Abstract
For the uropathogenic Escherichia coli strain 536 (O6:K15:H31), the DNA sequences of three pathogenicity islands (PAIs) (PAI I536 to PAI III536) and their flanking regions (about 270 kb) were determined to further characterize the virulence potential of this strain. PAI I536 to PAI III536 exhibit features typical of PAIs, such as (i) association with tRNA-encoding genes; (ii) G+C content differing from that of the host genome; (iii) flanking repeat structures; (iv) a mosaic-like structure comprising a multitude of functional, truncated, and nonfunctional putative open reading frames (ORFs) with known or unknown functions; and (v) the presence of many fragments of mobile genetic elements. PAI I536 to PAI III536 range between 68 and 102 kb in size. Although these islands contain several ORFs and known virulence determinants described for PAIs of other extraintestinal pathogenic E. coli (ExPEC) isolates, they also consist of as-yet-unidentified ORFs encoding putative virulence factors. The genetic structure of PAI IV536, which represents the core element of the so-called high-pathogenicity island encoding a siderophore system initially identified in pathogenic yersiniae, was further characterized by sample sequencing. For the first time, multiple PAI sequences (PAI I536 to PAI IV536) in uropathogenic E. coli were studied and their presence in several wild-type E. coli isolates was extensively investigated. The results obtained suggest that these PAIs or at least large fragments thereof are detectable in other pathogenic E. coli isolates. These results support our view that the acquisition of large DNA regions, such as PAIs, by horizontal gene transfer is an important factor for the evolution of bacterial pathogens.
Pathogenicity islands (PAIs), as a distinct type of genetic element, were described for the first time for uropathogenic Escherichia coli strain 536 (O6:K15:H31) (2, 17), which is one of the model organisms of extraintestinal pathogenic E. coli (ExPEC) used for studies on ExPEC pathogenesis and the evolution of bacterial pathogens. The PAI type of genetic elements is characterized by a large size (>10 kb), the presence of virulence-associated genes, frequent association with tRNA-encoding genes or other att sites for temperate bacteriophages, and a G+C content different from that of the rest of the chromosome. These elements are frequently flanked by repeat structures and carry many fragments of other mobile and accessory genetic elements, such as bacteriophages, plasmids, and insertion sequence (IS) elements. Some PAIs are unstable regions and can spontaneously disappear from the chromosome. Therefore, PAIs are considered to have evolved from mobile genetic elements by horizontal gene transfer. It can also be assumed that these DNA regions, since their acquisition, underwent and will continue to undergo further evolutionary changes, resulting in an ongoing evolution of bacterial pathogens (14-16).
In addition to the PAIs of E. coli strain 536, several PAIs have been characterized for other ExPEC strains, and for many of them at least partial DNA sequence information is available. Two PAIs have been identified for each of the uropathogenic isolates J96 and CFT073 (13, 30, 36) as well as for the sepsis isolate AL862 (23). One island has been described for the meningitis K1 isolate EV36 (5). Although some of these islands resemble each other, as they carry identical genes, they are markedly diverse with respect to size, genetic content and organization, chromosomal insertion site, and stability.
One aim of this study involved the characterization and sequence determination of already identified PAIs of uropathogenic E. coli strain 536 to obtain a detailed picture of these genetic elements. Additionally, to improve knowledge of the evolution and distribution of PAIs, the presence of PAI I536- to PAI IV536-specific sequences in different E. coli strains was investigated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A collection of 132 E. coli strains were used in this study and include the Institut für Molekulare Infektionsbiologie collection, which has already been used for the investigation of the flanking regions of determinants encoding S-family adhesins (8). Additional strains have been isolated during a long-term study of women with chronic urinary tract infections: 2A1, 5A1, 5A1U, 2A2, 16A2U, 16A3, 8B1, 16B1, 19B1, 8B2, 3B5, 1E1, 1E5, 8F4, 3N1, 3N2, and 3N5. All strains were grown in Luria-Bertani medium (33).
DNA techniques.
Isolation of plasmid DNA and recombinant DNA was performed as previously described (33).
PCR.
A description of the primers used in this study is available as supplementary material (http://www.uni-wuerzburg.de/infektionsbiologie). Screening for the presence of PAI I536- to PAI IV536-specific sequences was performed by PCR with primers specific for the individual region to be amplified. Chromosomal DNAs of E. coli strains 536 and MG1655 were used as positive and negative controls, respectively. DNA primers were purchased from MWG Biotech (Ebersberg, Germany). The Taq DNA polymerase used for the detection of genes in different E. coli strains was purchased from Qiagen (Hilden, Germany). Grouping into the main phylogenetic lineages of the ECOR strain collection was done by a triplex PCR described previously (6). The virulence assessment of the extraintestinal E. coli strains included a multiplex PCR specific for a set of typical virulence-associated genes of extraintestinal E. coli (20).
