Abstract
Chronic Pseudomonas aeruginosa lung infection is the major cause of morbidity and mortality in cystic fibrosis (CF) patients. One P. aeruginosa virulence factor unique to CF isolates is overproduction of alginate, phenotypically termed mucoidy. Mucoidy is the result of increased transcription from the algD gene and is activated by the transcriptional regulator AlgR. Mutations in algR result in a nonmucoid phenotype and loss of twitching motility. Additionally, AlgR controls transcription of algC, encoding a dual-function enzyme necessary for both lipopolysaccharide (LPS) and alginate production. Therefore, to determine the effect of algR on P. aeruginosa virulence, an algR mutant was examined for sensitivity to reactive oxygen intermediates, killing by phagocytes, systemic virulence, and the ability to maintain a murine lung infection. We found that P. aeruginosa PAO700 (algR::Gmr) was less lethal than PAO1, as tested in an acute septicemia infection mouse model, and was cleared more efficiently in a mouse pneumonia model. Additionally, the algR mutant (PAO700) was more sensitive to hypochlorite. However, PAO700 was more resistant to hydrogen peroxide and killed less readily in an acellular myeloperoxidase assay than PAO1. There was little difference in killing between PAO1 and PAO700 with macrophage-like J774 cells and human polymorhonuclear leukocytes. Two-dimensional gel analysis of P. aeruginosa algR mutant and wild-type protein extracts revealed 47 differentially regulated proteins, suggesting that AlgR plays both a positive role and a negative role in gene expression. Together, these results imply that AlgR is necessary for virulence and regulates genes in addition to the genes associated with alginate and LPS production and pilus function.
Pseudomonas aeruginosa is a ubiquitous, gram-negative, opportunistic pathogen that is capable of acute infection in neutropenic and burn patients (2). In cystic fibrosis (CF) patients, P. aeruginosa is the leading cause of morbidity and mortality due to its ability to establish a chronic infection (21). The dominant phenotype of P. aeruginosa associated with strains that chronically infect the respiratory tract of CF patients is overproduction of alginate (9, 13, 25, 58, 80). Alginate is an exopolysaccharide composed of a linear copolymer of β-d-mannuronic and α-l-guluronic acids (15, 34, 35). This excess production of alginate, phenotypically termed mucoidy, allows P. aeruginosa to evade phagocytosis by neutrophils and macrophages (59, 74). Furthermore, alginate insulates the bacterium from reactive oxygen intermediates (68-70) and hypochlorite generated by the phagocytic cells of the host (33). Since alginate overproduction is a hallmark phenotype of clinical P. aeruginosa isolates from CF patients, it has been studied extensively (25).
Several studies have examined the molecular mechanisms associated with the conversion of P. aeruginosa from a nonmucoid phenotype to an alginate-overproducing phenotype (25). Fyfe and Govan initially reported that mutations mapping to the late region of the P. aeruginosa chromosome (muc loci) are responsible for the conversion to mucoidy (19). Molecular characterization of the genes located in this region led to identification of five linked genes, algU (39, 42, 66, 67, 82) (algT [12, 23, 27]), mucA (5, 40, 64, 81), mucB (4, 40, 60) (algN [23]), mucC (4) (algM [43]), and mucD (4) (algY [43]). One of the major mechanisms that induce the constitutive mucoid phenotype of P. aeruginosa has been elucidated and directly involves the action of MucA (5, 41). Mutations in the mucA gene encoding an anti-sigma factor allow the release of the alternative sigma factor AlgU (AlgT, σ22) (60). AlgU (AlgT, σ22) is responsible for initiating transcription of the first committed step in the biosynthetic pathway of alginate, algD (39), itself (65), the heat shock sigma factor rpoH (64), and the transcriptional regulator algR (42). The transcriptional regulator AlgR positively activates algD transcription by binding to three different binding sites within the algD promoter (50, 51). Since AlgR shows homology to two-component transcriptional activators, investigators initially examined the role of phosphorylation in AlgR-dependent gene activation (11, 61). Although the AlgR protein was phosphorylated in vitro by CheA (11), it has recently been shown that phosphorylation may not play a role in the activation of AlgR-dependent activation of the algD promoter (37). Another gene associated with alginate production, algC, is also regulated by AlgR (83). The algC gene encodes a bifunctional enzyme that is involved in lipopolysaccharide (LPS) production (phosphoglucomutase activity) (8) and alginate production (phosphomannomutase activity) (83). Additionally, AlgC may be required for rhamnolipid production (53). AlgR binds to three sites within the algC promoter to regulate its transcription (18, 84). Transcriptional fusion studies of the algC promoter have shown that algC expression is reduced approximately fivefold in the absence of AlgR (83). Moreover, inactivation of the algR gene eliminates twitching motility, implying that algR has a role in the function of type IV fimbriae (77). Type IV pili have been shown to be required for P. aeruginosa attachment (6, 7) and biofilm formation (54). Taken together, these studies indicate that algR may play a more general role in the regulation of virulence in P. aeruginosa.
In this study, we examined the role that algR plays in stress responses to hypochlorite and hydrogen peroxide, interactions with murine macrophages and human neutrophils, and virulence, as tested in murine acute sepsis and inhalation pneumonia models.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are shown in Table 1. P. aeruginosa was grown on Pseudomonas isolation agar (PIA) (Difco) and in Luria-Bertani (LB) broth supplemented with 150 and 35 μg of gentamicin per ml, respectively, when required. PAO700, a derivative of PAO1 containing a chromosomally inactivated algR gene with a gentamicin cassette, was a generous gift from Vojo Deretic. Gene replacement was confirmed by Southern blotting (data not shown).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | algR+ Alg−wt | 28 |
| PAO700 | PAO1 algR::Gmr | V. Deretic |
| E. coli DH5α | lacZΔM15 recA1 | Bethesda Research Laboratories |
| Plasmids | ||
| pVDZ’2R | pVDZ’2 algR+ Tcr | This study |
| pDSL-1 | pVDZ’2 hemCD | This study |
| pCMR7 | pVDtac24 algR | 49 |
| pRK2013 | tra functions | 17 |
Alg−wt, nonmucoid wild type.
Nucleic acid manipulation and recombinant DNA methods and conjugation.
Construction of pVDZ'2R involved insertion of a BamHI fragment derived from pCMR7 containing the wild-type algR gene into plasmid pVDZ'2 (49). For construction of plasmid pDSL-1, a 2.1-kb fragment containing both hemC and hemD was generated by PCR by using the Expand High Fidelity PCR system (Roche). The hemCD 2.1-kb fragment was cloned into pCR2.1 (Invitrogen) and sequenced. The pCR2.1 plasmid containing both hemC and hemD was subsequently digested with BamHI and XbaI and then cloned into pVDZ'2, creating pDSL-1. Plasmids were introduced into P. aeruginosa by triparental conjugation with Escherichia coli DH5α harboring helper plasmid pRK2013 (17) and the plasmid to be introduced into P. aeruginosa, as described previously (31).
S1 nuclease protection assay.
The S1 nuclease protection assay was performed as previously described (65). P. aeruginosa strains were grown to an optical density at 600 nm (OD600) of 0.4. Briefly, bacterial cells were washed on ice in 50 mM Tris (pH 7.5) and lysed in 3.3% sodium dodecyl sulfate (SDS)-50 mM Tris (pH 7.5), and the total cellular RNA was isolated by centrifugation through a cushion of 5.7 M CsCl. S1 nuclease protection analysis was performed with single-stranded DNA probes uniformly labeled with 32P as previously described (65). Identical amounts of RNA (50 μg) were hybridized with equally distributed radiolabeled probe in each set of reactions. The hybridization probe was prepared from the M13 derivative M13hemC harboring the promoter region of P. aeruginosa hemC encompassing 732 bp (nucleotides 5922821 to 5922089 of the P. aeruginosa PAO1 genome). The M13hemC template was used with specific primer S303 (5′-AGGGTGACGGTCAAGCCGGG-3′; positions 96 to 115 with respect to the hemC initiation codon) to generate the single-stranded probe and sequencing ladder. S1 nuclease protection products were electrophoresed on sequencing gels (7.5% acrylamide, 8 M urea, 100 mM Tris, 100 mM boric acid, 2 mM EDTA; pH 8.3) along with the sequencing ladder produced by using the same template (M13hemC) and oligonucleotide primer (S303) that were used for preparing the S1 nuclease probes. Radioactive decay contributed to the presence of multiple bands observed in the 5′ end of the mRNA because of the uniform labeling of the single-stranded DNA probe and has been noted previously (32).
