Abstract
Proximity relationships within three doubly spin-labeled variants of the Na+/proline transporter PutP of Escherichia coli were studied by means of four-pulse double electron-electron resonance spectroscopy. The large value of 4.8 nm for the interspin distance determined between positions 107 in loop 4 and 223 in loop 7 strongly supports the idea of these positions being located on opposite sides of the membrane. Significant smaller values of between 1.8 and 2.5 nm were found for the average interspin distances between spin labels attached to the cytoplasmic loops 2 and 4 (position 37 and 107) and loops 2 and 6 (position 37 and 187). The large distance distribution widths visible in the pair correlation functions reveal a high flexibility of the studied loop regions. An increase of the distance between positions 37 and 187 upon Na+ binding suggests ligand-induced structural alterations of PutP. The results demonstrate that four-pulse double electron-electron resonance spectroscopy is a powerful means to investigate the structure and conformational changes of integral membrane proteins reconstituted in proteoliposomes.
INTRODUCTION
The knowledge of protein structure and dynamics is a basic prerequisite for the understanding of protein function. X-ray structural analysis and two-dimensional NMR spectroscopy provide deep insight into the structure-function relationship; however, the determination of the structure and conformational dynamics of membrane proteins and protein complexes is still a challenge. The combination of electron paramagnetic resonance (EPR) spectroscopy and site-directed spin labeling is emerging as an alternative technique (for reviews, see, e.g., Hubbell and Altenbach, 1994; Hubbell et al., 1998, 2000; Steinhoff, 2002). Here, natural amino acids at desired sites are replaced by cysteine residues which are then modified by nitroxide spin labels. This method has been successfully applied to define elements of the secondary and tertiary structure of membrane proteins, including solvent exposure (Hubbell et al., 1998), to determine the orientation and movements of individual segments of membrane proteins under physiological conditions or to characterize conformational changes that occur during protein function (Hubbell et al., 2000; Rink et al., 2000).
In the present report we apply and compare pulsed and continuous wave (cw) EPR methods to gather interresidue distance data and thereby local structural information of the membrane protein Na+/proline transporter PutP of Escherichia coli. The attachment of more than one spin label to a macromolecule leads to considerable changes of the cw EPR spectral shape if the interspin distance is <∼2 nm. These changes are due to the dipolar interactions between the individual spin labels and provide a measure for the interspin distance. In the range from 0.4 nm to 1.1 nm the intensities of the half-field transitions (Eaton et al., 1993) measured at low temperature (T < 200 K) provide a reliable distance measure. Peak height ratios, second moments, deconvolution based methods, and fitting algorithms of cw EPR spectra were shown to allow distance determination within the range of 0.8–2.2 nm (Likhtenshtein, 1976; Rabenstein and Shin, 1995; Steinhoff et al., 1997; Radzwill et al., 2001; Altenbach et al., 2001). These methods have been successfully applied to determine structural models of integral membrane proteins and protein complexes and to follow their conformational changes (see, e.g., Perozo et al., 1998; Mchaourab and Perozo, 2000; Wegener et al., 2001). In the distance range above 1.6 nm the uncertainty increases because the separation of the dipolar broadening contribution from the natural line width becomes increasingly difficult. For interspin distances exceeding 2 nm cw EPR is not applicable anymore; here pulse EPR techniques are a powerful approach. These techniques include pulse electron-electron double resonance techniques (Milov et al., 1981) such as four-pulse double electron-electron resonance (DEER) (Pannier et al., 2000), the 2 + 1 pulse sequence (Kurshev et al., 1989), multiple-quantum EPR (Borbat and Freed, 1999), and single-frequency techniques for refocusing electron-electron couplings (Jeschke et al., 2000). A comparison between the different methods for characterizing the distances between pairs of spin labels applied to the structurally well characterized, water-soluble protein human carbonic anhydrase II can be found in Persson et al. (2001).
