Abstract
We previously showed that Legionella pneumophila secretes, via its type II secretion system, phospholipase A activities that are distinguished by their specificity for certain phospholipids. In this study, we identified and characterized plaA, a gene encoding a phospholipase A that cleaves fatty acids from lysophospholipids. The plaA gene encoded a 309-amino-acid protein (PlaA) which had homology to a group of lipolytic enzymes containing the catalytic signature GDSL. In Escherichia coli, the cloned gene conferred trypsin-resistant hydrolysis of lysophosphatidylcholine and lysophosphatidylglycerol. An L. pneumophila plaA mutant was generated by allelic exchange. Although the mutant grew normally in standard buffered yeast extract broth, its culture supernatants lost greater than 80% of their ability to release fatty acids from lysophosphatidylcholine and lysophosphatidylglycerol, implying that PlaA is the major secreted lysophospholipase A of L. pneumophila. The mutant's reduced lipolytic activity was confirmed by growth on egg yolk agar and thin layer chromatography and was complemented by reintroduction of an intact copy of plaA. Overexpression of plaA completely protected L. pneumophila from the toxic effects of lysophosphatidylcholine, suggesting a role for PlaA in bacterial detoxification of lysophospholipids. The plaA mutant grew like the wild type in U937 cell macrophages and Hartmannella vermiformis amoebae, indicating that PlaA is not essential for intracellular infection of L. pneumophila. In the course of characterizing plaA, we discovered that wild-type legionellae secrete a phospholipid cholesterol acyltransferase activity, highlighting the spectrum of lipolytic enzymes produced by L. pneumophila.
Legionella pneumophila is a gram-negative organism which naturally multiplies within aquatic protozoa. Following inhalation by humans, the bacterium also infects lung macrophages and epithelial cells, causing Legionnaires' disease. L. pneumophila secretes a variety of potentially destructive enzymes, including a zinc metalloprotease, acid phosphatases, multiple lipases and phospholipases A (PLAs), a phospholipase C-like activity, and an RNase (2, 4, 13, 28, 34). These enzymatic activities, which are secreted via the bacterial type II secretion system, might hydrolyze major cell constituents and thus be important for the development of the disease (2, 28, 35, 45).
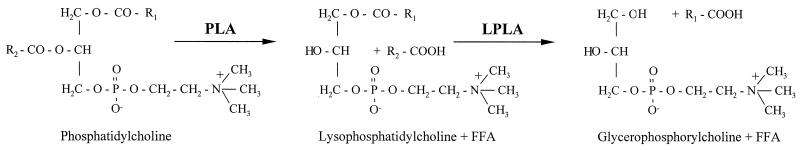
Recently, several secreted PLA activities have been found in the genus Legionella. Phospholipases are divided into several subgroups depending on their specificity for hydrolysis of ester bonds at different locations in the phospholipid molecule (49). PLAs cleave long-chain fatty acids from the glycerol backbone of phospholipid molecules, whereas phospholipases B release fatty acids both from the sn-1 and sn-2 positions of the glycerol backbone, and phospholipases C and D generate water-soluble compounds as well as 1,2-diacylglycerol or phosphatidic acid, respectively. PLAs can affect phospholipids and/or lysophospholipids having different polar head groups with varying degrees of specificity. Furthermore, they confer a positional specificity for the fatty acid in the sn-1 or sn-2 position and, moreover, for the length and saturation of the fatty acid bound in these positions. The first PLA activity of L. pneumophila hydrolyzes phospholipids containing both fatty acids producing lysophospholipids (25-27) (Fig. 1). The second PLA activity, a lysophospholipase A, preferentially liberates fatty acids from lysophospholipids having only one remaining fatty acid (25, 26, 28) (Fig. 1). Both PLA and lysophospholipase A activities are dependent upon the L. pneumophila type II secretion apparatus (2, 28, 46). Since Legionella type II protein secretion promotes intracellular survival and virulence and since PLAs are known to contribute to the pathogenesis of fungi and other bacteria (19, 20, 30, 35, 46, 50), we sought to characterize the genetic basis of L. pneumophila lysophospholipase A activity. Here, we identify the gene encoding the major lysophospholipase A of L. pneumophila and then describe its contribution to hydrolysis of lipids, detoxification of lysophosphatidylcholine, and intracellular infection of macrophages and amoebae.
FIG. 1.
Model of the two-step hydrolysis of phosphatidylcholine by secreted PLA and secreted lysophospholipase A (LPLA) activities of L. pneumophila.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Virulent L. pneumophila strain 130b (ATCC strain BAA-74, also known as Wadsworth or AA100) was used for mutagenesis of the Legionella plaA gene and later served as the wild-type control (22). Strains NU258 and NU259, direct derivatives of 130b, contain insertion mutations in the Legionella lspDE and lspG genes, respectively (46). The proA mutant AA200, another 130b derivative, is defective for expression of the L. pneumophila zinc metalloprotease (38). L. pneumophila was routinely grown on buffered charcoal-yeast extract (BCYE) agar for 2 days at 37°C (21). For detection of lipolytic activities, L. pneumophila was also grown on egg yolk agar, which contained the same constituents as BCYE agar except that 5% (vol/vol) egg yolk was added and starch was substituted for charcoal (6, 7, 14). For detection of proteolytic activity, bacterial strains were grown on casein agar, which contained the same ingredients as BCYE agar except that 1% (wt/vol) casein was added and charcoal was replaced by starch (54). In order to monitor extracellular growth in liquid media, L. pneumophila was cultured in buffered yeast extract (BYE) broth at 37°C with shaking at 250 rpm. Bacterial growth was monitored by determining the optical density of the culture at wavelength 660 nm (OD660; Beckman spectrophotometer DU500), following inoculation to an OD660 of 0.2 to 0.3. In order to assess L. pneumophila susceptibility to lysophosphatidylcholine, bacteria were grown in BYE broth until mid-log phase, and then 1-monopalmitoyllysophosphatidylcholine (MPLPC) was added to the cultures at final concentrations of 0.05, 0.1, and 0.2 mM. Growth of the bacteria was monitored by determination of the OD660, and after 4 or 16 h of incubation with the lipid, bacteria were plated onto BCYE for determination of CFU. Untreated controls were grown and plated in parallel to the treated cultures. Escherichia coli strains NovaBlue and DH5α, hosts for new recombinant plasmids, were grown in Luria-Bertani (LB) broth or agar (5). When appropriate, media were supplemented with antibiotics at final concentrations suitable for L. pneumophila (or E. coli) as follows: kanamycin, 25 μg/ml (50 μg/ml); chloramphenicol, 6 μg/ml (30 μg/ml); and ampicillin (only for E. coli; 100 μg/ml).
Preparation of culture supernatants and cell lysates.
Culture supernatants for assessment of hydrolytic activities were obtained at the end of exponential growth (i.e., OD660 of 2.2 to 2.3) by centrifugation for 5 min at 5,000 × g. Ten- or 20-fold-concentrated culture supernatants were prepared by isopropanol precipitation as described earlier (26). In short, 2 volumes of precooled isopropanol was added to 1 volume of culture supernatant, mixed, and incubated at −20°C for 10 min. The proteins were subsequently separated by centrifugation at 5,000 × g for 30 min at 4°C. The protein pellet was dissolved in the appropriate volume of 20 mM Tris-HCl (pH 7.5, 26°C). For the generation of cell lysates, bacteria from late exponential phase were pelleted by centrifugation as described above and then lysed by addition of a 1/20 volume of the original culture volume of 10 mg of lysozyme/ml and 1 μl of Triton X-100/ml at 37°C for 30 min. After repeated passage through a 26-gauge needle, the lysate was rediluted to its initial culture volume. Culture supernatants and cell lysates were tested immediately for enzymatic activities.
PCR and DNA sequence analysis.
