Abstract
The lytic outcome of natural infection by Chlamydia trachomatis was exploited to select CHO (Chinese hamster ovary) cells, following chemical mutagenesis, that were deficient in their ability to sustain productive chlamydial infection. Four distinct mutant cell phenotypes with defects in either attachment or postattachment mechanisms that are required for infection by C. trachomatis and Chlamydia pneumoniae were characterized.
Chlamydiae are obligate intracellular bacterial pathogens that cause a wide spectrum of diseases in humans. In the United States, genital tract infection with Chlamydia trachomatis is the most common sexually transmitted disease, often resulting in severe human reproductive pathology (1). In developing countries, ocular infection with C. trachomatis can lead to trachoma, the leading cause of preventable blindness (21). Chlamydia pneumoniae is an important cause of lower and upper respiratory tract infections in humans, and it has been implicated in the pathogenesis of atherosclerosis (17).
Strains of C. trachomatis are classified into two biovariants, the trachoma and lymphogranuloma venereum (LGV) biovars, each of which possesses distinct characteristics that make it pathophysiologic for the human host (18). Although both biovars share near-complete identity in genomic sequence and biology, they cause significantly different clinical syndromes. The trachoma biovar (serovars A to K) is primarily associated with infections of mucosal surfaces in the genital tract and in the eye, whereas the LGV biovar (serovars L1, L2, and L3) causes invasive infection in vivo and proliferates in the lymphatic tissues (18). Differences in tissue tropism and disease etiology between the biovars may also be reflected in their behavior in tissue culture systems. Studies of the two C. trachomatis biovars and their interaction with mammalian cells in culture have demonstrated a number of important physicochemical differences. Relative to that of the LGV biovar, attachment of the trachoma biovar to HeLa cells is markedly inhibited by mild heat treatment (14). Infectivity of both trachoma and LGV is significantly inhibited by the addition of exogenous heparin and heparan sulfate. However, LGV biovar attachment in the presence of heparin is significantly inhibited whereas trachoma biovar attachment is only moderately inhibited (4, 5, 25). Pretreatment of host cells with the polycation DEAE-dextran or poly-l-lysine enhances the cell association and infectivity of the trachoma biovar but not those of the LGV biovar (13, 14). C. trachomatis shares 92% of its genes and a high level of genome synteny with C. pneumoniae (11). C. pneumoniae also shares some biological properties with C. trachomatis in its interactions with host cells. Their properties for in vitro invasion of mammalian host cells are presumably similar, as infectivity of mammalian cells by C. pneumoniae is also competitively inhibited by the addition of exogenous heparin or heparan sulfate (24).
All chlamydiae share a unique developmental cycle consisting of two distinct forms, the infectious, metabolically inactive form, the elementary body (EB), which persists in the extracellular environment, and the noninfectious, metabolically active form, the reticulate body (RB), which reproduces in the intracellular environment (8). EBs that adhere to and are endocytosed by permissive host cells undergo morphological and physiologic differentiation to the RB form and, in that form, multiply within a specialized vacuole, termed an inclusion, within the host cell. Accumulating RBs differentiate back into EBs that are subsequently released into the extracellular environment following host cell lysis, thus completing the developmental cycle.
Considerably more is known about the later intracellular stages of infection than about the initial molecular interactions that mediate EB attachment to and entry into host cells. The complexity of the developmental cycle, combined with the lack of a method for genetic manipulation of chlamydiae, has posed a significant impediment in studying this process. Evidence has been presented to support receptor-mediated (microfilament-independent) endocytosis of chlamydiae into clathrin-coated pits, as well as microfilament-dependent uptake into non-clathrin-coated vesicles (9, 10, 16, 23). However, receptor binding is both saturable and highly sensitive to proteolytic treatment of the host cell surface (2, 22), suggesting that a protein component of the host plasma membrane is critical for this interaction. To date, no host cell receptor(s) to which chlamydiae bind has been identified, with the exception of a possible role for the estrogen receptor complex (6).
