Abstract
To link conformational transitions noted for DNA polymerases with kinetic results describing catalytic efficiency and fidelity, we investigate the role of key DNA polymerase β residues on subdomain motion through simulations of five single-residue mutants: Arg-283-Ala, Tyr-271-Ala, Asp-276-Val, Arg-258-Lys, and Arg-258-Ala. Since a movement toward a closed state was only observed for R258A, we suggest that Arg258 is crucial in modulating motion preceding chemistry. Analyses of protein/DNA interactions in the mutant active site indicate distinctive hydrogen bonding and van der Waals patterns arising from compensatory structural adjustments. By comparing closed mutant complexes with the wild-type enzyme, we interpret experimentally derived nucleotide binding affinities in molecular terms: R283A (decreased), Y271A (increased), D276V (increased), and R258A (decreased). Thus, compensatory interactions (e.g., in Y271A with adjacent residues Phe272, Asn279, and Arg283) increase the overall binding affinity for the incoming nucleotide although direct interactions may decrease. Together with energetic analyses, we predict that R258G might increase the rate of nucleotide insertion and maintain enzyme fidelity as R258A; D276L might increase the nucleotide binding affinity more than D276V; and R283A/K280A might decrease the nucleotide binding affinity and increase misinsertion more than R283A. The combined observations regarding key roles of specific residues (e.g., Arg258) and compensatory interactions echo the dual nature of polymerase active site, namely versatility (to accommodate various basepairs) and specificity (for preserving fidelity) and underscore an organized but pliant active site essential to enzyme function.
INTRODUCTION
DNA repair and replication processes are fundamental in maintaining genome integrity. The fidelity mechanisms by which DNA polymerases select the correct, but not incorrect, nucleotide require further elucidation at the level of atomic resolution. On the basis of extensive kinetic measurements and a large body of crystallographic structures for several DNA polymerases (e.g., Escherichia coli DNA polymerase I Klenow fragment (Kuchta et al., 1988; Dahlberg and Benkovic, 1991; Ollis et al., 1985; Beese et al., 1993), phage T7 DNA polymerase (Patel et al., 1991; Doublié et al., 1998, 1999), HIV-1 reverse transcriptase (Kati et al., 1992; Huang et al., 1998; Ding et al., 1998), phage T4 DNA polymerase (Frey et al., 1995), and DNA polymerase β (Beard et al., 2002a; Beard and Wilson, 2003; Vande Berg et al., 2001; Zhong et al., 1997, 1998; Kraynov et al., 1997; Ahn et al., 1997, 1998; Werneburg et al., 1996; Sawaya et al., 1997, 1994; Pelletier et al., 1994)), the overall pathway of incorporation of a correct 2′-deoxyribonucleoside 5′-triphosphate (dNTP) by a DNA polymerase has been proposed, as sketched in Fig. 1. The conformational closing (before chemistry, step 2 in Fig. 1) and opening (after chemistry, step 4 in Fig. 1) during each catalytic cycle (of increasing the primer strand by one nucleotide) have been suggested to be key players in the faithful incorporation of nucleotides via an “induced-fit” mechanism (Doublié et al., 1999; Sawaya et al., 1997; Li et al., 1998; Beard and Wilson, 1998). Through encasing the incoming dNTP and the complementary template nucleotide, polymerases generate a geometrically selective binding pocket for the nascent basepair (Echols and Goodman, 1991). The surface of the binding pocket for a correct incoming dNTP is comprised of polymerase residues making interactions with the DNA minor groove edge, van der Waals interactions with the sugar and base, and electrostatic interactions with the triphosphate moiety of the incoming nucleotide (Echols and Goodman, 1991; Kool et al., 2000; Goodman, 1997). In addition, the Watson-Crick edge of the templating base and the 3′-terminal nucleotide of the primer strand contribute interactions with the incoming nucleotide. Since this binding pocket is optimally formed for Watson-Crick basepairs, but not mispairs, the characterization of the dynamics of the interacting surfaces in the binding pocket is essential for detailing enzyme specificity.
FIGURE 1.
General pathway for nucleotide incorporation by DNA polymerases and corresponding crystallographic closed (A) and open (B) conformations of pol β/DNA complex as well as key protein residues in the active site in both the closed (C) and open structures (D). In the general pathway, the DNA polymerase binds primer/template DNA to produce a binary DNA polymerase complex, which subsequently binds a complementary dNTP to form a ternary substrate complex (step 1); the enzyme undergoes a subdomain motion forming a closed, catalytically competent state (step 2); after the chemical reaction of dNTP incorporation onto the DNA primer strand (step 3), the polymerase/product complex undergoes a second subdomain motion to result in an open state (step 4). The release of pyrophosphate (PPi) and DNA translocation (step 5) follow, after which the next reaction cycle can begin. Conformational comparisons for protein residues Asp190, Asp192, Phe272, Arg258, and Tyr296 in both crystallographic closed and open states of pol β are shown in (E). The coordinations of the nucleotide-binding Mg2+ (A) and the catalytic Mg2+ (B) are presented in (F).
DNA polymerase β (pol β) is a structurally and functionally attractive model to study polymerase mechanisms for efficient and faithful DNA synthesis. Pol β is the smallest eukaryotic cellular DNA polymerase characterized (Wilson, 1998) and lacks an accessory proofreading activity. It shares a common overall architecture for nucleotidyl transfer activity (Beard and Wilson, 2000) with other DNA polymerases: a hand-like shape consisting of thumb, palm, and fingers (Ollis et al., 1985). The function of the thumb and fingers subdomains of left-handed pol β are opposite those of right-handed DNA polymerases (Beard and Wilson, 2000). Three crystallographic complexes of human DNA pol β with substrates and products have been obtained, representing intermediates along pol β's DNA synthesis pathway in base excision repair (Sawaya et al., 1997): pol β/gapped DNA, pol β/gapped DNA/ddCTP, and pol β/nicked DNA. The thumb subdomain of pol β is ‘open’ in pol β/gapped DNA and pol β/nicked DNA, while it is ‘closed’ in pol β/gapped DNA/ddCTP (see Fig. 1, A and B). In the closed ternary complex, one Mg2+ (the nucleotide-binding ion) coordinates all three α, β, and γ-phosphates of the incoming ddCTP, and the other Mg2+ ion (the catalytic ion) is positioned between the α-phosphate of the incoming ddCTP and the 3′-terminus of the primer strand (see Fig. 1 F). Both magnesium ions are anchored to the active site by coordinating highly conserved acidic residues (i.e., Asp190, Asp192, and Asp256). This “two-metal-ion” mechanism operates in all DNA and RNA polymerases for nucleotide addition (Steitz, 1993).
To interpret effects of protein residues Arg283, Tyr271, Asp276, and Arg258 on the binding pocket (see Table 1), we examine conformational and energetic factors in dynamics simulations for five pol β mutants R283A, Y271A, D276V, R258A, and R258K. The residues selected for mutation are conserved in DNA pol β from Homo sapiens (human), Rattus norvegicus (rat), and Xenopus laevis (African clawed frog) (Reichenberger and Pfeiffer, 1998) and are known to affect polymerase fidelity, catalytic efficiency, and nucleotide binding efficiency in DNA synthesis (see Kunkel and Bebenek, 2000, for a summary; and Table 2, this article). For example, the alanine substitution for Arg283 of pol β decreases dramatically catalytic efficiency for correct, but not incorrect, nucleotide insertion, and leads to commensurate loss of fidelity (Beard et al., 2002a). The decreased efficiency in R283A is due to both a loss of nucleotide binding affinity and decrease in the rate of nucleotide insertion (Beard et al., 2002a, 1996; Ahn et al., 1997; Werneburg et al., 1996). The nucleotide binding affinity is increased in Y271A (Kraynov et al., 1997) and D276V (Vande Berg et al., 2001) although decreased in R258A (Menge et al., 1995) relative to the wild-type pol β. In addition, R258A exhibits an increased rate of nucleotide insertion with a negligible effect on polymerase fidelity (Menge et al., 1995; W. A. Beard, D. D. Shock, and S. H. Wilson, unpublished). Since primary effects of substituted residues cannot be intuited from crystal structures of wild-type enzymes, and only R283A has been crystallized (Beard et al., 1996), our dynamics simulations aim to expose kinetically relevant structural and dynamic features of the active site that may contribute to DNA synthesis efficiency, fidelity, and/or nucleotide binding affinity. By probing the influence of these enzyme modifications on the dynamic details of thumb subdomain conformational changes compared to wild-type (Yang et al., 2002a), we detail the roles of these key residues.
TABLE 1.