Western blotting.
P. aeruginosa strains PAO1, PAO700, PAO700/pCVDZ'2R and PAO1/pCMR7 were grown in LB broth at 37°C with aeration. Strains PAO700/pCVDZ'2R and PAO1/pCMR7 were grown in the presence of 50 μg of tetracycline per ml and 300 μg of carbenicillin per ml, respectively, to an OD600 of 0.2, isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 1 mM) was added, and the cultures were allowed to grow to an OD600 of 0.4. The cultures of all the strains were collected at an OD600 of 0.4, the OD600 used for the acellular, cellular, and animal virulence studies, by centrifugation (6,000 × g for 10 min). The bacteria were resuspended in 50 mM Tris-HCl (pH 8.0)-150 mM NaCl and then lysed by sonication. Total protein (15 μg) of each strain was loaded onto an SDS-10% polyacrylamide gel and then electroblotted onto a polyvinylidene difluoride membrane. The membrane was probed by using the anti-AlgR monoclonal antibody (11) with detection with a horseradish peroxidase-conjugated goat anti-mouse monoclonal antibody and an Opti-4CN substrate kit (Bio-Rad).
Oxidative stress killing assays and disk sensitivity assays.
The sensitivity of P. aeruginosa in suspension to sodium hypochlorite (NaOCl) was tested by the method of Learn et al. (33). P. aeruginosa was grown in 50 ml of LB broth to an OD600 of 0.4, centrifuged at 3,000 × g, and washed in 10 mM phosphate-buffered saline (PBS) (pH 7.4). The cells were resuspended in PBS-10 mM glucose (PBS-G) to an OD600 of 0.25 and allowed to rest for 30 min at 37°C. Approximately 2 × 108 CFU/ml was added to 9 ml of a 25 μM NaOCl solution in PBS-G and incubated at 37°C. Samples (1 ml) were removed, diluted serially, and plated onto LB medium at 0, 15, 30, and 45 min. To test the sensitivity of P. aeruginosa in suspension to H2O2, PAO1 (algR) and PAO700 (algR::Gmr) were grown to an OD600 of 0.4, washed in 10 mM PBS (pH 7.4), and resuspended in 10 ml of PBS-G to an OD600 of 0.25. The cells were allowed to rest for 30 min at 37°C. Approximately 2 × 108 CFU/ml was added to 9 ml of prewarmed PBS-G containing 15 mM (final concentration) H2O2 and incubated at 37°C. The numbers of CFU of surviving bacteria were determined by plating preparations on LB agar after serial dilution in PBS. Myeloperoxidase oxidative killing of P. aeruginosa was performed as previously described (80). Disk sensitivity assays were carried out as previously described (3, 42, 82).
Neutrophil- and macrophage-mediated bactericidal assays.
The procedures used for isolation of human neutrophils and the bactericidal assays were based on previously described methods (46). Briefly, heparinized blood was mixed by inversion with an equal volume of prewarmed 3% dextran (T500; Pharmacia) in 0.85% NaCl, and erythrocytes were sedimented by gravity for 18 min at room temperature. Leukocytes were collected by centrifugation at 500 × g for 10 min at 4°C, the pellets were resuspended in 40 ml of cold 0.85% saline, and a 10-ml Ficoll-Hypaque cushion was layered beneath the cell suspension. The solution containing 9.97% (wt/vol) Hypaque (sodium ditrizoate; Winthrop, New York, N.Y.) and 6.35% (wt/vol) Ficoll (T400; Pharmacia) had a density of 1.08 g/ml. After centrifugation at 250 × g for 40 min at 20°C, neutrophil pellets were resuspended in 10 ml of 0.2% NaCl and incubated for 20 s to hypotonically lyse the remaining erythrocytes. Immediately following the lysis of erythrocytes, 10 ml of 1.5% NaCl was added to balance the tonicity. Neutrophils were collected by centrifugation at 500 × g for 6 min at 4°C and resuspended in Hanks' buffered saline solution (HBSS) (pH 7.4). Neutrophils (3 × 107 cells) in HBSS were incubated with 3 × 108 CFU of P. aeruginosa in the presence of 10% heat-inactivated autologous human serum in a 10-ml (total volume) mixture. A 1-ml sample of cells for each time point was collected by centrifugation at 900 rpm (Sorvall RT6000 D) for 5 min and resuspended in an equal volume of cold sterile water to lyse the neutrophils before bacterial viability was determined.
Macrophage-mediated killing of P. aeruginosa was carried out as follows. Mouse macrophage cell line J774 (ATCC TIB-67) was grown in Dulbecco's modified Eagle's medium (DMEM) (low glucose; Cellgro) supplemented with 5 mM l-glutamine and 5% fetal bovine serum (HyClone) in a 5% CO2 atmosphere at 37°C. Mid-log-phase (OD600, 0.4) P. aeruginosa grown in LB broth at 37°C was collected by centrifugation at 9,200 × g and resuspended to an OD600 of 0.4 in PBS. Macrophage cells (5 × 107 cells) were incubated in DMEM containing 5% fetal bovine serum with 5 × 107 CFU of P. aeruginosa for 30 min at 37°C. After 30 min, the macrophages were washed three times with PBS to remove nonadherent P. aeruginosa. After the final wash, 1 ml of DMEM was added. Macrophages were harvested by removing the medium and then adding 1 ml of sterile water. Bacterial survival was determined by plating on LB medium.
Neutropenic and normal mouse model of fatal P. aeruginosa septicemia.
P. aeruginosa was grown in LB broth at 37°C to an OD600 of 0.4, collected by centrifugation at 3,000 × g, and washed twice in cold 1% protease peptone (Difco) in 10 mM PBS (pH 7.4). Infectious doses were determined at the time of infection by viable-cell (CFU) counting of serial dilutions plated on LB agar. For the neutropenic mouse model, 5- to 6-week-old C57BL/6 mice (Jackson Laboratory) were rendered neutropenic by three intraperitoneal injections of 200 μg of cyclophosphamide per g of body weight every other day as previously described (80). Two days following the final dose of cyclophosphamide, the mice were challenged by intraperitoneal injection of either wild-type P. aeruginosa PAO1 or PAO700 (algR::Gmr) in 0.2 ml of PBS. Mice in groups of five animals per dose were injected with inocula containing 102 or 103 CFU. For experiments with normal C57BL/6 mice, no cyclophosphamide was administered and the bacterial doses ranged from 106 to 108 CFU. The same route of injection, the intraperitoneal route, was used for all mice. The time required to cause mortality was recorded to determine the mean time to death.
Normal mouse model of P. aeruginosa pneumonia.