DNA sequencing and sequence analysis.
Overlapping cosmid clones covering the entire regions of PAI I536 to PAI III536 and their flanking regions of E. coli strain 536 were sequenced as follows. Small insert libraries (2 to 2.5 kb) were generated by mechanical shearing of cosmid DNA (29). After end repair with T4 polymerase, the fragments were ligated into the prepared pTZ19R vector. Isolated plasmids were sequenced from both ends by using dye-terminator chemistry and analyzed with ABI-377 automated DNA sequencers (Applied Biosystems, Weiterstadt, Germany). After assembly, the remaining gaps were closed by primer walking on the plasmid clones. The Phrap software implemented in the STADEN software package was used for assembly and editing of the sequence data (35). The genetic structure of PAI IV536 was confirmed by sample sequencing of the left and right borders and of several open reading frames (ORFs) located on PAI IV536. The resulting sequences were compared with those of the already published high-pathogenicity islands (HPIs) of Yersinia enterocolitica and Yersinia pseudotuberculosis (3, 4).
Homology searches were performed with the BLASTN, BLASTX, PSI-BLAST, and PHI-BLAST programs from the National Center for Biotechnology Information (1). Sequence annotation was performed by using Artemis software (32). Codon usage tables for PAI I536 to PAI IV536 were determined with Artemis and compared with codon usage tables for E. coli K-12, for different gram-negative and gram-positive bacterial pathogens, and for several bacteriophages of E. coli strain K-12, E. coli O157:H7 strain EDL933, Salmonella spp., and Shigella spp. (http://www.kazusa.or.jp/codon/cgi-bin).
Southern hybridization.
Several PCR results from the screening for PAI I536 to PAI IV536 sequences in different E. coli isolates were confirmed by Southern hybridization of EcoRI-digested chromosomal DNA from the investigated E. coli isolates. After agarose gel electrophoresis, the EcoRI-digested E. coli genomic DNA was transferred to Biodyne B nylon membranes (PALL, Roβdorf, Germany). The probes used for hybridization were obtained by PCR with the primer pairs used for PCR-based screening and the chromosomal DNA of E. coli strain MG1655 as a template. Hybridization and detection were carried out by using an enhanced chemiluminescence labeling and signal detection system (Amersham Pharmacia Biotech, Freiburg, Germany) according to the manufacturer's recommendations.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of PAI I536 to PAI III536 were submitted to the EMBL database under accession numbers AJ488511 (PAI I536), AJ494981 (PAI II536), and X16664 (PAI III536).
RESULTS AND DISCUSSION
Genetic features of PAIs of E. coli strain 536.
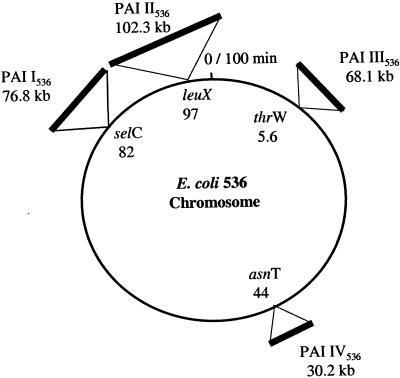
Three PAIs of E. coli strain 536 (PAI I536 to PAI III536) comprising about 270 kb were subcloned and sequenced. The genetic organization of PAI IV536, which represents the core element of the HPI initially described for Yersinia, was confirmed by PCR and by sample sequencing. The general characteristics of PAI I536 to PAI IV536 of uropathogenic E. coli strain 536 (e.g., chromosomal insertion site, association with tRNA-encoding genes, size, and encoded virulence factors) are compiled in Fig. 1 and Table1. The genetic organization of PAI I536 to PAI IV536 is depicted in Fig. 2. A detailed list of the known and putative ORFs identified on these islands is available as supplementary material (http://www.uni-wuerzburg.de/infektionsbiologie).
FIG. 1.
Comprehensive map of PAI I536 to PAI IV536 of uropathogenic E. coli strain 536. The map is based on the chromosome of E. coli strain MG1655. PAIs are indicated according to their chromosomal insertion sites next to tRNA-encoding genes.
FIG. 2.
Comparison of the genetic organization of PAI I536 to PAI IV536. Known or putative ORFs are grouped according to the following characteristics: blue, functional, known ORFs; green, truncated ORFs with a start codon and a stop codon; grey, as-yet-unidentified ORFs without homology on the DNA level. Nonfunctional ORFs (e.g., due to internal stop codons or frameshifts) are indicated by hatched symbols. ORF numbers are indicated below the corresponding ORF symbols. Functional or truncated tRNA-encoding genes are marked in red. Direct repeat (DR) structures flanking PAIs are indicated. Thick black lines below the PAIs represent regions of PAI I536 PAI IV536 which were detected by PCR. Several PAI-specific PCRs were grouped into PAI regions.
