Abstract
Based on a homology model of the Kv1.3 potassium channel, the recognitions of the six scorpion toxins, viz. agitoxin2, charybdotoxin, kaliotoxin, margatoxin, noxiustoxin, and Pandinus toxin, to the human Kv1.3 potassium channel have been investigated by using an approach of the Brownian dynamics (BD) simulation integrating molecular dynamics (MD) simulation. Reasonable three-dimensional structures of the toxin-channel complexes have been obtained employing BD simulations and triplet contact analyses. All of the available structures of the six scorpion toxins in the Research Collaboratory for Structural Bioinformatics Protein Data Bank determined by NMR were considered during the simulation, which indicated that the conformations of the toxin significantly affect both the molecular recognition and binding energy between the two proteins. BD simulations predicted that all the six scorpion toxins in this study use their β-sheets to bind to the extracellular entryway of the Kv1.3 channel, which is in line with the primary clues from the electrostatic interaction calculations and mutagenesis results. Additionally, the electrostatic interaction energies between the toxins and Kv1.3 channel correlate well with the binding affinities (−logKds), R2 = 0.603, suggesting that the electrostatic interaction is a dominant component for toxin-channel binding specificity. Most importantly, recognition residues and interaction contacts for the binding were identified. Lys-27 or Lys-28, residues Arg-24 or Arg-25 in the separate six toxins, and residues Tyr-400, Asp-402, His-404, Asp-386, and Gly-380 in each subunit of the Kv1.3 potassium channel, are the key residues for the toxin-channel recognitions. This is in agreement with the mutation results. MD simulations lasting 5 ns for the individual proteins and the toxin-channel complexes in a solvated lipid bilayer environment confirmed that the toxins are flexible and the channel is not flexible in the binding. The consistency between the results of the simulations and the experimental data indicated that our three-dimensional models of the toxin-channel complex are reasonable and can be used as a guide for future biological studies, such as the rational design of the blocking agents of the Kv1.3 channel and mutagenesis in both toxins and the Kv1.3 channel. Moreover, the simulation result demonstrates that the electrostatic interaction energies combined with the distribution frequencies from BD simulations might be used as criteria in ranking the binding configuration of a scorpion toxin to the Kv1.3 channel.
INTRODUCTION
Potassium channels are a ubiquitous family of membrane proteins that play critical roles in a wide variety of physiological processes, such as the regulation of muscle contraction, cell proliferation, insulin secretion, neuronal excitability (Wrisch and Grissmer, 2000; Cahalan et al., 2001; Kaczorowski and Garcia, 1999; Terlau and Stühmer, 1998). Increasing efforts are devoted to the structural and functional characterization of K+ channels. The best characterized K+ channels are derived from homologs of the Drosophila K+ channel genes including Shaker, Shab, Shaw, and Shal, which exist in mammalian tissues and were assigned as Kv1, Kv2, Kv3, and Kv4, respectively (Wrisch and Grissmer, 2000; Biggin et al., 2000; Lu et al., 2001; Zhou et al., 2001b). Each type of Kv channels may be divided into different subtypes (Wrisch and Grissmer, 2000). For the Kv1 channels, the voltage-gated potassium channel, Kv1.3, plays an important role in T-cell physiology because of its involvement in the regulation of membrane potential of cells, thereby influencing T-cell activation in vitro (Chandy et al., 1984; DeCoursey et al., 1984; Gulbis et al., 2000; Lewis and Cahalan, 1988). Immunosuppressants are used in the prevention of graft rejection following organ transplants, and in the management of autoimmune diseases. Clinically useful immunosuppressants might be developed by selectively targeting lymphocyte potassium channels (Beeton et al., 2001; Chandy et al., 2001; Wickenden, 2002). Immunosuppression via highly selective blockade of the T-cell potassium channels is attractive due to the functionally restricted tissue distribution of Kv1.3, its important role in T-cell activation, and the availability of specific and potent inhibitors of the channel. Accordingly, human Kv1.3 has been suggested to be an excellent target for modulating immune system functions by specific and potent blockers in the therapeutic management of stroke, epilepsy, and cardiac arrhythmias (Chandy et al., 1984; DeCoursey et al., 1984; Freudenthaler et al., 2002; Lewis and Cahalan, 1988). Therefore, it is of significance to study the action of drugs or other ligands (e.g., toxins) to these Kv1 channels.
Scorpion venoms are a rich source of ion channel modulators. The scorpion toxins have been useful molecules in probing the potassium channel structures and functions (Aiyar et al., 1996; Doyle et al., 1998; MacKinnon et al., 1988; Mourre et al., 1999; Peter et al., 2001; Thompson and Begenisich, 2000). To date, a variety of experimental strategies have defined functional domains within the Kv1 channels, and some thermodynamic mutant cycle analyses have been used to identify the specific residues in the S5–S6 linker region, which is a part of the scorpion-toxin receptor site. The homotetrameric arrangement of voltage-gated potassium channels was first revealed by the blocking analyses of charybdotoxin (ChTX, from Leiurus quinquestriatus) to wild-type and mutant channels (MacKinnon, 1991). Many potent polypeptide inhibitors of the lymphocyte potassium channels have been discovered in scorpion venom (Alessandri-Haber et al., 1999; Blanc et al., 1998; Garcia-Calvo et al., 1993; Gilquin et al., 2002; Goldstein et al., 1994). ChTX was the first polypeptide shown to block all Kv channels, inhibiting Kv1.3 with nanomolar affinity. Other scorpion toxins include noxiustoxin, kaliotoxin, margatoxin, agitoxin-2,hongotoxin, HsTx1, maurotoxin, and Pandinus toxins 1–3 (Pi1, Pi2, and Pi3) (Bontems et al., 1992; Fernandez et al., 1994; Garcia-Calvo et al., 1993; Goldstein et al., 1994; Krezel et al., 1995). In general, channel inhibitors can be pore blockers or gating modifiers (Goldstein et al., 1994; Harvey et al., 1995; Miller, 1995). Pore blockers bind to the channel in 1:1 stoichiometry and plug the pore of the channel impeding the flow of the ionic current. These toxins are small proteins that block the passage of K+ ions by binding at the pore entryway on the extracellular side of the channel, thereby inhibiting the ion flux. The interactions of toxins with potassium channels are among the strongest and most specific known in protein-protein complexes (MacKinnon et al., 1988).
However, many questions are still unresolved because of experimental difficulties and the lack of significant theoretical guidance. All drugs marketed that act on ion channels were discovered empirically rather than by molecular insight, and most of them have shown serious problems of safety and efficacy (Goldstein and Colatsky, 1996; Kaczorowski and Garcia, 1999). Therefore, computational simulation at the molecular level is a powerful tool in understanding electrophysiological experiments performed on wild-type and mutant channels. Our interest in the blockage mechanism of Kv1 channels stems from our efforts to design new ion channel blockers, with the eventual aim to develop new drugs for the treatment of diseases affecting both electrically excitable and nonexcitable tissues (Liu et al., 2003; Shen et al., 2003). However, no experimental data for the structure of scorpion toxins-Kv1.3 channel complexes have been reported.