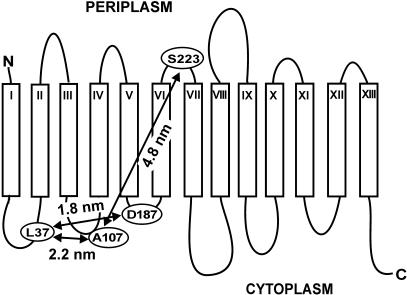
The aim of the present study is to apply DEER in combination with site-directed spin labeling for the determination of details of the so far unknown structure of the Na+/proline transporter PutP of E. coli. PutP is a member of the Na+/substrate symporter family (SSSF, TC 2.A.21) which currently comprises more than 200 similar proteins of pro- and eukaryotic origin (Reizer et al., 1994; Turk and Wright, 1997; Jung, 2001). These integral membrane proteins utilize the Na+ electrochemical gradient to drive the coupled uphill transport of a variety of substrates (sugars, amino acids, vitamins, osmolytes, ions, myo-inositol, urea, and water). For PutP of E. coli, gene fusion analyses, Cys accessibility studies, site-specific proteolysis, and site-directed spin labeling studies suggest a secondary structure model according to which PutP contains 13 transmembrane helices (TM) with the N-terminus located on the periplasmic side of the membrane and the C-terminus facing the cytoplasm (Jung et al., 1998a; Wegener et al., 2000) (compare also Fig. 1). As far as investigated, the 13-helix motif appears to be a special feature of the SSSF. Much attention has been paid to the identification of regions involved in the binding and translocation of Na+ and substrate (Jung, 2001; Pirch et al., 2002). Besides these studies, investigation of the structure and conformational dynamics of these transporters represents one of the current major challenges.
FIGURE 1.
13-Helix secondary structure model of the Na+/proline transporter of E. coli (PutP) according to Jung et al. (1998a). Putative transmembrane domains are represented as rectangles and numbered with Roman numerals; loops are numbered with Arabic numerals starting from the N-terminus. Amino acid positions which were spin labeled in our previous study are highlighted (boldface letters) (Wegener et al., 2000). Spin-labeled position of double Cys PutP derivatives, L37C/A107C, L37C/D187C, and A107C/S223C, investigated in this report are indicated by circles and connecting arrows.
Using site-directed spin labeling and EPR spectroscopy, this study provides for the first time information on distances between residue positions in the tertiary structure of PutP. In addition, the effect of PutP ligands on these distances is analyzed. The studies demonstrate that pulse DEER techniques can be used to investigate the structure and structural alterations of an integral membrane transport protein.
MATERIALS AND METHODS
Preparation and labeling of PutP
The generation of putP alleles encoding single Cys PutP molecules used in this study has already been described (Jung et al., 1998a). For overexpression, the putP alleles were cloned into plasmid pTrc99a (Amann et al., 1988) using restriction endonucleases NcoI and HindIII. The resulting plasmids were transformed into E. coli WG170 (F− trp lacZ rpsL thi Δ(putPA)101 proP219) (Stalmach et al., 1983). Cells were grown, membranes were prepared, and PutP was solubilized and purified by Ni-nitrilotriacetic acid affinity chromatography as described (Jung et al., 1998b). The eluted single Cys PutP molecules (in 50 mM KPi, pH 8.0, containing 300 mM KCl, 200 mM imidazole, 10% glycerol (v/v), and 0.04% β-D-dodecylmaltoside (w/v)) were reacted with methanethiosulfonate spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (Toronto Research Chemicals, Toronto) at molar spin label/protein ratio of 10:1 at 4°C for 3 h (hereafter we will refer to the spin-labeled residue as R1). Afterwards, unbound label was removed by dialysis against 50 mM KPi, pH 8.0, containing 10% glycerol and 0.04% β-D-dodecylmaltoside (w/v). The protein was reconstituted into proteoliposomes at a lipid/protein ratio of 20:1 (w/w) as described (Jung et al., 1998b). Finally, the proteoliposomes were washed twice with 50 mM KPi, pH 7.5, and resuspended in the same buffer to yield a PutP concentration of 30–80 μM. Proteoliposomes were frozen and stored in liquid N2 until use.
cw EPR measurements
cw EPR spectra were recorded with homemade X-Band EPR spectrometers equipped with a Bruker dielectric cavity for room-temperature (T ≅ 293 K) measurements or with an AEG TE-103 (Bruker, Ettlingen, Germany) cavity for low-temperature (T ≅ 170 K) measurements. The magnetic field was adjusted using a Bruker B-NM 12 B-field meter; microwave power was determined using a HP 430C bolometer. The PutP samples were loaded into quartz capillaries with an inner diameter of 1 mm. At room temperature 10 scans with a scan width of 12.8 mT (microwave power <0.6 mW, modulation amplitude <0.33 mT) were averaged. At 170 K 10 or 20 scans (12.8 mT scan width, microwave power <0.4 mW, modulation amplitude 0.23 mT) were averaged. After 12-bit AD conversion the data were processed using a personal computer.
cw EPR spectra simulations
cw EPR powder spectra were simulated based on the method described previously (Steinhoff et al., 1997). The motion of proteins is strongly restricted for temperatures <200 K. Below that temperature the apparent hyperfine splitting was found to deviate no more than 2% from the rigid limit value determined at 80 K and the EPR line shape resembles that of a powder spectrum. To account for the natural line shape the calculated powder stick spectrum is convoluted by a superposition of a Gaussian and a Lorentzian. Dipolar interaction is considered by additional convolution with a Pake pattern (Steinhoff et al., 1997; Radzwill et al., 2001).