PAO1 and PAO700 were grown in 200 ml of LB broth for 12 h. The bacteria were collected by centrifugation, washed in 10 ml of HBSS (Cellgro), and then resuspended in 5 ml of HBSS and combined. Fifteen female C57BL/6j mice were infected with both PAO1 and PAO700 by using a Glascol inhalation chamber (81). The parameters used for aerosol exposure were 30 min of nebulization, 15 min of cloud decay, and 2 min of decontamination (UV irradiation). To determine bacterial survival in the lungs, mice were euthanized at 0, 6, and 24 h postinfection; time zero was defined as the end of the inhalation cycle (after decontamination). The lungs of the mice were removed and homogenized in 1 ml of HBSS. The lung homogenates were serially diluted in HBSS and plated onto both PIA and PIA containing gentamicin (150 μg/ml) to determine the number of CFU of each strain per lung. The number of CFU of PAO700 was determined by determining the counts on the PIA plate containing gentamicin. The number of CFU of PAO1 was calculated by subtracting the number of CFU on the PIA plate containing gentamicin from the number of CFU on the PIA plate. The percentages of survival for both strains at 6 and 24 h were calculated with the following formula: (mean number of CFU from lungs of mice at each time point for each P. aeruginosa strain/mean number of CFU from lungs of mice at time zero for each P. aeruginosa strain) × 100.
Metabolic labeling of newly synthesized polypeptides and two-dimensional electrophoresis.
De novo-synthesized proteins were analyzed by using a previously described procedure (80). PAO1 and PAO700 were grown in LB broth to an OD600 of 0.4 at 37°C. A 10-ml aliquot of each culture was washed in M9 glucose medium with no amino acids added and then resuspended in M9 glucose medium supplemented with an 18-amino-acid mixture (without methionine and cysteine) at a concentration of 0.1% and incubated for 60 min at 37°C. The culture was labeled for 5 min with [35S]methionine and [35S]cysteine (Expre35S35S protein labeling mixture; 1,000 Ci/mmol; Perkin-Elmer) at a final concentration of 30 μCi/ml and centrifuged at 10,000 × g for 5 min at 4°C; then it was resuspended in 100 μl of lysis buffer (10 mM Tris-HCl [pH 8.0], 0.5% SDS), vortexed vigorously, heated to 70°C, and vortexed vigorously again. Ten microliters of nuclease buffer (50 mM MgCl2, 100 mM Tris-HCl [pH 7.0], 500 μg of RNase A per ml, 0.2 U of DNase I [Ambion] per μl) was added, and the samples were incubated for 30 min at 37°C. Total protein concentration was determined by the Bradford assay (Bio-Rad), and total incorporated 35S was determined by trichloroacetic acid precipitation. Two-dimensional gel electrophoresis was performed by the method of O'Farrell (52) by Kendrick Labs, Inc. (Madison, Wis.). Isoelectric focusing was carried out in glass tubes (inside diameter, 2.0 mm) by using 2% 4-8 ampholines (Gallard-Schlesinger Industries, Inc., Garden City, N.Y.) for 9,600 V · h. After equilibrium in SDS sample buffer (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, 0.0625 M Tris; pH 6.8), each tube gel was sealed to the top of a stacking gel that overlaid a 10% acrylamide slab gel (thickness, 0.75 mm). SDS slab gel electrophoresis was carried out for 4 h at 12.5 mA/gel. 14C-labeled molecular weight markers (Amersham Biosciences) were added to a well in agarose that sealed the tube gel to the slab gel. These markers appeared as bands at the basic edges of the autoradiographic films. After the gels were fixed in 50% methanol-10% acetic acid, they were dried onto filter paper with the acid edge to the left. Autoradiography was carried out by using Kodak BioMax film, and autoradiographs were developed after 6 days. Computerized comparisons were performed with duplicate gels and corresponding autoradiographic films. One film from each pair was scanned with a laser densitometer (model PDSI; Molecular Dynamics Inc, Sunnyvale, Calif.). The linearity of the scanner was checked prior to scanning with a calibrated neutral-density filter set (Melles Griot, Irvine, Calif.) so that all major spots and all changing spots were outlined, quantified, and matched on all the gels. In cases where protein spots were missing from some of the gels and present in others, a small area of background was outlined to facilitate matching. The general method of computerized analysis for these pairs included automatic spot finding and quantification, automatic background subtraction (mode of nonspot), and automatic spot matching in conjunction with detailed manual checking of the spot-finding and matching functions.
RESULTS
Inactivation of algR in P. aeruginosa causes increased sensitivity to hypochlorite.
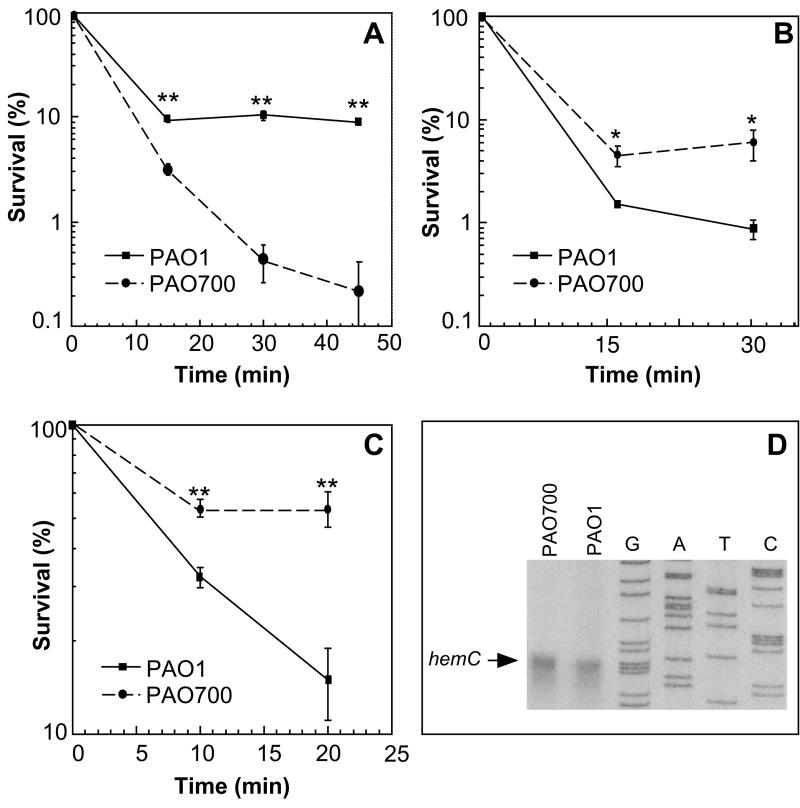
During chronic lung infections in CF patients, P. aeruginosa is exposed to the bactericidal products of polymorphonuclear leukocytes, high numbers of which are present in the lungs of CF patients (1, 30, 58, 72). One of the halogenated reactive oxygen intermediates generated by polymorphonuclear leukocytes with antipseudomonal activity is hypochlorous acid (48). In order to test whether algR may play a role in P. aeruginosa susceptibility to hypochlorite, we tested P. aeruginosa algR+ (PAO1) and algR::Gmr (PAO700) strains for resistance to this compound. The results of disk inhibition zone assays with different oxidants are shown in Table 2, and the rates of killing with hypochlorite in bacterial suspensions are shown in Fig. 1A. Both analyses may indicate that the algR mutant PAO700 had increased sensitivity to hypochlorite compared to the sensitivity of the parental algR+ strain PAO1. While the algR mutant cells showed increased sensitivity to hypochlorite, there was no significant difference in sensitivity to H2O2 between PAO1 and PAO700 (Table 2), as determined by the disk assay. PAO700's ability to resist hypochlorite was partially restored by introduction of algR-harboring plasmid pVDZ'2R (Table 2).
TABLE 2.
Sensitivity of P. aeruginosa to various reactive oxygen intermediatesa
| Strain (genotype) | pVDZ'2R (algR) | pDSL-1 (hemCD) | Diam (mm) of growth inhibition zone (mean ± SE)b
|
||
|---|---|---|---|---|---|
| NaOCl (5%) | Paraquat (2%) | H2O2 (3%) | |||
| PAO1 (algR+) | − | − | 15.3 ± 0.4 | 11.7 ± 0.6 | 9.0 ± 0.5 |
| PAO700 (algR::Gmr) | − | − | 23.0 ± 1.1 | 10.7 ± 0.6 | 8.7 ± 0.4 |
| PAO700 (algR::Gmr) | + | − | 19.3 ± 0.2 | NDc | ND |
| PAO700 (algR::Gmr) | − | + | 22.2 ± 1.9 | ND | ND |
All strains were grown in LB broth at 37°C to an OD600 of 0.4. The values are the averages of three measurements.