We sought to begin dissection of the interaction of chlamydiae with mammalian host cells by the selection of host cell mutants that were incapable of sustaining infection. Mutants were selected by exploiting the natural ability of chlamydiae to lyse its host cell at the terminal stage of the chlamydial developmental cycle. This experimental approach was based on the rationale that the selection process should result in mutants that have deficiencies in the attachment, uptake, or intracellular processes required for sustaining chlamydial growth. Coincidentally the same strategy was recently reported by Carabeo and Hackstadt (3), who characterized a single mutant CHO (Chinese hamster ovary) cell line that was markedly deficient for binding of LGV but not trachoma biovar strains. Our selection resulted in 12 mutant cell lines with four distinct properties for binding and infection by the two C. trachomatis biovars and C. pneumoniae. The production of host cell mutants that are resistant to chlamydial infection will provide the opportunity to better characterize host-dependent molecular bases for chlamydial infection and pathogenesis.
L929, HeLa 229, Hep-2, and CHO-K1 cells were routinely cultured in RPMI-1640 tissue culture medium (Gibco, Gaithersburg, Md.) supplemented with 10% fetal calf serum (HyClone, Logan, Utah), 0.1 mg of streptomycin sulfate (Sigma, St. Louis, Mo.) per ml, 0.1 mg of vancomycin hydrochloride (Sigma) per ml, and 1 mM glycine (Sigma). The growth medium was additionally supplemented with 1 μg of cycloheximide (Sigma) per ml upon infection with chlamydial strains. C. trachomatis L2/434/Bu was grown for 48 h in suspension cultures of L929 cells containing approximately 1 × 106 cells/ml. C. trachomatis D/UW-3/Cx was grown for 72 h on confluent HeLa 229 cell monolayers (∼5 × 107 cells) in T-150 tissue culture flasks (Costar, Corning, N.Y.). C. pneumoniae strain CWL-029 was grown for 72 h on confluent monolayers of Hep-2 cells in T-75 culture flasks (Costar). EBs were isolated from infected cell lines by sonication and purified by centrifugation through discontinuous Renografin (E. R. Squibb & Sons, Princeton, N.J.) density gradients as previously described (12). Purified EBs were washed with sterile Hank's balanced salt solution (HBSS) (Gibco), suspended in sterile, sucrose-phosphate-glutamic acid buffer, and frozen at −70°C until needed. Chlamydial EBs were enumerated as inclusion-forming units as previously described (5).
CHO-K1 cell monolayers were mutagenized by exposure to ethyl methane sulfonate (EMS) (Sigma) as previously described (7), with minor modifications. Briefly, 1 × 106 CHO-K1 cells were plated in three separate T-75 tissue culture flasks (Costar) and grown overnight at 37°C in a humidified, 5% CO2 environment. On the following day, the monolayers were washed three times with sterile HBSS. Fresh medium containing 300 μg of EMS per ml was added to each flask. CHO-K1 cell monolayers were incubated at 37°C in a humidified, 5% CO2 environment for an additional 24 h. The concentration of EMS that was used resulted in a survival rate that was approximately 30% of the level attained with unmutagenized the CHO-K1 cell line (data not shown). The mutagenized CHO cells (i.e., those derived from the parental CHO-K1 cell line) were washed three times with HBSS to remove the EMS, and fresh medium was added to each T-75 flask. Surviving CHO cells were grown for 7 days to allow mutant cell phenotypes to repopulate the flasks. During this period, the mutagenized cells were routinely washed three times with HBSS every 48 h to remove accumulated dead cell material.
Mutagenized CHO cells (∼5 × 106 cells) were inoculated with C. trachomatis serovar L2 at a multiplicity of infection of 10 and incubated for 2 h at room temperature. Unbound chlamydial EBs were then removed, and fresh medium was added to each flask. The Chlamydia-infected, mutagenized CHO cells were incubated and washed three times with HBSS on a daily basis to remove accumulated dead cell material. Surviving CHO cells were allowed to repopulate the tissue culture flasks to approximately 50% confluence and, again, inoculated with C. trachomatis L2 serovar to select for chlamydia-resistant phenotypes. Surviving mutant cells were trypinized and plated in a sterile, 96-well tissue culture plate (Costar). Individual cell lines were subjected to a final round of selection with C. trachomatis serovar L2, expanded, and formally cloned twice by limiting dilution (20). No inclusions were observed by immunofluorescence when the individual clonal lines were stained with a monoclonal antibody specific for C. trachomatis (data not shown). Moreover, PCR with primers for the ompA gene confirmed the lack of infected or persistently infected cells (data not shown).