Generally, PAI I536 to PAI III536 are mosaic-like structures consisting of many DNA fragments which show the highest homology on the nucleotide level to chromosomal regions of other pathogenic E. coli strains (e.g., O157:H7 strains EDL933 and Sakai and uropathogenic O6 strain CFT073) and of Shigella flexneri (she and SHI-2 PAIs). Many fragments of PAI I536 to PAI III536 are also highly homologous to regions of different virulence plasmids of E. coli (pColV, pB171, pO157, and pAPEC-1), Shigella spp. (pWR100 and pWR501), and Yersinia spp. (pMT1 and pYVe227). Other fragments represent as-yet-unidentified DNA sequences without homology on the DNA level. These different regions are interspersed by each other and consist of already known functional and nonfunctional or truncated ORFs, of as-yet-unidentified putative ORFs with homology on the amino acid level to already known proteins, or of putative ORFs with unknown functions. A marked fraction of ORFs located on these PAIs is derived from mobile accessory genetic elements, such as bacteriophages, plasmids, and IS elements. Interestingly, PAI IV536, which represents a broad-host-range PAI present in many different enterobacteria, comprises only functional ORFs. The ORFs detected on PAI I536 to PAI IV536 can be functionally grouped as shown in Table2.
In the following text, ORFs are designated in a numerical way according to their localization and order on a PAI (as indicated by the index), starting downstream of the tRNA-encoding gene, which serves as the chromosomal insertion site for the respective PAI. Consequently, the nonfunctional bacteriophage P4-like integrase-encoding ORF on PAI I536 which is located downstream of selC is designated ORF 1I-536.
Characteristics of PAI I536.
PAI I536 is associated with the tRNA-encoding gene selC. This island is 76,843 bp in size, is flanked by 16-bp direct repeats, and has a G+C content of 46%. The nonfunctional bacteriophage integrase gene (ORF 1I-536) immediately downstream of selC exhibits high homology to other intP4-like genes described for PAIs as well as to that of phage φR73 located at this tRNA-encoding gene. In addition to already known virulence-associated genes located on PAI I536, such as the alpha-hemolysin-encoding gene cluster, the annotation of PAI I536 sequences revealed two as-yet-unidentified putative adhesin determinants with no homology on the nucleotide level. The deduced amino acid sequences together with their genetic organization indicated that putative ORF 15I-536 to ORF 18I-536 and ORF 37I-536 to ORF 42I-536 represent gene clusters coding for F17- and CS12-like adhesins, respectively. Interestingly, the gene product encoded by putative ORF 18I-536 shows homology to the F17a fimbrial subunit as well as to the uroepithelial cell adherence protein UcaA of Proteus mirabilis (EMBL accession no. CAA54703), indicating that fimbriae containing this subunit may indeed be involved in urinary tract infections. It will be important to determine whether these putative determinants are expressed and how the encoded fimbrial adhesins, whose homologues have so far been described only for enterotoxigenic E. coli, contribute to the virulence of extraintestinal E. coli. Another adhesin-like protein may be encoded by putative ORF 47I-536, which is preceded by two putative ORFs (ORF 45I-536 and ORF 46I-536) which may encode an ATP-binding cassette transporter. These ORFs have in common the facts that they also show no homology on the DNA level but that the deduced amino acid sequences are homologous to proteins (NMB0586, NMB0587, and NMB0588) encoded by three adjacent genes in Neisseria meningitidis strain MC58. Other interesting ORFs which have not been described so far are putative ORF 2I-536 and ORF 3I-536. These overlapping ORFs are similar in size, and their deduced amino acid sequences (314 and 310 amino acids, respectively) are 43 and 39% identical to that of modification methylase NgoFVII of Neisseria gonorrhoeae, which consists of 374 amino acids. However, both ORFs show only 78% identity on the DNA level over the first 89 nucleotides, thus excluding the possibility of gene duplication. As the methylation status of chromosomal DNA is also regulated in response to different stimuli and affects gene expression, it will be interesting to investigate whether these modification methylase-encoding ORFs (in case they are expressed) also influence the gene expression of E. coli strain 536. The importance of the dam-encoded methylase for general gene expression and expression of virulence-associated genes in Salmonella and E. coli was previously reported (18, 19).
Characteristics of PAI II536.