In this study, a robust approach, integrating homology modeling, Brownian dynamics (BD), and long-time molecular dynamics (MD) simulations, has been employed for studying the association of scorpion toxins to the Kv1.3 channel. First, we constructed the three-dimensional (3D) structure model for the Kv1.3 potassium channel via homology modeling, taking the x-ray crystal structure of the KcsA potassium channel as a template. Then the docking feature of BD simulations (Cui et al., 2001, 2002; Fu et al., 2002; Northrup et al., 1999) and the structural refinement functionality of molecular mechanics were employed to localize the regions of binding, to identify the residues involved in complex formation, and to estimate the binding strength of the Kv1.3 channel with six scorpion toxins, viz. agitoxin2 from Leiurus quinquestriatus hebraeus (AgTX2), charybdotoxin from Leiurus quinquestriatus hebraeus (ChTX), kaliotoxin from Androctonus mauretanicus (KTX), margatoxin from Centruroides margaritatus (MgTX), noxiustoxin from Centruroides noxius (NTX), and Pandinus toxin 2 from Pandinus imperator (Pi2). Long-time MD simulations take the advantage of iteratively tracking the trajectory of conformational change, and therefore may capture the flexibilities of toxins and Kv1.3 channel during binding. So after the docked channel-toxin complexes were obtained, long-time MD simulations were carried out for the complexes embedded in the solvated palmitoyloleoylphosphatidylcholine (POPC) lipid bilayer.
SIMULATION MODELS AND METHODS
Models
The atomic coordinates of the six scorpion toxins, AgTX2 (Krezel et al., 1995), ChTX (Bontems et al., 1992), KTX (Fernandez et al., 1994), MgTX (Johnson et al., 1994), NTX (Dauplais et al., 1995), and Pi2 (Tenenholz et al., 1997), were obtained from the Brookhaven Protein Data Bank (PDB); their PDB entries are 1AGT, 1KTX, 1SXM, 1MTX, 2CRD, and 2PTA, respectively. The scorpion toxins have been previously divided into four subfamilies (Miller, 1995). Three of these subfamilies, including MgTX, NTX, and Pi2 toxins, possess the same Lys-27–Tyr-36 diad as in ChTX. Toxins forming the fourth subfamily, including KTX and AgTX2, have a threonine at position 36, and these two toxins uniquely possess a phenylalanine at position 24 or 25, located on the outer face of the β-sheet.
For the K+ channels, only the crystal structures of KcsA channel from Streptomyces lividans (PDB entry 1BL8) (Doyle et al., 1998; Zhou et al., 2001a) and the MthK channel from Methanobacterium thermoautotrophicum (PDB entry 1LNQ) (Jiang et al., 2002a,b) are available. Since currently the crystal structure of the human potassium channel Kv1.3 has not been determined, the three-dimensional model of the Kv1.3 channel was obtained by using the homology modeling based on the KcsA crystal structure. The eukaryotic structure of the voltage-dependent potassium channels shares a remarkable structure conservation of the channel pore with the prokaryotic potassium channel KcsA (Aiyar et al., 1996; Doyle et al., 1998; Biggin et al., 2000; Wrisch and Grissmer, 2000). Although the subunits of potassium channel KcsA contain only two transmembrane segments rather than the six transmembrane segments of the voltage-gated potassium channel, the amino acid sequences of these two proteins are very similar, especially their sequences in the pore region and extracellular entryway. Therefore, the x-ray structure of the KcsA channel was used as a template for constructing a 3D structural model of the Kv1.3.
Residues Arg-27, Ile-60, Arg-64, Glu-71, and Arg-117, missing from the current KcsA x-ray crystal structure, were added with the Biopolymer module encoded in Sybyl Release 6.8 (Tripos, St. Louis, MO). The 3D structural model of Kv1.3 channel was generated employing the Homology module of Insight II (Molecular Simulation, San Diego, CA) based on the corrected KcsA structure. The sequence alignment of KcsA (1BL8) with the Kv1.3 channel generated by CLUSTAL W (1.81) (Thompson et al., 1994) shows that the sequence identity between the two channels is 30.93%, and the similarity is 62.89% Fig. 1). The modeled side chains of the Kv1.3 channel were subjected to energy refinement using the adopted-basis Newton Raphson algorithm and the CHARMm22 force field as implemented in the Quanta program (Quanta Release 98, Molecular Simulations, San Diego, CA) to relieve possible steric clashes and overlaps. During the structure refinement the gradient tolerance was achieved to 0.05 kcal/(mol Å), and a distance-dependent dielectric constant of 4 was used to simulate the effect of solvent.
FIGURE 1.
Sequence alignments of of KcsA (1BL8) channel with the Kv1.3 channel generated by CLUSTAL W (Thompson et al., 1994). In the sequences, an asterisk indicates an identical or conserved residue, a colon indicates a conserved substitution, and a dot indicates a semiconserved substitution.
A model of POPC lipid bilayer was downloaded from http://moose.bio.ucalgary.ca/ (Tieleman et al., 1999). An equilibrated (by 1-ns NPT MD simulation) POPC lipid bilayer comprised of 128 lipid molecules was used as the starting point, with which a lipid bilayer consisting of 512 lipid molecules was constructed by extending the 128-lipid model in the xy plane two times aligned to the ±z axis. After 1 ns NPT MD simulation, the 512-lipid bilayer system was cut to an 80 × 80-nm square consisting of 251 lipid molecules, into which the Kv1.3 model was embedded.
Brownian dynamics simulations
The program package MacroDox version 3.2.2 (Northrup et al., 1999) was used to assign the titratable residues on proteins, solve the linearized Poisson-Boltzmann (PB) equation, and run the various Brownian dynamics simulations for the association between the scorpion toxins and the Kv1.3 channel. The BD algorithm for this program has been described in detail by Northrup et al. (1987, 1993, 1999). The CHARMm22 force field (MacKerell et al., 1998; Neria et al., 1996), which includes the charges of nonstandard residues such as pyroglutamic acid (PCA), was used to assign the charges of the Kv1.3 channel and the various scorpion toxins. The surface-accessibility-modified Tanford-Kirkwood (TK) method of Matthew (Matthew, 1985; Matthew and Gurd, 1986) produces results similar to those obtained with the Monte Carlo sampling and Hybrid Tanford/Roxby mean field methods encoded in the University of Houston Brownian Dynamics program (Davis et al., 1991; Madura et al., 1995) for the toxin-K+ channel interactions (Fu et al., 2002). Moreover, the main purpose of BD simulations in this study was to find reasonable binding configurations between the scorpion toxins and the Kv1.3 channel rather than to accurately calculate the binding constants. So the TK method encoded in MacroDox version 3.2.2 was employed to determine the protonation status of each titratable residue in the two proteins during the BD simulation at pH 7.0 and ionic strength 0.1 M. The six scorpion toxins all have three disulfide bonds, so the charges of the sulfur atoms of the cysteines involved in the disulfide bonds, i.e., Cys-18, Cys-14, Cys-18, Cys-28, Cys-33, and Cys-35 in AgTX2 and KTX; Cys-7, Cys-13, Cys-17, Cys-28, Cys-33, and Cys-35 in ChTX and Pi2; Cys-7, Cys-13, Cys-17, Cys-29, Cys-34, and Cys-36 in MgTX and NTX, were all zeroed out during the charge assignment. Although this test charge model was not as accurate as that of the effective charge method, it has been demonstrated in our previous work that the test charge model produced reasonable results for the interaction of protein-protein binding (Cui et al., 2001, 2002; Fu et al., 2002). Therefore, it was applied again in the present study. The TK-recommended partial charges were assigned to both the Kv1.3 channel and the scorpion toxins. The total charge is −10.896 e for the Kv1.3 channel. TK calculations indicated that the total charges of the toxin varied slightly from different conformations. It can be estimated that the total charges bared by AgTX are from 5.91 to 5.99 e, by ChTX are from 5.94 to 6.02 e, by KTX are from 4.94 to. 4.99 e, by MgTX are from 5.94 to 6.03 e, by NTX are from 6.89 to 7.02 e, and by Pi2 are from 6.96 to 7.08 e. After charge assignments, the electrostatic potentials of the Kv1.3 channel and the scorpion toxins were determined by numerically solving the linearized PB equation. Taking the above assigned charges as initial values, the PB equation was solved by the Warwicker-Watson method implemented in the MacroDox program. The protein interior dielectric constant and solvent dielectric constant were 4.0 and 78.3, respectively.