Immunofluorescence microscopy with Chlamydia-specific monoclonal antibodies was used to provide qualitative assessments of EB attachment to host cells plated on 12-mm-diameter coverslips. Serial dilutions of purified EBs were incubated with host cells for 30 min at 4°C, washed three times in HBSS, fixed with methanol, and stained with Chlamydia-specific monoclonal antibody (20). Quantitation of chlamydial infectivity was assessed by counting (in triplicate) the number of inclusion-forming units detected by immunofluorescence as previously described (19). Each cell line was tested in a minimum of three independent experiments.
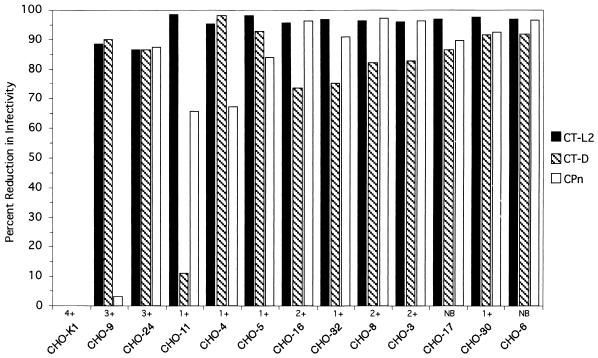
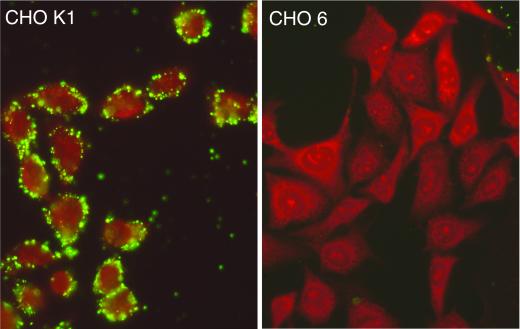
Most of the mutant cell lines (9 of 12) were resistant to infection by C. trachomatis (LGV and trachoma biovars) and by C. pneumoniae, although the resistance of the cell lines varied from 70 to 99% (Fig. 1). The majority of these cell lines (CHO-4, -5, -6, -11, -17, -30, and -32) showed little or no chlamydial EB attachment by direct fluorescence (Fig. 1). For example, cell lines such as CHO-6 were resistant to attachment by each of the three chlamydial strains (Fig. 2). Given that the trachoma biovar and the LGV biovar were both unable to attach or infect, this mutation is not directly related to differences in heparin-sensitive attachment between these biovars (4). This suggests that there is one host cell attachment receptor or cofactor that is utilized by each of these chlamydial biovars and species to mediate attachment. Alternatively, these data could be the result of more than one mutation affecting more than one receptor, although this seems unlikely because this phenotype was recovered in several independent clones.
FIG. 1.
Parallel assessments of infectivity of C. trachomatis serovar L2 (CT-L2), serovar D (CT-D), and C. pneumoniae (CPn) for each CHO cell mutant. The infectivity of each chlamydial strain for monolayers of each cell line was tested in triplicate. Approximately 48 h postinfection, chlamydial inclusions were detected by immunofluorescence and enumerated by counting inclusions. Similar data were obtained in at least three independent experiments. Qualitative estimates of attachment are indicated as follows: no binding (NB) or cellular-associated binding on a progressive numerical scale of 1+ to 4+, where 1+ represents approximately 10 EBs apparently associated with cell surfaces and 4+ represents many EBs bound with entire staining of the cells.
FIG. 2.
Immunofluorescence staining of C. trachomatis (L2) EBs attached to cells from the cell lines CHO-K1 and CHO-6. EBs were added to monolayers, incubated for 1 h at 4°C, fixed with methanol, and stained with a C. trachomatis-specific monoclonal antibody conjugated with fluorescein isothiocyanate.
A different phenotype was exhibited by CHO-24, which was 85% resistant to infection by all chlamydial strains tested but had attachment for EB similar to that of the parental cell line, CHO-K1. Thus, the CHO-24 cell line is unique in that it has a mutation that occurs after attachment, in a step of the infection process that is required for all the chlamydiae.