Genomic DNA of L. pneumophila was isolated as previously described (43). Primers plaa-d1 (5′-GCATCATCCAGCTTCTTGTC-3′) and plaa-e1 (5′-CTGGCTTCACAGACGCAACC-3′), based on the sequence found in the incomplete L. pneumophila database (http: //genome3.cpmc.colombia.edu/∼legion/), were used to amplify the plaA gene from strain 130b DNA. The 1,151-bp PCR product begins 355 bp upstream of plaA and ends at bp 796 within the open reading frame (ORF). To isolate an intact copy of plaA, the amplified fragment was labeled with digoxigenin (Boehringer Mannheim, Indianapolis, Ind.) and used as a probe to screen a 130b library by colony blot hybridization. The 130b genomic library consisted of 3- to 6-kb Sau3AI-restricted DNA cloned into pBR322 (31). A double-stranded sequence of cloned L. pneumophila DNA was determined using the BigDye terminator cycle sequencing mix (PE Applied Biosystems, Foster City, Calif.). Automated sequence analysis on an ABI Prism 373 DNA sequencer (Applied Biosystems) was performed at the Biotech Facility at Northwestern University Medical School, Chicago, Ill. Primers were obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa). Sequence database searches as well as protein alignments were performed by using the BLAST algorithm (1). The nucleotide sequence was also analyzed for promoters (44), and the predicted protein was analyzed with the SignalP program for a signal sequence (41).
Gene cloning and Legionella mutant construction.
To assist with ascribing function to plaA, several recombinant plasmids were derived from pAF1, a plaA-containing plasmid derived from our screen of the genomic library. First, a 2.3-kb BamHI/StuI fragment of pAF1 was subcloned into the BamHI/EcoRV sites of pBluescript II KS(+) (Stratagene, La Jolla, Calif.), yielding pAF2. Next, pAF2 was restricted with Bst98I in order to delete nucleotides 1078 to 1393 of plaA, treated with Klenow fragment, and ligated with a kanamycin resistance gene (Kmr) cassette from pVK3 (60), resulting in pAF3. Furthermore, a PCR product that contained only plaA was amplified using primers plaa-d1 and plaa-m1 (5′-ATAAGGACCATTGCGCTG-3′) and cloned into the T-tailed EcoRV site of pGEM-Teasy (Promega, Madison, Wis.), resulting in pAF7. Subsequently, the SalI and SphI sites of the pGEM-Teasy backbone were used to subclone plaA into the corresponding sites of pMMB207 (40), yielding pAF8. Plasmids were isolated from E. coli by alkaline lysis using the Midiprep kit from Bio-Rad Laboratories (Hercules, Calif.).
To isolate an L. pneumophila plaA mutant, pAF3 and allelic exchange were used to introduce a Kmr insertion mutation into the chromosome of strain 130b (35). Plasmid pAF3 was introduced into L. pneumophila by natural transformation (53). Based upon the observations that L. pneumophila transformation is correlated with type IV pilus expression and that pili are more prominent at 30°C than at 37°C (35, 53), modifications to the original transformation method were made (K. Allard and N. P. Cianciotto, unpublished observations). In detail, 130b bacteria were inoculated (i.e., OD660 = 0.2) into 2 ml of BYE broth contained within a polypropylene plastic tube, and then 5 μg of plasmid DNA/ml was added. After growing the culture for approximately 18 h at 30°C with moderate shaking until mid-exponential phase, the bacteria were plated on BCYE agar supplemented with kanamycin. PCR and Southern blot analysis were used to examine Kmr legionellae for the presence of the plaA mutation (2, 48). For generation of strains for complementation experiments, pMMB207 and pAF8 were introduced into wild-type and mutant L. pneumophila by electroporation as described previously (45).
Enzymatic assay for lipolytic activities.
Enzymatic activities were detected as described previously, with minor changes (2, 27, 28). In detail, 25 μl of different phospholipids or lipids were incubated with 25 μl of culture supernatant, cell lysate, or concentrated culture supernatant in a mixture containing 6 mM lipid substrate (3.4 mg of MPLPC/ml, 3.4 mg of 1-monopalmitoyllysophosphatidylglycerol [MPLPG]/ml, 2.2 mg of 1-monopalmitoylglycerol [1-MPG]/ml, 5 mg of 1,2-dipalmitoylphosphatidylglycerol [DPPG]/ml, 5 mg of 1,2-dipalmitoylphosphatidylcholine [DPPC]/ml), 3 mM NaN3, 0.5% (vol/vol) Triton X-100, and 20 mM Tris-HCl (pH 7.2). When concentrated culture supernatants were assessed for glycerophospholipid-cholesterol acyltransferase (GCAT) activity, 6 mM (0.25 mg/ml) cholesterol was added. All lipids, including thin-layer chromatography (TLC) standards, were obtained from Sigma Chemical (St. Louis, Mo.) or Avanti Polar Lipids, Inc. (Alabaster, Ala.). Prior to incubation, the lipid substrates were vortexed for 15 min at 37°C and then exposed to ultrasonication (Vibracell; Sonics and Materials Inc., Danbury, Conn.) three times for 20 s at an intensity of 5. In order to test whether the lysophospholipase A activity was trypsin resistant, we added 10 μl from a 10-mg/ml stock of trypsin (Sigma Chemical) in 20 mM Tris-HCl (pH 7.2) to the reaction mixtures. The incubations with bacterial products were performed at 37°C with continuous agitation at 250 rpm for 2.5 h in the case of unconcentrated L. pneumophila supernatants and lysates, for 18 h in the case of concentrated L. pneumophila supernatants, and for 5 h in the case of E. coli supernatants. Free fatty acids (FFA) were determined by means of the NEFA-C kit (WAKO Chemicals, Neuss, Germany) according to the instructions of the manufacturer. The assay was modified for the use of microtiter plates, i.e., 2 to 10 μl of the reaction mixture was analyzed following the addition of 50 μl of reagent A and 100 μl of reagent B. Depending upon the nature of the experiment, BYE broth, concentrated BYE broth, or LB broth was incubated and treated like the cultures and subsequently used as a negative control.
Lipid extraction and TLC.
For the detection of distinct polar and apolar lipids, reaction mixtures of lipids with concentrated culture supernatants, corresponding negative controls, or pieces from egg yolk agar were subjected to a lipid extraction (9, 27). The lower chloroform phase was subsequently used for separation of lipids by TLC. For detection of polar lipids, silica gel plates (Merck, Darmstadt, Germany) were developed in tanks containing a solvent mixture of chloroform-methanol-water in a ratio of 65:25:4 (vol/vol/vol) (27, 55). A mixture of petroleum ether-diethylether-glacial acetic acid in a ratio of 90:10:1 (vol/vol/vol) was used for separation of apolar lipids, including cholesterol esters (37). For visualization, silica plates were then stained with naphthol blue black (Aldrich Chemical Company, Milwaukee, Wis.) (42).
Intracellular infection of U937 cells and Hartmannella vermiformis amoebae.
U937, a human cell line that differentiates into macrophage-like cells upon treatment with phorbol esters, and H. vermiformis amoebae were used as hosts for in vitro infection by L. pneumophila (15, 16). The cell line and amoebae were maintained and infected as previously described (15, 16, 35). To assess intracellular growth of L. pneumophila, wells containing U937 cells or amoebae at a concentration of 106/ml and 105/ml, respectively, were infected with wild-type bacteria or isogenic mutants at a multiplicity of infection of 0.1. At various time points, the number of intracellular plus extracellular bacteria per well was determined by plating serial dilutions on BCYE agar (35). To measure the cytopathic effect of L. pneumophila strains on U937 cells, the ability of the infected monolayer to reduce alamar blue (Biosource International, Vacaville, Calif.) was determined (3, 52). Briefly, at various time points, the infected cells were thoroughly washed to eliminate the extracellular bacteria and then incubated with a mixture of medium and alamar blue at 10:1 (vol/vol) at 37°C for 3 h. After this time, the fluorescence (excitation, 540 nm; emission, 584 nm) was read in a Spectra Max Gemini fluorescence reader (Molecular Devices, Sunnyvale, Calif.).
Nucleotide sequence accession number.
The L. pneumophila 130b plaA sequence has been deposited in the GenBank database at the National Center for Biotechnology Information under accession number AF510106.
RESULTS
Identification of an L. pneumophila lysophospholipase A gene.
