Abstract
Reorientation of the regulatory domain of the myosin head is a feature of all current models of force generation in muscle. We have determined the orientation of the myosin regulatory light chain (RLC) using a spin-label bound rigidly and stereospecifically to the single Cys-154 of a mutant skeletal isoform. Labeled RLC was reconstituted into skeletal muscle fibers using a modified method that results in near-stoichiometric levels of RLC and fully functional muscle. Complex electron paramagnetic resonance spectra obtained in rigor necessitated the development of a novel decomposition technique. The strength of this method is that no specific model for a complex orientational distribution was presumed. The global analysis of a series of spectra, from fibers tilted with respect to the magnetic field, revealed two populations: one well-ordered (±15°) with the spin-label z axis parallel to actin, and a second population with a large distribution (±60°). A lack of order in relaxed or nonoverlap fibers demonstrated that regulatory domain ordering was defined by interaction with actin rather than the thick filament surface. No order was observed in the regulatory domain during isometric contraction, consistent with the substantial reorientation that occurs during force generation. For the first time, spin-label orientation has been interpreted in terms of the orientation of a labeled domain. A Monte Carlo conformational search technique was used to determine the orientation of the spin-label with respect to the protein. This in turn allows determination of the absolute orientation of the regulatory domain with respect to the actin axis. The comparison with the electron microscopy reconstructions verified the accuracy of the method; the electron paramagnetic resonance determined that axial orientation was within 10° of the electron microscopy model.
INTRODUCTION
Muscle contraction results from the cyclic interaction of the myosin head with actin. Models suggest that force generation and sliding of the thin and thick filaments is driven by an axial rotation or tilting of the myosin heads while bound to actin (Huxley, 1974, 1969; Reedy et al., 1965). Determination of the myosin head's structure in muscle fibers while in different states of the contractile cycle has proven difficult.
The myosin head consists of a globular catalytic domain that binds both actin and ATP. The catalytic domain extends into an elongated regulatory domain (Rayment et al., 1993a,b) which has two calmodulin-like subunits—the essential light chain (LC1 and LC3) and the regulatory light chain (RLC).
The current model of force production proposes that torque is generated in the catalytic domain, possibly by the rearrangement of the Switch I and II regions around the gamma phosphate of ATP, which is amplified by a large-scale reorientation of the regulatory domain. Rotation of the regulatory domain has been supported experimentally by a number of spectroscopic techniques: electron paramagnetic resonance (EPR) (Arata, 1990; Hambly et al., 1991, 1992), birefringence (Irving, 1993), fluorescence anisotropy (Allen et al., 1995; Irving et al., 1995; Ling et al., 1996), and fluorescence resonance energy transfer (Palm et al., 1999; Shih et al., 2000; Xiao et al., 1998, 2003; Xu and Root, 1998), as well as structural methods, electron microscopy (Jontes et al., 1995), and x ray (Dominguez et al., 1998; Houdusse and Cohen, 1996; Volkmann et al., 2000). The orientation of the RLC is, however, more complicated than these results would suggest, as numerous fluorescence anisotropy studies have reported the presence of more than one orientational population in all of the contractile cycle intermediate states (Bell et al., 2002; Sabido-David et al., 1998a,b) and in response to transient perturbations using either caged ATP/ADP or step-length changes of the fibers (Corrie et al., 1999; Hopkins et al., 1998; Irving et al., 1995). Unfortunately, fluorescence anisotropy does not resolve the various oriented populations. The observed anisotropy was modeled as either very broad distributions or a mixture of oriented and fully isotropic populations (Allen et al., 1995, 1996; Hopkins et al., 1998; Irving et al., 1995; Ling et al., 1996; Sabido-David et al., 1998b).
EPR is capable of resolving spin populations with different orientations as they resonate at different magnetic field strengths. Previous EPR studies of RLC have reported ordered and disordered populations (Arata, 1990; Hambly et al., 1991, 1992; Zhao et al., 1996). The origin of the disordered population was ascribed to non-specifically bound RLC or nonfunctional myosin heads. In this study we have improved the RLC exchange to achieve stoichiometric and functional reconstitution, thus ascertaining the observed heads are fully functional. The spectra of intrinsic rigor heads and myosin subfragment 1 (S1) heads infused into unlabeled muscle fibers were complex. A new EPR analysis method, which is independent of the physical model of orientational distribution, revealed the presence of two populations—one well-ordered, and the other with a broad, nonrandom distribution.
The ordered spin population was translated into the regulatory domain orientation, using molecular modeling and energy minimization methods, and found to be in excellent agreement with the orientation reported from electron microscopy (EM) for S1 heads decorating actin filaments (Holmes et al., 2003).
MATERIALS AND METHODS
Sample preparation
Psoas muscle fibers were obtained from freshly sacrificed New Zealand White rabbits as described in Fajer et al. (1988).
Myosin was prepared using the method of Tonomura et al. (1966) with modifications as described by Eads et al. (1984). Heavy meromyosin (HMM) was prepared by mild chymotryptic digestion, in the presence of Mg2+, from freshly prepared myosin (Wagner, 1982).
Myosin light chains were isolated from purified myosin using the guanidine-HCl method (Wagner, 1982) and separated on a Cibacron Blue 3GA column (Sigma, St. Louis, MI) according to Toste and Cooke (1979). RLC-Cys154, containing a single cysteine, was expressed in E. coli and purified on Q-Sepharose (Boey et al., 1994) and is referred to hereafter as RLC. TnC was purified according to Potter (1982).
RLC was labeled for 2 h by adding a fivefold excess of InVSL spin-label to 1 mg/mL protein solution at 22°C (Baumann et al., 2001). Free spin-label was removed by exhaustive dialysis. The final label/protein ratio was 0.9–1.0, determined by double integration of the EPR signal and protein concentration determination by BCA assay (Pierce, Rockford, IL).
Fiber exchange
The procedure used to extract RLC from skeletal muscle fibers and to reconstitute the fibers was based on methods developed by Moss and collaborators (Ling et al., 1996; Moss et al., 1982). Thin fiber bundles (4–8 single/double fibers) were dissected from larger glycerinated fiber bundles and rinsed in 150 mM KCl, 2.5 mM EDTA, 2.5 mM EGTA, 20 mM imidazole, pH 7 at 4°C for 5 min. The RLC was removed from muscle by incubation in 80 mM KCl, 10 mM EDTA, 10 mM CDTA, 1.5 mM TFP, 0.05% Triton X-100, 20 mM imidazole, pH 7.0 for 2 h at 4°C, with a change of solution after 1 h. Reconstitution consisted of sequential incubations for 45 min at 22°C in a relaxing solution (1 mM MgCl2, 7 mM EGTA, 5 mM MgATP, 20 mM imidazole, pH 7.0) containing 30 μM TnC, then 30 μM labeled RLC and finally 30 μM LC1/LC3. For EPR measurements the bundles were gathered and tied together using silk thread.
The efficiency of the exchange was evaluated on SDS PAGE gels. The integrated intensities of the protein bands were calibrated with serial dilutions of protein standards and the fiber samples were applied at two different concentrations to keep all intensities in the linear region of the gel. Light-chain concentrations were normalized to the concentration of actin which remains invariant during the exchange.