The CHO-9 cell line displayed a 90% reduction in infectivity with both C. trachomatis biovars, but there was little effect on the susceptibility of this cell line to C. pneumoniae infection. There was only a small apparent reduction in binding for the C. trachomatis biovars, as was observed with the CHO-24 cell line. These findings demonstrate that, while C. trachomatis and C. pneumoniae share an attachment mechanism, they have different biological requirements for postadherence interactions that lead to productive infection. The phenotype of the CHO-9 cell line suggests that a function after attachment, such as uptake or host modification after uptake, separates the pathways for these species.
The CHO-11 cell line was resistant to LGV infection (98% inhibition), showing a much greater susceptibility to trachoma biovar infection (12% inhibition). The phenotype of this cell line appears to be analogous to that reported by Carabeo and Hackstadt (3). The infectivity by C. pneumoniae for the CHO-11 cell line was inhibited by 65%. This suggests that the L2 serovar and the trachoma biovar differ in their respective needs for a host cell function at the attachment and/or invasion stage, as binding by the LGV biovar strain, measured by direct immunofluorescence, was minimal (data not shown). Carabeo and Hackstadt (3) concluded that the resistance to LGV biovar strains and sensitivity to trachoma biovar strains (serovars B or D) demonstrated by their cell line were likely due to utilization of different receptors and that this hypothesis is consistent with the differences in the susceptibilities of the biovars to heparan sulfate inhibition of attachment. This hypothesis requiring independent receptors is not consistent with the finding of mutant cell lines that are resistant to each of the chlamydial strains, demonstrating that there is a single primary receptor interaction that is also required for LGV infectivity. An alternative hypothesis for these data is that chlamydiae are utilizing a common pathway of entry that is composed of more than one subunit or structural modification of a subunit required for infection, with different requirements for each of the chlamydiae. While the LGV and trachoma biovars differ in their sensitivity to heparin inhibition for host cell adhesion (4), both biovars are markedly sensitive to heparin inhibition when infectivity is measured (5, 15). Thus, while a proportion of trachoma biovar EBs can adhere to host cells in the presence of heparin or heparan sulfate, the steps required for productive infection are nevertheless heparin dependent. The C. trachomatis-cell interactions described by Carabeo and Hackstadt (3) showed paradoxically that their mutant cell line recovered the ability to bind serovar L2 EBs following paraformaldehyde fixation and that this binding was inhibited by heparin. The CHO-11 cell line phenotype of partial resistance to C. pneumoniae supports a model of codependence in which a mutation for the LGV-specific component also is partially shared by C. pneumoniae. Together, these data suggest that the productive attachment processes of both biovars to a host receptor are intimately linked, as they are both sensitive to heparin inhibition. It should be noted that the selection of cell mutants was based solely on resistance to LGV infection; hence, cellular phenotypes with regard to the attachment and infectivity of other chlamydial strains will be limited to components that are either shared or not with those required for LGV infection. An additional implication is that these data are inconsistent with the notion that chlamydiae have multiple or alternative modes of entry, at least for epithelial cells.
Biochemical and genetic analysis of mammalian cell mutants resistant to chlamydial infection provides a new opportunity to define and dissect the steps of the chlamydial infection processes of adherence, entry, and inhibition of lysosomal fusion, as well as other host-dependent functions required for chlamydial growth. The fact that we and Carabeo and Hackstadt (3) independently used the same method that productively resulted in the isolation of host cell mutants resistant to infection by chlamydiae supports the conclusion that this is a robust experimental design to begin understanding the molecular bases of chlamydial infection of mammalian cells.
Acknowledgments
We thank E. Banta for outstanding technical support of this project and C. Fenner for editing the manuscript.
This research was supported by National Institutes of Health grants AI 42156 and AI32943.