Four-pulse DEER measurements
Four-pulse DEER measurements were performed using a Bruker Elexsys 580 spectrometer equipped with a 3-mm split-ring resonator under conditions of strong overcoupling (Q ≈ 100). The use of the 3-mm split-ring resonator increases sensitivity with respect to the 4-mm dielectric resonators used in our earlier work (Pannier et al., 2000) for the limited amount of protein samples and can provide a 2× larger microwave field amplitude at given incident power and quality factor. The measurements were performed at a temperature of 15 K to enhance sensitivity. With respect to measurements at 80 K, the number of accumulations in a given time decreases by a factor of 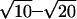 due to the increase in the longitudinal relaxation time at 15 K. This is slightly overcompensated by the gain in Boltzmann population difference, which is a factor of 5.3, so that any increase in the phase memory time Tm directly translates to a sensitivity advantage. Before insertion into the probehead, the samples were shock-frozen in liquid nitrogen to avoid crystallization of water. The pulse sequence (π/2)ν1-τ1-(π)ν1-t′-(π)ν2-τ1 + τ2 − t′-(π)ν1-τ2 -echo was used. (π/2)- and (π)-pulses had equal pulse lengths of 32 ns to assure equal excitation bandwidths. The interpulse delays were τ1 = 200 or 400 ns and τ2 = 1200 ns, except for the mutant A107R1/S223R1, where data for τ1 = 240 ns, τ2 = 2600 ns were also measured. The initial value of t′ was 80 ns and the increment 8 ns. A phase cycle +x/−x was applied to the first pulse and the two signals were subtracted. The pump frequency ν2 (typically 9.33 GHz) was set to the center of the resonator mode and the static magnetic field corresponded to pump excitation at the global maximum of the nitroxide spectrum. The observer frequency ν1 (typically 9.395 GHz) was set to the local maximum at the low-field edge of the spectrum. Accumulation times for the data sets varied between 14 and 32 h.
due to the increase in the longitudinal relaxation time at 15 K. This is slightly overcompensated by the gain in Boltzmann population difference, which is a factor of 5.3, so that any increase in the phase memory time Tm directly translates to a sensitivity advantage. Before insertion into the probehead, the samples were shock-frozen in liquid nitrogen to avoid crystallization of water. The pulse sequence (π/2)ν1-τ1-(π)ν1-t′-(π)ν2-τ1 + τ2 − t′-(π)ν1-τ2 -echo was used. (π/2)- and (π)-pulses had equal pulse lengths of 32 ns to assure equal excitation bandwidths. The interpulse delays were τ1 = 200 or 400 ns and τ2 = 1200 ns, except for the mutant A107R1/S223R1, where data for τ1 = 240 ns, τ2 = 2600 ns were also measured. The initial value of t′ was 80 ns and the increment 8 ns. A phase cycle +x/−x was applied to the first pulse and the two signals were subtracted. The pump frequency ν2 (typically 9.33 GHz) was set to the center of the resonator mode and the static magnetic field corresponded to pump excitation at the global maximum of the nitroxide spectrum. The observer frequency ν1 (typically 9.395 GHz) was set to the local maximum at the low-field edge of the spectrum. Accumulation times for the data sets varied between 14 and 32 h.
Four-pulse DEER data analysis
Distances or distance distributions were obtained from the dipolar time evolution data by procedures described by Jeschke and co-workers (Jeschke, 2002; Jeschke et al., 2002). Because of the limited signal-to-noise ratio for these protein samples, we tried to keep the number of fit parameters as small as possible. Model distance distributions consisting of a homogeneous background concentration chom and a single Gaussian peak characterized by its mean distance rm, the standard deviation σr, and an amplitude Ar were fitted to the data by minimization of the root mean square deviation using the shell factorization method (Jeschke et al., 2002) for signal simulation. Contributions of the Gaussian peak at distances r < 1.5 nm were dismissed, since spin pairs at such short distances cannot be excited by microwave pulses with pulse lengths tp ≥ 16 ns (Jeschke, 2002). The data were also processed by a model-free approach based on a cross talk-corrected approximate Pake transformation, mapping of the dipolar frequency distribution to a distance distribution, and distance-domain smoothing (Jeschke et al., 2002) with a Gaussian filter of 0.4 nm width.