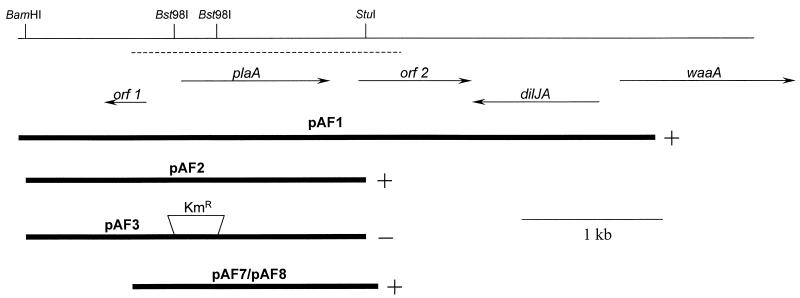
Recently, we determined the N-terminal sequence of a lysophospholipase A purified from culture supernatants of L. pneumophila strain 130b (28). The 17 N-terminal amino acids (i.e., TPLNNIVVFGDSLSDNG) were used in a BLAST search of the incomplete genome database of L. pneumophila Philadelphia strain 1 (http://genome3.cpmc.columbia.edu/∼legion/int_blast.html). One incomplete ORF encoded residues that matched exactly with the N-terminal sequence. Using primers based on the sequence of the putative lysophospholipase gene, we were able to PCR amplify a 1,151-bp fragment from 130b genomic DNA. Then, using labeled PCR product and colony blot hybridization screens, five positive clones were found in our L. pneumophila 130b genomic library. Sequencing of the 4.3-kb insertion in one of the recombinant plasmids, pAF1, confirmed the existence of an intact L. pneumophila lysophospholipase A gene, which we designated as plaA, for PLA gene A (Fig. 2).
FIG. 2.
The plaA locus in L. pneumophila and recombinant E. coli. The upper line represents a 5-kb region of the L. pneumophila chromosome that contains the lysophospholipase A gene (plaA), along with the location of relevant restriction enzyme sites. The dashed line represents the DNA region that we sequenced and have deposited in the GenBank database. The arrows below this line depict the relative location, size, and orientation of plaA and neighboring genes. The thick lines at the bottom of the figure represent the segments of Legionella DNA that were cloned into plasmid vectors. Plasmid pAF3 contained a Kmr gene cassette in place of the Legionella sequences that normally exist between the indicated Bst98I sites. The + and − symbols denote whether supernatants from the recombinant E. coli exhibited increased lysophospholipase A activity.
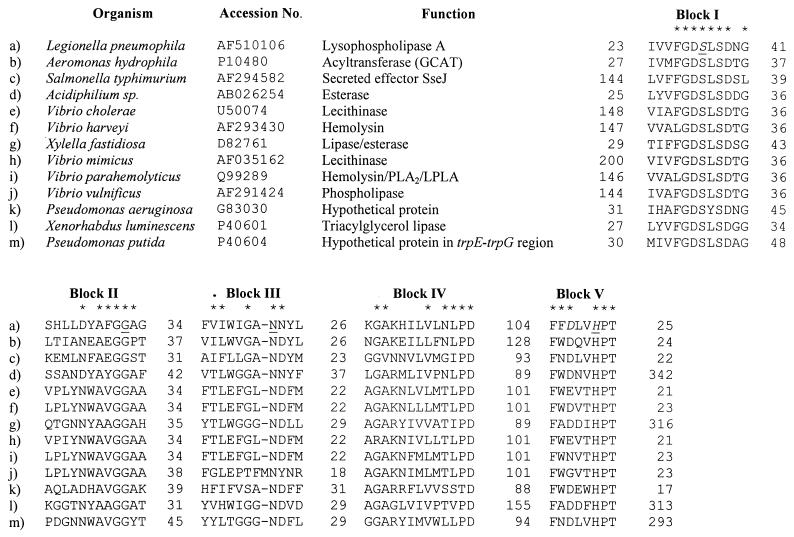
The 930-bp plaA sequence (GenBank accession no. AF510106) was predicted to encode a protein of 309 amino acids. The deduced PlaA sequence showed homology with numerous proteins, most of which are lipolytic enzymes. The closest overall homologies were with the GCAT of Aeromonas salmonicida (identity, 31%; similarity, 47%), phosphatidylcholine-sterol O-acyltransferase from Aeromonas hydrophila (identity, 31%; similarity, 47%), secreted effector J of Salmonella enterica serovar Typhimurium (identity, 29%; similarity, 47%), and the lecithinases of Vibrio cholerae and Vibrio mimicus (identity, 27%; similarity, 47 to 48%) (Fig. 3). These five enzymes and most of the PlaA homologs belong to a family of lipolytic proteins from prokaryotes as well as eukaryotes that contain a GDSL motif near their N termini (23, 57). Members of this group can be aligned along five blocks (I to V) of amino acid homology (57) (Fig. 3). We observed that this family of enzymes could be further classified into three subgroups, depending on the absence (group A) or presence of long N-terminal tails prior to block I (group B) or long C-terminal stretches after block V (group C). PlaA contains the GDSL motif close to its N terminus and possesses each of these blocks, suggesting that it is a member of this family (Fig. 3). L. pneumophila PlaA, like the GCAT protein of A. hydrophila, does not show any prolonged tails on either its N terminus or its C terminus and would therefore be a member of group A. After defining the three-dimensional structure of a GDSL-containing rhamnogalacturonan acetylesterase, Molgaard et al. identified a catalytic triad that is composed of the first serine in block I and the aspartic acid and histidine in block V (39). These authors further showed that only four amino acids (i.e., S, G, N, and H) are completely conserved among the GDSL enzymes, and they argued for this group of hydrolases to be designated as the SGNH hydrolase family (39). With its possession of SGNH (Fig. 3), L. pneumophila PlaA fulfills the proposed requirements of an SGNH hydrolase family member. PlaA was predicted to have an 18-amino-acid signal sequence, supporting our belief that it is a secreted enzyme (28, 46). The amino acids that occur after the predicted signal sequence are in exact agreement with the N-terminal amino acid sequence of the lysophospholipase A purified from L. pneumophila supernatants (28).
FIG. 3.
Sequence alignment of L. pneumophila PlaA with members of the GDSL family. Sequences of the 12 closest matches to PlaA are aligned along the five conserved blocks of the GDSL family. An asterisk designates those positions where an amino acid is conserved in at least six of the homologs. The amino acids comprising the putative catalytic triad of PlaA are shown in italics, and the residues conserved in the SGNH family are underlined. The digits before and after blocks I to V indicate the number of amino acid residues present before and after the conserved regions.
Two uncharacterized genes (i.e., orf 1 and orf 2), predicted to encode products with no significant homology to known proteins, flanked plaA (Fig. 2). Whereas the upstream orf 1 was oriented in the opposite direction from plaA, the downstream orf 2 was oriented in the same direction as plaA. We suspect that the plaA message is monocistronic, since plaA and orf 2 are separated by 192 bp and since −10 and −35 promoter sequences exist within the intergenic region (GenBank accession no. AF510106). The L. pneumophila genomic database (http://genome3.cpmc.columbia.edu/∼legion/int_blast.html) indicates that the dilJA gene, predicted to encode a chaperone, and the waaA gene, shown to encode a 3-deoxy-d-manno-oct-2-ulosonic acid transferase (10), lie downstream of orf 2 (Fig. 2).
Enzymatic activities of E. coli clones containing plaA.
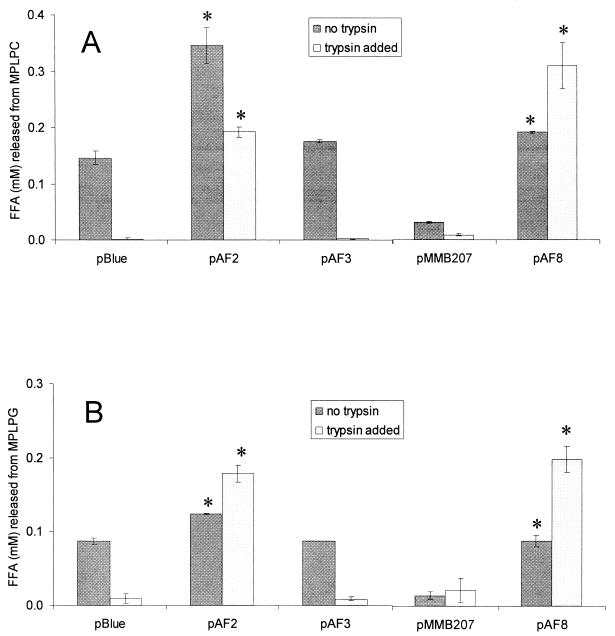
In order to confirm that plaA encodes a lysophospholipase A, we sought to examine secreted enzymatic activities associated with a set of recombinant E. coli clones. Toward that end, a 2.3-kb fragment containing plaA was subcloned from pAF1 into pBluescript KS(+), resulting in pAF2 (Fig. 2). Furthermore, the subcloned plaA was interrupted by deletion of the region between its two Bst98I sites, yielding pAF3 (Fig. 2). Finally, a 1.7-kb PCR fragment containing plaA, but no other complete ORF, was cloned into pGEM-Teasy and pMMB207, yielding pAF7 and pAF8, respectively (Fig. 2). Culture supernatants of E. coli clones containing either pAF1, pAF2, pAF7, or pAF8 released significantly more FFA from MPLPC, the known substrate of the L. pneumophila lysophospholipase A (28), than E. coli containing the corresponding vector (Fig. 4A and data not shown). Moreover, the clone harboring pAF3 and its inactivated plaA did not liberate increased amounts of FFA from MPLPC (Fig. 4A). Since L. pneumophila supernatants do not yield lysophosphatidylglycerol following exposure to lung surfactant (i.e., a source of phosphatidylglycerol) and since they release more FFA from DPPG than from DPPC (25, 26, 28), we hypothesized that lysophosphatidylglycerol might also be a substrate for PlaA. Therefore, we tested the supernatants from recombinant E. coli for the hydrolysis of MPLPG. As predicted, the E. coli clones containing pAF1, pAF2, pAF7, and pAF8, but not pAF3, were able to release more FFA from MPLPG (Fig. 4B and data not shown). Thus, PlaA confers activity towards both lysophosphatidylcholine and lysophosphatidylglycerol.