Comparison of residues Arg283, Tyr271, Asp276, and Arg258 interactions with DNA basepair in the active site and with other protein residues in both closed ternary and open binary nick crystal structures
| Key protein residue | Closed ternary* | Open nick binary* | ||
|---|---|---|---|---|
| Arg283 | Arg283:Cδ–T6:N3 | (3.50) | ||
| Arg283:NH1–T7:O4′ | (2.68) | Arg283:Cγ–Asn279:O | (3.04) | |
| Arg283:N–Asn279:O | (2.67) | |||
| Tyr271 | Tyr271:OH–P10:O2 | (2.66) | Tyr271:OH–P11:N2 | (2.29) |
| Tyr271:O–ddCTP:O3′ | (2.80) | Tyr271:O–Asn279:Oδ1 | (3.26) | |
| Tyr271:O–Asn279:Oδ1 | (2.75) | Tyr271:Cδ2–Glu295:Cβ | (3.00) | |
| Asp276 | Asp276:Cβ–ddCTP:C4 | (2.92) | ||
| Asp276:Oδ–Arg40:NH | (2.93) | Asp276:O–Asn279:N | (2.80) | |
| Asp276:O–Asn279:N | (3.61) | Asp276:O–Lys280:N | (3.03) | |
| Asp276:O–Lys280:N | (3.05) | |||
| Arg258 | Arg258:NH2–Glu295:Oɛ2 | (3.20) | Arg258:NH1–P11:C4′ | (2.93) |
| Arg258:NH2–Tyr296:OH | (2.92) | Arg258:NH2–Asp192:Cβ | (2.83) | |
The distance between two heavy atoms in each pair of interactions shown in parentheses in Å.
TABLE 2.
Experimental kinetic data for the wild–type pol β and mutants Arg283Ala, Tyr271Ala, Asp276Val, Arg258Ala, and Arg258Lys
| Pol β/DNA complex | Rate constants*kpol [s−1] | Fidelity† | Catalytic efficiency kpol/Kd [M−1 s−1] | dNTP binding affinity‡Kd[μM] | Reference | ||
|---|---|---|---|---|---|---|---|
| WT | 9.4–24 | 1600–51,000 | 125,000–1,100,000 | 8.6–108 | Ahn et al. (1997) | ||
| Arg-283-Ala | 0.048–0.83 | 150–6800 | 340–4900 | 61–170 | |||
| (decreased) | (decreased) | (decreased) | (increased) | ||||
| WT | 3.4–17 | 3900–51,000 | 180,000–510,000 | 6.7–66 | Kraynov et al. (1997) | ||
| Tyr-271-Ala | 0.58–4.1 | 3400–21,000 | 290,000–415,000 | 1.4–8.0 | |||
| (decreased) | (similar) | (similar) | (decreased) | ||||
| WT | 10.0 | N/A | 1,790,000 | 5.6 | Vande Berg et al. (2001) | ||
| Asp-276-Val | 6.3 | N/A | 10,500,000 | 0.6 | |||
| (G:dCTP) | (decreased) | (increased) | (decreased) | ||||
| kpol [s−1] | Fidelity§ (T:dGTP) | kcat/Km [M−1 s−1] | Kd[μM] (T:dATP) | Km[μM]¶ | kcat [s−1]¶ | ||
| WT | 5.3 | 4316 | 820,000 | 4.4 | 1.1 | 0.9 | |
| Arg-258-Ala | 18.0 | 8333 | 200,000 | 25 | 7.9 | 1.8 | W. A. Beard et al.(unpublished) |
| (increased) | (increased) | (decreased) | (increased) | (increased) | (increased) | ||
| Arg-258-Lys | N/A | N/A | 170,000 | N/A | 9.8 | 1.7 | W. A. Beard et al.(unpublished) |
| (T:dATP) | (decreased) | (increased) | (increased) |
Ranges exist for some data because a subset or all of 16 basepairs were experimentally investigated; single values refer to specific basepair for both mutant and wild-type (indicated in parentheses).
Rate constant kpol (step 2): rate of nucleotide incorporation for first–enzyme turnover.
Fidelity is 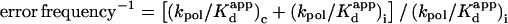 (c and i stand for correct and incorrect nucleotide incorporation, respectively).
(c and i stand for correct and incorrect nucleotide incorporation, respectively).
Kd[μM]: apparent equilibrium dissociation constant of dNTP; the larger the Kd value, the lower the dNTP binding affinity.
Fidelity is (kcat/Km)c/(kcat/Km)i (c and i stand for correct and incorrect nucleotide incorporation, respectively).
Km = Kd if kpol is the rate-limiting step; kcat measures the slowest step, or steps, in the reaction cycle during a steady-state assay.
We find that R258A approaches a closed state whereas mutants R283A, Y271A, D276V, and R258K move toward an open state by the end of the 8.1-ns trajectories. Analyses of local interaction networks help explain the increased rate of nucleotide insertion in R258A and decreased rates in the other mutants (relative to wild-type enzyme). Specifically, we unravel compensatory local interactions due to single-residue mutation in the active site both before and after chemistry and, together with energetic analyses, extrapolate effects of different mutants on enzyme fidelity, rate of nucleotide insertion, and/or nucleotide binding affinity relative to wild-type pol β: the rate of nucleotide insertion should be increased and fidelity maintained in R258G, as in R258A; nucleotide binding affinity should be increased in D276L more than D276V; and double mutant of R283A/K280A should decrease nucleotide binding affinity and increase misinsertion more than R283A. Such compensatory interactions and sensitivity, especially in the case of the Arg258 residue, underscore the highly organized but pliant active site essential to polymerase function.
METHODS
Systems setup
Intermediate models of mutants after chemistry for dynamics simulations
The wild-type pol β/DNA complex has been investigated by molecular dynamics (MD) simulations in our previous studies (Yang et al., 2002a). Two initial models were constructed for a closed form and an intermediate form between closed and open states after nucleotide incorporation. All MD simulations starting from the closed model failed to capture pol β's opening; however, simulations starting from the intermediate model approached an open state and captured important local side-chain motions in pol β's opening. Thus, simulations for the enzyme mutants here are started from this intermediate state, to facilitate comparison with the wild-type system.
The wild-type intermediate model was constructed as an average of the crystallographic closed, ternary complex (entry 1BPY) and open, binary nicked complex (entry 1BPZ) from the PDB/RCSB resource (Berman et al., 2000); see details in Yang et al. (2002a). On the basis of this intermediate model, we constructed five complexes of mutant pol β/DNA R283A, Y271A, D276V, R258K, and R258A by single residue replacement, leaving other protein residues and DNA base sequences unchanged. The generated steric clashes in all systems were removed by subsequent energy minimization and careful equilibration. The thumb subdomain in all initial mutant models is in a partially open state. The chemical product pyrophosphate (PPi) and the two specific Mg2+ ions in the active site were not considered in this work since they are absent in the crystallographic open structures, and to permit exact comparison with the simulations of our earlier work (Yang et al., 2002a,b); parallel work in our lab on the pre-chemistry structures is exploring the enzyme's selection specificity (Radhakrishnan and Schlick, 2004; K. Arora and T. Schlick, unpublished).
The five intermediate mutant models were similarly solvated in a periodic domain of face-centered cube and neutralized at a physiological ionic strength of 150 mM, as we did for the wild-type models in prior work (Yang et al., 2002a). This modeling produces five solvated intermediate mutant pol β/DNA complexes of R283A (41,958 atoms including 44 Na+ and 20 Cl−), Y271A (41,962 atoms including 44 Na+ and 21 Cl−), D276V (41,976 atoms including 43 Na+ and 21 Cl−), R258K (41,971 atoms including 44 Na+ and 21 Cl−), and R258A (41,958 atoms including 44 Na+ and 20 Cl−). Each system contains 11,830 solvated water molecules. We term the five solvated mutant models R283A, Y271A, D276V, R258K, and R258A herein. These models were each subjected to MD simulations over 8.1 ns. A 6-ns trajectory for the intermediate wild-type pol β/DNA complex (Yang et al., 2002b) is also analyzed for comparison.
Closed models of mutants before chemistry for calculating active-site electrostatic potentials and static protein/DNA interactions
On the basis of the crystallographic, closed ternary complex of pol β/DNA/ddCTP (Sawaya et al., 1997), the closed, ternary wild-type system is constructed by adding the missing protein residues 1–9 and an –OH group to the primer 3′-terminus and the C3′ atom of ddCTP in InsightII package, version 2000. Based on this closed wild-type system, the corresponding closed, ternary mutant systems R283A, Y271A, D276V, R258K, and R258A are constructed by single residue replacement, and subsequent energy minimization relaxes the structures. These closed wild-type and mutant systems before chemistry are employed to calculate the active-site electrostatic potential using the program QNIFFT (Sharp et al., 1990; Chin et al., 1999) and the interactions between the incoming 2′-deoxyribocytidine 5′-triphosphate (dCTP) and residues Arg283, Tyr271, Asp276, and Arg258 in wild-type, Ala283 in R283A, Ala271 in Y271A, Val276 in D276V, Lys258 in R258K, and Ala258 in R258A. These calculations could help explore the effect of key protein residues on polymerase active-site electrostatic environment and thus interpret the experimental data of nucleotide binding affinity observed for the mutants. The positions of residues Arg283, Tyr271, Asp276, and Arg258, with respect to the nascent basepair in the crystallographic closed and open structures, are presented in Fig. 1, C and D.