The Ermak-McCammon algorithm (Ermak and McCammon, 1978) was used to simulate the translational Brownian motion of two interacting proteins as the displacements Δr of the relative separation vector r between the centroids of the two proteins in a time step Δt according to the relation:
 |
(1) |
where D is the translational diffusion coefficient for the relative motion and assumed to be spatially isotropic; F is the systematic interparticle force, which is computed from the electrostatic field calculated before Brownian dynamics simulations; KBT is the Boltzmann constant times absolute temperature; and S is the stochastic component of the displacement arising from collisions of proteins with solvent molecules, which is generated by taking normally distributed random numbers obeying the relationship:
 |
(2) |
A similar equation governs the independent rotational Brownian motion of each particle, in which the force is replaced by a torque and D is replaced by an isotropic rotational diffusion coefficient Dir for each particle i.
Then BD simulations of various scorpion toxins binding to the Kv1.3 potassium channel were performed to identify the favorable complexes at pH 7.0 and ionic strength 0.1 M. For Brownian dynamics simulations of protein-protein interactions, the two proteins were treated as rigid bodies. The time step in the simulation was set at a default value of 10 ps. A spatial exclusion grid was consulted for every atom of the incoming molecule to avoid overlaps. The charges of the scorpion toxin were distributed onto the electrostatic potential grid of the Kv1.3 potassium channel; then the electrostatic interaction energies between the Kv1.3 potassium channel and the scorpion toxin were calculated by summing over the product of all of the above-assigned charges of the scorpion toxin and the corresponding electrostatic potential values generated by the Kv1.3 potassium channel. Trajectories were started with the scorpion toxin at a random position and orientation on the “b-surface” (a sphere with a radius of b; b = 76 Å in this study) centered on the Kv1.3 channel at which the forces due to the Kv1.3 channel are centrosymmetric. The center of mass of the scorpion toxin and the position of the oxygen atom of a water molecule in the selection filter of the x-ray crystal structure of KcsA were chosen to monitor the association during the BD simulations. The closest approach of the mobile scorpion toxin to the fixed receptor Kv1.3 potassium channel was recorded, and the trajectory was terminated when the mobile ligand escaped the “q-surface” (a sphere with a radius of q; q = 200 Å in this study).
All 17 NMR-determined structures of scorpion toxin AgTX2 in PDB entry 1AGT, 12 structures of ChTX in 2CRD, 11 structures of KTX in 1KTX, 23 structures of MgTX in 1MTX, 39 structures of NTX in 1SXM, and 20 structures of Pi2 in 2PTA, were docked with the Kv1.3 potassium channel, respectively, typically by running 10,000 trajectories for each Brownian dynamics simulation. The statistical analyses resulted in the number of occurrences for which each toxin amino acid residue formed intermolecular contacts to the channel in the complexes. Also, the occurrence frequencies of the intermolecular contacts between the key amino acid residues of the two proteins were obtained from the BD trajectories.
Molecular dynamics simulations
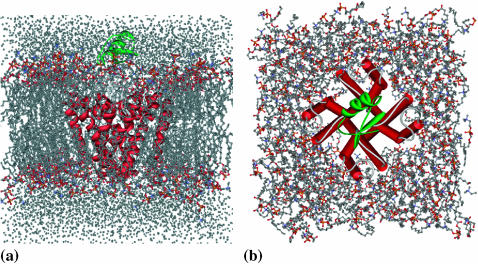
To measure the flexibility of the Kv1.3 channel in the environment of membrane, 5-ns MD simulations were performed on the Kv1.3 channel and its complexes with the scorpion toxins derived from the BD simulations. For each toxin-channel complex, the encounter configuration of the two proteins with lowest electrostatic interaction energy was used as the starting structure for MD simulation. Before MD simulations, the Kv1.3 channel or Kv1.3-toxin complex was incorporated into the POPC lipid bilayer model using the approach of Faraldo-Gomez et al. (2002). A hole was created on the POPC lipid bilayer model according to the surface shape of the Kv1.3 channel or its toxin complex, and then the Kv1.3 channel or its toxin complex was inserted into the hole. The lipid bilayer normally is oriented along the z axis, and the center of the bilayer is at the plane of z = 0. Kv1.3 was oriented with the pore along the z axis and the selectivity filter located near the upper layer of the membrane. Then the superstratum and underlayer of the protein-embedded lipid bilayer were respectively solvated by the SPC (Hermans et al., 1984) water model (Fig. 2, a and b), because the SPC model has been applied well in simulating lipid bilayer-water systems (Tieleman and Berendsen, 1998; Tieleman et al., 1999). The systems for MD simulations contain >40,000 atoms.
FIGURE 2.
Molecular graphics side views of the scorpion toxin AgTX and Kv1.3 potassium channel complex embedded in a POPC membrane solvated by water. These snapshots were taken near the beginning of the trajectory. (a) Side view of the system. (b) Top view of the system with water molecules hidden. The toxin was colored by cyan.
Molecular dynamics simulations were carried out using GROMACS version 3.14 (http://www.gromacs.org) (Berendsen et al., 1995; Lindahl et al., 2001) with GROMOS-96 force field (Lindahl et al., 2001; van Gunsteren et al., 1996). We used NPT conditions (i.e., constant number of particles, pressure, and temperature) in the simulation. A production simulation of 5 ns was run for the whole system after 1000 ps equilibration. The time step was 2 fs, with the LINCS algorithm (Hess et al., 1997) used to constrain bond lengths, and trajectory data were collected every 50 steps (0.1 ps). A constant pressure of 1 bar independently in all three directions was used, with a coupling constant of τp =1.0 ps. This allows the bilayer/protein area to adjust to its optimum value for the force field employed. Water, lipid, and protein were coupled separately to a temperature bath at 300 K, using a coupling constant τT = 0.1 ps. Particle-mesh Ewald method (Darden et al., 1993; Essmann et al., 1995) was used to calculate the long-range electrostatic interactions. Cutoff used for longer-range interactions was 0.9 nm for both the van der Waals interactions and the electrostatic interactions.
After the molecular dynamics simulation, energy minimization was again performed on the complexes to obtain the final binding complex structures. The details of the interaction of the scorpion toxins and the Kv1.3 potassium channel were analyzed using the LIGPLOT program (Wallace et al., 1995).
All calculations were carried out on a 64-CPU Silicon Graphics (Mountainview, CA) Origin3800 server and an O2 workstation.