FIG. 4.
Lysophospholipase A activity of E. coli containing L. pneumophila plaA. Culture supernatants of E. coli NovaBlue containing pBluescript KS(+) (pBlue) or its derivatives pAF2 or pAF3 as well as E. coli DH5α harboring pMMB207 or its derivative pAF8 were mixed with MPLPC (A) or MPLPG (B) in the presence or absence of trypsin and, after a 5-h incubation at 37°C, the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant and the amount released by uninoculated LB broth. The results represent the means ± standard deviations of duplicate cultures and are representative of three independent experiments. Asterisks denote significant differences in lysophospholipase activity between E. coli containing plaA and the respective vector control (P < 0.05; Student's t test).
Whereas some lipolytic enzymes are susceptible to inactivation by common proteases (8), the activated form of the GCAT of A. hydrophila, a homolog of PlaA (see above), is stable towards further proteolytic cleavage (11, 32). Hence, we were interested to test the effect of the serine protease trypsin on the cloned PlaA activity as well as the background lysophospholipase A activity of E. coli. As shown in Fig. 4, lysophospholipase activity in supernatants from E. coli clones containing either empty vector or the insertionally inactivated plaA was eliminated by the addition of trypsin. In contrast, the PlaA activity associated with pAF2 and pAF8 persisted after protease treatment (Fig. 4). Thus, L. pneumophila PlaA is a trypsin-resistant lysophospholipase A.
None of the culture supernatants cleaved the phospholipids DPPG and DPPC, which contain both fatty acids and are substrates for the L. pneumophila PLA (28) (data not shown). Taken together, these data confirm that L. pneumophila plaA specifically encodes a lysophospholipase A.
Isolation of L. pneumophila plaA mutants.
In order to determine the degree to which PlaA is responsible for the lysophospholipase A activity in Legionella supernatants, we constructed a set of L. pneumophila plaA mutants. More specifically, pAF3 (Fig. 2) and allelic exchange were used to introduce a Kmr cassette into the plaA gene of strain 130b. Two plaA mutants (i.e., NU270 and NU271) were obtained following two separate DNA transformations and allelic exchange selections. PCR and Southern blot analysis confirmed the mutations in plaA (data not shown). All of the following experiments, with the exception of the MPLPC sensitivity assays (see below), were performed with both NU270 and NU271 with comparable results, although for clarity only data for NU270 are presented. Thus, the phenotypes observed resulted directly or proximately from the mutation in plaA and not from spontaneous, second-site mutations.
To assess the importance of plaA for L. pneumophila extracellular growth, we compared, on three separate occasions, strains 130b and NU270 for their growth in BYE broth, the standard medium for culturing legionellae. As measured by the OD660 of the cultures, NU270 grew comparably to wild type throughout the logarithmic and stationary growth phases when incubated at 37°C with shaking (data not shown). Furthermore, the mutant grew normally on BCYE agar, the standard solid medium for culturing legionellae. Since L. pneumophila lysophospholipase A activity is dependent upon the lsp type II protein secretion system, and since wild-type and isogenic lspDE and lspG Legionella mutants have different colony morphologies (28, 46), we examined the colonial growth of NU270. After 4 days of growth on BCYE agar at 37°C and 10 days at room temperature, no differences in colony morphology were observed between the mutant and wild type (data not shown), indicating that the altered colony morphology of the secretion mutants is not due to a loss of PlaA. Thus, these data indicate that plaA is not required for normal extracellular growth in liquid or on solid bacteriological media.
Lipolytic activities of an L. pneumophila plaA mutant.
Growth of bacteria, including L. pneumophila, on egg yolk agar is often used to estimate secretion of lipolytic factors (6, 7, 14). PLAs generate clearing, due to their ability to produce lysophospholipids. Lysophospholipases A alleviate that clearing. Therefore, an L. pneumophila mutant lacking a lysophospholipase A should generate more clearing than the wild type. Indeed, NU270 produced more clearing on egg yolk agar than did strain 130b (data not shown). When the agar medium surrounding the bacterial growth was examined by TLC, we observed an enrichment of lysophosphatidylcholine associated with the mutant (data not shown). The phenotype of NU270 observed on egg yolk agar was fully complemented by a plasmid copy of plaA, indicating that alterations in clearing are due to the loss of plaA (data not shown). Taken together, these observations suggest a decrease in secreted lysophospholipase A activity in the L. pneumophila plaA mutant.
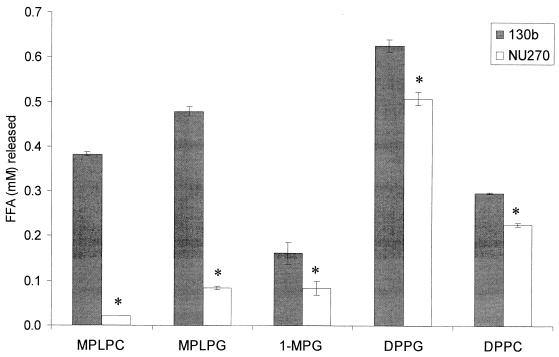
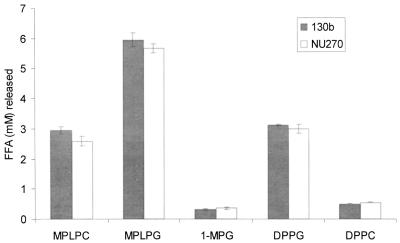
To more carefully assess the plaA mutant with respect to lipid hydrolysis, we tested culture supernatants for their ability to release FFA from MPLPC and MPLPG. Whereas hydrolysis of MPLPC by NU270 was reduced to less than 10% of wild-type activity, release of FFA from MPLPG was reduced to about 20% of normal levels (Fig. 5). These data indicate that PlaA is the major lysophospholipase A of L. pneumophila. The residual activities towards MPLPC and MPLPG suggest the presence of an additional, secreted lysophospholipase(s) A. Since partially purified PlaA had some activity against nonphospholipids (28), we next tested the relative ability of the mutant to hydrolyze 1-MPG. The hydrolysis of 1-MPG was reduced approximately 50% in NU270, confirming that PlaA is active against phospholipids and nonphospholipids (Fig. 5). The residual activity is likely due to other lipolytic enzymes, such as the recently described LipA lipase (4). Culture supernatants from the plaA mutant also had a modestly reduced release of FFA from DPPG and DPPC, two PLA substrates (Fig. 5). Since the plaA-containing E. coli clones did not liberate FFA from DPPG or DPPC (data not shown), this observation is likely due to an inability of mutant supernatants to further cleave those lysophospholipids produced by the action of the Legionella PLA. The ability of NU270 to fully release FFA from MPLPC, MPLPG, 1-MPG, DPPG, and DPPC was restored after trans complementation with plaA on pAF8 (Table 1). The activities of both the complemented wild type and NU270 against MPLPC and MPLPG were six- to ninefold higher than wild-type levels, a result that is likely due to multiple copies of plaA.
FIG. 5.