PAI II536 is associated with the tRNA-encoding gene leuX. This island is 102,200 bp in size, is flanked by 18-bp direct repeats, and has a G+C content of 46%. The functional bacteriophage integrase gene (ORF 1II-536) immediately downstream of leuX exhibits the highest homology to the intB gene of bacteriophage P4 located at this tRNA-encoding gene in E. coli K-12. Already known virulence determinants located on PAI II536 are the prf determinant (ORF 6II-536 to ORF 17II-536), which codes for the P-related fimbrial adhesin, and another alpha-hemolysin-encoding gene cluster (ORF 22II-536 to ORF 25II-536). Other putative virulence-associated genes present on this PAI are ORF 4II-536, which codes for the Hek adhesin described for E. coli (EMBL accession no. AY040859), and two putative ORFs (ORF 40II-536 and ORF 41II-536) without homology on the DNA level. The encoded gene product of ORF 40II-536 shows homology to filamentous hemagglutinin-like adhesins of Bordetella pertussis, Pseudomonas aeruginosa, and Yersinia pestis, and ORF 41II-536 shows homology to a conserved ORF which is located upstream of the hemagglutinin-encoding one and which is required for secretion of the adhesins. Another interesting as-yet-unidentified putative ORF is ORF 35II-536, whose encoded gene product shows homology to a fragment of modification methylase HgiDII of Herpetosiphon aurantiacus (EMBL accession no. P25265). The right-hand direct repeat structure is not immediately followed by E. coli K-12-specific sequences representing the conserved E. coli chromosomal backbone but by another 4-kb DNA region which is not present in E. coli K-12 strain MG1655. In this region, only one putative ORF coding for a hypothetical protein of E. coli O157:H7 strain Z5892 is located. This 4-kb DNA stretch is then followed by the E. coli K-12 chromosomal backbone starting with yjhS.
Characteristics of PAI III536.
PAI III536 is associated with the tRNA-encoding gene thrW. This island is 68,124 bp in size and has a G+C content of 47%. The functional bacteriophage integrase gene (ORF 1III-536) immediately downstream of thrW exhibits the highest homology to the int gene of bacteriophage SfX, which recognizes this tRNA-encoding gene as a chromosomal insertion site. This PAI is flanked by 47-bp direct repeats, as a truncated thrW gene located at the right-hand end of PAI III536 can, together with the functional copy of thrW at the left-hand end, serve as a direct repeat structure. However, approximately 7 kb of DNA is located between the truncated thrW gene at the right-hand end of PAI III536 and the transition into the E. coli K-12-like chromosomal backbone. This DNA stretch also represents foreign DNA, absent in E. coli strain MG1655, which contains an ORF with homology to the hemoglobin protease-encoding genes of pColV-K30 and pAPEC-1 (EMBL accession no. AJ223631 and AF218073, respectively). The Tsh hemoglobin protease has been shown to be involved in the virulence of, e.g., avian-pathogenic E. coli (10). As previously described (8), the right junction site of this composite structure includes sequences with homology to fragments of integrase genes of different S. flexneri and E. coli bacteriophages, followed by DNA sequences with homology to fragments of the insB and insA genes of the Iso IS1 element. These results demonstrate that the chromosomal region between thrW and yagU represents a composite structure of PAI III536 and other horizontally acquired sequences that could be considered to be flanked by sequences derived from Shigella bacteriophages which integrate into highly homologous attP sites next to the tRNA-encoding gene thrW. The occurrence of truncated tRNA-encoding genes within PAIs or DNA regions presumably acquired by horizontal gene transfer has also been observed for PAI II of CFT073 (30) and indicates the importance of these hot spots for chromosomal integration of foreign DNA sequences.
In addition to virulence determinants carried on PAI III536, such as the S fimbrial adhesin-encoding gene cluster sfa (ORF 17III-536 to ORF 25III-536) and the siderophore system-encoding gene cluster iro (ORF 27III-536 toORF 31III-536), sequence analysis of the entire region of PAI III536 revealed the presence of other known genes coding for virulence factors of E. coli; an example is ORF 52III-536, with homology to sap, which codes for an autotransporter-adhesin and which is located on the she PAI of S. flexneri 2a (EMBL accession no. AF200692). The as-yet-unidentified putative ORF 36III-536 encodes a HmuR-like heme receptor which has been described for Y. pestis (EMBL accession no. Q56989). Only short fragments of putative ORF 47III-536 and ORF 48III-536, coding for lysine decarboxylase (CadA) and cadaverine/lysine antiporter (CadB) homologues, respectively, show homology on the DNA level to the corresponding cadA and cadB genes of E. coli or Salmonella enterica serovar Typhimurium, indicating either their acquisition by horizontal gene transfer or the occurrence of multiple DNA rearrangement processes resulting in these cadA and cadB variants. Interestingly, ORFs with high homology to these genes are also located on PAI II of CFT073 (30). Whereas CadA and CadB have been proposed to be involved in acid tolerance mechanisms of Salmonella (12), the encoding genes are part of the so-called “black hole” in Shigella and enteroinvasive E. coli, which describes a large chromosomal deletion in these bacteria (25). The loss of this chromosomal region promotes the virulence of Shigella and enteroinvasive E. coli, as the product of lysine decarboxylase activity, cadaverine, blocks the action of Shigella enterotoxins (26).
