795C mutation on inactivation
To test this possibility, we used a pharmacological approach and studied the effects of DTT, a reducing reagent of disulfides in proteins, on channel inactivation for channel constructs in which we systematically varied residue 1795. We focused on the time course of the onset of inactivation and first compared the effects of DTT exposure on the onset kinetics of WT and Y1795C channels. As illustrated by the current traces in Fig. 3 A, DTT reversibly speeds the onset of inactivation of Y1795C channels, but has no effect on WT channels for which there is a Tyr at residue 1795. Fig. 3 B, which illustrates the effects of DTT on inactivation kinetics over a broad range of voltages, confirms this and, in addition, shows that DTT does not affect the kinetics of the onset of inactivation for channels in which the Tyr1795 is replaced by Ala (Y1795A (YA)). This result serves as a baseline against which the properties of mutant channels can be compared because it implies that, in the WT or Y1795A channel, it is not likely that disulfide bonds (and their disruption by DTT) contribute to these to biophysical properties of the channel. However, when Y1795C channels are treated with DTT, there is a significant speeding of the onset of inactivation, such that inactivation kinetics of DTT-treated Y1795C channels closely resemble the kinetics of WT channels (panel A, plot and inset). This is quantified in Fig. 3 C, which summarizes the effects of DTT treatment on inactivation kinetics measured at +10 mV. There is no significant effect of DTT on WT or YA channels (compare solid and open bars), but there is a significant decrease in t1/2 for YC channels, and t1/2 for YC channels + DTT is not significantly different from t1/2 of WT channels. These data provide evidence that disulfide bond formation may participate in some of the functional changes in channel gating induced by the LQT-3 mutation Y1795C. Interestingly, although not a focus of this study, we found no effect of DTT treatment on sustained current (Isus, bursting activity) of Y1795C channels (normalized Isus − DTT, 0.36 ± 0.04 (n = 14); + DTT, 0.3 ± 0.02, n = 3; p = not significant (NS)), and thus not all disease-associated function caused by the Y1795C mutation is likely to be mediated by disulfide bond formation.
FIGURE 3.
Evidence for disulfide bond modification of inactivation in Y1795C mutant channels. Cells expressing WT channel (○, •), A1795 (□, ▪), or C1795 (⋄, ♦) mutant channels were treated (solid symbols) or not treated (open symbols) with DTT. (A) The upper row shows current traces recorded from cells before, during, and after washout of exposure to DTT (50 mM) in the external bath. Currents are shown for WT (left) and YC channels (right) recorded at +10 mV. (B). The mean (± SE) time to half current decay (t1/2) is plotted versus test pulse voltage for each construct. The number of experiments was from six to nine for each construct. (C) The bars summarize t1/2 measured at +10 mV in the absence (open) and presence (solid) of DTT (10 mM). *p < 0.01 compared with untreated and with WT and YA; **NS versus WT.
Prediction of cysteine partners in disulfide bond by a mathematical model
Because the tertiary structure of the Na+ channel C-terminal domain has not yet been solved, we relied upon a model structure, previously reported by us, based on a remote relationship between the C-terminal domain and the structure of calmodulin. The model predicts that residue 1795 is almost adjacent to a cysteine molecule, residue C1850 (Fig. 4 A). Interestingly the residues are predicted to be in two different α-helices (H1 and H4; see Fig. 4, B and C), which cross in a region where two pairs of EF-hands are predicted to be packed against each other. The model also predicts that residue F1794, adjacent to Y1795 in the linear sequence, is far removed from C1850 (see Fig. 4 A), and thus the mutation F1794C would not be likely to form a disulfide bond with residue C1850 according to predictions of the model.
FIGURE 4.
Structural model of the C-terminal domain in the SCN5A Na+ channel. (A) The linear sequence of the proximal region of the C-terminal tail of SCN5A. The arrows indicate positions of F7194 (green), Y1795 (red), and C1850 (blue). (B) Model-predicted C-terminal structure. The N-terminal half (residues 1773–1863) of the SCN5A C-terminal domain was predicted to contain two EF-hand structural motifs that are packed to adopt a folding topology similar to the EF-hands in calmodulin (Cormier et al., 2002). The helix barrels H1-H2 show the first EF-hand structural motif in the model; the H3-H4 helix barrels show the second EF-hand structural motif. The three-dimensional ball-and-stick models show the predicted positions of the residues Y1795C and C1850 in the structural model. The sulfur atoms in the cysteines are colored in yellow. These two atoms are separated by 4 Å in the model structure. Residue F1794 that appears from the back of H1 helix is also shown in ball-and-stick model. (C) The helix wheel for H1 helix shows schematically the amphipathic nature of the H1 helix. The residues that are expected to be involved in the formation of a disulfide bond in the Y1795C mutant are shown in green. The ionizable residues in H1 helix are colored in red.