HMM reconstitution
A similar method was used for HMM as for fibers, except that RLC removal was accomplished in 1 h. The dissociated RLC was separated from the HMM on a Sephadex G-100 column (Sephadex, Uppsala, Sweden). The RLC-depleted HMM was incubated in 30-μM-labeled RLC in 1 mM MgCl2, 7 mM EGTA, 5 mM MgATP, 20 mM imidazole, pH 7 for 45 min at 22°C. The excess of labeled RLC was removed on the Sephadex G-100 column, and the HMM was dialyzed into rigor solution for infusion into skeletal muscle fibers.
S1 reconstitution
Papain S1 (10 μM) was incubated with 30 μM labeled RLC in 60 mM K-acetate, 2 mM ATP, 1 mM EGTA, 2 mM EDTA, 2 mM CDTA, 0.5 mM TFP, 20 mM MOPS, pH 6.7 for 20 min at 22°C. The RLC exchange was stopped by the addition of 4 mM MgCl2 and cooling to 4°C. The sample was clarified by centrifugation for 10 min at 2500 g in a Beckman microfuge (Beckman-Coulter, Fullerton, CA) and desalted on a G-25 column eluted with rigor buffer (130 mM K-propionate, 2 mM MgCl2, 1 mM EGTA, 1 mM NaN3, 20 mM MOPS, pH 7.0).
Stretching of muscle fiber beyond filament overlap
Six to eight exchanged fiber bundles consisting of 4–8 single fibers were tied together with silk thread within a 50-μL capillary. The bundle was slowly stretched at 0.33 mm/min using a small motor, while being perfused with a relaxing solution containing a force generation inhibitor (25 mM BDM, 5 mM MgATP, 1 mM EGTA, 3 mM MgCl2, 130 mM K-propionate, 20 mM sodium phosphate, pH 7.0). Sarcomere length was determined using laser diffraction (Uniphase 1507-0 HeNe laser, Uniphase, San Jose, CA). Muscle fibers stretched to ∼3.9 μm were held at this length in the EPR capillary by gluing the silk thread to the outside wall.
Circular dichroism
Ellipticity of RLC was measured using an AVIV Circular Dichroism Spectrometer Model 202 (AVIV, Lakewood, NJ). The percentage of helicity was calculated from the molar ellipticity,
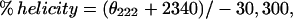 |
where θ222 is the molar ellipticity at 222 nm (Chen et al., 1972).
The use of mutated RLC and covalent modification with a spin probe may alter the secondary structure of the RLC. Circular dichroism (CD) spectroscopy was used to evaluate two effects: 1), whether the replacement of Cys-125 with an arginine residue in chicken skeletal RLC altered the RLC structure and 2), whether spin-labeling with InVSL altered the native RLC structure. The percentage of helical content between the native RLC and RLC-Cys154, as calculated according to Chen et al. (1972), changed by <2%, whereas the difference between labeled and unlabeled RLC was <1%.
Muscle mechanics
Muscle tension was determined on single fibers using a Cambridge Technology 400A force transducer (Cambridge Technology, Cambridge, MA). The extraction of endogenous RLC and the three-step reconstitution were accomplished in the well with the tension measured at 22°C. The temperature was adjusted to 4°C during RLC removal and raised to 22°C for the reconstitution.
SDS-PAGE gels of muscle fibers
Two to three milligrams of muscle fiber was dissolved in 50 μL of 0.55 M NaCl, 2% SDS, 5 mM DTT, finely minced, heated at 100°C for 3 min and sonicated 10 times for 2 s on ice with a Fisher Model 60 Sonic Dismembrator (Fisher, Pittsburgh, PA). The 15% gels were stained, scanned with a Canon D123OU personal scanner (Canon USA, Lake Success, NY), and analyzed using a LABVIEW (National Instruments, Austin, TX) program written in-house.
EPR spectroscopy
EPR experiments were carried out on a Bruker EMX X-band spectrometer (Bruker, Billerica, MA), using modified TM110 or TE102 cavities that allow variation of the fiber orientation with respect to the magnetic field as described in Fajer (1994a). The spectra were recorded with a microwave field strength (H1) of 0.03–0.08 G, a magnetic field modulation amplitude (Hm) of 2 G, and a modulation frequency of 50 kHz. EPR spectra were averaged and analyzed using spectral analysis software developed by our laboratory.
Molecular modeling
The structure of InVSL-modified cysteine was built using the Insight II (Accelrys, San Diego, CA) Builder module and added to the amino acid fragments library. The coordinates of the actomyosin complex obtained from EM reconstructions and the fitting of atomic structures of actin and S1 were from Holmes et al. (2003). The lowest energy/most probable position of the InVSL spin-label docked to Cys-154 of RLC was found utilizing Monte Carlo conformational searching of spin-label torsional space (Sale et al., 2002). The Monte Carlo search routine is implemented in CHARMM and uses the CHARMM19 force field for energy evaluations. The topology files and force fields for the spin-label have been developed as described in Sale et al. (2002) and are available from the authors. To find the orientation of the regulatory domain, with respect to actin, that was consistent with the EPR spectra, the whole regulatory domain was rotated around residue 783 in 5° increments around orthogonal axes. For each rotation, the orientation of the InVSL pπ orbital with respect to actin was calculated, stored in the database and searched for the conformer that would satisfy the observed spin-label orientation.
Spectral decomposition
EPR spectra of probes that do not move on the nanosecond timescale are a superposition of the resonances of spins at various angles with respect to the magnetic field, as
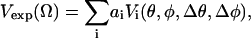 |
where the Vexp is experimental lineshape; Ω denotes orientational distribution of ensemble of spins; and Vi (θ, φ, Δθ, Δφ) is the individual lineshape (the basis set) of the spin packet i oriented at (θ,φ) with respect to the magnetic field H in polar coordinates and with an intrinsic dispersion of Δθ, Δφ. The dispersion here is assumed to be Gaussian but any other shape can be used for dispersion of the basis set spectra.
The resonance position of any single spin with the external magnetic field at angle θ with respect to the nitroxide pπ orbital and angle φ, its projection on the nitroxide plane, and the NO bond direction, is given by
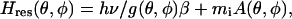 |
where mi is the nuclear spin number of the nitrogen, and β is the Bohr magneton. The g-tensor describing Zeeman interactions and the A-tensor of hyperfine interactions are given by their principal values (denoted by subscripts) and the orientation with respect to the field,
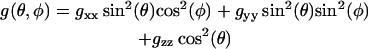 |
and
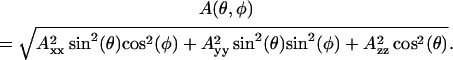 |
For randomly oriented samples, the individual spectra are summed over the isotropic distribution. For the macroscopically oriented samples, the spectra are summed over the orientational distribution of the label within the protein, distribution of the labeled domain within a sample, and over the distribution of the sample with respect to the magnetic field (Fajer, 1994a). These are three separate orientational distributions:
The orientation of the label with respect to the protein can be obtained from computational analysis of label rotamers in the known crystal structures (Sale et al., 2002) or from the combination of EPR and electron microscopy (Fajer, 1994a).
Orientation of the labeled domain is unknown, and is the object of this analysis.
Orientation of the sample with respect to the field is that of a cylinder (the myosin heads follow helical tracks on the thick filaments and the filaments themselves are azimuthally disordered), with the long axis parallel to the axis of a muscle fiber inclined to the magnetic field at an angle determined by the tilt of the experiment; in this case, 0°, 30°, and 90°.