RESULTS AND DISCUSSIONS
In our previous study the structural arrangement of the transmembrane helices TM II and TM III was determined by analyzing the residual mobility and accessibility of site-specifically attached nitroxide spin labels (Wegener et al., 2000). The results suggested that amino acid residues F45, S50, S54, W59, and M62 are part of TM II (cf. Fig. 1) whereas G39 and R40 on one side and A67 and G75 on the other side are located at membrane-water interfaces. TM III was suggested to be formed by residues S76 to G95. S71 and A107 were found to be located in water-exposed loops (loop3 and loop 4, respectively). Nitroxides side chains attached to positions 33 and 37 (loop 2) are on the cytoplasmic surface of the transporter but directed into an apolar environment. Binding of Na+ and/or proline to the transporter alters the mobility of the nitroxide group at positions 37 and 45. From these findings it was concluded that binding of ligands induces conformational alterations of PutP that involve at least part of TM II and the preceding cytoplasmic loop (loop L2). To further investigate the arrangement of cytoplasmic loops with respect to residue L37 the double Cys PutP derivatives L37C/A107C and L37C/D187C were prepared. EPR analyses of the spin-labeled samples were expected to provide information about distances between cytoplasmic loops L2 and L4 and between loops L2 and L6. In addition the doubly spin-labeled Cys PutP derivative A107C/S223C provides nitroxide positions on different sides of the lipid membrane (loops L4 and L7). According to the current model of PutP (Fig. 1) the interspin distance for this sample should exceed 4 nm.
cw EPR spectra (T = 293 K)
The room-temperature spectra of the single and double Cys PutP derivatives reacted with methane-thiosulfonate spin label are shown in Fig. 2. Differences in the S/N are due to different concentrations of the samples. The spectra of the single mutants L37R1 and A107R1 reveal intermediate motional restriction of the spin-label side chain as concluded from the apparent hyperfine splitting and the small line widths. This is in agreement with previous measurements and the proposed structure (Wegener et al., 2000) where these sites are located in the loops L2 and L4, respectively. In addition, accessibility measurements for water-soluble paramagnetic ions (Wegener et al., 2000) showed that the spin labels attached to positions 37 and 107 are close to the headgroup region of the bilayer or located in the water phase. The shape of the spectrum of the double mutant L37R1/A107R1 is in agreement with the superposition of the spectra of the two singly labeled samples (Fig. 2 b). The comparison of the spectra reveals that dipolar broadening is not observable in the cw EPR spectra of the double mutant at room temperature.
FIGURE 2.
Cw EPR spectra of the spin-labeled single (a) and double (b) Cys PutP derivatives recorded at room temperature (T ≅ 293 K). The narrow lines in the spectra of the single mutants L37R1 and A107R1 and the double mutants L37R1/A107R1 and L37R1/D187R1 are due to different (small) amounts of unbound spin label. In the top spectrum of b, we also show the integral weighted sum of the two single-labeled spectra of L37R1 and A107R1. This spectrum does not significantly differ from the spectrum of the double mutant L37R1/A107R1, especially no further broadening due to dipolar coupling is observable.
The spectra of the double Cys PutP derivatives L37R1/D187R1 and A107R1/S223R1 show more restricted motion of the nitroxide due to the fractions of the spin label located at positions 187 and 223, respectively: regarding the structural data (Jung et al., 1998a, Fig. 1) position 187 is located in the very short loop L6 which accounts for a more restricted motion of this nitroxide. S223 is located in the periplasmic loop L7. Information on tertiary interactions of this residue has not been available so far. Cysteine accessibility studies and analysis of the effect of ligand binding of fluorescence-labeled PutP-S223C implicates the residue in proline-induced conformational alterations (T. Pirch and H. Jung, unpublished information).
cw EPR spectra (T < 200 K)
In none of the low-temperature cw EPR spectra of the spin-labeled double Cys PutP derivatives shown in Fig. 3 does the dipolar interaction between the unpaired electrons of the nitroxides lead to a clearly visible dipolar line broadening. EPR spectra simulations with magnetic tensors and line width parameters determined from singly labeled samples show only minor deviations from the experimental data (not shown). Best fittings were obtained with interspin distance values equal to or exceeding (2.0 ± 0.2) nm with the distance distribution width fixed to 0.3 nm. We have to conclude that the average interspin distance values for the three samples must be larger than 1.8 nm. Although exact distance values are not obtainable for these samples by means of the cw EPR method the results provide a lower limit for the interresidue distance values.