Lipolytic activities of culture supernatants of wild-type and plaA mutant L. pneumophila. Culture supernatants from late log phase of BYE cultures of strains 130b and NU270 were incubated with MPLPC, MPLPG, 1-MPG, DPPG, or DPPC for 2.5 h at 37°C, and then the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant and the amount released by uninoculated BYE broth. The results represent the means ± standard deviations of triplicate cultures and are representative of two independent experiments. Asterisks denote significant differences in lipolytic activities between wild-type L. pneumophila and NU270 (P < 0.01 for MPLPC, MPLPG, DPPG, and DPPC hydrolysis, and P < 0.05 for hydrolysis of 1-MPG; Student's t test).
TABLE 1.
Genetic complementation of a lipolytic defect in the L. pneumophila plaA mutant
| Supernatant sample | FFA (mM) released froma:
|
||||
|---|---|---|---|---|---|
| MPLPC | MPLPG | 1-MPG | DPPG | DPPC | |
| 130b(pMMB207) | 0.437 ± 0.005 | 0.516 ± 0.018 | 0.169 ± 0.028 | 0.482 ± 0.011 | 0.201 ± 0.004 |
| 130b(pAF8) | 3.055 ± 0.112†b | 3.004 ± 0.138† | 0.468 ± 0.027† | 0.755 ± 0.138† | 0.323 ± 0.013† |
| NU270(pMMB207) | 0.020 ± 0.001 | 0.100 ± 0.011 | 0.081 ± 0.021 | 0.402 ± 0.011 | 0.161 ± 0.003 |
| NU270(pAF8) | 2.493 ± 0.062† | 2.849 ± 0.216† | 0.435 ± 0.029† | 0.654 ± 0.009† | 0.294 ± 0.005† |
Supernatants from late-log phase BYE cultures of strains 130b and NU270 containing pMMB207 or pAF8 were incubated with the indicated substrates for 2.5 h at 37°C, and subsequently the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant and the amount released by uninoculated BYE broth. The results represent the means ± standard deviations of triplicate cultures and are representative of two independent experiments.
A † denotes significant differences in lipolytic activity between the wild type or plaA mutant harboring the vector pMMB207 and the respective strains with the plaA-containing vector pAF8 (P < 0.05; Student's t test).
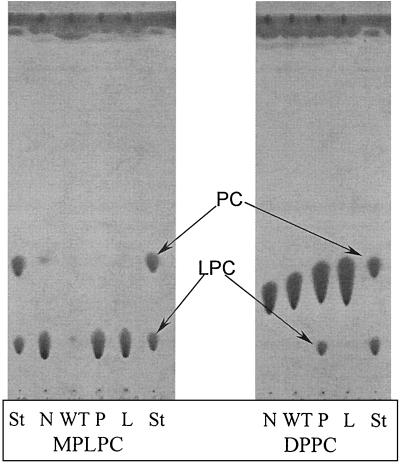
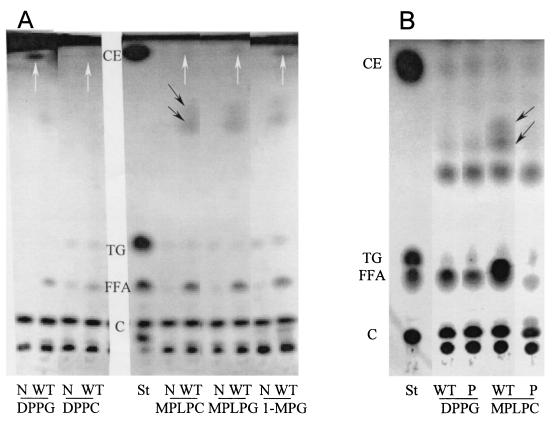
To confirm that the plaA mutant had altered lipolytic activities, we used TLC to examine the pattern of phospholipid cleavage caused by concentrated culture supernatants. In agreement with our FFA release data, strain 130b supernatants hydrolyzed more MPLPC and MPLPG than did those of NU270 (Fig. 6 and data not shown). Furthermore, the NU270 samples contained more lysophosphatidylcholine or lysophosphatidylglycerol following incubation with DPPC or DPPG, respectively (Fig. 6 and data not shown). These experiments also confirmed the reduced lysophospholipase A and PLA activities of L. pneumophila type II secretion mutants (Fig. 6 and data not shown). Taken together, the TLC experiments confirmed the significant loss of secreted lysophospholipase A activity that is associated with the plaA mutation in NU270.
FIG. 6.
TLC analysis of lipid hydrolysis by wild-type and mutant L. pneumophila. Tenfold-concentrated culture supernatants from late log phase of strains 130b (WT), NU270 (P), and an lspDE mutant (L) were incubated with MPLPC or DPPC for 18 h at 37°C. Subsequently, the lipids were extracted and separated by TLC. A mixture of concentrated BYE broth and the lipids was also incubated and served as a negative control (N). In the case of incubations with MPLPC (left panel), the samples were examined for the degradation of the lipid substrate and, in the cases of incubations with DPPC, for enrichment of lysophosphatidylcholine (LPC). For the qualitative identification of the lipid spots, lanes containing MPLPC and DPPC standards are marked St. The observations depicted here were made on at least two occasions.
With the isolation of a 130b mutant that does not express the major secreted lysophospholipase A, we were in a position to more clearly assess the prevalence of cell-associated lysophospholipase A activity. Therefore, we tested wild-type and mutant cell lysates for activities towards PlaA substrates MPLPC, MPLPG, and 1-MPG as well as PLA substrates DPPG and DPPC. High levels of activity against MPLPC, MPLPG, and 1-MPG were found in both strains (Fig. 7), implying the presence of a cell-associated lysophospholipase(s) A and perhaps lipase activity. Hydrolysis of DPPG and DPPC was also observed (Fig. 7), suggesting the presence of some unprocessed forms of the secreted PLA and/or an additional PLA(s) that is strictly linked to the bacterial cell.
FIG. 7.
Lipolytic activities of cell lysates of wild-type and plaA mutant L. pneumophila. Cell lysates from late log phase BYE cultures of strains 130b and NU270 were incubated with MPLPC, MPLPG, 1-MPG, DPPG, or DPPC for 2.5 h at 37°C. Subsequently, the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant and the amount released by uninoculated BYE broth that, like the cell samples, had been treated with lysozyme and Triton X-100. Results represent the means ± standard deviations of triplicate cultures and are representative of two independent experiments.
Susceptibility of L. pneumophila to cytotoxic lysophosphatidylcholine.
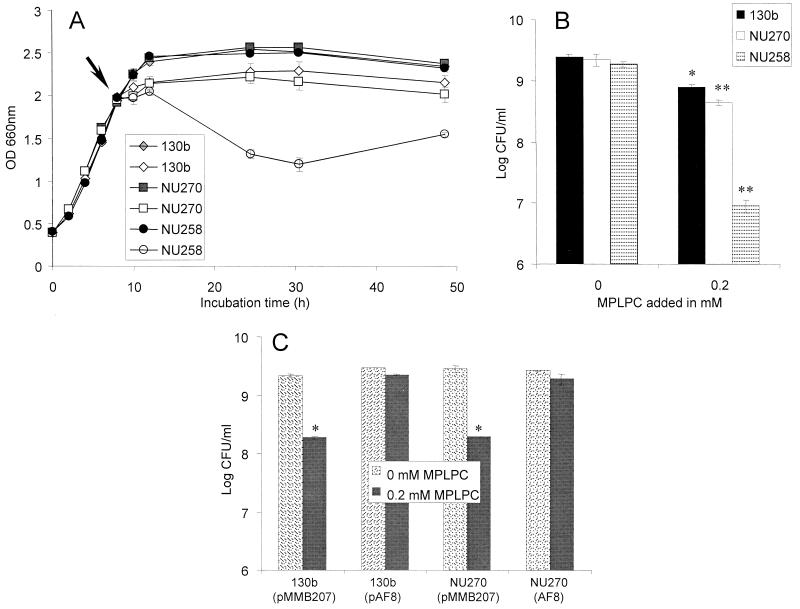
The PlaA substrate MPLPC is a potentially cytotoxic lipid (61) which can be generated by the action of host and pathogen PLAs on the membranes of eukaryotes and prokaryotes as well as lung surfactant (25, 33, 36, 56). Indeed, one of the suggested roles of lysophospholipases is the maintenance of lysophospholipids, such as MPLPC, at safe levels (29). For this reason, we examined whether wild-type and plaA mutant L. pneumophila differed in their susceptibility to lysophosphatidylcholine. More specifically, we added 0.05, 0.1, and 0.2 mM MPLPC to mid-log-phase cultures and then assessed changes in the viability of the bacteria. Previous experiments had shown that L. pneumophila lysophospholipase A secretion starts during mid-log phase and that wild-type cells grown in the presence of lung surfactant produce as much as 0.5 mM lysophosphatidylcholine (25, 26). Compared to an untreated control, wild-type bacteria showed reduced growth when exposed to 0.2 mM MPLPC (Fig. 8A). The viability difference between treated and untreated wild-type cultures was also confirmed when the cultures were plated for CFU determinations (Fig. 8B). These data show that lysophosphatidylcholine can be cytotoxic towards L. pneumophila. Mutant NU270 was only slightly more susceptible to lysophosphatidylcholine than the wild type (Fig. 8A and B), suggesting that PlaA is not critical for protection of L. pneumophila against MPLPC. Indeed, an L. pneumophila lspDE mutant was dramatically more sensitive to 0.2 mM MPLPC than was the wild type or the plaA mutant (Fig. 8A and B), indicating that other type II secreted factors can insure protection from toxic lysophosphatidylcholine. However, when plaA was introduced on a multicopy plasmid into the wild type or NU270, the bacteria were resistant to MPLPC (Fig. 8C), suggesting that PlaA may, under certain circumstances, play a role in detoxification of lysophosphatidylcholine.