The first step in the spectral decomposition is to create a library (database) of model spectra, each corresponding to a different θ, φ, and dispersion. In principle, there are four dimensions: θ, φ, Δθ, Δφ, and the exhaustive library should contain combinations of all angles and dispersions. Such a library results in nonunique solutions. For example, EPR lineshapes are not linear or singular with respect to orientation, and two different combinations of orientational parameters can yield very similar spectra. Therefore, we have limited the number of library spectra by increasing the step size between the grid points and by reducing the number of dimensions of the library, to generate unique spectra. The differences between the library spectra are defined by the signal/noise and resolution of the experimental spectrum. In practice, we discount the dimensions that do not determine the solution. For example, in the case presented here, φ and Δφ were found not to affect the solution, and were set to a constant value.
The experimental spectrum Vexp(Ω), is expressed as a linear combination of the library spectra using least-squares fitting to find the fraction of each orientational component. In each trial the library spectra are chosen at random and χ2 is calculated for each choice of orientational components: 50,000–500,000 random trials are sufficient to cover all the combinations of angular parameters. To avoid overfitting of the spectra or biasing toward a local minimum, the number of components was limited to 1–10 with random selection of the parameters from the library. The χ2 dependence on the number of components is biphasic; a sharp dropoff is followed by a plateau, when increasing the number of populations does not improve the fit significantly. Similar tracking of χ2 is performed as a function of the library size. The angle increment per library spectra is increased as long as there is no effect on the fit quality. Such a sparse library results in a more unique solution and faster decomposition. However, the fits to a single spectrum still generate many solutions, so global analysis of the sample tilt series was used to achieve a unique solution. Muscle fiber spectra are recorded parallel, perpendicular, and at 30° to the magnetic field, and all three spectra are analyzed simultaneously by concatenating into a single vector (Fajer, 1994a,b; Hustedt et al., 1997). The corresponding library consists of spectra (θ, φ, Δθ, Δφ) at three tilts concatenated into a single record. This computational strategy is implemented in MATLAB and is available from the authors.
RESULTS
Exchange of labeled RLC into skeletal muscle fibers
In the past, the relatively low levels of exchange of labeled light chains into skeletal muscle fibers resulted in ambiguity concerning functionality of the exchanged crossbridges. We therefore sought to increase the level of the exchanged light chains to near stoichiometry while preserving contractile function. Small bundles of fibers were used for the experiments and the usual exchange procedure was replaced by an extraction and three-step reconstitution that provided optimal levels of both stoichiometric exchange and functionality.
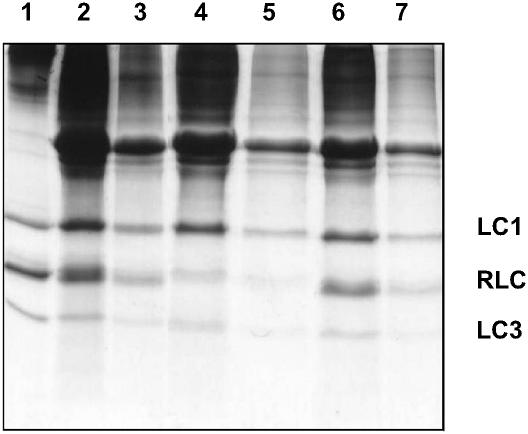
The relative protein levels at each of the extraction/reconstitution steps can be followed on the gel shown in Fig. 1 and are summarized in Table 1. The muscle fibers were depleted of 72 ± 4% endogenous RLC and reconstituted to 92 ± 6% of the original level; thus the labeled RLC represents 70 ± 10% of total RLC.
FIGURE 1.
SDS-PAGE gel of skeletal muscle fiber reconstitution with labeled RLC: Lane 1, purified myosin; Lanes 2 and 3, native fibers before RLC stripping, fourfold difference in loading; Lanes 4 and 5, fibers after RLC stripping, fourfold difference in loading; and Lanes 6 and 7, fibers after reconstitution with TnC, labeled RLC, and LC1/LC3 mixture, fourfold difference in loading.
TABLE 1.
Extent of RLC stripping and reconstitution, and active tension of fibers with exchanged RLC labeled at Cys-154
| Fiber sample
|
RLC content
|
LC1 content
|
LC3 content
|
TnC content
|
Active tension
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| %* ±SEM | n† | %* ±SEM | n† | %* ±SEM | n† | % ±SEM | n† | % ±SEM | n† | |
| Control | 100 | 5 | 100 | 5 | 100 | 5 | 100 | 3 | 100 | 2 |
| RLC stripped | 28 ± 4 | 5 | 96 ± 3 | 5 | 92 ± 6 | 5 | 17 ± 10 | 3 | 3 ± 1 | 4 |
| InVSL-RLC reconstituted | 92 ± 6 | 5 | 98 ± 5 | 5 | 97 ± 4 | 5 | 93 ± 10 | 3 | 91 ± 5 | 4 |
LC1, LC3, and RLC values were normalized to actin.
Where n is the number of independent samples.
Functionality of the exchanged fibers was assessed by measuring tension at submaximal activation levels. The tension at full activation is sensitive to the extent of damage to the contractile apparatus (91 ± 5%, corrected for the 26 ± 2% drop in tension of control fibers that is not uncommon in RLC exchanges; Sabido-David et al., 1998b), whereas tension at partial activation is inversely proportional to the amount of RLC present (Hoffman et al., 1990; Metzger and Moss, 1992; Moss et al., 1982).
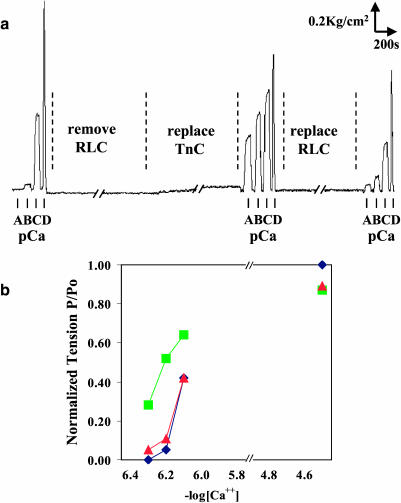
A tension trace of fibers taken during RLC removal and replacement is shown in Fig. 2 a. The isometric tension of the fibers was measured at submaximal levels of Ca2+, pCa 6.1, 6.2, 6.3, and at full activation, pCa 4.5. The increase of isometric tension with RLC removal was largest at pCa 6.2 (see Fig. 2 b), where the tension was 0.05 ± 0.01 of fully activated muscle before RLC removal, increased to 0.52 ± 0.02 upon RLC removal, and returned to 0.11 ± 0.01 on replacement with labeled RLC. The restoration of submaximal isometric tension to within 6% of its original value indicates that 87% of removed RLC has been replaced with labeled RLC at its original binding site. Thus we have achieved a near-stoichiometric and functional reconstitution.
FIGURE 2.
Fiber isometric tension at submaximal activation levels of calcium. (a) Tension trace of fiber before RLC removal, after RLC removal, and upon labeled RLC replacement. Isometric tension was taken at pCa of A = 6.3, B = 6.2, C = 6.1, and D = 4.5. (b) Normalized isometric fiber tension at submaximal activation levels of calcium; ♦ is before RLC removal, ▪ is with RLC removed, and ▴ is with RLC replaced.