Characteristics of PAI IV536.
PAI IV536 is associated with the tRNA-encoding gene asnT. This island represents the core element of the so-called HPI of pathogenic Yersinia species. It has been completely sequenced for several Yersinia strains (3, 4, 11). The left and right junction sites of PAI IV536 have also been sequenced before (34). We therefore verified the genetic organization of this PAI in E. coli strain 536 by sample sequencing of PCR products obtained with primers described elsewhere (22). The obtained sequences showed between 97 and 100% identity on the nucleotide level to other HPI-specific sequences of Y. pseudotuberculosis. Sequence determination of the left and right junction sites of PAI IV536 revealed only minor differences in the right-hand end of PAI IV536 compared to previous results. According to our sequence analysis, the 5′ end of PAI IV536 shows 98% identity to the sequence published earlier but comprises a 1,002-bp intergenic region between fyuA and Δb1978 instead of 1,226 bp, as described by Schubert and colleagues (34). According to these results, we conclude that by analogy to the HPI of Y. pseudotuberculosis, PAI IV536 is about 30.2 kb in size and has a G+C content of 57%. Flanking repeat structures are absent in PAI IV536, which contains the gene cluster required for biosynthesis of the siderophore system yersiniabactin.
Genes of mobile genetic elements present on PAI I536 to PAI IV536.
Besides the characteristic bacteriophage integrase-encoding genes located immediately downstream of the tRNA-encoding gene serving as a chromosomal insertion site for these PAIs, a considerable fraction of ORFs on PAI I536 to PAI III536 are remnants of various IS elements and transposons of different origins. The majority represent functional or nonfunctional transposase-encoding genes. Intact IS elements or transposons, however, have not been identified on these PAIs. Twenty-six fragments of IS elements have been detected on PAI I536 to PAI III536. They belong to different types of IS elements, including IS1, IS2, IS3, IS4, IS10, IS50R, IS100, IS110, IS629, IS630, IS679, IS911, IS1328, and IS1353. Many of the corresponding families of IS elements not only are restricted to enterobacteria but also are present among other bacteria (24). These IS elements are frequently located on virulence plasmids. Interestingly, at least one copy of each of the ORFs encoding the two subunits of the IS100 transposase is located on PAI I536 to PAI III536. Furthermore, several ORFs (copies of L0007 to L0010) of prophage CP-933 of E. coli O157:H7 strain EDL933 are always detectable on PAI I536 to PAI III536. Genes of prophage CP-933 are also present on PAIs of ExPEC strains CFT073 and AL862 (13, 23, 30). In addition, the putative ORF 63I-536 exhibits the highest homology to a fragment of an intron-associated reverse transcriptase/maturase-encoding ORF. The gene products of the adjacent putative ORF 52II-536 and ORF 53II-536, which share no sequence homology, are homologous to the quaternary ammonium compound resistance protein QacE of Brucella melitensis (EMBL accession no. AE009544) and E. coli (EMBL accession no. AF205943), respectively. In E. coli, qacE is known to be part of intron In53, a class 1 plasmid- and composite transposon-located integron (27). These data indicate that these PAIs in their present state result from repeated recombination events, including integration of different mobile genetic and other accessory genetic elements. PAI IV536 contains no remnants of IS elements.
Codon usage of ORFs located on PAI I536 to PAI IV536.
A comparison of codon usage tables for every PAI demonstrated that several codons occur with markedly different frequencies in these islands and in the chromosomal E. coli K-12-specific backbone. Details are available as supplementary material (http://www.uni-wuerzburg.de/infektionsbiologie). The codon usage of PAI I536 to PAI III536 differs from that of PAI IV536. The usage of codons such as ATA, ACA, AGA, and AGG is increased by at least a factor of 2 in these PAIs, whereas that of codon GCG is more than twofold lower than that in E. coli strain MG1655. This tendency is visible in several bacterial species, such as S. flexneri, Shigella sonnei, Salmonella enterica, Y. pseudotuberculosis, Y. enterocolitica, Staphylococcus aureus, and Bacillus subtilis, as well as in many bacteriophages of enterobacteria, such as 933W, ES18, P4, SfV, SfVI, and 7887. In PAI IV536, codons AGG and CCC are used at least twice as much as in E. coli strain MG1655, whereas the usage of codons ACT, AAA, and GGT is more than twofold lower in PAI IV536. These differences are indicative of the (more or less recent) acquisition of these PAIs by horizontal gene transfer.
Analysis of the presence of PAI I536- to PAI IV536-specific sequences in different E. coli isolates.