RESULTS AND DISCUSSION
Electrostatic potentials
The appearances of the electrostatic potentials on the surface of the Kv1.3 channel protein and the scorpion toxins generated by GRASP (Nicholls et al., 1991) are given in Fig. 3. The potential maps were calculated with a simplified Poisson-Boltzmann solver, as implemented and parameterized in Amber (Pearlman et al., 1995). As expected, the mouth of the Kv1.3 channel, which is located outside of the cell membrane, bears a large negative electrostatic potential that is centrosymmetric around the central axis of the Kv1.3 channel. The surfaces of the toxins, on the contrary, have large positive electrostatic potentials mainly formed by the side chains of positively charged residues such as arginine and lysine. This suggests that the toxins may associate with the entryway of the Kv1.3 channel using their positive patches. This conclusion was validated by the optimized toxin-channel complexes (see discussion below). The resultant dipole moment of Kv1.3 is oriented along the symmetry axis from the outer side to the inner side of the membrane. It primarily determines the orientations for the scorpion toxins binding to the channel. The formal charge on the Kv1.3 potassium channel is −12 e, and the formal charges on the toxins are 5 e, 6 e, 4 e, 6 e, 8 e, and 8 e for AgTX2, ChTX, KTX, MgTX, NTX, and Pi2, respectively. All the six toxins have one or two acidic residues in the region or near the region of the α-helix, and at least four basic residues in the three-stranded β-sheet, thus forming a dipole moment approximately in the direction from the α-helix region to the β-sheet region (Fig. 3). The orientation of the toxin is such that the side chain of a positively charged residue in a toxin probably protrudes into the Kv1.3 pore, interacting electrostatically with acidic residues (e.g., Asp-402) in the ion selective filter. Both the electrostatic potentials and dipoles show that the charge anisotropy is the driving force of the association of toxins to the Kv1.3 channel from the viewpoint of Coulombic interactions.
FIGURE 3.
Electrostatic potential contour maps for the Kv1.3 channel and the six scorpion toxins at an ionic strength of 0.1 M. The red contours represent isopotential surfaces where charge 1 e possesses electrostatic potential energy equal to −2.5 kT; the blue isopotential surfaces are for energy +2.5 kT. Arrows indicate the directions of the dipoles in the proteins. The picture was generated with the program GRASP (Nicholls et al., 1991).
BD-docked toxin-channel complexes
In the current BD simulation protocols, the proteins were treated as rigid bodies for simplicity, so the flexibility of the proteins was not considered. To overcome this shortcoming, we considered all of the available NMR conformations of the six scorpion toxins in solution when performing the BD simulations. For the Kv1.3 channel, because it is embedded in the membrane of the cell, it should not be substantially flexible (see the MD results below). In our previous BD studies (Cui et al., 2001, 2002; Fu et al., 2002), the simulations were run at an evaluated temperature, 500 K, to save simulation time. Higher temperature causes larger stochastic force and hence faster movement (longer distance) of the proteins per step, and may cause incomplete sampling of the association trajectory (docking conformational space). Thus, the current BD simulations were run at 300 K, to ensure that the docking procedure was able to explore extensively over all orientations of the toxin bound to the extracellular surface of the channel. Though computational cost was about 10 times longer than before, more encounter complexes were formed along with the BD trajectory. A typical BD trajectory lasted from 800 to 2000 ns, and the longest trajectory lasted >5000 ns.
The center of mass of the scorpion toxins and the position of the oxygen atom of a water molecule in the selection filter of the x-ray crystal structure of KcsA were chosen to monitor the association during the BD simulations. To avoid unfavorable complexes formed between two protein partners, the separation criterion defining an encounter complex was set to 30 Å. The distribution of the docked solutions of the two binding proteins is highly dependent on the distance criterion. The larger the distance used, the broader the distribution is. We used a large distance criterion to obtain more information about possible docking orientations from the BD simulations. This distance of 30 Å is large enough to get the most significant complexes from the simulations, as will be shown later.
To obtain favorable toxin-Kv1.3 channel complexes, we performed a detailed triplet contact analysis for each structure of the scorpion toxins interacting with the Kv1.3 channel. The most favorable triplet contacts in all complexes, satisfying the criterion of association (the distance between the two proteins is less than 30 Å), were obtained. We then analyzed the favorable triplet pairs between the scorpion toxins and the Kv1.3 channel using a new triplet contact distance, 5.5 Å. Among all the NMR conformations for each toxin, the top two structures that docked most favorably with the Kv1.3 channel, i.e., favorable clusters with lowest average electrostatic interaction energies and highest occurrence of the successfully docked complexes, were selected for further analyses. The closest triplet contacts for the top two structures of each scorpion toxin are listed in Table 1, indicating that the 7th and 13th NMR structures of AgTX2, the 1st and 6th structures of ChTX, the 4th and 11th structures of KTX, the 5th and 12th structures of MgTX, the 20th and 23rd structures of NTX, and the 3rd and 12th structures of Pi2 docked most favorably to the Kv1.3 channel.