EPR spectra of labeled RLC exchanged into skeletal muscle fibers
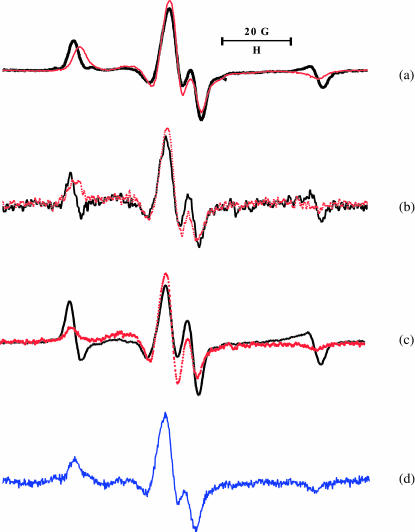
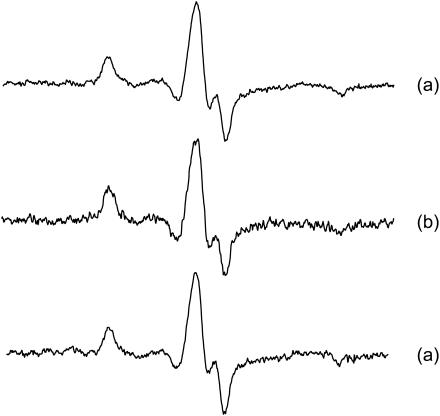
To study orientation and dynamics of labeled protein requires that the spin-label be ordered and immobilized on the surface of the protein. The label ordering is demonstrated by the spectral difference, or anisotropy, when the sample is rotated by 90° in the magnetic field. The tilt series spectra of rigor heads containing InVSL-labeled RLC for the intrinsic heads, exchanged HMM infused into unlabeled fibers, and S1 infused into fibers are shown in Fig. 3, a–c, respectively. The difference is most obvious in the low- and high-field region, which indicates ordering. Immobilization of the label is evident from the maximum splitting of random spectra of myofibrils, 2Aeff = 70.7 ± 0.1 G (Fig. 3 d), which compares closely to the rigid limit 70.9 ± 0.2 G obtained from labeled RLC cross-linked DITC-coated glass beads (Baumann et al., 2001).
FIGURE 3.
EPR spectra of: (a) skeletal muscle fibers with InVSL labeled RLC exchanged directly into the fiber, shown in rigor in the parallel orientation (black) and perpendicular orientation (red); (b) fibers decorated with HMM containing labeled RLC; (c) S1, labeled RLC, infused into a fiber; and (d) myofibrils with directly exchanged, labeled RLC.
The shift of the low-field peak and the positive intensity of the high-field peak in the parallel spectrum of muscle fibers (Fig. 3 a) indicates the presence of an ordered population of spins with their z axis close to the fiber axis, but there is also a second, less-ordered population contributing broader components to the spectrum, best represented by a part of the broad central peak.
Spectral decomposition
Magnetic parameters (g-tensor, hyperfine tensor, homogenous, and non-homogenous linewidth) were found by fitting myofibrillar spectra that have an isotropic orientational distribution (Fajer et al., 1990). Having determined these magnetic parameters, we created a library of spectra corresponding to various orientational distributions. The library of spectra were generated by varying the z axis of the nitroxide with respect to the fiber axis (θ), its projection on the x-y plane (φ), and the Gaussian dispersions around the two angles, constituting a four-dimensional grid. The figure of merit, χ2, between the experimental and best predicted spectra dropped twofold between one and two components, with a further 22% drop between two and three components, before leveling off (Table 2). The library was pruned from four dimensions to two dimensions by fixing φ to 45° and Δφ to 45°, since there was a <5% loss of χ2 by varying these parameters. The lower the number of library spectra that are used, without compromising the quality of the fit, the more unique is the solution. Therefore, we have progressively limited the number of spectra by increasing the grid interval for θ to 10°, with Gaussian disorder varying between 0° and 60° at 15° intervals. The final library that was found sufficient to fit experimental spectra with no increase of χ2 consisted of 50 spectra.
TABLE 2.
Decomposition of experimental spectra of labeled RLC-S1 in fibers
| Number of components | χ2 | θ | Δθ | Fraction (%) |
|---|---|---|---|---|
| One | 0.67 | 20 | 45 | 100 |
| Two | 0.27 | |||
| A | 0 | 15 | 24 | |
| B | 0–20* | 60 | 76 | |
| Three | 0.21 | |||
| A′ | 0 | 15 | 25 | |
| B′ | 0–10 | 60 | 50–68 | |
| C′ | 40–70* | 60 | 17–25 | |
| Four | 0.21 | |||
| A″ | 0–10* | 15 | 22–24 | |
| B″ | 0 | 60 | 56–58 | |
| C′ | 80 | 0 | 18 | |
| D″ | 20–30* | 0–15 | 1–5 |
The range of the solution from multiple decompositions with the same goodness of fit.
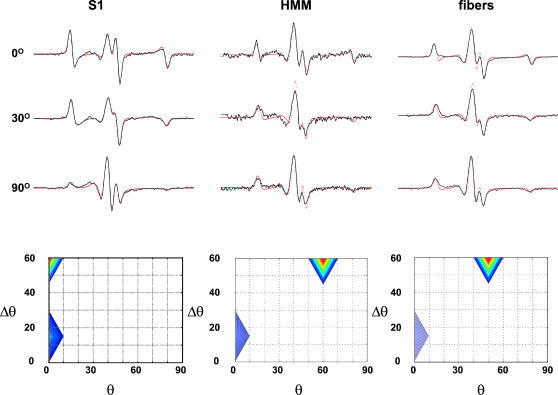
Furthermore, to decrease the ambiguity we have fitted globally all three tilt spectra obtained for each: S1 infused into unlabeled muscle fibers, HMM similarly infused, and muscle fibers whose native RLC was exchanged with labeled RLC (Fig. 4, top). The spectra of all the tilt series of the three samples are accounted for by only two populations. One population is better ordered than the other, Δθ = 15° versus Δθ = 50–60°. The ordered population is centered at 0° i.e., with the nitroxide z axis parallel to the magnetic field, whereas the less-ordered population is centered at θ = 20° for S1 and at θ = 60° for HMM and intrinsically labeled fibers. Note that the latter difference between S1 and HMM/fibers is not meaningful in view of the wide angular dispersion of this component. In infused S1, the ordered population represents ∼25% of total spins; this value decreases to 10% in HMM and intrinsic fibers.
FIGURE 4.
Top Global decomposition (red) of a tilt series of spectra (0°, 30°, 90°) of S1 decorated fibers (left), HMM infused into fibers (middle), and intrinsically labeled fibers (right). Only two components are required for a satisfactory fit, seen in the contour plot (bottom row). Bottom θ is the center of the distribution and Δθ is the dispersion, half-width at half-height. Blue, 0–30%; yellow, 30–60%; and red, >60%.
Origin of RLC ordering
The ordering of the regulatory domain in rigor might be due to interaction of the myosin head with the surface of the thick filament or the binding of the catalytic domain to the thin filament. The possible interaction with the thick filament surface is suggested by the high degree of order visualized by EM reconstruction (Offer and Knight, 1996) and fiber x-ray diffraction (Wray et al., 1978; Wray and Holmes, 1981). On the other hand, the catalytic domain in rigor fibers is known to be highly ordered by actin binding (Pollard et al., 1993; Thomas and Cooke, 1980). To test these alternative origins of labeled RLC ordering, the actomyosin interactions were abolished by relaxation and by stretching the fibers from a sarcomere length of 2.5 μm to 3.9 μm. The oriented population seen in rigor at resting sarcomere length disappears in the presence of ATP or as the fiber is stretched out-of-overlap (Fig. 5), which shows both spectra are random. Thus the labeled RLC orientation seen in rigor is dictated by crossbridge binding to actin.