Experimental validation of model-predicted tertiary structure
We thus tested both of these model-defined predictions and summarize the results of our experiments in Fig. 5. First, we replaced the Cys at position 1850 in the Y1795C construct with an Ala (C1805A_Y1795C) and then tested the effects of the reducing agent DTT on the kinetics of open state inactivation. If the disulfide bonds form between Cys1795 and Cys1850, then this substitution should prevent bond formation and ablate the effects of DTT on these gating parameters. Fig. 5 A shows that this is the case. Fig. 5 B shows that replacement of the Phe at position 1794 by Cys in the WT channel (with a Tyr at position 1795; construct F1794C + Y1795) does not induce sensitivity of inactivation kinetics to DTT. These results are consistent with a C-terminal structure in which residue 1795 is sufficiently close to residue 1850 to enable disulfide bond formation when both residues are Cys molecules and also with a structure for which residues 1850 and 1794 are separated physically by distances that preclude disulfide bond formation. The predictions of our model are consistent with these experimental results.
FIGURE 5.
Experimental tests of structural model predictions. The effects of DTT (10 mM) were measured on channels in which the possible cysteine pairs were probed by mutation. In each panel, the upper row illustrates currents measured (at +10 mV) in the absence and presence of DTT (10 mM). The lower row plots inactivation t1/2 versus test pulse voltage in the absence (open) and presence (solid) of DTT 10 mM. (A). The Cys at position 1850 was replaced by Ala, and Tyr1795 was replaced by Cys to generate construct C1850A_Y1795C. Plotted is mean ± SE data (n = 6 for each data set). (B) The Phe at position 1794 was replaced by Cys, and the Tyr at position 1795 was not changed to make construct F1794C_Y1795. Shown are mean ± SE data (n = 4–6 for each construct). At 10 mV, there is no significant difference between DTT-free or DTT-containing data (p > 0.3 in both cases).
DISCUSSION
Mutation Y1795C and pathophysiology: altered inactivation
Open state inactivation of voltage-gated Na+ channels is due to rapid block of the inner mouth of the channel pore by the cytoplasmic linker between domains III and IV that follows voltage-induced channel openings (Stuhmer et al., 1989; West et al., 1992; Vassilev et al., 1988, 1989; Catterall, 1995). Disruption of inactivation by inherited mutation of the III/IV linker (Bennett et al., 1995) or its docking sites, underlie channelopathies in skeletal muscle, neuronal tissue, and in the heart (Green et al., 1998; Lossin et al., 2002; Keating and Sanguinetti, 2001; Balser, 2002; Lerche et al., 2001; Moxley III, 2000; Steinlein and Noebels, 2000). The Y1795C mutation also disrupts inactivation, but not by direct alteration of the inactivation gate or its previously identified docking sites. This mutation, and several others associated both with LQT-3 and Brugada Syndrome, occur in an acid rich region of the C-terminal tail of the channel (An et al., 1998; Bezzina et al., 1999; Veldkamp et al., 2000; Akai et al., 2000; Wei et al., 1999; Ackerman et al., 2001; Rivolta et al., 2001; Baroudi and Chahine, 2000; Deschenes et al., 2000; Wehrens et al., 2000).