Based on the DNA sequences of PAI I536 to PAI IV536, we selected DNA primer combinations in order to detect specific sequences of these islands in a set of 132 different E. coli strains including many ExPEC and intestinal pathogenic E. coli isolates as well as several nonpathogenic E. coli strains (Fig. 2). Detailed results of this extensive screening for multiple PAIs, which complete recently published results on the presence of PAI III536 homologues in E. coli isolates (8), are available as supplementary material (http://www.uni-wuerzburg.de/infektionsbiologie). As shown in Table 3, PAI IV536-specific sequences showed a higher percentage of occurrence among the different E. coli strains tested in this study than sequences specific for PAI I536 to PAI III536. PAI III536-specific sequences were more frequently detectable than sequences specific for PAI II536. PAI I536-specific DNA regions were detected at the lowest frequency. PAI I536- to PAI III536-specific sequences were usually not detectable in intestinal pathogenic E. coli isolates, indicating that PAI I536 to PAI III536 represent a part of the ExPEC-specific gene pool. However, with a few exceptions due to local sequence differences, 17 to 28% of the intestinal pathogenic isolates were positive for all PAI IV536-specific PCRs. Between 67 and 92% of the ExPEC isolates were positive for all PAI IV536-specific PCRs, suggesting that they harbor the entire HPI core element. In contrast, the other three PAIs were not always completely detectable in some of the strains. In these instances, only certain PAI I536- to PAI III536-specific PCRs gave positive PCR results for the different strains used in this study.
TABLE 3.
Detection of PAI I536- to PAI IV536-specific sequences in different E. coli isolates
| E. coli strains (n)a | % Positive specific PCRs for the indicated region of PAI:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I536
|
II536
|
III536
|
IV536
|
|||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1 | 2 | 3 | 4 | |
| UTI (62) | 3.2 | 20.9 | 3.2 | 4.8 | 3.2 | 43.5 | 24.2 | 24.2 | 22.6 | 66.1 | 22.6 | 12.9 | 1.6 | 9.7 | 59.7 | 64.5 | 11.3 | 40.3 | 11.3 | 72.6 | 82.3 | 85.5 | 85.5 | 85.5 |
| Human and animal MNEC and SEPEC (28) | 0 | 14.3 | 0 | 0 | 0 | 32 | 25 | 7.1 | 21.4 | 35.7 | 25 | 10.7 | 0 | 7.1 | 7.1 | 46.4 | 7.1 | 28.6 | 7.1 | 64.3 | 85.7 | 89.3 | 89.3 | 89.3 |
| ECOR (28) | 7.1 | 14.3 | 3.6 | 7.1 | 7.1 | 21.4 | 17.9 | 7.1 | 16 | 47.4 | 5.3 | 0 | 0 | 0 | 35.7 | 39.3 | 0 | 32.1 | 0 | 53.6 | 75 | 78.6 | 78.6 | 78.6 |
| Diarrheagenic pathogens (18) | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 5.6 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 27.8 | 27.8 | 16.7 | 27.8 |
UTI, urinary tract infection; MNEC, meningitis-causing E. coli; SEPEC, sepsis-causing E. coli; ECOR, E. coli reference collection.
As already speculated, strains harboring determinants coding for members of the S family of fimbrial adhesins were shown to possess a common module of different forms of a PAI, depending on the different S adhesin types (8). The PCR screening based on the sequence of the complete region of PAI III536 confirmed this finding, as only some of the urinary tract infection isolates were positive for all PAI III536-specific PCRs. However, the chromosomal insertion site for their PAI III536 equivalent is so far unknown.
For some strains tested, all PAI I536-, PAI II536-, or PAI III536-specific sequences (with the above-mentioned exception for PAI III536) were detectable. Positive results in at least 13 specific PCRs for PAI I536 to PAI III536 strongly suggested that these strains contain the complete islands, as described for E. coli strain 536. Strains in which the complete PAIs of uropathogenic E. coli strain 536 have been detected are compiled in Table4. As shown by a previously described triplex PCR (6), all strains belong to major phylogenetic group B2, and the majority carry the same repertoire of virulence-associated genes as E. coli strain 536. The approximate genome sizes of these strains were determined by pulsed-field gel electrophoresis after CeuI digestion. The restriction patterns obtained indicated that, with the exception of those of strains RZ454 and RZ532 (4.6 Mb), the genome sizes are similar, ranging between 4.9 and 5.1 Mb. These results indicate that one or even multiple PAIs which have been described for uropathogenic E. coli strain 536 are also detectable in other ExPEC isolates. For the first time, we provide evidence by an extensive analysis of specific sequences of multiple PAIs in 132 different E. coli strains that identical PAIs, other than the HPI core element, most likely are present in different E. coli strains. This evidence indicates that these large chromosomal regions have been transferred by horizontal gene transfer.