FIGURE 5.
EPR spectra of skeletal muscle fibers with InVSL labeled RLC, stretched out-of-overlap: (a) in rigor, oriented parallel to the magnetic field; (b) in relaxation (in the presence of ATP, no Ca2+), parallel; and (c) relaxation, perpendicular.
The disordering observed within the larger component might arise due to several mechanisms: the helical mismatch of the myosin and actin filaments; the strain between the two heads originating from the common origin; a disordered label on the surface of the protein; or the inherent elasticity of the regulatory domain.
The first two mechanisms can be directly tested using isolated labeled HMM and S1 fragments infused into muscle fibers. The observation that twice as many heads are ordered in the absence of strain (S1) than in the presence (HMM, fibers) argues that a significant component of the disordered fraction in fibers arises from strain due to the common origin of the two heads. Helical mismatch is less likely to contribute to disorder as the HMM and fiber spectra are similar (Fig. 3).
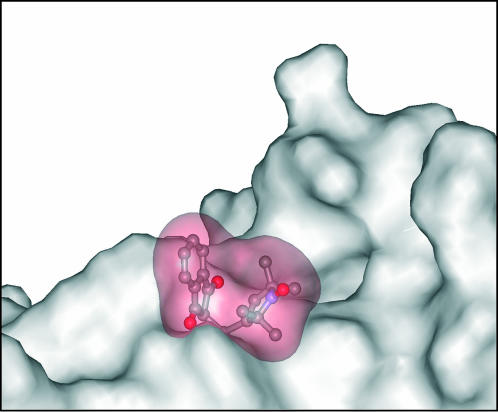
The third possible cause of a disordered fraction, non-stereospecific attachment of the spin-label to Cys-154, was explored using a Monte Carlo rotamer search of the spin-label attached to RLC (Sale et al., 2002). All single bonds between the protein backbone and the free radical were rotated at random, followed by energy minimization. The minimum that was found was isolated from other observed minima by 7 kT, reflecting steric hindrance imposed by the neighboring side chains (Fig. 6). Such a large energy difference demonstrates that the label environment constricts the orientation of the label to a well-defined value; i.e., the label does not have multiple orientations with respect to the labeled domain. This is in agreement with relative inaccessibility of the Cys-154 as the labeling in the folded state is negligible and RLC had to be unfolded in 5-M urea to be labeled.
FIGURE 6.
Surface rendering of the predicted conformation of the InVSL spin-label docked to Cys-154 of RLC. Note the steric constraints placed on the label orientation by the adjacent side chains.
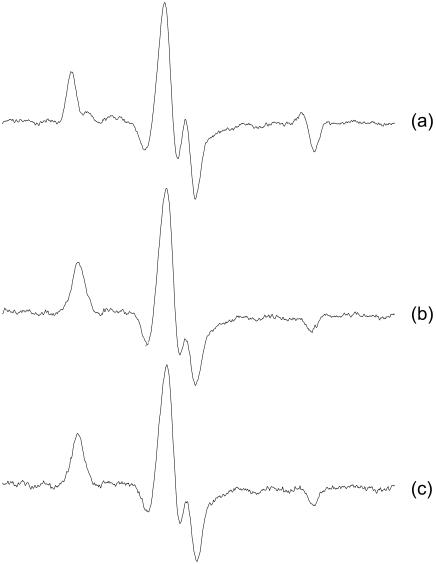
Isometric contraction
The orientation of the labeled RLC was also investigated in isometric contraction. Fig. 7 shows the EPR lineshapes of fibers parallel to the magnetic field perfused with rigor solution, in the presence of 5 mM MgATP (relaxation) and in isometric contraction in the presence of Ca2+. To avoid a possible rigor core—depletion of MgATP within the fiber bundle—we have used a backup solution consisting of creatine phosphate and creatine phosphokinase (Adhikari and Fajer, 1996). The spectra of relaxed and activated fibers have identical lineshape. The ordered fraction visible in the rigor spectrum is not present in the isometric contraction and no additional population is observed. Superficially, this seems to be at odds with the results of Baker et al. (1998), who have observed a shift of orientational populations in scallop muscle between contraction and relaxation. However, the same authors reported an “almost undetectable” shift (<4%) for the rabbit skeletal fibers. The difference between scallop and rabbit results was originally thought to be due to non-specifically bound RLC, however, our reconstitution method is specific and confirms this basic difference between the two species.
FIGURE 7.
EPR spectra of muscle fibers with labeled RLC exchanged directly into the fiber at full overlap, parallel to the field: (a) rigor, (b) relaxation, and (c) isometric contraction.
Regulatory domain modeling
A well-defined probe orientation implies stereospecific attachment of the reporting label on the RLC surface, allowing interpretation of the probe orientation in terms of protein orientation. Steric restrictions imposed by neighboring side chains orient the InVSL spin-label on the RLC surface, alleviating problems associated with probe floppiness, that have been observed in previous EPR and fluorescence studies (Baker et al., 1998; Corrie et al., 1999; Hambly et al., 1991; Ling et al., 1996).
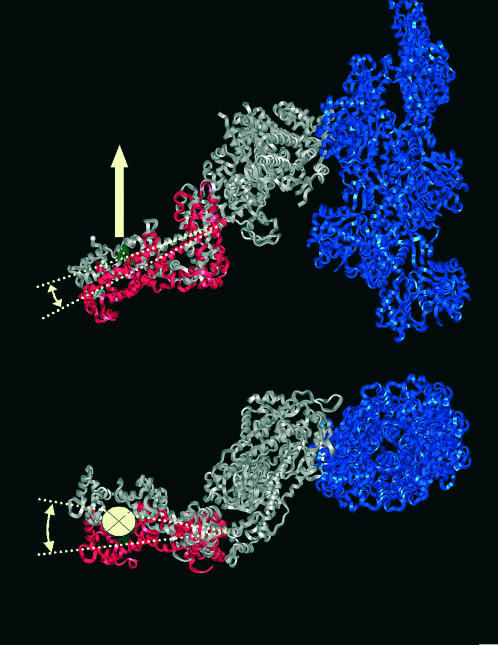
The modeling was accomplished in two steps. First the orientation of the probe was determined using a Monte Carlo conformational search method, to find the lowest energy spin-label conformer (Sale et al., 2002). Once the probe orientation was found, the regulatory domain was rotated around three orthogonal axes with their origin at residue 783. The result was a family of regulatory domain structures with all possible orientations. For each of the positions the angle between the label nitroxide z axis and actin was calculated and compared to the experimentally found value. A unique structure that places the z axis parallel to the actin filament and which does not have a steric clash with actin is shown in Fig. 8 (red). Comparison with the EM reconstruction (gray) of Holmes et al. (2003) reveals a small (8°) axial rotation and a 22° twist around the actin filament axis. Note that the largest difference is in the azimuthal direction, where the resolution of the EM reconstructions is the least well-resolved. The excellent agreement between EPR and EM underscores that the protein orientation detected by this extrinsic probe, under physiological conditions, in a force-producing fiber, complements the characterization obtained under the more harsh conditions used for EM. To the best of our knowledge, this is the first attempt to interpret EPR spectra in terms of the absolute orientation of a labeled protein domain within protein complex.
FIGURE 8.
Orientation of regulatory domain derived from EPR spectra (red). The z axis of the nitroxide probe (white arrow) is oriented parallel to the actin filament (blue). Axial view (top), end-on view along the axis of the actin filament (bottom). In gray the S1 structure as determined by Holmes et al. (2003) from fits of atomic models of actin and S1 to EM reconstructions.