Minimization, equilibration, and dynamics protocol
Energy minimizations, equilibrations, and dynamics simulations for all five intermediate mutant systems (after chemistry) were performed using the program CHARMM (Brooks et al., 1983; MacKerell and Banavali, 2000) and the all-atom version 26a2 force field (Chemistry Department, Harvard University, Cambridge, MA). Each system was minimized using the steepest-descent method for 10,000 steps followed by adapted-basis Newton-Raphson (Brooks et al., 1983; Schlick, 1992) for 20,000 steps. Each system was then equilibrated for 30 ps at room temperature by the stochastic LN integrator (Schlick et al., 1997; Barth and Schlick, 1998a,b; Schlick, 2001) before dynamics production runs. (See Yang et al., 2002a, for a thorough examination of the stability and reliability of LN integrator for large macromolecular systems in terms of thermodynamic, structural, and dynamic properties compared to single-timestep Langevin as well as Newtonian, i.e., Velocity Verlet, propagators.) The LN triple-timestep protocol of Δτ/Δtm/Δt = 1/2/150 fs and all other parameters for dynamics simulations here are the same as those used in our initial work (Yang et al., 2002a).
RESULTS
Thumb moves to a closed state in the R258A simulation whereas it moves slightly toward an open state in other mutant simulations
The α-helix N (thumb subdomain) defines the closed and open states for pol β. Using averaged dynamic structures over the last 1.5 ns of each mutant simulation compared to the crystallographic closed and open complexes (Fig. 2), we rank the degrees of opening for pol β mutants as Y271A ≈ R258K ≈ R283A > D276V, whereas α-helix N in R258A moves toward a closed state similar to the crystallographic closed structure.
FIGURE 2.
Comparison of α-helix N between simulated mutant systems and the crystallographic closed (red) and open (green) structures from two perspectives. All structures are superimposed according to the Cα atoms of the catalytic palm subdomain. The colored ribbons—orange, cyan, pink, gray, and blue—represent, respectively, the simulated mutants R283A, Y271A, D276V, R258K, and R258A. The nascent DNA basepair (P11–T6) in the crystallographic open structure is also presented to indicate the DNA position in the pol β/DNA complex.
As shown by the time evolution of the root mean-square deviations (RMSD) of α-helix N Cα atoms relative to those of both closed and open crystallographic structures (Fig. 3), the minimum RMSD of R258A relative to the closed crystallographic structure (1.13 Å) indicates that R258A has approached a closed state that is not superimposable with the closed crystallographic structure. The average RMSDs for the five pol β mutants over the last 1.5 ns of the simulations with respect to the crystallographic open structure are 2.79 Å for Y271A, 2.86 Å for R258K, 2.89 Å for R283A, 3.33 Å for D276V, and 6.62 Å for R258A, all much larger than the minimum RMSD observed in the wild-type simulation (0.8 Å) (Yang et al., 2002a).
FIGURE 3.
Time evolution of the RMS deviation of the α-helix N Cα atoms in all mutant trajectories relative to those in the crystallographic closed (red) and open (green) structures. All systems are superimposed by the palm Cα atoms.
Phe272 ring flips in trajectories of R283A, D276V, and R258A, but not in Y271A and R258K trajectories
We have suggested, based on prior simulations, a possible sequence of events in (wild-type) pol β opening: Phe272 aromatic ring flip, thumb subdomain opening, and Arg258 side-chain rotation toward Asp192 (Yang et al., 2002a). More recent simulations of pol β complexed to mismatched basepairs at the primer terminus (Yang et al., 2002b) and ongoing novel simulation applications (Radhakrishnan and Schlick, 2004) support this scenario of events. Fig. 4 presents the conformations of key protein residues (Asp190, Asp192, Arg258, or mutated Arg258, Phe272, and Tyr296) in the averaged dynamic structure over the last 1.5 ns for mutants compared to positions observed in the crystallographic closed and open forms (see also Fig. 1 E), and Fig. 5 follows the Phe272 ring flips by evolution of two dihedral angles (χ1 = C–Cα–Cβ–Cγ, measuring rotation of the entire phenyl ring, and χ2 = Cα–Cβ–Cγ–Cδ, showing inclination of the ring plane) in both wild-type and mutant simulations. We observe that Phe272 flips in R283A (at 2.3 ns), D276V (at 4.9 ns), and R258A (at 0.2 ns) trajectories, but not in Y271A and R258K trajectories. The side chain of residue 258 does not rotate toward Asp192 in any mutant simulation.
FIGURE 4.
Conformational comparison for key protein residues in the active site in the crystallographic closed structure (red), crystallographic open nick structure (green), and in the simulated mutants R283A (orange), Y271A (cyan), D276V (pink), R258K (gray), and R258A (blue).
FIGURE 5.
Time evolution of the two dihedral angles associated with the Phe272 ring flip motion in all mutant simulations over 8.1 ns and a wild-type simulation over 6 ns. The two dihedral angles are defined by the following atom quadruplets: C–Cα–Cβ–Cγ for χ1 (red) and Cα–Cβ–Cγ–Cδ for χ2 (blue). Average values for the two dihedral angles during the characteristic ranges are also presented for all trajectories.
The rapid Phe272 flip in R258A likely reflects the additional space introduced by exchange of the large arginine with alanine. Lysine, however, at the same position (258), decreases flexibility and delays the side-chain flipping (see Figs. 4 and 5). The flip repression in Y271A may be explained by the disruption of interactions between Tyr271 and the newly incorporated primer nucleotide via several hydrogen bonds, as we observed previously (Yang et al., 2002a). Still, we suggest that Y271A compensates this interaction loss by employing the main-chain carboxyl of the adjacent residue, Phe272, to intermittently hydrogen-bond with the 3′-OH of the newly incorporated nucleotide (see Discussion).
Tyr296 is also very flexible in R258A (Figs. 4 and 6): dihedral angle C–Cα–Cβ–Cγ of Tyr296, which measures the ring's flexibility, jumps from ∼170° to 75° at ∼6.3 ns; this angle remains at ∼170° during other mutant and wild-type simulations (Yang et al., 2002b). This can be explained by loss of the Tyr296/Arg258 hydrogen bond in R258A, an interaction that prevents Arg258 from interfering with the ability of Asp192 to ligand the two essential Mg2+ ions in the closed state (Sawaya et al., 1997).
FIGURE 6.
Time evolution of a dihedral angle in residue Tyr296 indicating the ring flip in the R258A simulation. This dihedral angle χ1 is defined by atoms C–Cα–Cβ–Cγ in Tyr296. Average values for χ1 in two characteristic ranges are also given.
Protein/DNA interactions show distinctive patterns in mutants
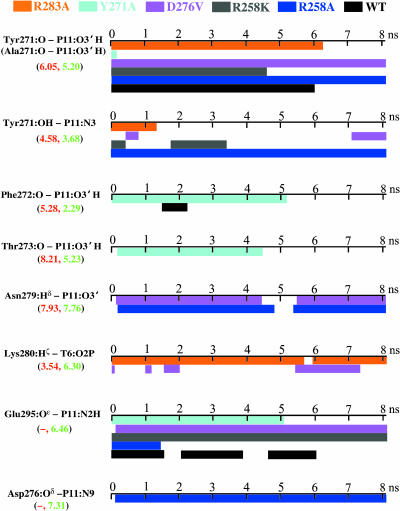
Arg283, Tyr271, and Asp276 are known to influence pol β's fidelity, catalytic efficiency, and nucleotide binding affinity (Vande Berg et al., 2001; Kraynov et al., 1997; Beard et al., 1996) by contributing hydrogen-bond donors (Tyr271 and Arg283) and van der Waals interactions (Asp276) with nascent basepairs in the closed ternary complex (Sawaya et al., 1997). Our analyses of hydrogen bonds and van der Waals contacts between the protein and DNA in the active site present distinctive interaction patterns in the mutants (Fig. 7). For example, hydrogen bonds of Phe272:O, Thr273:O with P11:O3′H exist during most of the Y271A simulation while these interactions are not present in the other mutant simulations. The Asn279:Hδ/P11:O3′ hydrogen bond forms only in R258A and D276V trajectories. Lys280 is the only protein residue hydrogen-bonding to the templating nucleotide T6 (the Watson-Crick partner for P11), and this hydrogen bond maintains throughout R283A trajectory and only transiently forms in D276V trajectory. We also note that Asp276:Oδ makes van der Waals contacts with P11:N9 (distances ≤ 3.5 Å) only in R258A simulation. In addition, the Asp276:Oδ1/Arg40:Hη hydrogen bond maintains during most of the R258A simulation (consistent with thumb closing).