FIGURE 3.
Low-temperature (T ≅ 170 K) cw EPR spectra of the spin-labeled double Cys PutP derivatives (solid line). For comparison we show also simulated EPR powder spectra according to the method described by Steinhoff et al. (1997) (dotted line). The values of the g- and A-tensor components were taken from Wegener et al. (2001): gxx = 2.0087, gyy = 2.0066, gzz = 2.0026, Axx = 0.52 mT, Ayy = 0.45 mT. The spectra were convoluted with a field-independent line-shape function, composed of a superposition of 48% Lorentzian and 52% Gaussian of 0.36 and 0.26 mT widths, respectively. The results of the fitting procedure show that an average interspin distance of 2.0 nm with a distance distribution width of 0.3 nm reproduce the experimental spectra in a proper way. In addition the value of the hyperfine splitting Azz was fitted to values of Azz,L37R1/A107R1 = 3.49 mT, Azz,L37R1/D187R1 = 3.47 mT, and Azz,A107R1/S223R1 = 3.48 mT, respectively.
DEER time-domain experiments
DEER time-domain data were acquired for the singly labeled mutant L37R1 and the three doubly labeled mutants L37R1/D187R1, L37R1/A107R1, and A107R1/S223R1. The normalized signals V(t)/V(t = 0) data are plotted in Fig. 4 as a function of the dipolar evolution time t = t′ − τ1. For the control experiment on the singly labeled mutant L37R1 we expect an exponential decay of the signal governed by the homogeneous distribution of protein molecules in the sample. Indeed, the data (Fig. 4 a) can be fitted by an exponential decay corresponding to an approximate local protein concentration of 1.8 mM. In contrast, the signal for the doubly spin-labeled mutant L37R1/D187R1 deviates strongly from an exponential decay (Fig. 4 b). The deviation is due to a fast-decaying contribution close to t = 0 (arrow) that corresponds to preferred distances between the two labels shorter than 2.2 nm. To characterize the distance distribution in more detail and get an estimate for the reliability of the data, we also present results of two different data analysis procedures (Fig. 5, a and b). By using a model-free direct transformation of the data with the cross talk-corrected approximate Pake transformation (Jeschke et al., 2002) we obtain a distance distribution that is rather broad and may have a maximum at distances <1.8 nm that are not accessible by our experiments (solid line in Fig. 5 a). However, the cw EPR measurements show that the distribution function must significantly decrease for interspin distances <1.8 nm. A fit of a distance distribution consisting of a Gaussian peak (distance between the two labels in one protein molecule) and a homogeneous background (intermolecular distances) provides a distance distribution that is very close to the distribution obtained by direct transformation (solid line in Fig. 5 b). The extracted distance distribution did not significantly change when microwave pulses with lengths of 16 ns instead of 32 ns were used. Fits for several fixed mean distances rm and variation of σr, Ar, and chom show that the data can still be described satisfactorily with mean distances up to ∼2.2 nm, but that the quality of the fit deteriorates strongly for rm > 2.2 nm (data not shown). The asymmetric width of the distribution was estimated to approach 0.8 nm, which suggests that the loop regions are very poorly ordered.
FIGURE 4.
Four-pulse DEER dipolar time evolution data for spin-labeled PutP derivatives. (a) L37R1, (b) L37R1/D187R1, (c) L37R1/D187R1 in the presence of 50 mM NaCl, (d) L37R1/D187R1 in the presence of 50 mM NaCl and 2 mM proline, (e) L37R1/A107R1, and (f) A107R1/S223R1.
FIGURE 5.
Extraction of distance distributions by direct transformation of the data to pair correlation functions (a and c) and fits of a Gaussian peak and homogeneous background (b and d) (y axes in arbitrary units). (a and b) L37R1/D187R1 in the absence (solid line) and presence (dashed line) of 50 mM NaCl. (c and d) L37R1/D187R1 (solid line) and L37R1/A107R1 (dashed line) in the absence of NaCl.