TABLE 1.
Possible triplet contacts between structures of scorpion toxins and Kv1.3 channel at an ionic strength of 0.1 M
| Toxins | Structures | Triplets (Kv1.3 channel-toxin residues) | Eelec | F | ||
|---|---|---|---|---|---|---|
| AgTX2 | 7 | D402(A)-K32 | D402(D)-K27 | D386(A)-R31 | −9.11 | 162 |
| D402(D)-K27 | D402(B)-G1 | D386(A)-R31 | −9.09 | 169 | ||
| D402(A)-K32 | D402(D)-K27 | D402(B)-G1 | −9.04 | 186 | ||
| D402(B)-K32 | D402(C)-G1 | D386(B)-R31 | −9.01 | 120 | ||
| D402(A)-G1 | D402(B)-K27 | D386(B)-R31 | −9.69 | 155 | ||
| 13 | D402(A)-R24 | D402(D)-K27 | D376(D)-K19 | −9.08 | 306 | |
| D402(A)-K27 | D402(D)-G1 | D386(A)-R24 | −9.11 | 287 | ||
| D402(D)-K27 | D376(D)-K19 | D386(A)-K16 | −9.24 | 291 | ||
| D402(B)-R24 | D402(C)-G1 | D376(D)-R31 | −9.28 | 102 | ||
| ChTX | 1 | D402(C)-R25 | H404(A)-S37 | D386(B)-K11 | −8.25 | 263 |
| D386(A)-R34 | D402(C)-R25 | H404(A)-S37 | −8.23 | 216 | ||
| D402(C)-K27 | D402(B)-R25 | D402(D)-K11 | −9.05 | 149 | ||
| D402(A)-R34 | H404(A)-K27 | D386(B)-K31 | −8.29 | 254 | ||
| D402(C)-R34 | D402(B)-R25 | H404(C)-S37 | −9.06 | 281 | ||
| D402(A)-K27 | D402(C)-R25 | H404(A)-S37 | −8.23 | 210 | ||
| 6 | D402(A)-R34 | H404(B)-S37 | D386(A)-K31 | −8.58 | 187 | |
| D402(B)-Q1 | H404(B)-S37 | D376(A)-K32 | −8.77 | 139 | ||
| D402(A)-R34 | D402(B)-R25 | H404(B)-S37 | −8.41 | 174 | ||
| KTX | 4 | D402(A)-R24 | D402(D)-K27 | D386(A)-K7 | −10.92 | 455 |
| D402(B)-K16 | D402(D)-K27 | D386(A)-K7 | −10.90 | 393 | ||
| D402(A)-K16 | D402(B)-K7 | H404(C)-K27 | −10.47 | 331 | ||
| D402(A)-R24 | D402(B)-K16 | D386(A)-K7 | −10.92 | 256 | ||
| D402(A)-R24 | D402(B)-K16 | D402(D)-K27 | −10.92 | 255 | ||
| D402(A)-K32 | D402(B)- K7 | D402(C)-K27 | −10.44 | 241 | ||
| D402(A)-K27 | D402(D)-K16 | D402(C)-K7 | −10.45 | 486 | ||
| 11 | D402(A)-R24 | D402(B)-K16 | D402(D)-R31 | −11.23 | 452 | |
| D402(A)-R31 | D402(D)-K16 | D402(C)-R24 | −10.96 | 393 | ||
| D402(A)-R24 | D402(D)-R31 | D376(D)-K7 | −11.29 | 376 | ||
| D402(A)-K16 | D402(B)-K7 | D402(C)-R31 | −10.89 | 365 | ||
| MgTX | 5 | D402(A)-K18 | D402(D)-K6 | D376(D)-K11 | −10.22 | 283 |
| D402(D)-K18 | D402(B)-K6 | D376(B)-K11 | −10.07 | 199 | ||
| D402(A)-K11 | D402(D)-K6 | D376(B)-K33 | −10.27 | 146 | ||
| D402(A)-K28 | D402(D)-K35 | D402(C)-K11 | −10.85 | 102 | ||
| D402(A)-K35 | D402(C)-K28 | D402(B)-K11 | −10.32 | 186 | ||
| 12 | D402(A)-K18 | D402(D)-K11 | D402(B)-K6 | −11.49 | 275 | |
| D402(D)-K18 | D402(C)-K6 | D402(B)-K11 | −10.81 | 129 | ||
| D402(A)-K6 | D402(C)-K11 | D402(B)-K18 | −11.15 | 206 | ||
| D402(A)-K11 | D402(C)-K18 | D386(D)-K6 | −10.64 | 146 | ||
| D402(C)-K11 | D402(B)-K18 | D386(A)-K6 | −10.15 | 206 | ||
| NTX | 20 | D402(A)-K35 | D402(C)-K28 | D402(B)-K11 | −9.36 | 467 |
| D402(C)-K35 | D402(B)-K28 | D402(D)-K11 | −9.22 | 420 | ||
| D402(A)-K28 | D402(C)-K11 | D402(D)-K35 | −8.55 | 312 | ||
| D402(A)-K35 | D402(C)-K28 | D376(C)-K18 | −8.48 | 164 | ||
| D402(A)-K11 | D402(B)-K35 | D402(D)-K28 | −8.52 | 327 | ||
| 23 | D402(A)-K35 | D402(B)-K11 | D402(C)-K28 | −9.19 | 473 | |
| D402(C)-K35 | D402(B)-K28 | D402(D)-K11 | −8.33 | 367 | ||
| D402(A)-K11 | D402(B)-K35 | D402(D)-K28 | −8.41 | 235 | ||
| D402(A)-K35 | D402(B)-K28 | D402(D)-K11 | −8.42 | 209 | ||
| D402(B)-K28 | D402(C)-K35 | D402(D)-K11 | −8.33 | 167 | ||
| Pi2 | 3 | D402(D)-T4 | D402(C)-K11 | D386(B)-K19 | −12.68 | 386 |
| D402(B)-K18 | D402(D)-T4 | D386(B)-K19 | −12.65 | 253 | ||
| D402(A)-R31 | D402(D)-T4 | D386(B)-K19 | −12.60 | 338 | ||
| D402(B)-K18 | D402(C)-K11 | D386(B)-K19 | −12.59 | 241 | ||
| D402(B)-K18 | D402(A)-R31 | D386(B)-K19 | −12.62 | 282 | ||
| D402(B)-K18 | D402(A)-R31 | D402(D)-T4 | −12.65 | 253 | ||
| D402(B)-K18 | D402(A)-R31 | D402(C)-K11 | −12.65 | 213 | ||
| 12 | D402(B)-K19 | D402(C)-K11 | D386(B)-K18 | −12.37 | 499 | |
| D402(B)-T4 | D402(C)-K34 | D386(A)-K38 | −13.58 | 265 | ||
| D402(B)-K19 | D402(C)-K11 | D386(A)-R31 | −12.59 | 223 | ||
| D402(C)-K11 | D386(B)-K18 | D386(A)-R31 | −12.61 | 444 | ||
| D402(C)-K19 | H404(A)-E20 | D386(B)-K38 | −13.30 | 429 | ||
| D402(B)-K19 | D402(A)-R31 | D386(B)-K18 | −12.56 | 465 | ||
| D402(B)-K19 | D402(C)-K11 | H404(B)-E20 | −12.37 | 238 | ||
| D402(B)-K19 | D402(A)-R31 | H404(B)-E20 | −12.28 | 256 | ||
Eelec, average electrostatic interaction energy between the two proteins (kcal/mol); F, occurrence of successfully docked complexes out of 10,000 attempts.
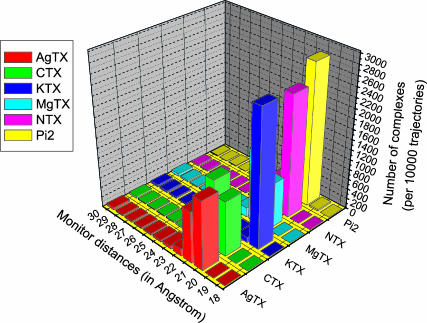
Complexes between a scorpion toxin structure and the Kv1.3 channel in which a set of selected monitor atoms in both proteins are all within 30 Å were identified as favorable complexes. Clusters of similarly docked complexes between the toxin structures and the Kv1.3 channel were identified. The highest populated clusters are the most favorably docked complexes as listed in Table 1. The distributions of the distances between the scorpion toxins and the Kv1.3 channel, i.e., the population of clusters, are shown in Fig. 4, indicating that the largest distributions are the ones in which the proteins are at the distances between 19 Å and 22 Å. This finding supports the selected interaction criterion for the formation of encounter complexes between the scorpion toxins and the potassium channel.
FIGURE 4.
Distribution of distances between the two monitors for all complexes of the most favorably docked structures of the scorpion toxins associating with the Kv1.3 channel. The monitor distances <30 Å were recorded.