FIGURE 7.
Time evolution of some important hydrogen bonds and van der Waals contacts between protein and DNA nascent basepair for dynamics simulations of R283A (orange), Y271A (cyan), D276V (pink), R258K (gray), and R258A (blue) over 8.1 ns, and of wild-type (black) over 6 ns. The values in parentheses indicate the distance (in Å) between two heavy atoms in each pair of corresponding interactions in the closed (red) and open (green) states of the crystallographic pol β/DNA complexes; and the hyphen (–) indicates not applicable.
Thus, each mutant has distinctive hydrogen bonding and/or van der Waals interaction patterns between protein and DNA in the active site, all differing from those in the wild-type. These differences may arise from compensatory interactions induced by single residue mutation (see Discussion). Furthermore, our analyses show that residues Tyr271, Asp276, Asn279, Lys280, and/or Glu295 interact with DNA nascent basepair, consistent with their suggested roles in pol β fidelity and catalytic efficiency in DNA synthesis (Vande Berg et al., 2001; Kraynov et al., 1997; Ahn et al., 1997, 1998; Werneburg et al., 1996; Sawaya et al., 1994; Pelletier et al., 1994; Beard et al., 1996).
Negative electrostatic environment in the active site traps a counterion Na+, a possible surrogate for missing Mg2+
We find in all mutant simulations that one sodium counterion is trapped in the active site by coordinating conserved aspartate(s) and several dynamic water molecules (Fig. 8). This Na+ may represent a partial electrostatic surrogate for the missing Mg2+ ions. Specifically, in R283A, a trapped Na+ interacts with both Asp192 and Asp256 during the last 1.8 ns of the simulation. For D276V, the trapped Na+ coordinates Asp256, occasionally Asp190, during most of the simulation, in a manner similar to that in wild-type (Yang et al., 2002a). In R258K and Y271A simulations, the trapped Na+ only coordinates Asp190. For R258A, the Na+ coordinates both Asp190 and Asp192 during the first third of the simulation, and then it moves to coordinate all three aspartates (Asp190, Asp192, Asp256) for the remainder of the simulation, which coincides with that of the catalytic Mg2+ in the crystallographic closed structure (Sawaya et al., 1997). This suggests a role for the catalytic Mg2+ in maintaining the enzyme in a closed state, in which key functional groups are assembled for nucleotide incorporation before chemistry. In addition, our observed Na+ coordination pattern further supports a closed state reached by R258A after simulation.
FIGURE 8.
(A) The sodium counterion probability density of >6% in the active site for mutant and wild-type enzyme simulations. The densities are accumulated on a 1 Å cubic lattice over 500 snapshots for the mutants and 200 snapshots for the wild-type, and sampled at a frequency of 3 ps during 6.6–8.1 ns for each mutant trajectory and during 4.4–5.0 ns for the wild-type trajectory. (B) Typical snapshots for mutant and wild-type systems to show sodium ion coordination with key protein residues Asp190, Asp192, Asp256, and several dynamic water molecules in the active site.
Interactions between mutated residues and the incoming dCTP reflect the effect of single-residue mutation on nucleotide binding affinity
We further investigate the effect of single-residue mutation on active-site electrostatic environment by comparing electrostatic potentials and interactions between the incoming nucleotide and mutated residue in closed ternary wild-type and mutants. This analysis may help interpret the varying nucleotide binding affinities among these mutants in Table 2 (Vande Berg et al., 2001; Kraynov et al., 1997; Beard et al., 1996; Menge et al., 1995). We present in Fig. 9 the active-site electrostatic potentials and in Fig. 10 the total, electrostatic, and van der Waals interaction energies between dCTP and protein residue (at positions 283, 271, 276, or 258) in the closed ternary complex of wild-type pol β/DNA/dCTP and corresponding mutants.
FIGURE 9.
Local electrostatic potential around the incoming dCTP (green) and the complementary template base dG (yellow) in closed, ternary complexes of the wild-type and five mutants R283A, Y271A, D276V, R258K, and R258A. For clarity, the other DNA primer/template residues are not shown. The positions of 40, 258, 271, 279, 280, and 283 in each potential map are indicated.
FIGURE 10.
Comparison of interaction energies between the incoming dCTP and protein residues selected for mutation in the ternary, closed wild-type pol β/DNA/dCTP complex and the corresponding mutants of R283A, Y271A, D276V, R258A, and R258K. In each pair of energy bars, the left bar represents energy in wild-type (WT) and the right bar represents energy in mutant.
Observed local variations in Fig. 9 include negative electrostatic potential at positions 258 for R258A and 283 for R283A instead of positive in wild-type. In addition, the alanine substitution for arginine increases the negative electrostatic potential at positions 271, 276, 279, and 280 in both R283A and R258A, and decreases the positive potential at position 258 in R283A and position 283 in R258A. A positive potential is observed at position 276 in D276V instead of negative in wild-type. We also note more negative potential at positions 279 and 283 in D276V relative to the wild-type. The alanine substitution for Tyr271 in Y271A increases positive potential at position 271, and somewhat increases negative potential at positions 279 and 283. The lysine replacement of Arg258 decreases positive potential at position 258, and enhances negative electrostatic potential at positions 271, 276, and 283. Thus, as expected, single-residue mutation for key protein residue alters electrostatic potentials of the mutated and neighboring residues, and thus the active-site electrostatic environment.
Since the incoming nucleotide (dCTP here) contains a planar base and a high, negatively-charged triphosphate group (charge of −4), a large, positive protein side chain makes strong van der Waals and electrostatic interactions with nucleotide, whereas a small, negative side chain has weaker interactions. Indeed, when the large, positively-charged arginine is substituted by small, nonpolar alanine in R283A and R258A, both van der Waals and electrostatic interactions between dCTP and residue 283 or 258 decrease, especially the electrostatic interactions. This decrease is consistent with, and may help explain, the decreased nucleotide binding affinity in R283A (∼30-fold diminished; Beard et al., 1996) and R258A (∼5–10-fold diminished; Menge et al., 1995; W. A. Beard, D. D. Shock, and S. H. Wilson, unpublished).
Similarly, substitutions of Tyr271 with alanine in Y271A and of Arg258 with the relatively smaller lysine in R258K also decrease the electrostatic and van der Waals interactions between dCTP and residue 271 or 258. However, this decreased interaction is at variance with the increased nucleotide binding affinity in Y271A (approximately four-to-fivefold increased; Kraynov et al., 1997). This discrepancy may be reconciled in part with enhanced interactions between certain residues (i.e., Phe272, Asn279, and Arg283) spatially adjacent to residue 271 and the dCTP that compensate for loss of the dCTP/residue 271 interaction, and thereby increase the nucleotide binding affinity in Y271A. Indeed, we determine that interactions of dCTP with Phe272, Asn279, and Arg283 in Y271A increase by 0.74, 3.94, and 1.27 kcal/mol, respectively, relative to those in the wild-type, which also agrees with the increased negative potential at residues 279 and 283 (Fig. 9).
When the negatively-charged aspartate is replaced by nonpolar valine in D276V, the interaction between Val276 and dCTP greatly increases, mostly from the attractive electrostatic interaction between hydrogen atoms of residue 276 and triphosphate group of dCTP (−17.59 kcal/mol for wild-type and −37.98 kcal/mol for D276V). This increase is consistent with D276V's increased nucleotide binding affinity (approximately ninefold; Vande Berg et al., 2001). We also observe that interactions between dCTP and residues Asn279, Arg283 increase in D276V relative to those in wild-type (3.90, 1.14 kcal/mol for Asn279 and Arg283 increased, respectively).
DISCUSSION
A kinetic and structural survey of DNA polymerases from various families displaying divergent fidelities indicates that polymerase specificity is primarily determined by the efficiency by which the polymerase inserts the correct nucleotide (this is inferred from the fact that DNA polymerases insert incorrect nucleotides with similar efficiencies; Beard et al., 2002a). Furthermore, the efficiency of correct insertion appears to be primarily governed by electrostatic environment of the incoming nucleotide and its effects on the binding pocket geometry (Beard and Wilson, 2003). Accordingly, a better understanding of the contribution of protein side chains (especially their charge effects) on dynamics of the nascent basepair binding pocket is a prerequisite to an atomic-level dynamics description of DNA polymerase fidelity. In our study, we have selectively replaced charged side chains situated at or near the binding pocket and compared dynamics simulations of mutant systems to the wild-type dynamics (Yang et al., 2002a). The effect of these single-residue mutations on electrostatic environment of the incoming nucleotide was also investigated. These dynamic and energetic details are used to generate atomic-level insights into enzyme fidelity, rate of nucleotide insertion, and/or nucleotide binding affinity in mutants, and offer testable predictions for different mutants.