After addition of sodium chloride solution to the L37R1/D187R1 sample (total concentration of NaCl 50 mM) we find a strong increase of the proton modulations (fast oscillations marked by the bar in Fig. 4 c). This indicates a change in the environment of at least one of the two spin labels, which may be due to either better accessibility to water or a rearrangement of the protein structure that brings other side groups in closer contact with the nitroxide group. Furthermore, the average distance between the spin labels becomes larger as is manifest in the broadening of the feature close to t = 0. The increase in the mean distance between the two residues is also clearly revealed in the distance distributions shown in Fig. 5, a and b, although the maximum might still be <1.8 nm. Addition of a proline solution to the sample that already contains NaCl (total concentration of proline 2 mM) did not lead to further significant changes in the time-domain signal (Fig. 4 d) or the distance distribution (data not shown).
The data for the doubly labeled mutant L37R1/A107R1 (Fig. 4 e) are also significantly nonexponential. Here the distance distribution extends to longer distances than for the mutant L37R1/D187R1 in the absence of NaCl (Fig. 5, c and d). We may safely conclude that the mean distance between the two labels is <2.5 nm. Addition of NaCl solution causes only a slight increase in the proton modulations for the mutant L37R1/A107R1 (data not shown) and no significant change in the distance distribution. The dipolar evolution is also essentially unchanged after subsequent addition of proline solution.
Yet a different behavior is observed for the doubly labeled mutant A107R1/S223R1 (Fig. 4 f). Again the data are significantly nonexponential, but in this case a slow oscillation corresponding to longer distances is observed, which is superimposed with strong proton modulations even in the absence of NaCl. To measure the larger distance with sufficient confidence we increased τ2 to 2.6 μs (Fig. 6). The best fit (solid noiseless line) corresponds to a distance of 4.8 nm in agreement with the expectations for two residues on different sides of the membrane. The width of the Gaussian peak of 0.9 nm again indicates sizeable disorder in the loop regions, but note that this parameter is not very precisely defined at the given maximum dipolar evolution time t and signal-to-noise ratio. Fits with Gaussian peaks at several fixed mean distances and variation of σr, Ar, and chom reveal that the absolute error of this distance is smaller than 0.8 nm.
FIGURE 6.
Four-pulse DEER dipolar time evolution data for the doubly spin-labeled mutant A107R1/S223R1 (noisy data represented by wavy line) and fit by a distance distribution consisting of a single Gaussian peak at 4.8 nm with a width of 0.9 nm and homogeneous background (smooth solid line).
Implications for the structural model of PutP
The only information on tertiary interactions within a member of the SSSF available so far has recently been gained by chemical cross-linking of splits of the sodium/galactose transporter of Vibrio parahaemolyticus (vSGLT) (Xie et al., 2000). The studies suggest that the hydrophilic loops between TMs IV and V and between X and XI are within 8 Å of each other. The current study provides first information on proximity relationships within PutP of E. coli and allows for the first time a determination of long-range distances within the transporter molecule. The DEER data are in good agreement with the recently proposed secondary structure model of PutP (Jung et al., 1998a; Wegener et al., 2000) (compare Fig. 7 and Fig. 1). The large mean distance (4.8 nm) between positions 107 and 223 strongly supports the idea of these positions being located on opposite sides of the membrane, whereas the much shorter distances determined between positions 37, 107, and 187 are in agreement with a location on the same side of the membrane. In addition, the large widths of distribution of distances between the different positions arise most likely from locations on flexible loop regions of the transporter. The shortest distance (∼1.8 nm) was determined between positions 37 and 187 located on the cytoplasmic loops between TMs I and II and TMs V and VI, respectively (Fig. 7). Although L37 itself is not important for PutP function, R40 in its vicinity plays a crucial role in coupling of Na+ and proline transport (Quick et al., 1999). Similarly, also D187 is important for an efficient coupling of the transport of both ligands (Quick and Jung, 1998). Both residues are highly conserved within the SSSF. Taken together with the DEER measurements, it can be speculated that R40 and D187 interact with each other during the transport cycle. Clearly, further analyses are required to substantiate this idea.
FIGURE 7.
Secondary structure model of PutP of E. coli with the spin-labeled sites highlighted (compare to Fig. 1). Given are the mean distances between these sites as determined by four-pulse DEER.