Growth inhibition zones around 6-mm-diameter disks soaked with different compounds were measured after overnight incubation. The differences in sensitivity to NaOCl for PAO1 and PAO700 and for PAO700 and PAO700/pVDZ'2R as determined by (Fischer's protected-least-significant-difference test) were significant (P < 0.0001).
ND, not determined.
FIG. 1.
NaOCl (A), hydrogen peroxide (B), and human myeloperoxidase-H2O2-Cl− system (C) sensitivity of P. aeruginosa PAO1 and PAO700 (algR:: Gmr) and S1 nuclease protection analysis of hemC (D). PAO1, wild-type algR+ strain; PAO700, algR::Gmr mutant of PAO1. (A) PAO1 and PAO700 (108 CFU each) were incubated in the presence of 25 μM NaOCl. Survival is expressed as the fraction (percentage) of the initial cell input that survived the treatment. The P values (as determined by a t test) were 1.1 × 10−4, 6.5 × 10−4, and 1.3 × 10−4 for the 15-, 30-, and 45-min time points, respectively. Two asterisks indicate that the P value was <0.01. (B) PAO1 and PAO700 (108 CFU each) were incubated in the presence of 14.7 mM H2O2. The P values (as determined by a t test) were 4 × 10−2 and 1 × 10−2 for the 15- and 30-min time points, respectively. An asterisk indicates that the P value was <0.05. (C) PAO1 and PAO700 (108 CFU each) were incubated in the presence of a complete myeloperoxidase-glucose oxidase system. Survival is expressed as the fraction (percentage) of the initial cell input that survived the treatment. The P values (as determined by a t test) were 2.4 × 10−3 and 6.3 × 10−3 for the 10- and 20-min time points, respectively. Two asterisks indicate that the P value was <0.01. The bars indicate standard errors, and each point is the average for three separate experiments. (D) hemC transcription is not affected in PAO700. Equal amounts of total RNA from the two strains grown to an OD600 of 0.4 were used for S1 nuclease protection analyses as described in Materials and Methods. Lanes G, A, T, and C, sequencing lanes. The arrow indicates the position of the 5′ end of the hemC transcript.
Immediately downstream of algR are two genes, hemC and hemD, that are involved in heme production in P. aeruginosa. An E. coli hemC mutant has low catalase activity, indicating that heme production is adversely affected (45). In order to determine if the gentamicin cassette inserted into algR had polar effects on the downstream genes hemC and hemD, these genes were cloned into pVDZ'2, and the resulting plasmid, pDSL-1, was conjugated into PAO700. The algR mutant strain harboring pDSL-1 was tested in the hypochlorite and hydrogen peroxide plate assays. There was not a significant difference between PAO700 and PAO700 containing plasmid pDSL-1(hemCD) after exposure to hypochlorite (Table 2). In addition, an S1 nuclease protection assay examining hemC transcription in PAO1 and PAO700 revealed no difference in the hemC transcript between the two strains (Fig. 1D). These results suggest that the sensitivities to hypochlorite observed are a direct result of genes regulated by AlgR.
Inactivation of algR causes decreased sensitivity of P. aeruginosa to coupled human myeloperoxidase-glucose oxidase bactericidal system.
One of the primary mechanisms of bacterial killing utilized by neutrophils in vivo is the generation and release of hypochlorite by the enzyme myeloperoxidase (48). In order to determine the susceptibility of algR strain PAO700 to killing by this means, an in vitro human myeloperoxidase-glucose oxidase bactericidal system was tested with PAO1 (algR+) and PAO700 (algR::Gmr). Interestingly, P. aeruginosa PAO700 (algR::Gmr) was more resistant to killing by a human myeloperoxidase-glucose oxidase bactericidal system than the algR-containing strain PAO1 was (Fig. 1C). This result was not consistent with the results obtained in the hypochlorite assays. Hydrogen peroxide is also generated by glucose oxidase in the myeloperoxidase-glucose oxidase bactericidal system, and PAO700 resistance to this compound may override the hypochlorite effects observed previously. In order to examine this possibility, PAO1 and PAO700 sensitivities to 15 mM H2O2 in broth cultures were tested. Inactivation of algR resulted in an eightfold increase in P. aeruginosa resistance to hydrogen peroxide (Fig. 1B) in this assay. The survival values for PAO1 and PAO700 were significantly different at the 10- and 20-min time points (P < 2.4 × 10−3 and P < 6.3 × 10−3, respectively, as determined by a t test).
Inactivation of algR does not reduce P. aeruginosa survival in cellular bactericidal systems.
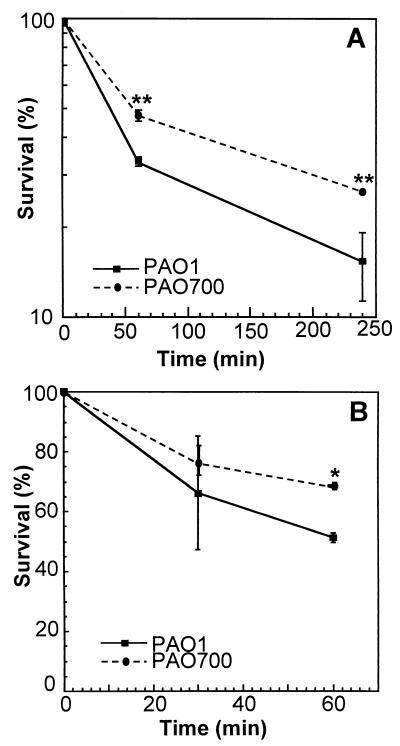
Since PAO700 (algR::Gmr) was more sensitive to hypochlorite and more resistant to hydrogen peroxide acellular modes of killing than PAO1 (algR+), we tested the viability of the algR strain PAO700 in macrophages. Initially, we tested P. aeruginosa PAO1 and the algR mutant PAO700 with the immortalized murine macrophage-like cell line J774. There was a slight difference in survival between PAO1 (8%) and PAO700 (18%) after 4 h (Fig. 2A), suggesting that AlgR may regulate genes necessary for survival after exposure to macrophages. In order to examine this relationship further, we examined the sensitivities of PAO1 and PAO700 to elicited mouse peritoneal and bone marrow-derived macrophages. There was no statistical difference between the sensitivities of PAO1 (algR+)and PAO700 (algR::Gmr) to killing by murine peritoneal or bone marrow-derived macrophages (data not shown). These results may indicate that algR does not play a role in the sensitivity of P. aeruginosa to killing or recognition by macrophages.
FIG. 2.
Differential sensitivities of algR+ (PAO1) and algR::Gmr (PAO700) P. aeruginosa strains to the murine macrophage-like cell line J774 (A) and primary human neutrophils (B). Levels of survival are as defined in the legend to Fig. 1. (A) Macrophages (5 × 106 cells) were infected at a multiplicity of infection of 1 and incubated for 1 and 4 h. The P values (as determined by a t test) were 2.2 × 10−3 and 1.5 × 10−3 for 60 and 240 min, respectively. Two asterisks indicate that the P value was <0.01. (B) Freshly prepared peripheral blood neutrophils (3 × 107 cells) were incubated with PAO1 and PAO700 (1 × 108 CFU) for 15 and 30 min. The P value (as determined by a t test) was 4.5 × 10−2 for 30 min. An asterisk indicates that the P value was <0.05.
Since neutrophils have been cited as the main host response cells for clearance of P. aeruginosa and since no difference was observed with macrophages, we next tested the sensitivities of PAO1 and PAO700 to human neutrophils. There was no statistical significance between PAO1 and PAO700 after 30 min of exposure to human neutrophils. However, after 1 h, algR mutant strain PAO700 was approximately 10% more resistant to killing by human neutrophils (Fig. 2B). This result is consistent with the results of the myeloperoxidase-glucose oxidase assays and may indicate that AlgR represses genes required for P. aeruginosa to survive exposure to primary host response cells.