PAIs and their importance for the evolution of ExPEC.
The results of this study increase knowledge on the variability of the genetic structures of PAIs in ExPEC. PAI IV536 represents a broad-host-range PAI also present in many other E. coli pathotypes and different enterobacteria and differs with respect to G+C content and genetic organization from PAI I536 to PAI III536, which have similar G+C contents and structural features and which have so far been detected only in ExPEC. However, the latter islands share several ORFs which are also present on many PAIs of other ExPEC isolates, such as bacteriophage CP-933-related sequences. This fact may be indicative of continously ongoing DNA rearrangements among PAI sequences resulting in evolution in ExPEC strains of different PAI variants deriving from an ancestral PAI which also consisted of phage CP-933. Although many PAIs of ExPEC strains superficially resemble each other with respect to the presence and/or genetic linkage of certain virulence determinants, there is nevertheless considerable variability in PAI composition and structural organization (8, 13, 21, 23, 30, 31, 36). The majority of homologous DNA sequences present on PAIs, such as fragments of IS elements and other accessory genetic elements, allow recombination and consequently PAI rearrangements and deletion and/or acquisition of foreign DNA that has been acquired by horizontal gene transfer. This feature facilitates rapid and continuous DNA rearrangements and remodeling of PAIs. In addition, some PAIs of ExPEC represent unstable genetic elements which can be deleted from the chromosome (2, 9, 28). It is assumed that such deleted PAIs, or at least large parts thereof, could be transferred to suitable recipients and thus contribute to the ongoing evolution of pathogenic bacteria (7, 15, 28).
In this study, we provide evidence that large regions of PAI I536 to PAI III536 can be detected in different ExPEC isolates, thus arguing for en bloc acquisition of these DNA regions by horizontal gene transfer. In summary, the study of the genetic organization of PAIs of uropathogenic E. coli strain 536 revealed the existence of several putative virulence determinants so far unknown in E. coli. A comparison of PAI I536 to PAI III536 with other PAIs of ExPEC supports a model in which PAI mobilization and horizontal gene transfer as well as continous reorganization of PAIs by recombination, for example, mediated by multiple fragments of accessory DNA elements present on one or multiple PAIs, contribute to the ongoing evolution of ExPEC variants.
TABLE 1.
Main features of PAI I536 to PAI IV536 of E. coli strain 536
| PAI | Chromosomal insertion site (tRNA) | Size (kb) | G+C content (%) | Known or putative virulence factor(s) | Integrase gene | Direct repeats (bp) |
|---|---|---|---|---|---|---|
| I536 | selC | 76.8 | 46 | Alpha-hemolysin, F17-like fimbriae,a CS12-like fimbriaea | P4-likeb | 16 |
| II536 | leuX | 102.2 | 46 | Hek adhesin, P-related fimbriae, alpha-hemolysin, hemagglutinin-like adhesina | P4 | 18 |
| III536 | thrW | 68.1 | 47 | S fimbriae, iro siderophore system, HmuR-like heme receptor,a Sap adhesin, Tsh-like hemoglobin proteasec | SfX | 47 |
| IV536 | asnT | 30.2 | 57 | Yersiniabactin siderophore system | P4-likeb |
Putative ORF or operon shows no homology on the DNA level but does with respect to the deduced amino acid sequence and genetic structure.
Integrase-encoding gene is nonfunctional due to internal stop codons or a mutated start codon.
Tsh-like hemoglobin protease-encoding gene is located in the 7-kb region outside of PAI III536, which is absent from the E. coli K-12 strain MG1655 genome.
TABLE 2.
Functional categories of known and putative ORFs located on PAI I536 to PAI IV536
| Known or putative function | No. of ORFs that are:
|
||
|---|---|---|---|
| Intact | Truncated | Nonfunctionala | |
| Related to mobile genetic elements | 14 | 33 | 3 |
| Toxin | 8 | ||
| Adhesin | 37 | 2 | |
| Iron acquisition | 14 | 3 | |
| Other virulence-associated function | 1 | 3 | |
| Enzymes | 2 | 10 | |
| Regulation | 3 | 5 | |
| Other functions | 7 | 17 | 2 |
| Hypothetical or unknownb | 31 | 48 | 2 |
| Without homology | 4 | ||
Due to internal stop codons or frameshifts.
Putative ORFs or their encoded products show homology on the DNA or protein level.
TABLE 4.