Most importantly, the EPR measurements provide first estimates on directions of ligand-induced movements of amino acid positions of PutP relative to each other. Previous analyses of the influence of ligand binding on cysteine accessibility to sulfhydryl reagents, on fluorescence emission spectra, and on nitroxide mobility of single-labeled PutP molecules has identified amino acid positions involved in ligand-induced structural changes (Wegener et al., 2000; Jung, 2001; Pirch et al., 2002). In this study, the increase in proton modulations observed in the DEER measurements upon Na+ binding to PutP-L37R1/D187R1 suggests that one or both positions are exposed to a different environment most likely via a Na+-induced conformational change. An involvement of positions 37 and 187 in Na+-induced structural alterations is also suggested by the observed increase of the mean distance between these positions. In contrast to the 37/187 pair, Na+ and proline did not significantly influence the DEER signal of PutP-L37R1/A107R1. Ligand-induced distance changes in PutP-A107R1/S223R1 were not measured because the overall distance between positions 107 and 223 is too large and the DEER signal is approximately eightfold less sensitive to distance alterations than at shorter distances, as discussed, e.g., for the 37/187 pair.
In conclusion, the studies demonstrate that pulse DEER techniques can be used to investigate the structure and structural alterations of integral membrane proteins. Four-pulse DEER has successfully been applied to obtain for the first time information on distances between amino acid positions in the tertiary structure of the Na+ proline transporter PutP. In addition, it is shown that Na+ binding leads to alterations of these distances.
Acknowledgments
We thank M. Pannier for help in the initial stages of this work and C. Bauer for technical assistance.
This work was financially supported by the Deutsche Forschungsgemeinschaft (SFB 431-P10 (H.J.) and 431-P18 (H.J.S.)).
References
- Altenbach, C., K. J. Oh, R. Trabanino, K. Hideg, and W. Hubbell. 2001. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 40:15471–15482. [DOI] [PubMed] [Google Scholar]
- Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 69:301–315. [DOI] [PubMed] [Google Scholar]
- Borbat, P. P., and J. H. Freed. 1999. Multiple-quantum ESR and distance measurements. Chem. Phys. Lett. 313:145–154. [Google Scholar]
- Eaton, S. S., K. M. More, B. M. Sawant, and G. R. Eaton. 1993. Use of the EPR half-field transition to determine the interspin distance and the orientation of the interspin vector in systems with two unpaired electrons. J. Am. Chem. Soc. 105:6560–6567. [Google Scholar]
- Hubbell, W. L., and C. Altenbach. 1994. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol. 4:566–573. [Google Scholar]
- Hubbell, W. L., D. S. Cafiso, and C. Altenbach. 2000. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7:735–739. [DOI] [PubMed] [Google Scholar]
- Hubbell, W. L., A. Gross, R. Langen, and M. A. Lietzow. 1998. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 8:649–656. [DOI] [PubMed] [Google Scholar]
- Jeschke, G. 2002. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 3:927–932. [DOI] [PubMed] [Google Scholar]
- Jeschke, G., A. Koch, U. Jonas, and A. Godt. 2002. Direct conversion of EPR dipolar time evolution data to distance distributions. J. Magn. Reson. 155:72–82. [DOI] [PubMed] [Google Scholar]
- Jeschke, G., M. Pannier, A. Godt, and H. W. Spiess. 2000. Dipolar spectroscopy and spin alignment in electron paramagnetic resonance. Chem. Phys. Lett. 331:243–252. [Google Scholar]
- Jung, H. 2001. Towards the molecular mechanism of Na+/solute symport in prokaryotes. Biochim. Biophys. Acta. 1505:131–143. [DOI] [PubMed] [Google Scholar]
- Jung, H., R. Rübenhagen, S. Tebbe, K. Leifker, N. Tholema, M. Quick, and R. Schmid. 1998a. Topology of the Na+/proline transporter of Escherichia coli. J. Biol. Chem. 273:26400–26407. [DOI] [PubMed] [Google Scholar]
- Jung, H., S. Tebbe, R. Schmid, and K. Jung. 1998b. Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry. 37:11083–11088. [DOI] [PubMed] [Google Scholar]
- Kurshev, V. V., A. M. Raitsimring, and Y. D. Tsvetkov. 1989. Selection of dipolar interaction by the 2+1 pulse train ESE. J. Magn. Reson. 81:441–454. [Google Scholar]
- Likhtenshtein, G. I. 1976. Spin Labeling Methods in Molecular Biology. John Wiley, New York.