Inactivation of algR causes decreased systemic virulence of P. aeruginosa in neutropenic and normal C57BL/6 mouse models of fatal Pseudomonas septicemia.
The algR strain PAO700 was differentially sensitive and resistant to in vitro oxidative stresses but did not show sensitivity to macrophage killing. In order to test if algR plays a role in the overall virulence of the organism, wild-type strain PAO1 and algR insertionally inactivated strain PAO700 were tested in a systemic virulence model of fatal Pseudomonas septicemia. Initially, we determined the 50% lethal doses of PAO1 (algR+) and PAO700 (algR::Gmr) in the neutropenic mouse model of P. aeruginosa sepsis. C57BL/6j mice were rendered neutropenic by cyclophosphamide treatment and challenged with P. aeruginosa by intraperitoneal injection. There was no difference between the 50% lethal doses of the two strains in neutropenic mice (Table 3).
TABLE 3.
Mortalities of neutropenic C57BL/6j mice challenged with PAO1 and PAO700
| P. aeruginosa strain (genotype) | Infectious dose (CFU)a | Mortalityb | Mean time to death (h) |
|---|---|---|---|
| PAO1 (algR+) | 1.0 × 102 | 4/5 | 42.4 |
| 1.0 × 103 | 4/5 | 37.9 | |
| PAO700 (algR) | 1.0 × 102 | 4/5 | 34.9 |
| 1.0 × 103 | 5/5 | 27.9 |
The actual infectious doses administered were determined by plating at the time of injection.
Mortality is expressed as the number of mice that died/the total number of mice tested. Mice were challenged by intraperitoneal injection, and deaths occurred within 48 h postinjection; no additional fatalities were observed after that.
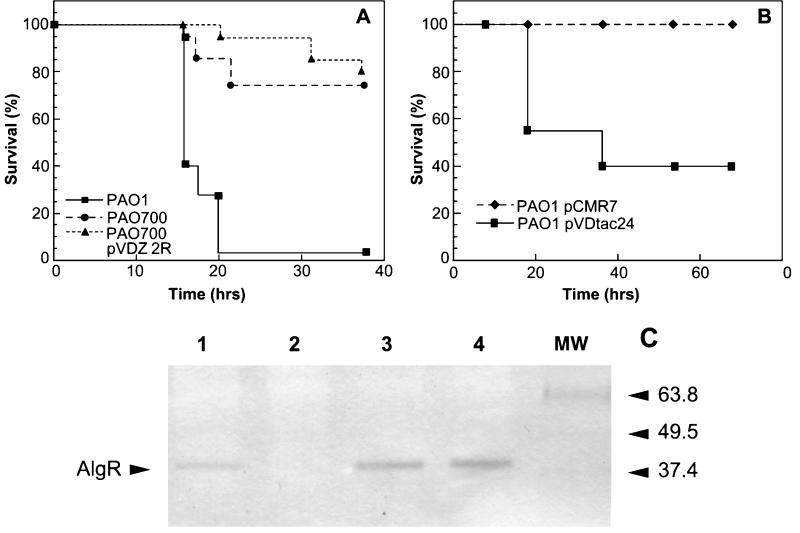
The decreased number of neutrophils may have inhibited the ability of the mice to clear the infection from the peritoneum. In order to assess the virulence in a normal mouse, intraperitoneal challenges with 1 × 106 CFU of PAO1 and 1 × 106 CFU of PAO700 were performed. The mice infected with PAO700 exhibited 74% survival, whereas none of the mice infected with PAO1 survived (Fig. 3A). Attempts to complement virulence by reintroduction of the algR gene into PAO700 resulted in a further decrease in virulence, indicating that control of the AlgR intracellular concentration is critical for P. aeruginosa virulence (Fig. 3A). This led to the question of whether we could suppress virulence altogether by overexpressing algR alone. PAO1 harboring the algR-overexpressing plasmid pCMR-7 was completely avirulent compared to PAO1 after infection with 1 × 106 CFU (Fig. 3B). Additionally, Western blot analysis of cell extracts of PAO1, PAO700, PAO700 harboring plasmid pVDZ'2R, and PAO1 containing plasmid pCMR7 with anti-AlgR grown to the same OD600 (0.4) that was used in the infections revealed that AlgR production increased in PAO700 harboring pVDZ'2 and PAO1 harboring pCMR-7 compared to AlgR production in PAO1 (Fig. 3C). These results may indicate that AlgR plays a role in negatively and positively regulating virulence factors for P. aeruginosa and that increased AlgR expression levels can suppress virulence.
FIG. 3.
Survival curves for PAO1 (algR+), PAO700 (algR::Gmr), and PAO700 containing pVDZ'2R (A) and for PAO1/pCMR7 and PAO1/pVDtac24 (B) and Western blot analysis of cell extracts of PAO1, PAO700, PAO1 with plasmid pVDZ'2R, and PAO1 harboring plasmid pCMR-7 with anti-AlgR (C). (A and B) C57BL/6 mice were infected by intraperitoneal injection with 1.5 × 106 CFU of each strain and monitored for 48 to 72 h. PAO1, algR+; PAO700, algR mutant of PAO1; PAO700 pVDZ 2R, PAO700 harboring plasmid pVDZ'2 with algR; PAO1 pCMR7, PAO1 (algR+) harboring plasmid pVDtac24 containing algR; PAO1 pVDtac24, PAO1 (algR+) with vector pVDtac24. Survival is expressed as the fraction (percentage) of the total number of mice (20) in each group that survived. (C) Western blot analysis of cell extracts of PAO1 (lane 1), PAO700 (lane 2), PAO700 containing plasmid pVDZ'2R (lane 3), and PAO1 with AlgR overexpression plasmid pCMR-7 (lane 4). Lane MW contained molecular weight markers. All cell extracts examined were from the stage of growth (OD600, 0.4) that was used in the virulence studies whose results are shown in panels A and B.
P. aeruginosa PAO700 (algR::Gmr) is cleared more readily from murine lungs.
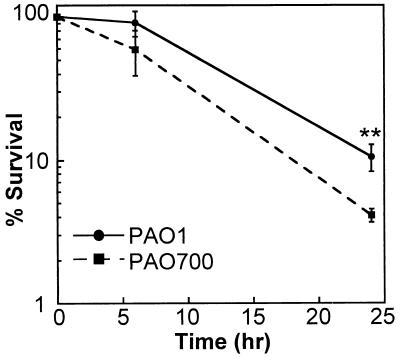
CF patients are colonized by both mucoid and nonmucoid strains of P. aeruginosa. Since algR mutant strain PAO700 displayed reduced virulence in a murine systemic model, we tested this strain for its ability to survive in murine lungs and compared it to parental strain PAO1 in a coinfection pneumonia model. We introduced 5 × 106 cells of each strain (PAO1 and PAO700) simultaneously into the lungs of C57BL/6j mice by inhalation and monitored the bacterial clearance by determining the numbers of CFU in lung homogenates from animals at 6 and 24 h. There was no statistical significance between the numbers of the two P. aeruginosa strains found in the lungs 6 h postinfection. However, at 24 h, there were significantly fewer PAO700 cells (3.3 × 104 cells) than PAO1 cells (2.5 × 105 cells) (Fig. 4). These results may indicate that the P. aeruginosa algR mutant strain is more readily cleared from mouse lungs than PAO1.
FIG. 4.
Clearance of PAO1 and PAO700 (algR::Gmr) following coinfection. Fifteen female C57BL/6 mice were infected with a mixed culture of PAO1 and PAO700 (algR::Gmr) that was aerosolized by using a Glascol aerosolization chamber (see Materials and Methods). Five mice were sacrificed at 0, 6, and 24 h postinfection. The lungs were removed, and the bacteria were quantified. The percentage of survival was determined as follows: (number of CFU of bacteria for each mouse/average number of CFU for mice at time zero) × 100. The P value (as determined by a t test) was 3.7 × 10−3 for 24 h. Two asterisks indicate that the P value was <0.01.