Mechanistic basis for altered function: insight from molecular modeling
How might inherited C-terminal mutations, and in particular the Y1795C LQT-3 mutation, alter channel inactivation? Without a solved structure for Nav1.5 (SCN5A), it is difficult to reconcile the functional changes caused by the Y1795C mutation with information that is based solely on its cDNA sequence. However, the experimental results presented in this work provide insight into the plausible structure of the C-terminal domain, and thus can be used to understand how this mutation may alter channel inactivation. In a previous study, we constructed a molecular model for the first half of the C-terminal domain with a remote homologous structural template calmodulin. Based on this structural model, the distance between the Cα atoms of Y1795 and C1850 is estimated to be 6.8 Å. In general, a disulfide bond involves two cysteines for which the average Cα distance is 7 Å. Y1795C and C1850 are thus expected to be in range to form a disulfide bridge, provided that the molecular model is reasonably accurate (see Fig. 4). We have demonstrated with CD spectroscopy that the first half of the C-terminal domain is indeed consistent with the calmodulin structure in that the C-terminal domain forms a structure comprised of mostly α-helices (Cormier et al., 2002). In this work, we have further confirmed that the predicted head-to-tail distance for part of the model structure with 56 consecutive residues is in range to form a disulfide bond between C1850 and C1795. This head-to-tail distance is extremely sensitive to the tertiary structure of the intervening residues; any structural discrepancies in the orientation and packing of the helices between these two residues would introduce a large range of variation in this head-to-tail distance. The implication of the observed disulfide bond formation is that the tertiary structure for the C-terminal domain that consists of a pair of EF-hand structural motifs must be a reasonable model for the first half of the C-terminal domain.
The structural model has provided insights into the question as to how a disulfide bond in the C-terminal domain might introduce such a drastic changes in inactivation kinetics involving the III-IV loop and the pore domain. Mutational analysis revealed distinct effects of the Y1795C mutation that are particularly significant at positive voltages. At voltages positive to 0 mV, the kinetics of the onset of open state inactivation are significantly and uniquely slowed by the Y1795C mutation independent of test pulse voltage. Intuitively, this voltage independent effect may involve intradomain interactions, as the disulfide bond formation observed in this work. Two speculative models may be considered that involve the C-domain in the inactivation mechanism: 1), the C-terminal domain competes with the III-IV loop binding site in the opened pore domain, such that the III-IV loop can not effectively bind to the pore domain to inactive (block) the channel; or 2), the C-terminal domain interacts with the III-IV loop to hinder the interaction of the III-IV loop with the opened pore domain. A C-terminal domain stabilized by the interhelix disulfide bond can enhance both mechanisms and in turn slow down the inactivation kinetics. Moreover, the fact that polar residues in position 1795 speed up the kinetics of the inactivation is consistent with the possibility that regions near the mutation site may be close to the interaction interface of the C-terminal domain. This region is hydrophobic (see Fig. 4 B) and is likely to interact with the hydrophobic surface exposed in the opened pore domain or the isoleucine-phenylalanine-methionine hydrophobic motif in the III-IV loop. The experimental work so far has not provided direct evidence to support either of the models above. Since the III-IV loop is highly positively charged, the second model also suggests that long range electrostatic interactions involving the negative charge near the mutation site (see Fig. 4 B) can significantly affect the interactions and in turn the kinetics of the inactivation mechanism. These possibilities, which need to be tested with further experimental work, offer a structural basis for life threatening arrhythmias that accompany inherited mutations of the C-terminal domain of the human heart Na+ channel.