DISCUSSION
The improvement in the reconstitution technique of RLC into muscle fibers, combined with the novel spectral decomposition technique, allowed us to: 1), quantify and parameterize the complex orientational population and 2), determine for the first time the absolute orientation of a labeled domain in a macromolecular protein system using EPR.
The EPR spectra of the labeled RLC in either intrinsic or infused heads all revealed that up to a quarter of heads contain highly ordered probes, whereas the remainder is less ordered. The improved reconstitution resulted in the near-stoichiometric incorporation of labeled RLC into muscle fibers without functional impairment. Therefore, both the highly ordered and less ordered populations observed here represent functional heads. The ordering was found to be dependent on binding to actin, since no order is observed in fibers stretched beyond overlap or in relaxed fibers. Some of the disorder can be attributed to the strain between the heads as the ordered fraction decreases in HMM as compared to S1.
Reconstitution
A common problem in the interpretation of spectroscopic data from the regulatory domain of myosin has been the relatively low level of labeling (15%) of the myosin head in skeletal muscle (Arata, 1990; Hambly et al., 1991, 1992). Moreover, there has always been concern that the labeled heads may be functionally impaired by the exchange procedure. The lower level of exchange can be somewhat increased (∼50%) by using smooth muscle RLC to make smooth/skeletal chimeras (Ling et al., 1996). Alternatively, one can use invertebrate muscle (scallop) that has a higher exchange efficiency (∼75%), but which exhibits different functional characteristics compared to skeletal muscle (Baker et al., 1998). Therefore the conclusions obtained from previous experiments with low level exchange or utilizing cross-species chimeras may not reflect the behavior of the skeletal muscle crossbridge.
In this study our method of reconstitution incorporates 70% of labeled skeletal RLC into skeletal muscle, comparing well with chicken gizzard RLC in scallop fibers. This high level of incorporation results in 40% of crossbridges having both heads exchanged with labeled RLC. The suppression of tension at submaximal Ca2+ levels after removal of RLC, with a full recovery after addition of stoichiometric amounts of labeled RLC, argues strongly that: 1), the reconstituted RLC binds to the same sites as the original light chain and 2), labeled RLC substitutes well for unlabeled light chain. Therefore, the high level of exchange combined with the return of function to normal makes us confident that the observed EPR spectra are representative of native skeletal RLC.
RLC orientation
A number of previous spectroscopic studies have identified multiple, but broadly distributed, populations of optical and magnetic probes in rigor. Fluorescence polarization anisotropy of labeled RLC in muscle fibers was consistent with half of the probes being isotropically distributed and half being oriented at a well-defined angle (Ling et al., 1996). This analysis was not a result of multiple orientations of the probe with respect to the protein, as bifunctional probes with two attachment sites determining the orientation of the probe also showed a similar distribution (Corrie et al., 1999). Although fluorescence cannot distinguish between broad homogeneous populations and a mixture of ordered and disordered species, EPR studies, which resolve orientations along the magnetic field axis, identified similar components. The original EPR studies of RLC orientation observed between two and three populations of various degrees of order and mobility in skeletal muscle (Arata, 1990; Hambly et al., 1991; Zhao et al., 1996). Subsequently, Thomas and collaborators saw two relatively broad components in scallop muscle in rigor and relaxation (Baker et al., 1998).
In all these studies, there was at least one substantial component that was somewhat ordered in rigor; however, that degree of ordering was obscured by probe mobility resulting in motional averaging. Our finding that ∼25% of probes represent an ordered, yet immobilized, population allows for an accurate estimate of its orientation and distribution, θ = 0° with a narrow dispersion of 15°, using either a standard Gaussian model or the decomposition method developed here.
The orientation of both populations is defined by their binding to actin, since stretching the fiber to zero-overlap abolishes ordering. Similarly detachment of the heads by ATP (relaxation) also eliminates both ordered fractions. This is in general agreement with other studies on skeletal muscle using fluorescence (Allen et al., 1995, 1996; Hopkins et al., 1998; Irving et al., 1995; Ling et al., 1996; Sabido-David et al., 1998b) and EPR (Hambly et al., 1991, 1992; Zhao et al., 1996). Only in scallop muscle does some ordering persist in relaxation, which might represent a species-specific characteristic of invertebrate muscle (Baker et al., 1998). In contraction, we do not observe any ordered rigor component, again consistent with all the other studies. Even in scallop little anisotropy was detected between the parallel and perpendicular spectra during contraction, which suggests that no ordered population exists in this state (Baker et al., 1998).
It is unclear why the broad population exists in rigor. The simplest explanation would be that strain imposed by either the helical mismatch of the thick and thin filaments or the common origin of the two heads of each myosin distorts the regulatory domain. The helical mismatch scenario seems unlikely as the infused HMM, which is not constrained by the thick filament, has a similar distribution as the intrinsically labeled fibers. On the other hand, the common origin scenario has some experimental support, as the fraction of ordered heads is twice as large for single heads compared to double-headed myosin. However, since 75% of heads are contributing to the broad population in S1, the above cannot be the sole reason for the disorder.
Alternative explanations for the broad population are that the labeled RLC adopts several conformations, or that the spin-label is non-stereospecifically bound to the RLC. Our data argue against both alternatives, inasmuch as there is a high level of exchange and functionality, and the spin-label simulations showed a well-defined, minimum energy conformation. However, further experimental verification of label stereospecificity will be provided by bifunctional spin probes whose orientation and mobility are restricted by two attachment sites. These experiments are currently underway in our laboratory.
Although we have no direct proof, all our data points to a large conformational heterogeneity of the regulatory domain. We propose that the broad orientational distribution we have observed is a consequence of regulatory domain flexibility. Such flexibility has been observed by electrical birefringence (Highsmith and Eden, 1990), electron paramagnetic resonance (Adhikari et al., 1999), fluorescence resonance energy transfer (Palm et al., 1999; Xiao et al., 1998; Xu and Root, 1998), phosphorescence (Brown et al., 2001), and electron microscopy (Burgess et al., 2002). Similar disorder has been seen in insect flight muscle that is generally better ordered than skeletal muscle (Chen et al., 2002). These EM reconstruction data show that C–Cα bond of residue 154 of RLC is distributed over 60° with respect to the actin filament.
In summary, we have developed a reconstitution method that is stoichiometric, functional, and suitable for spectroscopic studies of skeletal muscle. The spectral decomposition developed here indicates the presence of two populations: one very well ordered, the other more disordered. The ordering of the regulatory domain is determined by actin binding, demonstrated by the absence of any order during relaxation, contraction, or in fibers stretched beyond overlap. The ordered population reports on the orientation of the regulatory domain with precision. Detailed computational analysis of the spectra allowed us to find the orientation of the regulatory domain with respect to actin filament in rigor, which is in excellent agreement with cryo-EM reconstructions.
Acknowledgments
We thank Drs. Hui Li and Louise Brown for many discussions and technical help, Dr. Rasmus Schröder for sharing the coordinates of the actomyosin complex before publication, Dr. Kalman Hideg for providing us with indane/dione spin-label, Dr. Ewa Bienkiewicz for help with circular dichroism spectroscopy, and Dr. David Budil for discussions about spectral decomposition.