Altered conformational motions in mutants help explain observed changes in the rate of nucleotide insertion, kpol
Structural (Arndt et al., 2001), kinetic (Vande Berg et al., 2001), and computational (Yang et al., 2002a,b) data have suggested that the large subdomain movement per se in the polymerase cycle (Fig. 1) is relatively rapid. Although our natural dynamics simulations occur over short time frames relative to events that limit catalytic cycling, previous accelerated sampling (high-temperature simulations and targeted molecular dynamics simulations; Yang et al., 2002a,b) and current novel pathway methodology applications (Radhakrishnan and Schlick, 2004; Arora and Schlick, 2003) suggest that Arg258 side-chain rotation could be slow enough to be kinetically significant. Such slow rearrangements involving the Arg258 rotation might similarly limit pol β's closing before chemistry. When an incorrect nucleotide binds to pol β, these slow, local rearrangements would discourage incorrect nucleotide insertion and thereby play a role in polymerase selectivity.
Our dynamics simulations for the mutant pol β/DNA complexes offer additional details of the molecular changes that may occur during thumb subdomain motions. The thumb moves toward an ‘open’ state in R258K, Y271A, D276V, and R283A simulations, but still remains more ‘closed’ than the crystallographic open conformation (Sawaya et al., 1997). In contrast, the thumb moves toward a ‘closed’ state in R258A simulation, similar to the crystallographic closed structure (Sawaya et al., 1997). Thus, the subtle residue changes in the vicinity of the polymerase active site affect the subdomain motion and may indicate that the equilibrium constant for this structural transition is near unity.
Since Asp192 forms a salt-bridge with Arg258 in the open ‘inactive’ conformation while it coordinates both the catalytic and nucleotide-binding Mg2+ in the closed ‘active’ conformation, removing the positive charge on residue 258 would be expected to promote the active closed conformation because Asp192 would be free to coordinate both Mg2+ ions required for catalysis. Indeed, we note a favored closed conformation for the simulated R258A. When the positive charge at this position is maintained in the enzyme (i.e., wild-type, R258K, Y271A, D276V, and R283A), our simulations indicate accordingly that the thumb subdomain opens somewhat. Since kinetic data have indicated that polymerase closing before chemistry and opening after chemistry have similar half-lives (Dahlberg and Benkovic, 1991; Patel et al., 1991; Zhong et al., 1997), our simulations after chemistry might suggest that the alanine substitution for Arg258 (R258A) facilitates the enzyme closing whereas the other single-residue mutations somewhat delay the enzyme closing before chemistry. A facilitated closing in R258A agrees with its increased rate of nucleotide insertion (W. A. Beard et al., unpublished) whereas inhibited closing in the other mutants is consistent with their decreased rates (Vande Berg et al., 2001; Kraynov et al., 1997; Ahn et al., 1997; Beard et al., 1996) relative to the wild-type enzyme (Table 2). The single-residue mutation in the active site may alter the enzyme's conformational closing and/or affect the chemistry step (if rate-limiting), and thus change the rate of nucleotide insertion, kpol.
In addition, our suggested important role of Arg258 rotation in polymerase closing (Yang et al., 2002a) gains support from parallel studies on generating pol β's closing reaction kinetics profile before chemistry using transition path sampling (Radhakrishnan and Schlick, 2004). Since these results indicate that the Arg258 rotation has a relatively high free energy barrier of ∼19 ± 3 kBT in the closing pathway, this closing motion may be facilitated by replacement of arginine with alanine, since the latter may reduce this energy barrier.
Because it was also found that R258A does not have significantly altered fidelity with respect to the wild-type enzyme (W. A. Beard et al., unpublished), both correct and incorrect insertions are likely influenced similarly by the alanine substitution. (Recall that fidelity is defined as the reciprocal of the misinsertion frequency, namely, 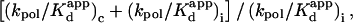 where c and i refer to correct and incorrect nucleotide incorporation, respectively; see footnote in Table 2.) This leads us to suggest that a replacement of Arg258 by the small, nonpolar glycine (R258G) will similarly increase the rate of nucleotide insertion and affect enzyme fidelity negligibly.
where c and i refer to correct and incorrect nucleotide incorporation, respectively; see footnote in Table 2.) This leads us to suggest that a replacement of Arg258 by the small, nonpolar glycine (R258G) will similarly increase the rate of nucleotide insertion and affect enzyme fidelity negligibly.
Interactions of mutated residue with incoming nucleotide and resulting induced compensatory interactions help explain altered nucleotide binding affinity in mutants
Below, we discuss compensatory protein/DNA interactions in the active site observed in mutants both before and after chemistry. Furthermore, in each closed, ternary mutant system before chemistry, we relate the nucleotide binding affinity to the interaction between incoming nucleotide and mutated residue as well as induced compensatory interactions. We then offer some testable hypotheses regarding effects of different mutants on the nucleotide binding affinity.
In the Y271A simulation, the alanine substitution disrupts the hydrogen bond of Tyr271 with the base of the newly incorporated nucleotide (P11) in the active site. However, the hydrogen bonds between P11:O3′H and main-chain oxygens of Phe272 and Thr273, adjacent to residue 271, are maintained during most of the Y271A trajectory (see Fig. 7). In contrast, these hydrogen bonds are seldom observed in the other mutant simulations. These compensatory interactions might help stabilize the newly incorporated primer nucleotide. Similarly, in the closed ternary Y271A complex before chemistry, interactions between certain active-site residues (i.e., Phe272, Asn279, and Arg283) and dCTP may compensate for the loss of interaction between residue 271 and dCTP (Fig. 10) due to the alanine substitution, and thereby help account for the mutant's moderate increase in nucleotide binding affinity (Kraynov et al., 1997).
In the closed, ternary D276V mutant, the increased interaction between Val276 and dCTP agrees well with the increased nucleotide binding affinity relative to wild-type (Vande Berg et al., 2001). We find that the increased Val276/dCTP interaction mostly stems from the increased electrostatic interaction (∼20 kcal/mol) between the partially positively-charged hydrogen atoms of Val276 and the negatively-charged triphosphate group of dCTP, rather than the slightly increased van der Waals interaction (∼0.25 kcal/mol) between Val276 and the base of dCTP, as proposed previously (Vande Berg et al., 2001). These results lead us to hypothesize that a leucine substitution for Asp276 (D276L) will increase the nucleotide binding affinity more than D276V since leucine has a larger size and more hydrogen atoms than valine. Furthermore, the enhanced interactions between dCTP and residues 279, 283 in the closed, ternary D276V complex may also contribute to the nucleotide binding affinity. In addition, we note that the hydrogen bond between Asn279:Hδ and P11:O3′ is maintained mostly throughout the D276V simulation (after chemistry), and this may help stabilize the newly incorporated DNA primer terminus. This is consistent with the experimental observation that the D276V mutant exhibits a nonproductive DNA binding mode where the primer terminus is stabilized in the polymerase active site (Vande Berg et al., 2001).
Arg283 does not directly interact with the incoming nucleotide in either the crystallographic open or closed forms of pol β. However, in the closed conformation, Arg283 makes van der Waals contacts with the minor groove edge of the templating base and is hydrogen-bonded to the sugar of the preceding template nucleotide (Table 1). Substituting the large arginine side chain with the small alanine alleviates steric clashes with specific non-Watson-Crick basepairs in the binding pocket and therefore alters specificity (Osheroff et al., 1999). Indeed, our calculated interaction between residue 283 and the templating base decreases ∼14 kcal/mol in the closed, ternary R283A before chemistry relative to that in the wild-type. The decreased interaction between residue 283 and dCTP in our calculation may help explain the decreased nucleotide binding affinity in R283A (∼30-fold) relative to the wild-type (Beard et al., 1996). Residue Lys280, like Arg283, hydrogen-bonds with the template base in a closed state, and substitution of Lys280 with smaller side chains reveals loss of binding affinity for the incoming nucleotide with template purines (Beard et al., 2002b). We observe that the Lys280:Hδ/T6:O2P hydrogen bond is maintained throughout the R283A simulation after chemistry; and the interaction between Lys280 and dCTP increases 0.3 kcal/mol in the closed, ternary R283A before chemistry relative to that in the wild-type. These results suggest that Lys280 can help stabilize the newly incorporated primer base (after chemistry) or the incoming nucleotide (before chemistry) in the R283A mutant. Therefore, we hypothesize that the double mutant of R283A/K280A could further decrease the nucleotide binding affinity with a corresponding loss of fidelity.