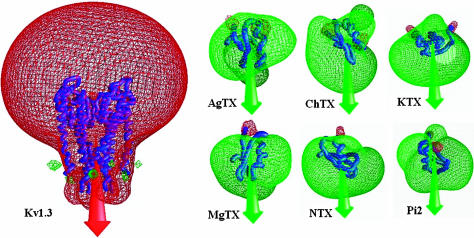
Using BD simulations followed by a modified triplet contact analysis, we identified the favorable complexes formed between the scorpion toxins and the Kv1.3 channel. The BD trajectories of the most favorably docked structures of the scorpion toxins gave >3000 candidate complexes in 10,000 BD trajectories. The distribution of the centers of mass of AgTX2 around the Kv1.3 channel is presented in Fig. 5 as an example, which shows that AgTX2 is located at the extracellular entryway of the Kv1.3 channel. Also, the residues of toxins occurring most frequently in the triplet contacts of the favorable complexes formed between the scorpion toxins and the Kv1.3 channel in the 10,000 BD trajectories are listed in Table 2. From Tables 1 and 2 it can be noticed that Lys-27 (or Lys-28), Arg-24 (or Arg-25) in the six toxins, and Asp-402, His-404, Asp-386, and Asp-376 in each subunit of the Kv1.3 potassium channel are the key residues for the toxin-channel triplet contacts. This is in agreement with the electrostatic potential calculations. In all of the resultant candidate complexes of AgTX2 and Kv1.3, the side chain of Lys-27 in AgTX2 is located in the pore near the outer binding site, demonstrating that such configurations were sterically possible. Nonetheless, the orientation of the toxin is widely distributed around the pore axis with a slightly higher occurrence observed for the complexes in whichArg-24 of the toxin was near Asp-402 from one of the four channel subunits. Configuration with lowest binding energies was selected from the BD trajectories to be the final docked toxin-channel complex. If configurations had the same lowest binding energy, the one with minimum triplet contact distances was selected to be the final docked complex.
FIGURE 5.
Distribution of the structures of AgTX formed encounter complex around the extracelluar mouth of the Kv1.3 channel. The Kv1.3 channel is represented as Cα trace. Each ball represents the center of mass of the toxin in an encounter snapshot with the Kv1.3 channel.
TABLE 2.
Most frequent residues (ordered from left to right) of the toxins occurred in the triplet contacts during the binding of the toxins to the potassium channel
| Toxin | Residues | ||||||
|---|---|---|---|---|---|---|---|
| AgTX2 | Lys-27 | Arg-24 | Lys-32 | Arg-31 | Gly-1 | Lys-16 | Lys-19 |
| ChTX | Lys-27 | Arg-25 | Lys-31 | Arg-34 | Lys-32 | Lys-11 | Ser-37 |
| KTX | Lys-27 | Arg-24 | Lys-32 | Lys-16 | Lys-7 | Asp-20 | Arg-31 |
| MgTX | Lys-28 | Lys-33 | Lys-18 | Thr-1 | Lys-35 | His-39 | Lys-11 |
| NTX | Lys-28 | Lys-33 | Lys-18 | Lys-15 | Lys-6 | Lys-11 | Glu-19 |
| Pi2 | Lys-27 | Lys-32 | Lys-34 | Arg-31 | Lys-19 | Lys-18 | Lys-11 |
Electrostatic interaction energies
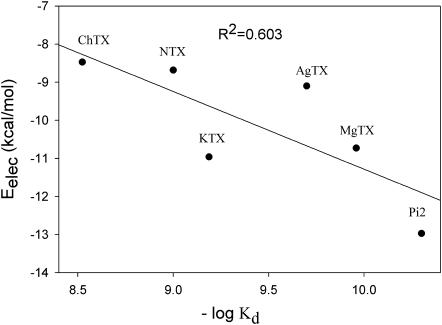
The electrostatic interaction energies between the scorpion toxins and Kv1.3 channel were calculated using the following process. The charges of the toxins were assigned onto the electrostatic potential grids of the Kv1.3 channel, and then the electrostatic interaction energies were calculated by summing all the above assigned charges of the toxins times the corresponding electrostatic potential values at the grids of the Kv1.3 channel. The calculation results are listed in Table 1. The average electrostatic interaction energies between the six scorpion toxins and the Kv1.3 channel range from −6.8 to −13.0 kcal/mol. The electrostatic interaction energies of different conformations of the same scorpion toxin to the channel fluctuate within 2 kcal/mol. For example, the average electrostatic interaction energies for the 1st, 7th, 13th, and 17th structures in the AgTX NMR entry are −7.45 kcal/mol, −8.34 kcal/mol, −8.79 kcal/mol, and −7.88 kcal/mol, respectively. This indicates that the conformations of the toxins indeed affect the docking results. The lowest electrostatic interaction energies (most favorably docked conformations) are −9.10 kcal/mol for AgTX2, −8.47 kcal/mol for ChTX, −10.96 kcal/mol for KTX, −10.73 kcal/mol for MgTX, −8.68 kcal/mol for NTX, and −12.97 kcal/mol for Pi2. The experimental dissociation constant (Kd values) of Kv1.3 channel protein to the six toxins are 3 nM (ChTX), 1 nM (NTX), 110 pM (MgTX), 650 pM (KTX), 200 pM (AgTX2), and 50 pM (Pi2) (Chandy et al., 2001). The electrostatic interaction energies between the scorpion toxins and Kv1.3 channel correlate well with the –logKd, R2 = 0.603 (see Fig. 6). This indicates that the electrostatic interaction is a dominant component for toxin-channel binding specificity. Moreover, the simulation result demonstrates that the electrostatic interaction energies combining with the distribution frequencies from BD simulations might be used as criteria in ranking the binding configuration of a scorpion toxin to the Kv1.3 channel.
FIGURE 6.
Correlation between the electrostatic interaction energies and the values of −logKd of the six scorpion toxins to Kv1.3 channel.
Flexibilities of Kv1.3 channel and scorpion toxins
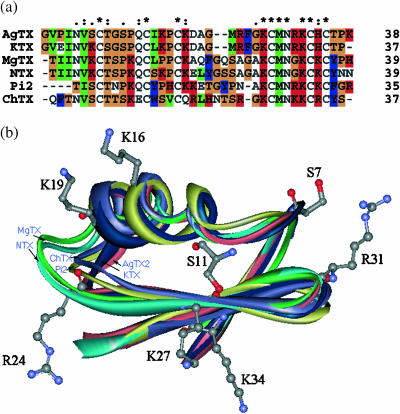
To measure the flexibilities of the Kv1.3 channel and the scorpion toxins in the binding, conventional MD simulations were performed for the Kv1.3 channel and its complexes with toxins in a solvated lipid bilayer mimicking the plasma membrane environment. As will be discussed below, the six toxins are highly homologous in sequence (Fig. 7 a), and the high-resolution NMR structures (Blanc et al., 1997, 1998; Fajloun et al., 2000; Harvey et al., 1995; Krezel et al., 1995; Romi et al., 1993) demonstrated that their 3D structures share the same overall conformation and adopt a similar folding pattern (Fig. 7 b). Therefore, we just took AgTX2 as an example to address the flexibility of the toxins in binding to the Kv1.3 channel.
FIGURE 7.
(a) Sequence alignments of the six scorpion toxins generated by CLUSTAL W (Thompson et al., 1994). In the sequences, an asterisk indicates an identical or conserved residue, a colon indicates a conserved substitution, and a dot indicates a semiconserved substitution. (b) Structure alignment of the six scorpion toxins with some of the AgTX2 residues shown.