FIG. 8.
Effect of MPLPC on the viability of wild-type and mutant L. pneumophila. Strains 130b, plaA mutant NU270, and lspDE mutant NU258 were inoculated into BYE broth at an OD660 of 0.2 to 0.3 and grown at 37°C with shaking. (A) When the cultures reached mid-log phase (arrow), 0.2 mM MPLPC (⋄, □, ○) or medium control (♦, ▪, •) was added. Bacterial growth was monitored by recording the cultures' OD660. (B) After 16 h of incubation with MPLPC, cultures were serially diluted and plated on BCYE agar for determination of CFU. Wild-type and plaA mutant L. pneumophila containing pMMB207 or pAF8 were treated as described above. (C) Bacteria were plated for determination of CFU after 4 h of incubation with 0.2 mM MPLPC. Data represent the means ± standard deviations of triplicate cultures and are representative of two independent experiments. One asterisk designates significant differences between L. pneumophila cultures untreated versus treated with MPLPC (P < 0.05; Student's t test). An additional asterisk denotes significant differences between the wild type and the plaA mutant or between the wild type and the lspDE mutant (P < 0.05; Student's t test).
Influence of the L. pneumophila zinc metalloprotease on secreted lipolytic activities.
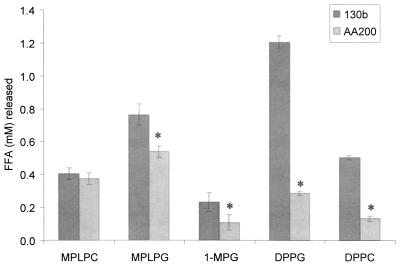
The PlaA-like GCAT of A. hydrophila is activated by a serine protease, which removes a small peptide between cysteine residues C225 and C281 (11, 32). This cleavage event occurs after the removal of the protein's 18-amino-acid signal peptide, and the resulting disulfide bridge formed between the cysteines renders the GCAT relatively resistant toward trypsin treatment (11, 17, 59). Since PlaA has two cysteines (i.e., C225 and C254) in locations similar to those of the Aeromonas cysteines and is trypsin resistant, we were interested in whether the type II-secreted zinc metalloprotease of L. pneumophila is involved in PlaA activation. Therefore, we compared culture supernatants from strain 130b and its isogenic proA mutant AA200 for their ability to release FFA from different lipid substrates. Although the strains showed no difference in FFA liberation from MPLPC, the major substrate of PlaA, the protease mutant released less FFA from MPLPG, 1-MPG, DPPG, and DPPC (Fig. 9). These data indicate that the L. pneumophila metalloprotease has no role in PlaA processing but may promote activation of the Legionella PLA.
FIG. 9.
Lipolytic activities of culture supernatants of wild-type and proA mutant L. pneumophila. Culture supernatants from log-phase cultures of strains 130b and proA mutant AA200 were incubated with MPLPC, MPLPG, 1-MPG, DPPG, or DPPC for 2.5 h at 37°C. Subsequently, the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant versus the amount released by uninoculated BYE broth. Results represent the means ± standard deviations of four cultures and are representative of two independent experiments. Asterisks denote significant differences in lipolytic activities between wild-type L. pneumophila and the proA mutant (P < 0.05; Student's t test).
Identification of an L. pneumophila GCAT activity.
Since PlaA shared homology and trypsin resistance with the GCAT from Aeromonas, we sought to determine if PlaA can transfer fatty acids from phospholipids to cholesterol. Since it was not known whether L. pneumophila possesses this activity, we first examined wild-type legionellae for GCAT activity. Toward that end, we incubated concentrated culture supernatants of strain 130b with cholesterol-lipid mixtures containing DPPG, DPPC, MPLPC, MPLPG, or 1-MPG and subsequently analyzed the lipid extracts for cholesterol esters by TLC. Most obvious, the wild type transferred fatty acids from DPPG to cholesterol, showing that L. pneumophila indeed confers GCAT activity (Fig. 10A). Transfer of fatty acids from DPPC, MPLPC, and MPLPG to cholesterol was less prominent (Fig. 10A), suggesting that phosphatidylglycerol is the preferred fatty acyl donor for L. pneumophila GCAT. Interestingly, cholesterol esters were also generated from mixtures of wild-type supernatants, cholesterol, and the nonphospholipid 1-MPG (Fig. 10A), indicating that L. pneumophila acyltransferase activity also accepts fatty acyl donors without an intramolecular phosphate. Next, to find out whether PlaA acts as a GCAT, we examined concentrated culture supernatants of NU270 for a loss of acyltransferase activity. However, cholesterol esters were observed in comparable quantities for the wild type and the plaA mutant (Fig. 10B and data not shown), suggesting that factors other than PlaA confer L. pneumophila GCAT activity. When concentrated culture supernatants of wild-type L. pneumophila were incubated with lipid-cholesterol mixtures, unknown apolar compounds were detected (Fig. 10). Interestingly, these compounds were absent in MPLPC-cholesterol incubations with the plaA mutant (Fig. 10B), suggesting that PlaA is involved in their generation. These compounds were not derivatives of cholesterol, since they were found when cholesterol was excluded from the reactions (data not shown).
FIG. 10.
GCAT activity of culture supernatants from wild-type and plaA mutant L. pneumophila. (A) Twenty-fold-concentrated culture supernatants of strains 130b (WT) incubated with mixtures of DPPG, DPPC, MPLPC, MPLPG, or 1-MPG with cholesterol. (B) Twenty-fold-concentrated culture supernatants of strains 130b (WT) and NU270 (P) incubated with mixtures of DPPG or MPLPC with cholesterol for 18 h at 37°C. Subsequently, the lipids were extracted and separated by TLC. A mixture of concentrated BYE broth and the lipids served as a negative control (N). The samples were examined for generation of cholesterol esters as a measure of GCAT activity. Lanes containing cholesterol palmitate (CE), tripalmitoylglycerol (TG), palmitic acid (FFA), and cholesterol (C) standards are marked St. The white arrows in panel A indicate cholesterol esters formed by L. pneumophila GCAT activity. The black arrows indicate the position of unknown polar compounds produced by L. pneumophila concentrated supernatants. Data are representative of two independent experiments.
Intracellular infection by an L. pneumophila plaA mutant.