Recall that we observe several protein/DNA hydrogen bonds in the R258A and R258K trajectories, such as Tyr271:O/P11:O3′H in both mutants, Tyr271:OH/P11:N3 and Asn279:Hδ/P11:O3′ in R258A, and Glu295:Oɛ/P11:N2H in R258K. These interactions help stabilize the newly incorporated primer base in the simulated mutants. In the closed, ternary complexes of R258A and R258K, the alanine and lysine substitutions for Arg258 both increase negative electrostatic potential at position 283, and this leads to increased interaction between Arg283 and the incoming dCTP. Such compensatory interactions indicate the high sensitivity of the enzyme system to the active-site electrostatic environment. The modified polymerases exhibit different and multifaceted conformational pathways in terms of their nature, sequence, and rate of local structural changes.
CONCLUSION
Taken together, our findings provide structural and dynamics insights arising from each mutant system regarding the enzyme's rate of nucleotide insertion, fidelity, and/or nucleotide binding affinity. The preference for a closed form of the simulated R258A suggests a facilitated enzyme closing before chemistry in this mutant, whereas the tendency for an open form of the simulated R283A, Y271A, D276V, and R258K mutants suggests a somewhat delayed enzyme closing before chemistry. These observations tie well with kinetic measurements regarding the increased rate of nucleotide insertion in R258A and decreased rates in the other mutants relative to the wild-type enzyme. We thus hypothesize that R258G should similarly facilitate the enzyme closing before chemistry, increase the nucleotide insertion rate, and maintain fidelity relative to wild-type pol β.
Analyses of active-site protein/DNA interactions in dynamics mutant simulations (after chemistry) indicate distinctive hydrogen-bonding and van der Waals patterns arising from compensatory interactions. The increased or decreased interactions between the incoming nucleotide and mutated residues as well as the induced compensatory interactions in closed, ternary mutant systems (before chemistry) could help explain their altered nucleotide binding affinity with respect to the wild-type enzyme. These results lead us to suggest effects of other mutants on the nucleotide binding affinity. For example, leucine substitution for Asp276 (D276L) may increase the nucleotide binding affinity more than D276V, and the double mutant of R283A/K280A may decrease the nucleotide binding affinity and also increase misinsertion more than R283A.
Such sensitivity to very localized changes in key residues and specific compensatory interactions underscore the precise, highly organized active site that has evolved in DNA polymerases to preserve their vital function and also their pliant, flexible nature in compensating for local changes. This highly cooperative environment may be essential to direct the system to the chemical reaction of nucleotide incorporation. Further studies are underway to delineate this delicate orchestration of events in the enzyme reaction profile. Experiments to test our hypotheses regarding different mutants will also be of great interest.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
The work was supported by National Science Foundation grant ASC-9318159 and National Institutes of Health grant R01 GM55164 to T.S., and National Institutes of Health grants CA75449 and CA28038 to S.B. Acknowledgment is made to the donors of the American Chemical Society Petroleum Research Fund for partial support of this research (award PRF39115-AC4 to T.S.). Computations were supported by the National Computational Science Alliance under MCA99S021N and utilized the NCSA SGI Origin2000.
APPENDIX A
Sequence homology studies
It is instructive to review the literature regarding homology studies outside of the pol β group. This Appendix shows two or three conserved active-site residues in several DNA polymerases belonging to different families, and certain conserved protein residues among some DNA polymerases in the X family.
On the basis of sequence homologies (Delarue et al., 1990; Ito and Braithwaite, 1991; Braithwaite and Ito, 1993) and crystallographic structure analyses (Joyce and Steitz, 1994), DNA polymerases have been grouped into five families, e.g., A, B, C, D, and X. Prototypes for each family are E. coli DNA polymerase I (family A), DNA polymerase II (family B), DNA polymerase III α-subunit (family C), archeal polymerases (family D) (Cann and Ishino, 1999), and pol β and terminal transferase (family X). Several newly discovered DNA polymerases known for low fidelity synthesis on undamaged DNA and ability to bypass DNA lesions in vitro are associated with the Y family of DNA polymerases (Ohmori et al., 2001). Overall, there is very little similarity between the amino acid sequences of DNA polymerases in the different families, except for a few acidic residues coordinated with two functional metal ions located in the palm subdomain (Delarue et al., 1990).
Amino acid sequence comparisons have suggested three conserved carboxylate-containing residues in the active sites of all classes of polymerases (Singh and Modak, 1998). However, only two aspartate residues are thought to be structurally conserved among Klenow fragment, HIV-1 RT, RB69 pol α polymerase (Wang et al., 1997), and the third ones are variable. In Table A1, the three active-site residues coordinated with the metal ions are listed for some representative DNA polymerases: pol β (Sawaya et al., 1997, 1994; Pelletier et al., 1994), Klenow fragment of E. coli DNA polymerase I (Polesky et al., 1990), Taq DNA polymerase (Klentaq1; Li et al., 1998), T7 DNA polymerase (Doublié et al., 1998; Doublié and Ellenberger, 1998), Bacteriophage RB69 (Wang et al., 1997), Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4; family Y; Ling et al., 2001), and HIV-1 RT (Huang et al., 1998; Kohlstaedt et al., 1992; Larder et al., 1987, 1989); the third residues that are not absolutely conserved are underlined. The conservation of the metal-binding site in these highly divergent DNA polymerases underscores the importance of the metal ions in assisting nucleotide polymerization.
TABLE A1.
Conserved residues among some representative DNA polymerases from different families
| DNA polymerases | Carboxylate triads | Reference | ||
|---|---|---|---|---|
| DNA pol β | Asp190 | Asp192 | Asp256 | Sawaya et al. (1994, 1997); Pelletier et al. (1994) |
| Klenow fragment | Asp705 | Asp882 | Asp883* | Polesky et al. (1990) |
| Klentaq | Asp610 | Asp785 | Glu786* | Li et al. (1998) |
| T7 DNA polymerase | Asp475 | Asp654 | Glu655* | Doublié and Ellenberger (1998); Doublié et al. (1998) |
| Bacteriophage RB69 gp43 | Asp411 | Asp621* | Asp623 | Wang et al. (1997) |
| P2 DNA polymerase IV (Dpo4) | Asp7 | Asp105 | Glu106 | Ling et al., 2001 |
| HIV RT | Asp110 | Asp185 | Asp186* | Huang et al. (1998); Kohlstaedt et al. (1992); Larder et al. (1987, 1989) |
The underlined residues are not absolutely conserved and are variable in the polymerase.
Despite the nonhomology among different families of DNA polymerases, members of family X, which pol β belongs to, share some invariant residues including the strictly conserved aspartates in the active site. On the basis of sequence alignments (Oliveros et al., 1997; García-Diaz et al., 2000; Maciejewski et al., 2001; Showalter et al., 2001), the conserved residues are classified as metal ligands, DNA primer/template ligands, dNTP ligands, and others. In Table A2, we list these for some representative DNA polymerases in family X (pol β, ASFV polymerase X, human TdT, Yeast polymerase IV, mouse polymerase λ, and human polymerase μ). The pol β residues mutated in this article and their corresponding residues in other family X members are also indicated. Among family X members, each includes a catalytic triad of aspartates involved in metal binding. Mutations of residues Asp190, Asp192, and Asp256 in pol β have resulted in severe loss of catalytic activity (Date et al., 1991; Menge et al., 1995). Invariant residues Lys234, Arg254, Arg283, and Tyr296 in pol β are involved in DNA binding. Lys234 and Arg254 in pol β, which are both located in the palm subdomain and hydrogen-bonded to the nascent basepairs (Pelletier et al., 1994), are invariant except Lys234 in human pol μ (Arg387) and Arg254 in ASFV polymerase X (Gln98). Arg283, which locates in the α-helix N of the thumb subdomain and hydrogen-bonds to the templating base, is invariant in all family X members. Tyr296, located on a loop in the thumb subdomain and interacting with the backbone phosphates of the template strand, aligns with Tyr140 of ASFV polymerase X and is always substituted for a histidine residue in other members. The generally conserved residues Lys234 and Arg283 of pol β are proposed to break up the water structures in the minor groove of the template-primer upon complex formation, as the first step in the B-DNA to A-DNA transition at the pol β active site (Pelletier et al., 1996).
TABLE A2.