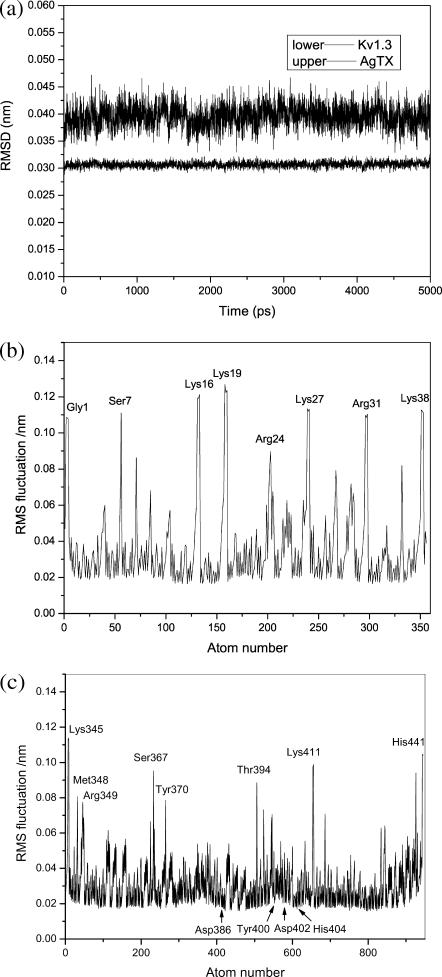
In general, for the Kv1.3 alone, the root mean-square deviations (RMSDs) of the channel backbone atoms relative to the starting structure (3D model of the Kv1.3 constructed by homology modeling) in the 5-ns MD trajectory fluctuate from 0.029 to 0.032 nm (Fig. 8 a). This indicates that the structure of the Kv1.3 channel is not so flexible embedded in the lipid bilayer on a nanosecond timescale, and it is reasonable that the flexibility of the Kv1.3 channel was not considered in the BD simulations. The RMSDs of the scorpion toxins fluctuate between 0.035 and 0.045 nm (Fig. 8 a), indicating that the structures of the scorpion toxins are more flexible than that of the Kv1.3 channel. Therefore, the conformational changes of the toxins were considered in the BD simulations.
FIGURE 8.
Flexibilities of the AgTX2 and Kv1.3 channel in their binding deduced from the 5-ns MD simulations. (a) RMSDs of Kv1.3 and AgTX2 in their separated state. (b) RMS fluctuations of the atoms of AgTX2 in the complex. (c) RMS fluctuations of the atoms of Kv1.3 (only shows one monomer) in the complex.
The MD simulation on the AgTX2-Kv1.3 complex addressed the most flexible residues in the two-protein binding. The atomic RMS fluctuation (RMSF) of AgTX2 indicated that the fluctuation scales of most of the atoms of the toxin are small, ranging from 0.02 to 0.06 nm; however, the RMSFs of positively charged residues, Lys-16, Lys-19, Arg-24, Lys-27, Arg-31, and Lys-38, involved in the toxin-Kv1.3 binding are relatively large (Fig. 8 b). For the Kv1.3 channel, the RMSFs of most atoms are less than 0.04 nm (Fig. 8 c); the RMSFs of the residues playing an important role in the toxin binding, such as Gly-380, Asp-386, Tyr-400, Asp-402, and His-404, are only ∼0.02 nm, and the residues of the Kv1.3 channel with large RMSFs, such as Tyr-370, Thr-394, Lys-411, and residues at the ends, are not involved in the toxin-channel binding. This demonstrates again that the flexibilities of the toxins should be considered in the toxin-channel binding simulations, whereas the flexibility of the Kv1.3 channel plays a less important role for the toxin recognition.
Contacts between the toxins and the channel
During the MD simulations, the hydrogen bonds between these pairs kept on breaking and forming, and the average number of hydrogen bonds formed between the toxin and the channel is ∼2–3 (data not shown), which implies triplet contact analysis is quite necessary and sufficient when determining the toxin-channel complexes in BD simulation. The most frequently formed hydrogen bonds between the potassium channel and the toxins were listed in Table 3.
TABLE 3.
Possible hydrogen bonds formed between the scorpion toxins and the Kv1.3 channel in the toxin-channel complex
| Scorpion toxin
|
Kv1.3 channel
|
||||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Distance (Å) | |
| AgTX2 | Arg-31 | Nη2 | Asp-386(B) | OΩ1 | 2.94 |
| Arg-31 | Nη1 | Asp-386(B) | Oδ1 | 3.09 | |
| Lys-27 | Nζ | Tyr-400(A) | O | 3.19 | |
| Arg-24 | Nη2 | Asp-402(A) | O | 2.92 | |
| ChTX | Ser-37 | Oγ | Ser-378(D) | O | 2.93 |
| Arg-34 | Nη1 | Gly-401(B) | O | 3.25 | |
| Lys-31 | Nζ | Gly-380(B) | O | 2.62 | |
| Lys-27 | Nζ | Asp-402(B) | Oδ2 | 3.34 | |
| KTX | Arg-31 | Nη1 | Asp-386(B) | Oδ1 | 2.44 |
| Asn-30 | N | Asp-402(B) | O | 2.61 | |
| Arg-24 | Nη1 | Asp-402(A) | Oδ1 | 2.96 | |
| Lys-27 | Nζ | Tyr-400(A) | O | 2.36 | |
| MgTX | Lys-33 | Nζ | Asp-386(A) | Oδ1 | 2.91 |
| Asn-31 | Nδ2 | Val-406(B) | O | 2.81 | |
| Lys-28 | Nζ | His-404(B) | Nδ1 | 3.15 | |
| Thr-1 | Oζ1 | Asp-402(C) | Nɛ2 | 2.74 | |
| NTX | Asn-39 | Nδ2 | Asp-402(D) | O | 2.89 |
| Tyr-37 | Oη | Gly-401(B) | O | 3.02 | |
| Asn-31 | Nδ2 | Asp-402(C) | O | 2.83 | |
| Lys-28 | Nζ | Tyr-400(A) | O | 2.87 | |
| Pi2 | Lys-34 | Nζ | Asp-402(C) | Oδ1 | 2.65 |
| Lys-32 | Nζ | His-404(A) | Nɛ2 | 2.84 | |
| Asn-30 | N | Asp-402(A) | O | 2.89 | |
| Lys-27 | Nζ | Tyr-400(D) | O | 2.72 | |
| Asn-25 | O | His-404(B) | Nɛ2 | 3.15 | |
| Tyr-14 | Oη | Asp-402(D) | Oδ1 | 3.07 | |
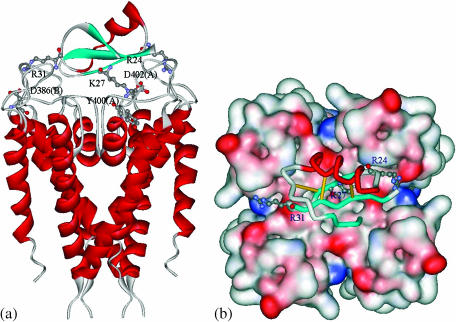
After MD simulations, we performed an energy minimization for each toxin-channel complex to achieve an energy tolerance of 0.05 kcal/(mol × Å) using the adopted-basis Newton-Raphson algorithm and the CHARMm22 force-field as implemented in the Quanta program (1998 Release; Molecular Simulations). Taking AgTX2 as an example, the optimized structure of the complex is shown in Fig. 9. Structurally, the toxins use their β-sheet to associate with the outer pore of the Kv1.3 channel. The optimized structures of the complexes have favorable electrostatic interactions between the two proteins. For example, the flat β-sheet face of AgTX2 has a patch containing five positively charged residues, which covers ∼200 Å2 of the AgTX2 surface; there are twelve negatively charged residues at the outer pore area of the Kv1.3 channel, i.e., Asp-386(A–D), Asp-402(A–D) and His-404(A–D), interacting favorably with the positive patch through hydrogen bonding and electrostatic interactions. Numerous Kv1.3 residues are involved in hydrophobic contacts with the flat β-sheet face of the toxins, including Ser-11, Phe-25, Met-29, Asn-30, His-34, and Thr-36, as shown in Table 4 and Fig. 9.