Characterization of different ExPEC isolates presumably containing complete PAIs of E. coli strain 536 or large fragments thereof
| Strain | Serotypea | Sourceb | Genome size (Mb)c | Characteristicsd | Presence of entire PAIe
|
||
|---|---|---|---|---|---|---|---|
| I536 | II536 | III536 | |||||
| 536 | O6:K15:H31 | UTI | 4.9 | malX papAH papC papEF papGIII fimH fyuA sfalfocDE sfaS hlyA kpsMTK5 | + | + | + |
| ECOR 52 | ND | Feces | 5 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc focG cnf1 iutA hlyA kpsMTK5 | + | − | − |
| ECOR 60 | ND | UTI | 5.1 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc focG cnfl iutA hlyA rfc kpsMTK5 | + | − | − |
| RZ479 | O6:K+:H− | UTI | 5.1 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 hlyA kpsMTK5 | + | + | + |
| 2980 | O18ac:K5 | Feces | 5 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS hlyA kpsMTK5 | − | + | − |
| HK24 | ND | Sepsis | 5 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 hlyA kpsMTK5 | − | + | + |
| HK8 | ND | Sepsis | 5 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 iutA hlyA kpsMTK5 | − | − | + |
| RZ422 | O6:K14:H− | UTI | 4.9 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc focG cnf1 cdtB traT hlyA kpsMTK5 | − | − | + |
| RZ451 | O6:K18/22:H31 | UTI | 5 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 traT hlyA kpsMTK5 | − | − | + |
| RZ505 | O6:K14:H | UTI | 5.1 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 hlyA kpsMTK5 | − | − | + |
| RZ532 | O6:K+:H31 | UTI | 4.6 | malX papAH papC papEF papGII/III fimH fyuA sfalfoc sfaS cnf1 hlyA kpsMTK5 | − | − | + |
ND, not determined.
UTI, urinary tract infection.
As determined by pulsed-field gel electrophoresis after CeuI digestion.
As determined by multiplex PCR; designation of PCR products is according to McCormick et al. (26).
Chromosomal insertion site of PAI III536 homologues is unknown. +, present; −, not present.
Acknowledgments
The work of the group in Würzburg, Germany, was supported by the SFB479, the Fonds der Chemischen Industrie, the Bayerische Forschungsstiftung, and the FAZIT-Stiftung. The Labor für Genomanalysen received support from the Forschungsmittel des Landes Niedersachsen.
We thank G. Balling and B. Middendorf (Würzburg, Germany) for sequence determination and analysis of the left and right borders of PAI IV536 as well as M. Schultheiss and B. Plaschke (Würzburg, Germany) for excellent technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cieslewicz, M., and E. Vimr. 1997. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol. Microbiol. 26:237-249. [DOI] [PubMed] [Google Scholar]
- 6.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrindt, U., and J. Hacker. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4:550-557. [DOI] [PubMed] [Google Scholar]
- 8.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrindt, U., U. Hentschel, J. B. Kaper, and J. Hacker. 2002. Genome plasticity in pathogenic and nonpathogenic enterobacteria. Curr. Top. Microbiol. Immunol. 264:157-175. [PubMed] [Google Scholar]
- 10.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletions of 102 kb of chromosomal DNA, which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 12.Foster, J. W., Y. K. Park, I. S. Bang, K. Karem, H. Betts, H. K. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140:341-352. [DOI] [PubMed] [Google Scholar]
- 13.Guyer, D. M., J.-S. Kao, and H. L. T. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli strain CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 15.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, J., G. Blum-Oehler, B. Janke, G. Nagy, and W. Goebel. 1999. Pathogenicity islands of extraintestinal Escherichia coli, p. 59-76. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands, plasmids, and other mobile elements. ASM Press, Washington, D.C.
- 17.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale, W. B., M. W. van der Woude, B. A. Braaten, and D. A. Low. 1998. Regulation of uropathogenic Escherichia coli adhesin expression by DNA methylation. Mol. Genet. Metab. 65:191-196. [DOI] [PubMed] [Google Scholar]
- 19.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. R., T. T. O'Bryan, M. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalioui, L., and C. Le Bouguénec. 2001. afa-8 gene cluster is carried by a pathogenicity island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. Black holes and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, B. A., M. I. Fernandez, A. M. Siber, and A. T. Maurelli. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell. Microbiol. 1:143-155. [DOI] [PubMed] [Google Scholar]
- 27.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 29.Oefner, P., S. P. Hunicke-Smith, L. Chiang, F. Dietrich, J. Mulligan, and R. W. Davis. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko, D. A., J. A. Phillips, X. Li, and H. L. T. Mobley. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. 184:1041-1049. [DOI] [PubMed] [Google Scholar]
- 31.Redford, P., and R. A. Welch. 2002. Extraintestinal Escherichia coli as a model system for the study of pathogenicity islands. Curr. Top. Microbiol. Immunol. 264:15-30. [PubMed] [Google Scholar]
- 32.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schubert, S., A. Rakin, D. Fischer, J. Sorsa, and J. Heesemann. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409-414. [DOI] [PubMed] [Google Scholar]
- 35.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 36.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]