Conserved residues in representative members of family X DNA polymerases
| Conserved residues
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA polymerases | Metal ligands | DNA ligands | dNTP ligands | Salt-bridges | Others | Mutated residues | |||||||||||||
| DNA pol β* | Asp190 | Asp192 | Asp256 | Lys234 | Arg254 | Arg283 | Tyr296 | Arg183 | Phe272 | Gly274 | Gly179/Phe272 | Arg182/Glu316 | Tyr327 | Pro330 | Arg333 | Arg258 | Tyr271 | Asp276 | Arg283 |
| ASFV Pol X† | Asp49 | Asp51 | Asp100 | Lys85 | Gln98 | Arg127 | Tyr140 | Arg42 | Phe116 | Gly118 | Gly38/Phe116 | Arg41/Glu156 | Tyr167 | Pro170 | Arg173 | Phe102 | His115 | Val120 | Arg127 |
| Human TdT‡ | Asp343 | Asp345 | Asp433 | Lys402 | Arg431 | Arg459 | His473 | Arg336 | Trp449 | Gly451 | Gly332/Trp449 | Arg335/Glu489 | Tyr500 | Pro503 | Arg506 | Val435 | Gly448 | Arg453 | Arg459 |
| Yeast Pol IV§ | Asp467 | Asp469 | Asp502 | Lys480 | Arg500 | Arg529 | His542 | Arg360 | Tyr518 | Gly520 | Gly356/Tyr518 | Asn359/Glu556 | Tyr567 | Pro570 | Arg573 | Phe504 | His517 | Lys522 | Arg529 |
| Mouse Pol λ¶ | Asp425 | Asp427 | Asp488 | Lys470 | Arg486 | Arg515 | His528 | Arg418 | Phe504 | Gly506 | Gly414/Phe504 | Arg417/Glu554 | Tyr565 | Pro568 | Arg571 | Ile490 | Tyr503 | Ala508 | Arg515 |
| Human Pol μ‖ | Asp330 | Asp332 | Asp418 | Arg387 | Arg416 | Arg445 | His459 | Arg323 | Trp434 | Gly346 | Gly319/Trp434 | Arg322/Glu475 | Tyr486 | Pro489 | Arg492 | Val420 | Gly433 | Lys438 | Arg445 |
Residues Phe272 and Gly274 are located at the end of α-helix M interact by van der Waals contacts with the sugar moiety (C2′ and C3′ carbons) of the nucleotide, which is suggested to participate in nucleotide selectivity of DNA over RNA (Pelletier et al., 1994). The Gly274 residue is invariant in all sequences aligned here. Arg183, located in the α-helix K of the palm subdomain and hydrogen-bonded to the negatively-charged phosphate moiety of the nucleotide, is invariant in all family X members here. The cis-peptide bond found between Gly274 and Ser275 generates a dramatic bend between α-helices M and N of the thumb subdomain and is critical during catalytic cycling (Beard et al., 2002a).
A salt-bridge between pol β residue Arg182 (located in α-helix K) and Glu316 (located in α-helix O) stabilizes the open thumb position. On the other hand, the closed thumb position is stabilized by a hydrogen bond between Gly179 and Phe272. These four residues are invariant or highly conserved in all members of the pol X family. Pol β residues Tyr327, Pro330, and Arg333 located at the C-terminus of the thumb are invariant in all the family X DNA polymerases here. These C-terminal residues could contribute to the dynamics of the thumb movement.
There is only one reported natural variant of human DNA pol β, in which residue 295 has Glu instead of Lys, and this variant exhibits an inhibitory effect in an in vitro base excision repair assay and might play some role in carcinogenesis of the gastric mucosa (Iwanaga et al., 1999). In sum, the pol β residues Arg258, Tyr271, Asp276, and Arg283 chosen here for mutation are not conserved, except for Arg283.
Abbreviations used: pol β, DNA polymerase β; dNTP, 2′-deoxyribonucleoside 5′-triphosphate; ddCTP, 2′, 3′-dideoxyribocytidine 5′-triphosphate; PPi, pyrophosphate.
References
- Ahn, J., V. S. Kraynov, X. Zhong, B. G. Werneburg, and M.-D. Tsai. 1998. DNA polymerase β: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem. J. 331:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, J., B. G. Werneburg, and M.-D. Tsai. 1997. DNA polymerase β: structure-fidelity relationship from pre-steady-state kinetic analyses of all possible correct and incorrect basepairs for wild type and R283A mutant. Biochemistry. 36:1100–1107. [DOI] [PubMed] [Google Scholar]
- Arndt, J. W., W. Gong, X. Zhong, A. K. Showalter, J. Liu, C. A. Dunlap, Z. Lin, C. Paxson, M.-D. Tsai, and M. K. Chan. 2001. Insight into the catalytic mechanism of DNA polymerase β: structures of intermediate complexes. Biochemistry. 40:5368–5375. [DOI] [PubMed] [Google Scholar]
- Arora, K., and T. Schlick. 2003. Deoxyadenosine sugar puckering pathway simulated by the stochastic difference equation algorithm. Chem. Phys. Lett. 378:1–8. [Google Scholar]
- Barth, E., and T. Schlick. 1998a. Overcoming stability limitation in biomolecular dynamics. I. Combining force splitting via extrapolation with Langevin dynamics in LN. J. Chem. Phys. 109:1617–1632. [Google Scholar]
- Barth, E., and T. Schlick. 1998b. Extrapolation versus impulse in multiple-timestepping schemes. II. Linear analysis and applications to Newtonian and Langevin dynamics. J. Chem. Phys. 109:1633–1642. [Google Scholar]
- Beard, W. A., W. P. Osheroff, R. Prasad, M. R. Sawaya, M. Jaju, T. G. Wood, J. Kraut, T. A. Kunkel, and S. H. Wilson. 1996. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase β. J. Biol. Chem. 271:12141–12144. [DOI] [PubMed] [Google Scholar]
- Beard, W. A., D. D. Shock, B. J. Vande Berg, and S. H. Wilson. 2002a. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 49:47393–47398. [DOI] [PubMed] [Google Scholar]
- Beard, W. A., D. D. Shock, X.-P. Yang, S. F. DeLauder, and S. H. Wilson. 2002b. Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 277:8235–8242. [DOI] [PubMed] [Google Scholar]
- Beard, W. A., and S. H. Wilson. 1998. Structural insights into DNA polymerase β fidelity: hold tight if you want it right. Chem. Biol. 5:R7–R13. [DOI] [PubMed] [Google Scholar]
- Beard, W. A., and S. H. Wilson. 2000. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat. Res. 460:231–244. [DOI] [PubMed] [Google Scholar]
- Beard, W. A., and S. H. Wilson. 2003. Structural insights into the origins of DNA polymerase fidelity. Structure. 11:489–496. [DOI] [PubMed] [Google Scholar]
- Beese, L. S., V. Derbyshire, and T. A. Steitz. 1993. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 260:352–355. [DOI] [PubMed] [Google Scholar]
- Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187–217. [Google Scholar]
- Cann, I. K. O., and Y. Ishino. 1999. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 152:1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, K., K. A. Sharp, B. Honig, and A. M. Pyle. 1999. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 6:1055–1061. [DOI] [PubMed] [Google Scholar]
- Dahlberg, M. E., and S. J. Benkovic. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 30:4835–4843. [DOI] [PubMed] [Google Scholar]
- Date, T., S. Yamamoto, K. Tanihara, Y. Nishimoto, and A. Matsukage. 1991. Aspartic acid residues at positions 190 and 192 of rat DNA polymerase β are involved in primer binding. Biochemistry. 30:5286–5292. [DOI] [PubMed] [Google Scholar]
- Delarue, M., O. Poch, N. Tordo, D. Moras, and P. Argos. 1990. An attempt to unify the structure of polymerases. Protein Eng. 3:461–467. [DOI] [PubMed] [Google Scholar]
- Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, Jr., A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284:1095–1111. [DOI] [PubMed] [Google Scholar]
- Doublié, S., and T. Ellenberger. 1998. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 8:704–712. [DOI] [PubMed] [Google Scholar]
- Doublié, S., M. R. Sawaya, and T. Ellenberger. 1999. An open and closed case for all polymerases. Structure. 7:R31–R35. [DOI] [PubMed] [Google Scholar]
- Doublié, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 391:251–258. [DOI] [PubMed] [Google Scholar]
- Echols, H., and M. F. Goodman. 1991. Fidelity mechanism in DNA replication. Annu. Rev. Biochem. 60:477–511. [DOI] [PubMed] [Google Scholar]
- Frey, M. W., L. C. Sowers, D. P. Millar, and S. J. Benkovic. 1995. The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase. Biochemistry. 34:9185–9192. [DOI] [PubMed] [Google Scholar]
- García-Díaz, M., O. Domínguez, L. A. López-Fernández, L. T. de Lera, M. L. Saníger, J. F. Ruiz, M. Párraga, M. J. García-Ortiz, T. Kirchhoff, J. del Mazo, A. Bernad, and L. Blanco. 2000. DNA polymerase lambda (pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 301:851–867. [DOI] [PubMed] [Google Scholar]
- Goodman, M. F. 1997. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. USA. 94:10493–10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 282:1669–1675. [DOI] [PubMed] [Google Scholar]