M. Tateyama and H. Liu contributed equally to this work.
References
- Abriel, H., C. Cabo, X. H. Wehrens, I. Rivolta, H. K. Motoike, M. Memmi, C. Napolitano, S. G. Priori, and R. S. Kass. 2001. Novel arrhythmogenic mechanism revealed by a long-qt syndrome mutation in the cardiac Na(+) channel. Circ. Res. 88:740–745. [DOI] [PubMed] [Google Scholar]
- Ackerman, M. J., B. L. Siu, W. Q. Sturner, D. J. Tester, C. R. Valdivia, J. C. Makielski, and J. A. Towbin. 2001. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 286:2264–2269. [DOI] [PubMed] [Google Scholar]
- Akai, J., N. Makita, H. Sakurada, N. Shirai, K. Ueda, A. Kitabatake, K. Nakazawa, A. Kimura, and M. Hiraoka. 2000. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 479:29–34. [DOI] [PubMed] [Google Scholar]
- An, R. H., X. L. Wang, B. Kerem, J. Benhorin, A. Medina, M. Goldmit, and R. S. Kass. 1998. Novel LQT-3 mutation affects Na+ channel activity through interactions between alpha- and beta1-subunits. Circ. Res. 83:141–146. [DOI] [PubMed] [Google Scholar]
- Balser, J. R. 2002. Inherited sodium channelopathies: models for acquired arrhythmias? Am. J. Physiol. Heart Circ. Physiol. 282:H1175–H1180. [DOI] [PubMed] [Google Scholar]
- Baroudi, G., and M. Chahine. 2000. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 487:224–228. [DOI] [PubMed] [Google Scholar]
- Bennett, P. B., K. Yazawa, N. Makita, and A. L. George. 1995. Molecular mechanism for an inherited cardiac arrhythmia. Nature (Lond.). 376:683–685. [DOI] [PubMed] [Google Scholar]
- Bezzina, C., M. W. Veldkamp, M. P. van den Berg, A. V. Postma, M. B. Rook, J. W. Viersma, I. M. van Langen, G. Tan-Sindhunata, M. T. Bink-Boelkens, A. H. van Der Hout, M. M. Mannens, and A. A. Wilde. 1999. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 85:1206–1213. [DOI] [PubMed] [Google Scholar]
- Catterall, W. A. 1995. Structure and function of voltage-gated ion channels. Annu. Rev. Biochem. 64:493–531. [DOI] [PubMed] [Google Scholar]
- Clancy, C. E., and R. S. Kass. 2002. Defective cardiac ion channels: from mutations to clinical syndromes. J. Clin. Invest. 110:1075–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, C. E., M. Tateyama, and R. S. Kass. 2002. Insights into the molecular mechanisms of bradycardia-triggered arrhythmias in long QT-3 syndrome. J. Clin. Invest. 110:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier, J. W., I. Rivolta, M. Tateyama, A. S. Yang, and R. S. Kass. 2002. Secondary structure of the human cardiac Na+ channel C terminus. Evidence for a role of helical structures in modulation of channel inactivation. J. Biol. Chem. 277:9233–9241. [DOI] [PubMed] [Google Scholar]
- Deschenes, I., G. Baroudi, M. Berthet, I. Barde, T. Chalvidan, I. Denjoy, P. Guicheney, and M. Chahine. 2000. Electrophysiological characterization of SCN5A mutations causing long QT (E1784K) and Brugada (R1512W and R1432G) syndromes. Cardiovasc. Res. 46:55–65. [DOI] [PubMed] [Google Scholar]
- Green, D. S., A. L. George, Jr., and S. C. Cannon. 1998. Human sodium channel gating defects caused by missense mutations in S6 segments associated with myotonia: S804F and V1293I. J. Physiol. 510:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch, T. J. 2000. Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Rev. Neurosci. 1:21–30. [DOI] [PubMed] [Google Scholar]
- Jentsch, T. J., B. C. Schroeder, C. Kubisch, T. Friedrich, and V. Stein. 2000. Pathophysiology of KCNQ channels: neonatal epilepsy and progressive deafness. Epilepsia. 41:1068–1069. [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott, K., and F. Lehmann-Horn. 2001. Human muscle voltage-gated ion channels and hereditary disease. Curr. Opin. Pharmacol. 1:280–287. [DOI] [PubMed] [Google Scholar]
- Keating, M. T., and M. C. Sanguinetti. 2001. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 104:569–580. [DOI] [PubMed] [Google Scholar]
- Kullmann, D. M. 2002. The neuronal channelopathies. Brain. 125:1177–1195. [DOI] [PubMed] [Google Scholar]
- Lerche, H., K. Jurkat-Rott, and F. Lehmann-Horn. 2001. Ion channels and epilepsy. Am. J. Med. Genet. 106:146–159. [DOI] [PubMed] [Google Scholar]