Research was sponsored by National Science Foundation grant No. IBN-98-08708, National High Magnetic Field Laboratory In-House grant No. 5024, American Heart Association grant No. 9950424N, and National Heart Foundation of Australia grant No. G 01S 0424 (to B.H.)
Ken Sale's current address is Biosystems Research Department, Sandia National Laboratories, Livermore, CA 94551.
Abbreviations used: ADP, adenosine 5′ diphosphate; ATP, adenosine 5′ triphosphate; BDM, butadione monoxime; CDTA, trans-1,2-diaminocyclo-hexane-n,n,n′,n′-tetraacetic acid; DMF, n,n′-dimethyl formamide; DTT, dithiothreitol; EDTA, ethylenediamine tetraacetic acid aminoethyl ether; EGTA, ethylene-glycol-bis-n,n,n′,n′-tetraacetic acid; HMM, heavy meromyosin; InVSL, 2-(-Oxyl-2,2,5,5-tetramethyl-3-pyrrolin-3-methynyl)indane-1,3-dione; LC1, skeletal light chain 1; RLC-Cys154, recombinant light chain 2 with single cysteine at position 154; LC3, skeletal light chain 3; MOPS, 3-(n-morpholino)propane-sulfonic acid; PMSF, phenylmethylsulfonylfluoride; RLC, regulatory light chain; S1, myosin subfragment 1; TFP, trifluoperazine; TRIS, TRIS (hydroxymethyl)-aminomethane.
References
- Adhikari, B., J. Somerset, J. T. Stull, and P. G. Fajer. 1999. Dynamic modulation of the regulatory domain of myosin heads by pH, ionic strength, and RLC phosphorylation in synthetic myosin filaments. Biochemistry. 38:3127–3132. [DOI] [PubMed] [Google Scholar]
- Adhikari, B. B., and P. G. Fajer. 1996. Myosin head orientation and mobility during isometric contraction: effects of osmotic compression. Biophys. J. 70:1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, T. S., D. C. Sabido, N. Ling, M. Irving, and Y. E. Goldman. 1995. Transients of fluorescence polarization in skeletal muscle fibers labeled with rhodamine on the regulatory light chain. Biophys. J. 68:81S–86S. [PMC free article] [PubMed] [Google Scholar]
- Allen, T. S., N. Ling, M. Irving, and Y. E. Goldman. 1996. Orientation changes in myosin regulatory light chains following photorelease of ATP in skinned muscle fibers. Biophys. J. 70:1847–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata, T. 1990. Orientation of spin-labeled light chain 2 of myosin heads in muscle fibers. J. Mol. Biol. 214:471–478. [DOI] [PubMed] [Google Scholar]
- Baker, J. E., I. Brust-Mascher, S. Ramachandran, L. E. LaConte, and D. D. Thomas. 1998. A large and distinct rotation of the myosin light chain domain occurs upon muscle contraction. Proc. Natl. Acad. Sci. USA. 95:2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, B. A., B. D. Hambly, K. Hideg, and P. G. Fajer. 2001. The regulatory domain of the myosin head behaves as a rigid lever. Biochemistry. 40:7868–7873. [DOI] [PubMed] [Google Scholar]
- Bell, M. G., R. E. Dale, U. A. van der Heide, and Y. E. Goldman. 2002. Polarized fluorescence depletion reports orientation distribution and rotational dynamics of muscle cross-bridges. Biophys. J. 83:1050–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boey, W., W. Huang, B. Bennetts, J. Sparrow,, C. G. dos Remedios, and B. D. Hambly. 1994. Fluorescence resonance energy transfer within the regulatory light chain of myosin. Eur. J. Biochem. 219:603–610. [DOI] [PubMed] [Google Scholar]
- Brown, L. J., N. Klonis, W. H. Sawyer, P. G. Fajer, and B. D. Hambly. 2001. Independent movement of the regulatory and catalytic domains of myosin heads revealed by phosphorescence anisotropy. Biochemistry. 40:8283–8291. [DOI] [PubMed] [Google Scholar]
- Burgess, S., M. Walker, F. Wang, J. R. Sellers, H. D. White, P. J. Knight, and J. Trinick. 2002. The pre-power stroke conformation of myosin V. J. Cell Biol. 159:983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. F., H. Winkler, M. K. Reedy, M. C. Reedy, and K. A. Taylor. 2002. Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle. J. Struct. Biol. 138:92–104. [DOI] [PubMed] [Google Scholar]
- Chen, Y. H., J. T. Yang, and H. M. Martinez. 1972. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 11:4120–4131. [DOI] [PubMed] [Google Scholar]
- Corrie, J. E., B. D. Brandmeier, R. E. Ferguson, D. R. Trentham, J. Kendrick-Jones, S. C. Hopkins, U. A. van der Heide, Y. E. Goldman, C. Sabido-David, R. E. Dale, S. Criddle, and M. Irving. 1999. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature. 400:425–430. [DOI] [PubMed] [Google Scholar]
- Dominguez, R., Y. Freyzon, K. M. Trybus, and C. Cohen. 1998. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 94:559–571. [DOI] [PubMed] [Google Scholar]
- Eads, T. M., D. D. Thomas, and R. H. Austin. 1984. Microsecond rotational motions of eosin-labeled myosin measured by time-resolved anisotropy of absorption and phosphorescence. J. Mol. Biol. 179:55–81. [DOI] [PubMed] [Google Scholar]
- Fajer, P. G., E. A. Fajer, N. J. Brunsvold, and D. D. Thomas. 1988. Effects of AMPPNP on the orientation and rotational dynamics of spin-labeled muscle cross-bridges. Biophys. J. 53:513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajer, P. G., E. A. Fajer, and D. D. Thomas. 1990. Myosin heads have a broad orientational distribution during isometric muscle contraction: time-resolved EPR studies using caged ATP. Proc. Natl. Acad. Sci. USA. 87:5538–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajer, P. G. 1994a. Determination of spin-label orientation within the myosin head. Proc. Natl. Acad. Sci. USA. 91:937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajer, P. G. 1994b. Method for the determination of myosin head orientation from EPR spectra. Biophys. J. 66:2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambly, B. D., K. Franks, and R. Cooke. 1991. Orientation of spin-labeled light chain-2 exchanged onto myosin cross-bridge in glycerinated muscle fibers. Biophys. J. 59:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambly, B. D., F. Kathleen, and R. Cooke. 1992. Paramagnetic probes attached to a light chain on the myosin head are highly disordered in active muscle fibers. Biophys. J. 63:1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highsmith, S., and D. Eden. 1990. Ligand-induced myosin subfragment 1 global conformational change. Biochemistry. 29:4087–4093. [DOI] [PubMed] [Google Scholar]
- Hoffman, P. A., J. M. Metzger, M. L. Greaser, and R. L. Moss. 1990. Effects of partial extraction of light chain 2 on the Ca2+ sensitivities of isometric tension, stiffness, and velocity of shortening in skinned skeletal muscle fibers. J. Gen. Physiol. 95:477–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, K. C., I. Angert, F. J. Kull, W. Jahn, and R. R. Schroder. 2003. Electron cryomicroscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 425:423–427. [DOI] [PubMed] [Google Scholar]