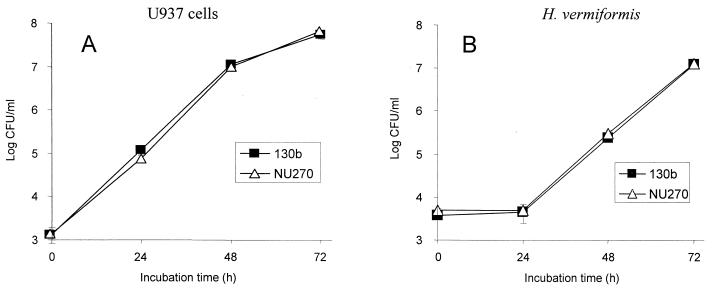
Since the type II protein secretion system of L. pneumophila promotes infection of macrophages and protozoa and since lysophospholipase A activity is dependent on type II secretion (46), we were interested in determining the role of plaA in intracellular infection. Toward that end, we first compared the ability of strains 130b and NU270 to infect monolayers of U937 cells, a standard model for L. pneumophila macrophage infection. Strains 130b and NU270 behaved similarly, showing a typical pattern of intracellular growth in which bacterial numbers increased from 100,000- to 1,000,000-fold by 72 h postinoculation (Fig. 11A) (2, 35). Furthermore, NU270 did not seem to have a defect in entry or attachment to the macrophages, since the numbers of bacteria recovered after the uptake period (0 h) were comparable for the wild type and the mutant (Fig. 11A). It is well known that L. pneumophila replication leads to death of the macrophage (45, 46). However, strains 130b and NU270 produced comparable effects on host cell viability (data not shown), indicating no major role for plaA in this in vitro model of cytopathogenicity. To determine the role of PlaA in amoeba infection, H. vermiformis cultures were inoculated with strains 130b and NU270 and the number of bacteria was recorded at different times. As in the macrophage infection system, comparable numbers of the wild type and mutant were recovered from the amoeba cocultures (Fig. 11B). Taken together, these data indicate that plaA is not required for intracellular infection by L. pneumophila.
FIG. 11.
Intracellular infection by wild-type and plaA mutant L. pneumophila. Strains 130b and NU270 were used to infect monolayers of U937 macrophages (A) or cultures of H. vermiformis amoebae (B) at a multiplicity of infection of 0.1. At 0, 24, 48, and 72 h postinoculation, the numbers of bacteria were quantitated by plating aliquots on BCYE agar. Results represent the means ± standard deviations of triplicate samples and are representative of two independent experiments.
DISCUSSION
In this study, we identified the gene for the major L. pneumophila lysophospholipase A, an enzyme that cleaves fatty acids from lysophospholipids but not phospholipids containing both fatty acids. The substrates for PlaA are produced, at least in part, through the action of a secreted PLA (Fig. 1) recently identified (27). Since the plaA mutant showed residual (i.e., ca. 10%) lysophospholipase A activity in its culture supernatants, we conclude that L. pneumophila secretes lysophospholipases A in addition to PlaA. Furthermore, we found that L. pneumophila possesses cell-associated lysophospholipase A, as well as PLA, activities that are distinct from PlaA. In support of these data, we found two predicted homologs of PlaA in the unfinished L. pneumophila genome database (unpublished observations). Because the plaA mutant also had a reduced ability to release FFA from the nonphospholipid 1-MPG, we believe that PlaA also possesses lipase activity. This observation is in good agreement with the fact that PlaA-containing fractions obtained from L. pneumophila supernatants release FFA from both MPLPC and 1-MPG (28). The cleavage of both phospholipids and non-phosphate-containing lipids has also been described for staphylococcal (phospho)lipases (51).
Based upon its predicted amino acid sequence, L. pneumophila PlaA is a new member of the GDSL and SGNH hydrolase families (39, 57). The best-characterized lipolytic protein in the GDSL group is the Aeromonas GCAT, an enzyme that will also behave as a lysophospholipase or phospholipase, in the absence of an acceptor molecule (12). We observed that L. pneumophila supernatants do contain a GCAT activity. However, the plaA mutant was not lacking this secreted activity, suggesting that L. pneumophila PlaA, unlike its Aeromonas homolog, is not acting as a cholesterol acyltransferase. Since the Legionella GCAT activity, like that of A. salmonicida (13), transferred fatty acids from DPPG, it may be identical with the secreted, DPPG- and DPPC-cleaving PLA of L. pneumophila (26, 27, 28). Further investigations are necessary to elucidate the structural determinants for the L. pneumophila acyltransferase activity and to understand why only some GDSL proteins act as acyltransferases.
Since the predicted size of the translated plaA sequence is 34.5 kDa but purified PlaA migrates as a 28- or 29-kDa protein upon reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (28), we suspect that PlaA is proteolytically modified. The first cleavage of PlaA is undoubtedly the removal of its 18-amino-acid signal sequence by leader peptidase, an event that is a prerequisite for type II secretion. Following upon the Aeromonas GCAT example (11), the second modification may represent a restriction of the protein between cysteines at positions 225 and 281, a process that would yield a mature protein of about 29 kDa. Because the Legionella metalloprotease mutant was normal for lysophospholipase A activity, the final stage of PlaA maturation and the relevant protease remain to be determined. Interestingly, though, the culture supernatants of the proA mutant showed reductions in PLA activity and, to a lesser extent, MPLPG-specific lysophospholipase A and lipase activities. These data suggest that some of the virulence attenuation observed with the protease mutant (38) might be due to a loss of lipolytic activities.
One of the suggested functions of lysophospholipases is protection from high levels of cytotoxic lysophosphatidylcholine (29, 61). Indeed, increased expression of plaA afforded L. pneumophila greater survival in the presence of MPLPC. However, since an lsp mutant, unlike the plaA mutant, was hypersensitive to MPLPC, we believe that other type II secreted factors are most critical for L. pneumophila resistance to lysophosphatidylcholine. The other secreted lysophospholipase A activity and the newly discovered cell-associated activity are candidate effectors of lysophospholipid detoxification. Interestingly, it has been shown that gram-negative bacteria are more resistant to lysophosphatidylcholine than gram-positive bacteria (18, 58). In support of these data, when we searched the finished and unfinished bacterial genome databases we only found evidence for GDSL protein homologs in gram-negative organisms (unpublished observations).
Recently, lysophospholipases A and members of the GDSL family have been associated with virulence and/or potentially pathogenic activities. For example, a lysophospholipase A activity of the fungus Cryptococcus neoformans was implicated in intracellular replication and in virulence in both mouse inhalational and rabbit meningitis models (19). Furthermore, a pldA mutant of Campylobacter coli that is reduced in lysophospholipase A and PLA activity is defective for lysis of erythrocytes (30). Finally, an sseJ mutant of S. enterica serovar Typhimurium is mildly attenuated for systemic virulence in mice, and the SseJ protein, a member of the GDSL family (see Fig. 3), appears to be involved in the formation of tubular extensions of the Salmonella-containing vacuole (47). On the other hand, GCAT loss in A. salmonicida produces no decrease in virulence in Atlantic salmon (59), and the absence of Lec phospholipase, another GDSL protein, does not diminish fluid accumulation by V. cholerae in rabbit ileal loops (24).
The L. pneumophila plaA mutant revealed no defect in intracellular multiplication within U937 cell macrophages and H. vermiformis amoebae, suggesting that PlaA is not critical for intracellular infection. However, it is possible that the other Legionella lysophospholipases A compensate for the lack of PlaA, necessitating a need to generate and examine mutants defective for multiple GDSL proteins. Furthermore, it is conceivable that PlaA plays a role in extracellular events and, thus, examination of plaA mutants in an animal model is of interest. Additional targets for future investigation involve identifying the type II exoenzymes that promote L. pneumophila intracellular infection and resistance to cytotoxic lysophosphatidylcholine, the enzyme(s) that is responsible for Legionella GCAT activity, and the function of the other lysophospholipase A activities and GDSL proteins that are encoded by L. pneumophila.
Acknowledgments
We thank the past and present members of the Cianciotto laboratory, Markus Koerber Abdelhak Belmadani, Frauke Fehrmann, and Anatoly Mayburd for helpful discussions and comments. We are grateful to Lucy Tompkins and Paul Edelstein for providing the isogenic proA mutant. We acknowledge Joseph Vogel and colleagues for their initial description of low-temperature transformation as presented at scientific meetings.
A. Flieger was supported by grant Fl359 1-1 from “Deutsche Forschungsgemeinschaft.” This work was further funded by NIH grant AI 43987 awarded to N.P.C.