- Ito, J., and D. K. Braithwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga, A., M. Ouchida, K. Miyazaki, K. Hori, and T. Mukai. 1999. Functional mutation of DNA polymerase β found in human gastric cancer—inability of the base excision repair in vitro. Mutat. Res. 435:121–128. [DOI] [PubMed] [Google Scholar]
- Joyce, C. M., and T. A. Steitz. 1994. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 63:777–822. [DOI] [PubMed] [Google Scholar]
- Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988–25997. [PubMed] [Google Scholar]
- Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structures at 3.5 Å of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 256:1783–1790. [DOI] [PubMed] [Google Scholar]
- Kool, E. T., J. C. Morales, and K. M. Guckian. 2000. Mimicking the structure and function of DNA: insight into DNA stability and replication. Angew. Chem. Int. Ed. 39:990–1009. [DOI] [PubMed] [Google Scholar]
- Kraynov, V. S., B. G. Werneburg, X. Zhong, H. Lee, J. Ahn, and M.-D. Tsai. 1997. DNA polymerase β: analysis of the contributions of tyrosine-271 and asparagine-279 to substrate specificity and fidelity of DNA replication by pre-steady-state kinetics. Biochem. J. 323:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta, R. D., P. Benkovic, and S. J. Benkovic. 1988. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 27:6716–6725. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497–529. [DOI] [PubMed] [Google Scholar]
- Larder, B. A., S. D. Kemp, and D. J. Purifoy. 1989. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc. Natl. Acad. Sci. USA. 86:4803–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder, B. A., D. J. Purifoy, K. L. Powell, and G. Darby. 1987. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 327:716–717. [DOI] [PubMed] [Google Scholar]
- Li, Y., S. Korolev, and G. Waksman. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17:7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, H., F. Boudsocq, R. Woodgate, and W. Yang. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 107:91–102. [DOI] [PubMed] [Google Scholar]
- Maciejewski, M. W., R. Shin, B. Pan, A. Marintchev, A. Denninger, M. A. Mullen, K. Chen, M. R. Gryk, and G. P. Mullen. 2001. Solution structure of a viral DNA repair polymerase. Nat. Struct. Biol. 8:936–941. [DOI] [PubMed] [Google Scholar]
- MacKerell, A. D., Jr., and N. K. Banavali. 2000. All-atom empirical force field for nucleic acids. II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comp. Chem. 21:105–120. [Google Scholar]
- Menge, K. L., Z. Hostomsky, B. R. Nodes, G. O. Hudson, S. Rahmati, E. W. Moomaw, R. J. Almassy, and Z. Hostomska. 1995. Structure-function analysis of the mammalian DNA polymerase β active site: role of aspartic acid 256, arginine 254 and arginine 258 in nucleotidyl transfer. Biochemistry. 34:15934–15942. [DOI] [PubMed] [Google Scholar]
- Ohmori, H., E. C. Friedberg, R. P. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkle, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell. 8:7–8. [DOI] [PubMed] [Google Scholar]
- Oliveros, M., R. J. Yúñez, M. L. Salas, J. Salas, E. Viñuela, and L. Blanco. 1997. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J. Biol. Chem. 272:30899–30910. [DOI] [PubMed] [Google Scholar]
- Ollis, D. L., P. Brick, R. Hamlin, N. G. Xuong, and T. A. Steitz. 1985. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 313:762–766. [DOI] [PubMed] [Google Scholar]
- Osheroff, W. P., W. A. Beard, S. H. Wilson, and T. A. Kunkel. 1999. Base substitution specificity of DNA polymerase β depends on interactions in the DNA minor groove. J. Biol. Chem. 274:20749–20752. [DOI] [PubMed] [Google Scholar]
- Patel, S. S., I. Wong, and K. A. Johnson. 1991. Pre-steady-state kinetic analysis of processive DNA replication inducing complete characterization of an exonuclease-deficient mutant. Biochemistry. 30:511–525. [DOI] [PubMed] [Google Scholar]
- Pelletier, H., M. R. Sawaya, A. Kumar, S. H. Wilson, and J. Kraut. 1994. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 264:1891–1903. [PubMed] [Google Scholar]
- Pelletier, H., M. R. Sawaya, W. Wolfle, S. H. Wilson, and J. Kraut. 1996. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 35:12762–12777. [DOI] [PubMed] [Google Scholar]
- Polesky, A. H., T. A. Steitz, N. D. Grindley, and C. M. Joyce. 1990. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J. Biol. Chem. 265:14579–14591. [PubMed] [Google Scholar]
- Radhakrishnan, R., and T. Schlick. 2003. Orchestration of cooperative events in DNA synthesis and repair mechanism unraveled by transition path sampling of DNA polymerase β's closing. Proc. Natl. Acad. Sci. USA. 101:5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberger, S., and P. Pfeiffer. 1998. Cloning, purification and characterization of DNA polymerase β from Xenopus laevis. Eur. J. Biochem. 251:81–90. [DOI] [PubMed] [Google Scholar]
- Sawaya, M. R., H. Pelletier, A. Kumar, S. H. Wilson, and J. Kraut. 1994. Crystal structures of rat DNA polymerase β: evidence for a common polymerase mechanism. Science. 264:1930–1935. [DOI] [PubMed] [Google Scholar]
- Sawaya, M. R., R. Prasad, S. H. Wilson, J. Kraut, and H. Pelletier. 1997. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 36:11205–11215. [DOI] [PubMed] [Google Scholar]
- Schlick, T. 1992. Optimization methods in computational chemistry. In Reviews in Computational Chemistry, Vol. 3. K. B. Lipkowitz, and D. B. Boyd, editors. VCH Publishers, New York, NY. 1–71.
- Schlick, T. 2001. Time-trimming tricks for dynamic simulations: splitting force updates to reduce computational work. Structure. 9:R45–R53. [DOI] [PubMed] [Google Scholar]
- Schlick, T., E. Barth, and M. Mandziuk. 1997. Biomolecular dynamics at long timesteps: bridging the timescale gap between simulation and experimentation. Annu. Rev. Biophys. Biomol. Struct. 26:181–222. [DOI] [PubMed] [Google Scholar]
- Sharp, K. A., B. Honig, and S. C. Harvey. 1990. Electrical potential of transfer RNAs: codon-anticodon recognition. Biochemistry. 29:340–346. [DOI] [PubMed] [Google Scholar]
- Showalter, A. K., I. L. Byeon, M. Su, and M. Tsai. 2001. Solution structure of a viral DNA polymerase X and evidence for a mutagenic function. Nat. Struct. Biol. 8:942–946. [DOI] [PubMed] [Google Scholar]
- Singh, K., and M. J. Modak. 1998. A unified DNA- and dNTP-binding mode for DNA polymerases. Trends Biochem. Sci. 23:277–281. [DOI] [PubMed] [Google Scholar]
- Steitz, T. A. 1993. DNA- and RNA-dependent DNA polymerases. Curr. Opin. Struct. Biol. 3:31–38. [Google Scholar]
- Vande Berg, B. J., W. A. Beard, and S. H. Wilson. 2001. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β. J. Biol. Chem. 276:3408–3416. [DOI] [PubMed] [Google Scholar]
- Wang, J., A. K. M. A. Sattar, C. C. Wang, J. D. Karam, W. H. Konigsberg, and T. A. Steitz. 1997. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 89:1087–1099. [DOI] [PubMed] [Google Scholar]
- Werneburg, B. G., J. Ahn, X. Zhong, R. J. Hondal, V. S. Kraynov, and M.-D. Tsai. 1996. DNA polymerase β: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry. 35:7041–7050. [DOI] [PubMed] [Google Scholar]
- Wilson, S. H. 1998. Mammalian base excision repair and DNA polymerase β. Mutat. Res. 407:203–215. [DOI] [PubMed] [Google Scholar]
- Yang, L., W. A. Beard, S. H. Wilson, S. Broyde, and T. Schlick. 2002a. Polymerase β simulations suggest that Arg258 rotation is a slow step rather than large subdomain motion per se. J. Mol. Biol. 317:651–671. [DOI] [PubMed] [Google Scholar]
- Yang, L., W. A. Beard, S. H. Wilson, B. Roux, S. Broyde, and T. Schlick. 2002b. Local deformations revealed by dynamics simulations of DNA polymerase β with DNA mismatches at the primer terminus. J. Mol. Biol. 321:459–478. [DOI] [PubMed] [Google Scholar]
- Zhong, X., S. S. Patel, and M.-D. Tsai. 1998. DNA polymerase β. IV. Dissecting the functional roles of the two metal ions with CrIIIdTTP. J. Am. Chem. Soc. 120:235–236. [Google Scholar]
- Zhong, X., S. S. Patel, B. G. Werneburg, and M.-D. Tsai. 1997. DNA polymerase β: multiple conformational changes in the mechanism of catalysis. Biochemistry. 36:11891–11900. [DOI] [PubMed] [Google Scholar]