- Hopkins, S. C., C. Sabido-David, J. E. Corrie, M. Irving, and Y. E. Goldman. 1998. Fluorescence polarization transients from rhodamine isomers on the myosin regulatory light chain in skeletal muscle fibers. Biophys. J. 74:3093–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse, A., and C. Cohen. 1996. Structure of the regulatory domain of scallop myosin at 2Å resolution: implications for regulation. Structure. 4:21–32. [DOI] [PubMed] [Google Scholar]
- Hustedt, E. J., A. I. Smirnov, C. F. Laub, C. E. Cobb, and A. H. Beth. 1997. Molecular distances from dipolar coupled spin-labels: the global analysis of multifrequency continuous wave electron paramagnetic resonance data. Biophys. J. 72:1861–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, A. F. 1974. Muscular contraction. J. Physiol. 243:1–43. [PMC free article] [PubMed] [Google Scholar]
- Huxley, H. E. 1969. The mechanism of muscular contraction. Science. 164:1356–1365. [PubMed] [Google Scholar]
- Irving, M. 1993. Birefringence changes associated with isometric contraction and rapid shortening steps in frog skeletal muscle fibres. J. Physiol. 472:127–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving, M., T. S. Allen, C. Sabido-David, J. Craik, B. D. Brandmeier, J. Kendrick-Jones, J. E. Corrie, D. R. Trentham, and Y. E. Goldman. 1995. Tilting of the light-chain region of myosin during step length changes and active force generation in skeletal muscle. Nature. 375:688–691. [DOI] [PubMed] [Google Scholar]
- Jontes, J. D., E. M. Wilson-Kubalek, and R. A. Milligan. 1995. A 32° tail swing in brush border myosin I on ADP release. Nature. 378:751–753. [DOI] [PubMed] [Google Scholar]
- Ling, N., C. Shrimpton, J. Sleep, J. Kendrick-Jones, and M. Irving. 1996. Fluorescent probes of the orientation of myosin regulatory light chains in relaxed, rigor, and contracting muscle. Biophys. J. 70:1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, J. M., and R. L. Moss. 1992. Myosin light chain 2 modulates calcium-sensitive cross-bridge transitions in vertebrate skeletal muscle. Biophys. J. 63:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, R. L., G. G. Giulian, and M. L. Greaser. 1982. Physiological effects accompanying the removal of myosin LC2 from skinned skeletal muscle fibers. J. Biol. Chem. 257:8588–8591. [PubMed] [Google Scholar]
- Offer, G., and P. Knight. 1996. The structure of the head-tail junction of the myosin molecule. J. Mol. Biol. 256:407–416. [DOI] [PubMed] [Google Scholar]
- Palm, T., K. Sale, L. Brown, H. Li, B. Hambly, and P. G. Fajer. 1999. Intradomain distances in the regulatory domain of the myosin head in prepower and postpower stroke states: fluorescence energy transfer. Biochemistry. 38:13026–13034. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., D. Bhandari, P. Maupin, D. Wachsstock, A. G. Weeds, and H. G. Zot. 1993. Direct visualization by electron microscopy of the weakly bound intermediates in the actomyosin adenosine triphosphatase cycle. Biophys. J. 64:454–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, J. D. 1982. Preparation of troponin and its subunits. Methods Enzymol. 85B:241–263. [DOI] [PubMed] [Google Scholar]
- Rayment, I., H. M. Holden, M. Whittaker, C. B. Yohn, M. Lorenz, K. C. Holmes, and R. A. Milligan. 1993a. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 261:58–65. [DOI] [PubMed] [Google Scholar]
- Rayment, I., W. R. Rypniewski, K. Schmidt-Base, R. Smith, D. R. Tomchick, M. M. Benning, D. A. Winkelmann, G. Wesenberg, and H. M. Holden. 1993b. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 261:50–65. [DOI] [PubMed] [Google Scholar]
- Reedy, M. K., K. C. Holmes, and R. T. Tregear. 1965. Induced changes in orientation of the cross-bridges of glycerinated insect flight muscle. Nature. 207:1276–1279. [DOI] [PubMed] [Google Scholar]
- Sabido-David, C., B. D. Brandmeier, J. Craik, J. E. Corrie, D. R. Trentham, and M. Irving. 1998a. Steady-state fluorescence polarization studies of the orientation of myosin regulatory light chains in single skeletal muscle fibers using pure isomers of iodoacetamidotetramethylrhodamine. Biophys. J. 74:3083–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabido-David, C., S. C. Hopkins, L. D. Saraswat, S. Lowey, Y. E. Goldman, and M. Irving. 1998b. Orientation changes of fluorescent probes at five sites on the myosin regulatory light chain during contraction of single skeletal muscle fibres. J. Mol. Biol. 279:387–402. [DOI] [PubMed] [Google Scholar]
- Sale, K., C. Sar, K. A. Sharp, K. Hideg, and P. G. Fajer. 2002. Structural determination of spin-label immobilization and orientation: a Monte Carlo minimization approach. J. Magn. Reson. 156:104–112. [DOI] [PubMed] [Google Scholar]
- Shih, W. M., Z. Gryczynski, J. R. Lakowicz, and J. A. Spudich. 2000. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell. 102:683–694. [DOI] [PubMed] [Google Scholar]
- Thomas, D. D., and R. Cooke. 1980. Orientation of spin-labeled myosin heads in glycerinated muscle fibers. Biophys. J. 32:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonomura, Y., P. Appel, and M. Morales. 1966. On the molecular weight of myosin II. Biochemistry. 5:515–521. [DOI] [PubMed] [Google Scholar]
- Toste, A. P., and R. Cooke. 1979. Interactions of contractile proteins with free and immobilized Cibracron Blue F3GA. Anal. Biochem. 95:317–328. [DOI] [PubMed] [Google Scholar]
- Volkmann, N., D. Hanein, G. Ouyang, K. M. Trybus, D. J. DeRosier, and S. Lowey. 2000. Evidence for cleft closure in actomyosin upon ADP release. Nat. Struct. Biol. 7:1147–1155. [DOI] [PubMed] [Google Scholar]
- Wagner, P. D. 1982. Preparation and fractionation of myosin light chains and exchange of the essential light chains. Methods Enzymol. 85:72–81. [DOI] [PubMed] [Google Scholar]
- Wray, J., P. Vibert, and C. Cohen. 1978. Actin filaments in muscle: pattern of myosin and tropomyosin/troponin attachments. J. Mol. Biol. 124:501–521. [DOI] [PubMed] [Google Scholar]
- Wray, J. S., and K. C. Holmes. 1981. X-ray diffraction studies of muscle. Annu. Rev. Physiol. 43:553–565. [DOI] [PubMed] [Google Scholar]
- Xiao, M., H. Li, G. E. Snyder, R. Cooke, R. G. Yount, and P. R. Selvin. 1998. Conformational changes between the active-site and regulatory light chain of myosin as determined by luminescence resonance energy transfer: the effect of nucleotides and actin. Proc. Natl. Acad. Sci. USA. 95:15309–15314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, M., J. G. Reifenberger, A. L. Wells, C. Baldacchino, L. Q. Chen, P. Ge, H. L. Sweeney, and P. R. Selvin. 2003. An actin-dependent conformational change in myosin. Nat. Struct. Biol. 10:402–408. [DOI] [PubMed] [Google Scholar]
- Xu, J., and D. D. Root. 1998. Domain motion between the regulatory light chain and the nucleotide site in skeletal myosin. J. Struct. Biol. 123:150–161. [DOI] [PubMed] [Google Scholar]
- Zhao, L., J. Gollub, and R. Cooke. 1996. Orientation of paramagnetic probes attached to gizzard regulatory light chain bound to myosin heads in rabbit skeletal muscle. Biochemistry. 35:10158–10165. [DOI] [PubMed] [Google Scholar]