- Mchaourab, H. S., and E. Perozo. 2000. Determination of protein folds and conformational dynamics using spin-labeling EPR spectroscopy. In Distance Measurements in Biological Systems by EPR. L. Berliner, S. S. Eaton, and G. R. Eaton, editors. Kluwer, New York.
- Milov, A. D., K. M. Salikhov, and M. D. Shirov. 1981. Application of ELDOR in electron-spin echo for paramagnetic center space distribution in solids. Fiz. Tverd. Tela. 23:975–982. [Google Scholar]
- Pannier, M., S. Veit, A. Godt, G. Jeschke, and H. W. Spiess. 2000. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142:331–340. [DOI] [PubMed] [Google Scholar]
- Perozo, E., D. M. Cortes, and L. G. Cuello. 1998. Three-dimensional architecture of a K+ channel: implications for the mechanism of ion channel gating. Nat. Struct. Biol. 5:459–469. [DOI] [PubMed] [Google Scholar]
- Persson, M., J. R. Harbridge, P. Hammarstrom, R. Mitri, L. G. Martensson, U. Carlsson, G. R. Eaton, and S. S. Eaton. 2001. Comparison of electron paramagnetic resonance methods to determine distances between spin labels on human carbonic anhydrase II. Biophys. J. 80:2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirch, T., M. Quick, M. Nietschke, M. Langkamp, and H. Jung. 2002. Sites important for Na+ and substrate binding in the Na+/proline transporter of Escherichia coli, a member of the Na+/solute symporter family. J. Biol. Chem. 277:8790–8796. [DOI] [PubMed] [Google Scholar]
- Quick, M., and H. Jung. 1998. A conserved aspartate residue, Asp187, is important for Na+-dependent proline binding and transport by the Na+/proline transporter of Escherichia coli. Biochemistry. 37:13800–13806. [DOI] [PubMed] [Google Scholar]
- Quick, M., S. Stölting, and H. Jung. 1999. Role of conserved Arg40 and Arg117 in the Na+/proline transporter of Escherichia coli. Biochemistry. 38:13523–13529. [DOI] [PubMed] [Google Scholar]
- Rabenstein, M. D., and Y. K. Shin. 1995. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. USA. 92:8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzwill, N., K. Gerwert, and H. J. Steinhoff. 2001. Time-resolved detection of transient movement of helices F and G in doubly spin-labeled bacteriorhodopsin. Biophys. J. 80:2856–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer, J., A. Reizer, and M. H. J. Saier. 1994. A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta. 1197:133–166. [DOI] [PubMed] [Google Scholar]
- Rink, T., M. Pfeiffer, D. Oesterhelt, K. Gerwert, and H. J. Steinhoff. 2000. Unraveling photoexcited conformational changes of bacteriorhodopsin by time resolved electron paramagnetic resonance spectroscopy. Biophys. J. 78:1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmach, M. E., S. Grothe, and J. M. Wood. 1983. Two proline porters in Escherichia coli K-12. J. Bacteriol. 156:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff, H.-J. 2002. Methods for study of protein dynamics and protein-protein interaction in protein-ubiquitination by electron paramagnetic resonance spectroscopy. Front. Biosci. 7:c97–110. [DOI] [PubMed] [Google Scholar]
- Steinhoff, H. J., N. Radzwill, W. Thevis, V. Lenz, D. Brandenburg, A. Antson, G. Dodson, and A. Wollmer. 1997. Determination of interspin distances between spin labels attached to insulin: comparison of electron paramagnetic resonance data with the x-ray structure. Biophys. J. 73:3287–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk, E., and E. M. Wright. 1997. Membrane topology motifs in the SGLT cotransporter family. J. Membr. Biol. 159:1–20. [DOI] [PubMed] [Google Scholar]
- Wegener, A. A., J. P. Klare, M. Engelhard, and H. J. Steinhoff. 2001. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20:5312–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener, C., S. Tebbe, H. J. Steinhoff, and H. Jung. 2000. Spin labeling analysis of structure and dynamics of the Na+/proline transporter of Escherichia coli. Biochemistry. 39:4831–4837. [DOI] [PubMed] [Google Scholar]
- Xie, Z., E. Turk, and E. M. Wright. 2000. Characterization of the Vibrio parahaemolyticus Na +/glucose cotransporter. A bacterial member of the sodium/glucose transporter (SGLT) family. J. Biol. Chem. 275:25959–25964. [DOI] [PubMed] [Google Scholar]