Inactivation of P. aeruginosa PAO1 algR eliminates twitching motility.
There is at least one possible explanation for the reduced virulence of algR mutant strain PAO700. Mutations in algR result in a loss of twitching motility in P. aeruginosa (77). P. aeruginosa strains that lack pili have been shown to be less virulent than and exhibit reduced adherence to human and murine respiratory tract epithelia compared to parental strain PAO1 (6, 7, 44). In order to test these observations, twitching motility assays were performed with PAO1, PAO700, and PAO700 harboring the algR plasmid pVDZ'2R (Table 4). Insertional inactivation of algR resulted in no observable twitching motility when the standard plate assay was used. The introduction of pVDZ'2R harboring algR into PAO700 resulted in an increase in the twitching motility to nearly wild-type levels.
TABLE 4.
Twitching motility assay with PAO1, PAO700, and PAO700/pVDZ’2R
| P. aeruginosa strain (genotype) | pVDZ’2R (algR) | Twitching zone (mm) (mean ± SE)a |
|---|---|---|
| PAO1 (algR+) | − | 19.9 ± 1.8 |
| PAO700 (algR::Gmr) | − | NTb |
| PAO700 (algR::Gmr) | + | 15.5 ± 3.8 |
Twitching motility assays were performed after 48 h of growth at 37°C on 0.3% LB agar. Bacteria were inoculated by stabbing LB agar with a needle. The zones of twitching were measured on the bottoms of petri dishes after staining with Coomassie blue.
NT, no twitching motility observed.
Expression of a subset of polypeptides is increased in algR P. aeruginosa cells.
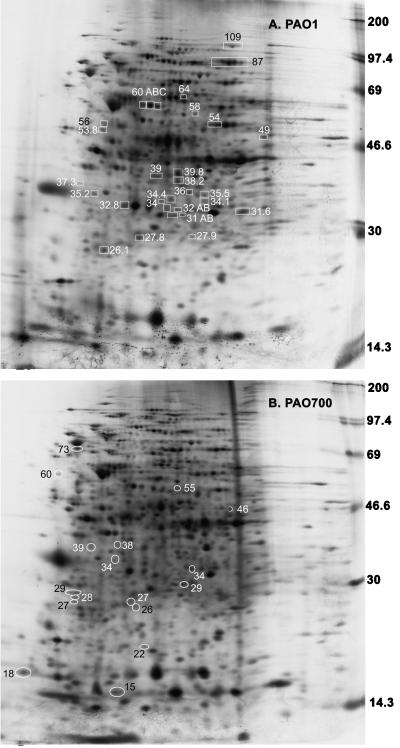
Since algR encodes a transcriptional regulator and its inactivation resulted in decreased virulence while AlgR overexpression led to a complete loss of virulence, we decided to examine profiles of metabolically labeled proteins in algR+ and algR::Gmr strains (Fig. 5). Figure 5 shows logarithmic-growth-stage-specific protein expression in PAO1 and PAO700 at the points used in the virulence studies. The patterns of labeled proteins on two-dimensional gels indicated that at least 30 peptides were downregulated or absent in PAO700 (Fig. 5B) compared to the peptides in PAO1 (Fig. 5A). Conversely, approximately 17 proteins were present only in PAO700 (Fig. 5B). Additionally, the steady-state expression of PAO1 and the steady-state expression of PAO700 at the same stage of logarithmic growth were compared. These comparisons revealed that there were at least 24 differentially expressed proteins; 20 proteins were upregulated by AlgR, and 4 proteins were downregulated by AlgR (data not shown). The finding that several proteins can be induced in algR::Gmr P. aeruginosa may suggest a role not previously appreciated for algR as a negative regulator of gene expression in P. aeruginosa.
FIG. 5.
Two-dimensional polyacrylamide gel electrophoresis comparison of P. aeruginosa PAO1 and PAO700 (algR::Gmr) proteins. P. aeruginosa PAO1 and PAO700 were each grown to an OD600 of 0.4. The cells were lysed as described in Materials and Methods. Isoelectric focusing was performed from left (acid) to right (basic). The boxes indicate proteins that were threefold or more lower on the PAO1 two-dimensional gel than on the PAO700 two-dimensional gel. The ovals indicate proteins that were present only on the PAO700 two-dimensional gel.
DISCUSSION
In this study, the effects of algR inactivation on P. aeruginosa virulence and sensitivity to acellular and cellular microbicidal systems were analyzed. AlgR was discovered to be a transcriptional activator of algD and to be required for the production of alginate (10). AlgR binds to three different sites within the algD promoter and interacts with RNA polymerase associated with AlgU (AlgT, σ22) to promote its transcription (18, 29, 49-51). AlgR also activates algC expression in a similar manner by interacting with three binding sites within its promoter region (18, 83). AlgR aids in the persistence of CF infections by activating the alginate biosynthetic pathway, which results in an excess production of alginate. The role of AlgR has been extended to the regulation of type IV pili, and an algR mutation resulted in a loss of twitching motility (77). Our data indicate that AlgR may play a role in regulating protection against oxidants (e.g., hypochlorite) relative to the infectious process. The algR promoter is regulated by AlgU (AlgT, σ22), which has been shown to regulate oxidative stress in P. aeruginosa (80). We have shown that some of the oxidative stress response of AlgU (AlgT, σ22) may be partially due to effects on genes regulated by AlgR, as PAO700 (algR::Gmr) was sensitive to hypochlorite. The partial complementation effects shown in Table 2 may have been due to the slight overexpression of algR shown in Fig. 3C due to copy number effects of pVDZ'2R. P. aeruginosa seems to be very sensitive to the level of AlgR in the cell. In fact, we have observed that pVDZ'2R complementation of PAO568 algR (mucA2) restores approximately 80% of the wild-type levels of alginate production (data not shown), the same level of complementation which we observed in the hypochlorite disk assays (Table 2). Interestingly, the algR null mutant was more resistant to hydrogen peroxide, the converse of algU mutant PAO6852 (80). Infiltration of neutrophils into the airways of chronically infected CF patients is well known. The sensitivity of the algR mutant strain to hypochlorite indicates that AlgR may control genes involved in counteracting damage caused by reactive chloride compounds. This may partially explain the ability of P. aeruginosa to resist clearance by neutrophils, which release hypochlorite in phagosomes and to the external milieu. It is reasonable to assume that upregulation of algR-dependent genes could be beneficial and provide some protective advantage to P. aeruginosa.
In contrast to the results obtained in acellular assays, which indicated that algR has a role in the defense against hypochlorite, only small differences in susceptibility to the bactericidal action of phagocytic cells were noted in our experiments. Granulocytes, which play a role in the control of P. aeruginosa infections, kill microorganisms by oxygen-dependent and oxygen-independent pathways (38, 48, 73). Oxygen-independent pathways are sufficient to kill P. aeruginosa, and this may explain the lack of effect seen in the cellular assays (48).
The experiments involving the intraperitoneally challenged neutropenic and normal mice indicate that algR activity in wild-type P. aeruginosa may be required for the processes associated with mortality in acute systemic disease. The degree of disparity between the survival of the mice infected with PAO1 and the survival of the mice infected with PAO700 could not have been predicted based on the in vitro data. The effects of the algR mutant on P. aeruginosa virulence observed may be explained by changes in the concentrations of one or more of many Pseudomonas virulence factors. There is one known factor directly affected by AlgR which may explain the reduced virulence observed: nonfunctional pili (Table 4). The loss of twitching motility displayed by an algR mutant may result in the inability of this mutant to form a biofilm (54). The inability to form a biofilm may interfere with quorum sensing, which controls many virulence factors (26, 56, 57, 62, 63, 71, 78, 79).