- Liu, H., M. Tateyama, C. E. Clancy, H. Abriel, and R. S. Kass. 2002. Channel openings are necessary but not sufficient for use-dependent block of cardiac Na(+) channels by flecainide: evidence from the analysis of disease-linked mutations. J. Gen. Physiol. 120:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin, C., D. W. Wang, T. H. Rhodes, C. G. Vanoye, and A. L. George, Jr. 2002. Molecular basis of an inherited epilepsy. Neuron. 34:877–884. [DOI] [PubMed] [Google Scholar]
- Marban, E. 2002. Cardiac channelopathies. Nature. 415:213–218. [DOI] [PubMed] [Google Scholar]
- Miller, C. 2000a. Ion channel surprises: prokaryotes do it again! Neuron. 25:7–9. [DOI] [PubMed] [Google Scholar]
- Miller, C. 2000b. Ion channels: doing hard chemistry with hard ions. Curr. Opin. Chem. Biol. 4:148–151. [DOI] [PubMed] [Google Scholar]
- Moxley III,, R. T. 2000. Channelopathies. Curr. Treat. Options Neurol. 2:31–47. [DOI] [PubMed] [Google Scholar]
- Nassir, F., Y. Xie, and N. O. Davidson. 2003. Apolipoprotein[a] secretion from hepatoma cells is regulated in a size-dependent mannor by alterations in disulfide bond formation. J. Lipid Res. 44:816–827. [DOI] [PubMed] [Google Scholar]
- Premkumar, L. S., F. Qin, and A. Auerbach. 1997. Subconductance states of a mutant NMDA receptor channel kinetics, calcium, and voltage dependence. J. Gen. Physiol. 109:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, F., A. Auerbach, and F. Sachs. 1996. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 70:264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta, I., H. Abriel, M. Tateyama, H. Liu, M. Memmi, P. Vardas, C. Napolitano, S. G. Priori, and R. S. Kass. 2001. Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J. Biol. Chem. 276:30623–30630. [DOI] [PubMed] [Google Scholar]
- Rivolta, I., C. E. Clancy, M. Tateyama, H. Liu, S. G. Priori, and R. S. Kass. 2002. A novel SCN5A mutation associated with long QT-3: altered inactivation kinetics and channel dysfunction. Physiol. Genomics. 10:191–197. [DOI] [PubMed] [Google Scholar]
- Schwake, M., T. Friedrich, and T. J. Jentsch. 2001. An internalization signal in ClC-5, an endosomal Cl-channel mutated in dent's disease. J. Biol. Chem. 276:12049–12054. [DOI] [PubMed] [Google Scholar]
- Steinlein, O. K., and J. L. Noebels. 2000. Ion channels and epilepsy in man and mouse. Curr. Opin. Genet. Dev. 10:286–291. [DOI] [PubMed] [Google Scholar]
- Stuhmer, W., F. Conti, H. Suzuki, X. Wang, M. Noda, N. Yahagi, H. Kubo, and S. Numa. 1989. Structural parts involved in activation and inactivation of the sodium channel. Nature (Lond.). 339:597–603. [DOI] [PubMed] [Google Scholar]
- Vassilev, P. M., T. Scheuer, and W. A. Catterall. 1988. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 241:1658–1661. [DOI] [PubMed] [Google Scholar]
- Vassilev, P., T. Scheuer, and W. A. Catterall. 1989. Inhibition of inactivation of single sodium channels by a site-directed antibody. Proc. Natl. Acad. Sci. USA. 86:8147–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldkamp, M. W., P. C. Viswanathan, C. Bezzina, A. Baartscheer, A. A. Wilde, and J. R. Balser. 2000. Two distinct congenital arrhythmias evoked by a multidysfunctional Na(+) channel. Circ. Res. 86:E91–E97. [DOI] [PubMed] [Google Scholar]
- Wehrens, X. H., H. Abriel, C. Cabo, J. Benhorin, and R. S. Kass. 2000. Arrhythmogenic mechanism of an LQT-3 mutation of the human heart Na(+) channel alpha-subunit: a computational analysis. Circulation. 102:584–590. [DOI] [PubMed] [Google Scholar]
- Wei, J., D. W. Wang, M. Alings, F. Fish, M. Wathen, D. M. Roden, and A. L. George, Jr. 1999. Congenital long-QT syndrome caused by a novel mutation in a conserved acidic domain of the cardiac Na+ channel. Circulation. 99:3165–3171. [DOI] [PubMed] [Google Scholar]
- West, J. W., D. E. Patton, T. Scheuer, Y. Wang, A. L. Goldin, and W. A. Catterall. 1992. A cluster of hydrophobic amino acid residues required for fast Na(+)-channel inactivation. Proc. Natl. Acad. Sci. USA. 89:10910–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, A. S. 2002. Structure-dependent sequence alignment for remotely related proteins. Bioinformatics. 18:1658–1665. [DOI] [PubMed] [Google Scholar]
- Yang, A. S., and B. Honig. 2000. An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance. J. Mol. Biol. 301:665–678. [DOI] [PubMed] [Google Scholar]
- Yang, A. S., and L. Y. Wang. 2002. Local structure-based sequence profile database for local and global protein structure predictions. Bioinformatics. 18:1650–1657. [DOI] [PubMed] [Google Scholar]