Editor: D. L. Burns
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon, V., S. Kurtz, and N. P. Cianciotto. 2001. The Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragon, V., S. Kurtz, O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 6.Baine, W. B. 1985. Cytolytic and phospholipase C activity in Legionella species. J. Gen. Microbiol. 131:1383-1391. [DOI] [PubMed] [Google Scholar]
- 7.Baine, W. B. 1988. A phospholipase C from the Dallas 1E strain of Legionella pneumophila serogroup 5: purification and characterization of conditions of optimal activity with an artificial substrate. J. Gen. Microbiol. 134:489-498. [DOI] [PubMed] [Google Scholar]
- 8.Bernback, S., O. Hernell, and L. Blackberg. 1987. Bovine pregastric lipase: a model for the human enzyme with respect to properties relevant to its site of action. Biochim. Biophys. Acta 922:206-213. [DOI] [PubMed] [Google Scholar]
- 9.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 10.Brabetz, W., C. E. Schirmer, and H. Brade. 2000. 3-Deoxy-d-manno-oct-2-ulosonic acid (Kdo) transferase of Legionella pneumophila transfers two Kdo residues to a structurally different lipid A precursor of Escherichia coli. J. Bacteriol. 182:4654-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumlik, M. J., F. G. van der Goot, K. R. Wong, and J. T. Buckley. 1997. The disulfide bond in the Aeromonas hydrophila lipase/acyltransferase stabilizes the structure but is not required for secretion or activity. J. Bacteriol. 179:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckley, J. T., L. N. Halasa, and S. MacIntyre. 1982. Purification and partial characterization of a bacterial phospholipid:cholesterol acyltransferase. J. Biol. Chem. 257:3320-3325. [PubMed] [Google Scholar]
- 13.Buckley, J. T. 1982. Substrate specificity of bacterial glycerophospholipid:cholesterol acyltransferase. Biochemistry 21:6699-6703. [DOI] [PubMed] [Google Scholar]
- 14.Chrisope, G. L., C. W. Fox, and R. T. Marshall. 1976. Lecithin agar for detection of microbial phospholipases. Appl. Environ. Microbiol. 31:784-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, G. B. Toews, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman, G., and P. W. Whitby. 1993. A comparison of the amino acid sequence of the serine protease of the fish pathogen Aeromonas salmonicida subsp. salmonicida with those of other subtilisin-type enzymes relative to their substrate-binding sites. J. Gen. Microbiol. 139:245-249. [DOI] [PubMed] [Google Scholar]
- 18.Coonrod, J. D., and K. Yoneda. 1983. Detection and partial characterization of antibacterial factor(s) in alveolar lining material of rats. J. Clin. Investig. 71:129-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166-175. [DOI] [PubMed] [Google Scholar]
- 20.Dorrell, N., M. C. Martino, R. A. Stabler, S. J. Ward, Z. W. Zhang, A. A. McColm, M. J. Farthing, and B. W. Wren. 1999. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology 117:1098-1104. [DOI] [PubMed] [Google Scholar]
- 21.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farn, J. L., R. A. Strugnell, P. A. Hoyne, W. P. Michalski, and J. M. Tennent. 2001. Molecular characterization of a secreted enzyme with phospholipase B activity from Moraxella bovis. J. Bacteriol. 183:6717-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiore, A. E., J. M. Michalski, R. G. Russell, C. L. Sears, and J. B. Kaper. 1997. Cloning, characterization, and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect. Immun. 65:3112-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flieger, A., S. Gong, M. Faigle, H. A. Mayer, U. Kehrer, J. Muβotter, P. Bartmann, and B. Neumeister. 2000. Phospholipase A secreted by Legionella pneumophila destroys alveolar surfactant phospholipids. FEMS Microbiol. Lett. 188:129-133. [DOI] [PubMed] [Google Scholar]
- 26.Flieger, A., S. Gong, M. Faigle, H. Northoff, and B. Neumeister. 2001. In vitro secretion kinetics of proteins from Legionella pneumophila in comparison to proteins from non-pneumophila species. Microbiology 147:3127-3134. [DOI] [PubMed] [Google Scholar]
- 27.Flieger, A., S. Gong, M. Faigle, M. Deeg, P. Bartmann, and B. Neumeister. 2000. Novel phospholipase A activity secreted by Legionella species. J. Bacteriol. 182:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flieger, A., S. Gong, M. Faigle, S. Stevanovic, N. P. Cianciotto, and B. Neumeister. 2001. Novel lysophospholipase A secreted by Legionella pneumophila. J. Bacteriol. 183:2121-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garsetti, D., F. Holtsberg, M. R. Steiner, R. W. Egan, and M. A. Clark. 1992. Butyric acid-induced differentiation of HL-60 cells increases the expression of a single lysophospholipase. Biochem. J. 288:831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant, K. A., I. U. Belandia, N. Dekker, P. T. Richardson, and S. F. Park. 1997. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect. Immun. 65:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickey, E. K., and N. P. Cianciotto. 1994. Cloning and sequencing of the Legionella pneumophila fur gene. Gene 143:117-121. [DOI] [PubMed] [Google Scholar]
- 32.Hilton, S., W. D. McCubbin, C. M. Kay, and J. T. Buckley. 1990. Purification and spectral study of a microbial fatty acyltransferase: activation by limited proteolysis. Biochemistry 29:9072-9078. [DOI] [PubMed] [Google Scholar]
- 33.Holm, B. A., L. Keicher, M. Y. Liu, J. Sokolowski, and G. Enhorning. 1991. Inhibition of pulmonary surfactant function by phospholipases. J. Appl. Physiol. 71:317-321. [DOI] [PubMed] [Google Scholar]
- 34.Keen, M. G., and P. S. Hoffman. 1989. Characterization of a Legionella pneumophila extracellular protease exhibiting hemolytic and cytotoxic activities. Infect. Immun. 57:732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Lara, I. M., and O. Geiger. 2001. Novel pathway for phosphatidylcholine biosynthesis in bacteria associated with eukaryotes. J. Biotechnol. 91:211-221. [DOI] [PubMed] [Google Scholar]
- 37.MacIntyre, S., and J. T. Buckley. 1978. Presence of glycerophospholipid:cholesterol acyltransferase and phospholipase in culture supernatant of Aeromonas hydrophila. J. Bacteriol. 135:402-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 39.Molgaard, A., S. Kauppinen, and S. Larsen. 2000. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure Fold. Des. 8:373-383. [DOI] [PubMed] [Google Scholar]
- 40.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 42.Plekhanov, A. Y. 1999. Rapid staining of lipids on thin-layer chromatograms with amido black 10B and other water-soluble stains. Anal. Biochem. 271:186-187. [DOI] [PubMed] [Google Scholar]
- 43.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition, p. 737-738. In L. Hunter and T. E. Klein (ed.), Biocomputing. Proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co., Singapore.
- 45.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schmiel, D. H., and V. L. Miller. 1999. Bacterial phospholipases and pathogenesis. Microbes Infect. 1:1103-1112. [DOI] [PubMed] [Google Scholar]
- 50.Schmiel, D. H., E. Wagar, L. Karamanou, D. Weeks, and V. L. Miller. 1998. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect. Immun. 66:3941-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simons, J. W., F. Gotz, M. R. Egmond, and H. M. Verheij. 1998. Biochemical properties of staphylococcal (phospho)lipases. Chem. Phys. Lipids 93:27-37. [DOI] [PubMed] [Google Scholar]
- 52.Stone, B. J., and Y. A. Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szeto, L., and H. A. Shuman. 1990. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect. Immun. 58:2585-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Touchstone, J. C., S. S. Levin, M. F. Dobbins, L. Matthews, P. C. Beers, and S. G. Gabbe. 1983. (3-sn-Phosphatidyl)cholines (lecithins) in amniotic fluid. Clin. Chem. 29:1951-1954. [PubMed] [Google Scholar]
- 56.Touqui, L., and L. Arbibe. 1999. A role for phospholipase A2 in ARDS pathogenesis. Mol. Med. Today 5:244-249. [DOI] [PubMed] [Google Scholar]
- 57.Upton, C., and J. T. Buckley. 1995. A new family of lipolytic enzymes? Trends Biochem. Sci. 20:178-179. [DOI] [PubMed] [Google Scholar]
- 58.Van Rensburg, C. E., G. K. Joone, J. F. O'Sullivan, and R. Anderson. 1992. Antimicrobial activities of clofazimine and B669 are mediated by lysophospholipids. Antimicrob. Agents Chemother. 36:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vipond, R., I. R. Bricknell, E. Durant, T. J. Bowden, A. E. Ellis, M. Smith, and S. MacIntyre. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 66:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weltzien, H. U. 1979. Cytolytic and membrane-perturbing properties of lysophosphatidylcholine. Biochim. Biophys. Acta 559:259-287. [DOI] [PubMed] [Google Scholar]