It is also possible that an algR knockout results in reduced algC expression. Zielinski et al. showed that algC expression is reduced 5.7-fold in an algR background through studies of transcriptional fusion of the algC promoter to lacZ (83). It has been reported that a PAO1 algC::Tetr mutant strain is avirulent in neonatal mouse pneumonia and burned mouse models of P. aeruginosa virulence. These studies also showed that inactivation of algC results in serum sensitivity and phage susceptibility (22, 76). A recent study also noted that mutations in algC resulted in reduced virulence in the Caenorhabditis elegans infection model (20). We have observed that PAO700 (algR::Gmr) does not display an altered LPS pattern, which is in agreement with the findings obtained by Olvera et al. (53) when they examined another algR PAO1 strain, CDM1/1 (data not shown). Additionally, there was no difference in serum sensitivity between PAO1 and PAO700 (data not shown). Our results obtained with PAO700 exhibiting reduced virulence in the septicemia model agree with these previous reports but indicate that LPS expression may not be affected in algR mutant strain PAO700. Nonfunctional pili may explain the reduced virulence of the algR mutant observed in this study.
As mentioned previously, our attempt to complement algR in trans in PAO700 suggests that the cellular concentration of AlgR is critical. This is the first study to report that AlgR may have the ability to repress transcription. Even though pVDZ'2R is maintained in P. aeruginosa at a relatively low level, there is still more than one copy of the plasmid per cell, which leads to a higher concentration of AlgR in the cell (Fig. 3C). Western blot analysis with anti-AlgR revealed that the intracellular concentrations of AlgR were indeed elevated in PAO700 harboring plasmid pVDZ'2R. We hypothesized that the higher AlgR concentration could affect the transcription of AlgR-regulated genes necessary for virulence. By overexpressing AlgR from plasmid pCMR7, we were able to show that AlgR can indeed suppress virulence in P. aeruginosa PAO1. These results indicate that AlgR may be capable of acting as a repressor on some genes when AlgR levels are elevated. This possibility is intriguing in light of AlgR's role in alginate production. When P. aeruginosa is mucoid, AlgU (AlgT, σ22) is available for initiating transcription of algR, causing an increase in the cellular AlgR concentration. This raises the possibility that Pseudomonas may use the intracellular AlgR concentration as one mechanism to repress virulence factors when it displays the mucoid phenotype in the unique environment of the CF lung. Studies have shown that mucoid strains of P. aeruginosa are unpiliated and lose flagella and that the LPS becomes rough (14, 16, 24, 36, 55). It is possible that in the conversion to mucoidy the energy used for production of extracellular structures (pili, flagella, and LPS) is redirected to produce alginate. AlgR may act as the switch to downregulate the genes. AlgR may also act indirectly through other transcriptional regulators to obtain the same result.
Currently, except for algD and algC, we do not know which genes are directly regulated by AlgR. Since the extent of AlgR regulation is unknown, we can only speculate as to which genes are responsible for the decrease in virulence seen in the algR knockout. A cursory examination of the P. aeruginosa genome by using algD and algC AlgR binding sites as search parameters revealed that approximately 79 open reading frames contain high-affinity AlgR binding sites. These can be roughly broken down into the following eight different categories based on homology: (i) alginate, (ii) antibiotic resistance, (iii) motility, (iv) transcription, (v) pyrimidine or purine metabolism, (vi) carbon metabolism, (vii) iron acquisition, and (viii) stress response. The largest category appears to be transcription, indicating that AlgR may be involved in regulatory cascades in P. aeruginosa. It has recently been recognized based on the genome sequence that P. aeruginosa contains 118 two-component transcriptional regulators, and the range of genes controlled by these two-component regulators is just beginning to be appreciated (75). Based on the steady-state protein expression shown by two-dimensional SDS-polyacrylamide gel electrophoresis analysis, the number of genes that are regulated by AlgR appears to be at least 24, but stage-specific expression implies that there may be as many as 47 such genes. The growth conditions used in this analysis (rich medium with aerobic aeration) do not account for the P. aeruginosa stress response, which may activate transcription of other AlgR-dependent genes. Thus, AlgR appears to play a much larger role in P. aeruginosa transcriptional regulation than previously appreciated. Based on the results of our systemic virulence studies, some of the 24 to 47 genes may be important for systemic virulence. Therefore, this study indicates that AlgR has more than one role in the overall virulence and pathogenesis of this organism. The number of transcriptional regulators that contain high-affinity AlgR binding sites may explain some of the pleiotropic effects observed with the algR mutant in this study. We have also observed that PAO700 produces approximately three times more pyoverdine in LB and iron-limiting media, indicating that AlgR has a negative regulatory role in pyoverdine production (data not shown). It has been shown that pyoverdine is required for virulence in a burned mouse model and that it is required for acquisition of iron from the host (47). However, a P. aeruginosa mutant exhibiting an increase in pyoverdine production has not previously been examined for virulence. One possibility for AlgR involvement in pyoverdine regulation is through pvdA that contains an AlgR consensus sequence that matches 9 of 10 residues of the algD RB1 binding site. Additionally, two other iron acquisition open reading frames, PA3268 and PA1909, encoding putative TonB-dependent homologues, may be involved in the increased pyoverdine phenotype observed. Future studies should elucidate the mechanism by which AlgR controls virulence in P. aeruginosa.
Acknowledgments
We thank Vojo Deretic for providing P. aeruginosa strain PAO700 and the anti-AlgR antibody.
This work was supported by grant SCHURR97ZO from the Cystic Fibrosis Foundation and by grant LEQSF-199-02-RD-A-42 from the Louisiana Board of Regents Support Fund. This research was also supported in part by a grant from the W. M. Keck Foundation of Los Angeles
Editor: J. T. Barbieri
REFERENCES
- 1.Barton, A. D., K. Ryder, R. V. Lourenco, W. Dralle, and S. G. Weiss. 1976. Inflammatory reaction and airway damage in cystic fibrosis. J. Lab. Clin. Med. 88:423-426. [PubMed] [Google Scholar]
- 2.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Ladeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, J. C., J. M. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, J. C., M. J. Schurr, H. Yu, D. W. Rowen, and V. Deretic. 1997. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology 143:3473-3480. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comolli, J. C., L. L. Waite, K. E. Mostov, and J. N. Engel. 1999. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect. Immun. 67:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne, M. J., K. S. Russell, C. L. Coyle, and J. B. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic, V. 1996. Molecular biology of mucoidy in Pseudomonas aeruginosa, p. 223-244. In J. A. Dodge, D. J. H. Brock, and J. H. Widdicombe (ed.), Cystic fibrosis current topics, vol. 3. J. Wiley and Sons, Chichester, United Kingdom.