Editor: D. L. Burns
REFERENCES
- 1.Anonymous.1997. Chlamydia trachomatis genital infections—United States, 1995. Morb. Mortal. Wkly. Rep. 46:193-198. [PubMed] [Google Scholar]
- 2.Byrne, G. I., and J. W. Moulder. 1978. Parasite-specified phagocytosis of Chlamydia psittaci and Chlamydia trachomatis by L and HeLa cells. Infect. Immun. 19:598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carabeo, R. A., and T. Hackstadt. 2001. Isolation and characterization of a mutant Chinese hamster ovary cell line that is resistant to Chlamydia trachomatis infection at a novel step in the attachment process. Infect. Immun. 69:5899-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J. C., and R. S. Stephens. 1997. Chlamydia trachomatis glycosaminoglycan-dependent and independent attachment to eukaryotic cells. Microbial. Path. 22:23-30. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. C., and R. S. Stephens. 1994. Trachoma and LGV biovars of Chlamydia trachomatis share the same glycosaminoglycan-dependent mechanism for infection of eukaryotic cells. Mol. Microbiol. 11:501-507. [DOI] [PubMed] [Google Scholar]
- 6.Davis, C. H., J. E. Raulston, and P. B. Wyrick. 2002. Protein disulfide isomerase, a component of the estrogen receptor complex is associated with Chlamydia trachomatis serovar E attached to human endometrial epithelial cells. Infect. Immun. 70:3413-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman, M. M. 1987. Chinese hamster ovary cells. Methods Enzymol. 151:3-8. [DOI] [PubMed] [Google Scholar]
- 8.Hatch, T. P. 1999. Developmental biology, p. 29-67. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 9.Hodinka, R. L., C. H. Davis, J. Choong, and P. B. Wyrick. 1988. Ultrastructural study of endocytosis of Chlamydia trachomatis by McCoy cells. Infect. Immun. 56:1456-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodinka, R. L., and P. B. Wyrick. 1986. Ultrastructural study of mode of entry of Chlamydia psittaci into L-929 cells. Infect. Immun. 54:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 12.Koehler, J. E., R. R. Burgess, N. E. Thompson, and R. S. Stephens. 1990. Chlamydia trachomatis RNA polymerase major σ subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli σ70 and Bacillus subtilis σ43. J. Biol. Chem. 265:13206-13214. [PubMed] [Google Scholar]
- 13.Kuo, C., S. Wang, and J. T. Grayston. 1972. Differentiation of TRIC and LGV organisms based on enhancement of infectivity by DEAE-dextran in cell culture. J. Infect. Dis. 125:313-317. [DOI] [PubMed] [Google Scholar]
- 14.Kuo, C. C., and T. Grayston. 1976. Interaction of Chlamydia trachomatis organisms and HeLa 229 cells. Infect. Immun. 13:1103-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo, C. C., S. P. Wang, and J. T. Grayston. 1973. Effect of polycations, polyanions and neuraminidase on the infectivity of trachoma-inclusion conjunctivitis and lymphogranuloma venereum organisms in HeLa cells: sialic acid residues as possible receptors for trachoma-inclusion conjunctivitis. Infect. Immun. 8:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds, D. J., and J. H. Pearce. 1990. Characterization of the cytochalasin D-resistant (pinocytic) mechanisms of endocytosis utilized by chlamydiae. Infect. Immun. 58:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed]
- 18.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 19.Stephens, R. S., C. C. Kuo, and M. R. Tam. 1982. Sensitivity of immunofluorescence with monoclonal antibodies for detection of Chlamydia trachomatis inclusions in cell culture. J. Clin. Microbiol. 16:4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens, R. S., M. R. Tam, C. C. Kuo, and R. C. Nowinski. 1982. Monoclonal antibodies to Chlamydia trachomatis: antibody specificities and antigen characterization. J. Immunol. 128:1083-1089. [PubMed] [Google Scholar]
- 21.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W. H. O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 22.Vretou, E., P. C. Goswami, and S. K. Bose. 1989. Adherence of multiple serovars of Chlamydia trachomatis to a common receptor on HeLa and McCoy cells is mediated by thermolabile protein(s). J. Gen. Microbiol. 135:3229-3237. [DOI] [PubMed] [Google Scholar]
- 23.Ward, M. E., and A. Murray. 1984. Control mechanisms governing the infectivity of Chlamydia trachomatis for HeLa cells: mechanisms of endocytosis. J. Gen. Microbiol. 130:1765-1780. [DOI] [PubMed] [Google Scholar]
- 24.Wuppermann, F. N., J. H. Hegemann, and C. A. Jantos. 2001. Heparan sulfate-like glycosaminoglycan is a cellular receptor for Chlamydia pneumoniae. J. Infect. Dis. 184:181-187. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, J. P., and R. S. Stephens. 1992. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell 69:861-869. [DOI] [PubMed] [Google Scholar]