- 10.Deretic, V., R. Dikshit, W. M. Konyecsni, A. M. Chakrabarty, and T. K. Misra. 1989. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171:1278-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deretic, V., J. H. J. Leveau, C. D. Mohr, and N. S. Hibler. 1992. In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol. Microbiol. 6:2761-2767. [DOI] [PubMed] [Google Scholar]
- 12.DeVries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doggett, R. G., G. M. Harrison, R. N. Stillwell, and E. S. Wallis. 1966. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J. Pediatr. 68:215-221. [Google Scholar]
- 14.Drake, D., and T. C. Montie. 1988. Flagella, motility and invasive virulence of Pseudomonas aeruginosa. J. Gen. Microbiol. 134:43-52. [DOI] [PubMed] [Google Scholar]
- 15.Evans, L. R., and A. Linker. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara, S., N. A. Zielinski, and A. M. Chakrabarty. 1993. Enhancer-like activity of A1gR1-binding site in alginate gene activation: positional, orientational, and sequence specificity. J. Bacteriol. 175:5452-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fyfe, J. A. M., and J. R. W. Govan. 1980. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J. Gen. Microbiol. 119:443-450. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilligan, P. H. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 4:35-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, J. B., M. J. Coyne, Jr., A. N. Neely, and I. A. Holder. 1995. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect. Immun. 63:4166-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg, J. B., W. L. Gorman, J. Flynn, and D. E. Ohman. 1993. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J. Bacteriol. 175:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg, J. B., and G. B. Pler. 1996. Pseudomonas aeruginosa lipopolysaccharides and pathogenesis. Trends Microbiol. 4:490-494. [DOI] [PubMed] [Google Scholar]
- 25.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 27.Hershberger, C. D., R. W. Ye, M. R. Parsek, Z.-D. Xie, and A. M. Chakrabarty. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative σ factor (σE). Proc. Natl. Acad. Sci. USA 92:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 29.Kato, J., and A. M. Chakrabarty. 1991. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 88:1760-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch, C., and N. Høiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 31.Konyecsni, W. M., and V. Deretic. 1988. Broad-host-range plasmid and M13 bacteriophage-derived vectors for promoter analysis in Escherichia coli and Pseudomonas aeruginosa. Gene 74:375-386. [DOI] [PubMed] [Google Scholar]
- 32.Konyecsni, W. M., and V. Deretic. 1990. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J. Bacteriol. 172:2511-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Learn, D. B., E. P. Brestel, and S. Seetharama. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 55:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linker, A., and R. S. Jones. 1966. A new polysaccharide resembling alginic acid isolated from pseudomonads. J. Biol. Chem. 241:3845-3851. [PubMed] [Google Scholar]
- 35.Linker, A., and R. S. Jones. 1964. A polysaccharide resembling alginic acid from a Pseudomonas micro-organism. Nature 204:187-188. [DOI] [PubMed] [Google Scholar]
- 36.Luzar, M. A., M. J. Thomassen, and T. C. Montie. 1985. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect. Immun. 50:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, S., U. Selvaraj, D. E. Ohman, R. Quarless, D. J. Hassett, and D. J. Wozniak. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandell, G. L. 1974. Bactericidal activity of aerobic and anaerobic polymorphonuclear neutrophils. Infect. Immun. 9:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, D. W., B. W. Holloway, and V. Deretic. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, D. W., M. J. Schurr, M. H. Mudd, and V. Deretic. 1993. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol. Microbiol. 9:497-506. [DOI] [PubMed] [Google Scholar]
- 41.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. W. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to stress response. J. Bacteriol. 176:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147-155. [DOI] [PubMed] [Google Scholar]
- 45.McConville, M. L., and H. P. Charles. 1979. Mutants of Escherichia coli K12 accumulating porphobilinogen: a new locus, hemC. J. Gen. Microbiol. 111:193-200. [DOI] [PubMed] [Google Scholar]
- 46.Metcalf, J. A., J. I. Gallin, W. M. Nauseef, and R. Root. 1986. Laboratory manual of neutrophil function. Raven Press, New York, N.Y.
- 47.Meyer, J.-M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizgerd, J. P., and J. D. Brain. 1995. Reactive oxygen species in the killing of Pseudomonas aeruginosa by human leukocytes. Curr. Microbiol. 31:124-128. [DOI] [PubMed] [Google Scholar]
- 49.Mohr, C. D., N. S. Hibler, and V. Deretic. 1991. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol. 173:5136-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr, C. D., J. H. J. Leveau, D. P. Krieg, N. S. Hibler, and V. Deretic. 1992. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 174:6624-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohr, C. D., D. W. Martin, W. M. Konyecsni, J. R. Govan, S. Lory, and V. Deretic. 1990. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172:6576-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 53.Olvera, C., J. B. Goldberg, R. Sanchez, and G. Soberon-Chavez. 1999. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179:85-90. [DOI] [PubMed] [Google Scholar]
- 54.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 55.Pasloske, B. L., A. M. Joffe, Q. Sun, K. Volpel, W. Paranchych, F. Eftekhar, and D. P. Speert. 1988. Serial isolates of Pseudomonas aeruginosa from a cystic fibrosis patient have identical pilin sequences. Infect. Immun. 56:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 57.Pearson, J. P., M. Feldman, B. H. Iglewski, and A. Prince. 2000. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68:4331-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedersen, S. S. 1992. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS 100(Suppl. 28):1-79. [PubMed] [Google Scholar]
- 59.Pederson, S. S., N. Høiby, and F. Espersen. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowen, D. W., and V. Deretic. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36:314-327. [DOI] [PubMed] [Google Scholar]
- 61.Roychoudhury, S., K. Sakai, D. Schlictman, and A. M. Chakrabarty. 1992. Signal transduction in exopolysaccharide alginate synthesis: phosphorylation of the response regulator AlgR1 in Pseudomonas aeruginosa and Escherichia coli. Gene 112:45-51. [DOI] [PubMed] [Google Scholar]
- 62.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 63.Rumbaugh, K. P., J. A. Griswold, B. H. Iglewski, and A. N. Hamood. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67:5854-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schurr, M. J., and V. Deretic. 1997. Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24:411-420. [DOI] [PubMed] [Google Scholar]
- 65.Schurr, M. J., H. Yu, J. C. Boucher, N. S. Hibler, and V. Deretic. 1995. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (σE) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J. Bacteriol. 177:5670-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, N. S. Hibler, and V. Deretic. 1995. Biochemical characterization and posttranslational modification of AlgU, a regulator of stress response in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 216:874-880. [DOI] [PubMed] [Google Scholar]
- 68.Simpson, J. A., S. E. Smith, and R. T. Dean. 1988. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J. Gen. Microbiol. 134:29-36. [DOI] [PubMed] [Google Scholar]
- 69.Simpson, J. A., S. E. Smith, and R. T. Dean. 1993. Alginate may accumulate in cystic fibrosis lung because the enzymatic and free radical capacities of phagocytic cells are inadequate for its degradation. Biochem. Mol. Biol. Int. 30:1021-1034. [PubMed] [Google Scholar]
- 70.Simpson, J. A., S. E. Smith, and R. T. Dean. 1989. Scavenging by alginate of free radicals released by macrophages. Free Radic. Biol. Med. 6:347-353. [DOI] [PubMed] [Google Scholar]
- 71.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 72.Speert, D. P. 1994. Pseudomonas aeruginosa infections in patients with cystic fibrosis, p. 183-236. In A. L. Baltch and R. P. Smith (ed.), Pseudomonas aeruginosa infections and treatment. Marcel Dekker, Inc., New York, N.Y.
- 73.Speert, D. P., M. Bond, R. C. Woodman, and J. T. Curnutte. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524-1531. [DOI] [PubMed] [Google Scholar]
- 74.Speert, D. P., and S. Gordon. 1992. Phagocytosis of unopsonized Pseudomonas aeruginosa by murine macrophages is a two-step process requiring glucose. J. Clin. Investig. 90:1085-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 76.Tang, H. B., E. DiMango, R. Bryan, M. Gambello, B. H. Iglewski, J. B. Goldberg, and A. Prince. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitchurch, C. B., R. A. Alm, and J. S. Mattick. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 93:9839-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winzer, K., and P. Williams. 2001. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291:131-143. [DOI] [PubMed] [Google Scholar]
- 79.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, A. Heydorn, K. Mathee, C. Moser, L. Eberl, S. Molin, N. Hoiby, and M. Givskov. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 146:2481-2493. [DOI] [PubMed] [Google Scholar]
- 80.Yu, H., J. C. Boucher, N. S. Hibler, and V. Deretic. 1996. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (σE). Infect. Immun. 64:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu, H., M. Hanes, C. E. Chrisp, J. C. Boucher, and V. Deretic. 1998. Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect. Immun. 66:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli σE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]
- 84.Zielinski, N. A., R. Maharaj, S. Roychoudhury, C. E. Danganan, W. Hendrickson, and A. M. Chakrabarty. 1992. Alginate synthesis in Pseudomonas aeruginosa: environmental regulation of the algC promoter. J. Bacteriol. 174:7680-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]