FIGURE 9.
Optimized structure of the AgTX2-kv1.3 channel complex. (a) Side view of the structure. The two proteins were represented as ribbon structure. R24, K27, and R31 in AgTX2 formed three hydrogen bonds with D402, Y400, and D386 in Kv1.3, respectively. (b) Top view of the structure. The molecular surface of Kv1.3 channel is colored by electrostatic potential (red, negative; blue, positive, white, uncharged). The toxin is represented as Cα trace, and the important residues in binding are represented as ball-and-stick model.
TABLE 4.
Possible hydrophobic contacts between the scorpion toxins and Kv1.3 channel in the toxin-channel complex
| Scorpion toxin
|
Kv1.3 channel
|
||||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Distance (Å) | |
| AgTX2 | Ser-11 | Cβ | His-404(C) | Cɛ1 | 3.60 |
| Met-29 | Cγ | His-404(B) | Cɛ1 | 3.65 | |
| Asn-30 | Cγ | Met-403(B) | Cɛ | 3.86 | |
| Gly-10 | Cα | Ser-378(C) | C | 3.88 | |
| Phe-25 | Cδ1 | His-404(A) | Cɛ1 | 3.38 | |
| Thr-36 | Cγ2 | Asp-402(A) | Cβ | 3.73 | |
| Asn-30 | Cγ | Gly-380(B) | C | 3.86 | |
| ChTX | Ser-37 | Cβ | Ser-378(D) | C | 3.82 |
| Tyr-36 | Cβ | His-404(D) | Cɛ1 | 3.67 | |
| Lys-31 | Cɛ | Met-403(B) | Cɛ | 3.70 | |
| Met-29 | Cδ | Asp-402(B) | Cγ | 3.02 | |
| Met-29 | C | His-404(C) | Cɛ1 | 3.51 | |
| Lys-27 | Cδ | Gly-401(C) | C | 3.18 | |
| KTX | Asn-30 | Cγ | Gly-380(B) | Cα | 3.62 |
| Asn-30 | Cβ | Met-403(B) | Sδ | 3.78 | |
| Met-29 | Sδ | Gly-401(B) | C | 3.74 | |
| Met-29 | Cα | Asp-402(B) | C | 3.79 | |
| Lys-27 | Cɛ | Gly-401(D) | Cα | 3.86 | |
| Phe-25 | Cδ1 | His-404(A) | Cɛ1 | 3.50 | |
| MgTX | His-39 | C | His-404(C) | Cδ2 | 3.43 |
| Pro-38 | Cγ | Ser-379(C) | Cβ | 3.57 | |
| Tyr-37 | Cɛ1 | Gly-401(C) | C | 3.67 | |
| Lys-35 | Cɛ | Gly-401(D) | Cα | 3.57 | |
| Lys-33 | Cɛ | Gly-480(A) | Cα | 3.76 | |
| Met-30 | Cγ | His-404(D) | Cγ | 2.25 | |
| Met-30 | Cγ | Asp-402(A) | Cγ | 3.58 | |
| Lys-28 | Cδ | His-404(B) | Cɛ1 | 3.51 | |
| Ile-2 | Cδ1 | Met-403(A) | C | 3.30 | |
| NTX | Asn-39 | Cγ | Ser-479(D) | Cβ | 3.74 |
| Tyr-37 | Cδ1 | His-404(B) | Cɛ1 | 3.62 | |
| Met-30 | Cγ | His-404(A) | Cɛ1 | 3.37 | |
| Lys-28 | Cɛ | His-404(D) | Cɛ1 | 3.32 | |
| Ile-2 | Cδ1 | His-404(C) | Cɛ1 | 3.22 | |
| Thr-1 | Cγ2 | Ser-379(B) | Cβ | 3.66 | |
| Pi2 | Phe-36 | Cɛ2 | His-404(B) | Cɛ1 | 3.53 |
| Arg-31 | Cɛ | Gly-380(A) | Cα | 3.53 | |
| Asn-30 | Cγ | Ser-379(A) | Cβ | 3.75 | |
| Met-29 | Cɛ | Gly-401(C) | C | 3.87 | |
| Lys-27 | Cδ | Gly-401(D) | C | 3.86 | |
| Pro-10 | Cγ | His-404(D) | Cɛ1 | 3.59 | |
| Pro-10 | Cδ | Ser-379(D) | Cα | 3.58 | |
The high-resolution NMR structures of these toxins (Blanc et al., 1997, 1998; Fajloun et al., 2000; Harvey et al., 1995; Krezel et al., 1995; Romi et al., 1993) have demonstrated that they share the same overall conformation and adopt a similar folding pattern (Fig. 7 b); the RMSD of the structural superposition between two toxins ranges from 0.51 to 1.45 Å. The global structures of the toxins consist of a three-stranded antiparallel β-sheet and an α-helix, which are stabilized by three disulfide bonds. Although the 3D folding of the six toxins is similar, it is important to note that their specificity and affinities depend on the residues situated at their external surface and especially the surface contact with the channel protein. Small changes, either in the Kv1.3 channel or in the toxins, could drastically change their binding affinities. For example, site-specific mutants (P10S, S14W, A25R, A25Delta) of NTX inhibited Kv1.3 channel with Kd values of 30, 0.6, 112, and 166 nM, respectively (Mullmann et al., 2001), and mutants (H404A, H404Y, D402N, and Y400V) of Kv1.3 (Aiyar et al., 1996) resulted in KTX inhibiting the channel with Kd values of 0.72, 0.03, 0.38, and 2.5 nM, respectively. In contrast, these same NTX mutants had no effect on maxi-K channel activity with estimated Kd values exceeding 1 mM.
The principal toxin-Kv1.3 channel interactions derived from the refined structures were analyzed using the LIGPLOT program (Wallace et al., 1995). The hydrogen bonds and the hydrophobic contacts present in the refined complexes are listed in Tables 3 and 4, respectively. The refined 3D models clearly indicate the important residue pairs for the binding, which cover most of the key residues that were experimentally verified as important for scorpion toxin-potassium channel binding (Aiyar et al., 1996; DeCoursey et al., 1984; Freudenthaler et al., 2002; Harvey et al., 1995; Hopkins, 1998; Miller 1995; Naranjo and Miller, 1996). For example, the conserved toxin residue Lys-27 (or Lys-28 in NTX and MgTX,) protrudes into the pore, interacting with residues Tyr-400, Asp-402, or His-404 in the signature sequence GYGD of the potassium channel via hydrogen bonding and electrostatic and hydrophobic interactions. These interaction features are in agreement with the mutation experiments. Mutations of Arg-24-Ala, Lys-27-Met, and Asn-30-Ala decreased the binding of AgTX2 to the Shaker channel by 128.8-, 685.7-, and 696.1-fold, respectively (Gross and MacKinnon, 1996). Replacement of Asp-402 in Kv1.3 resulted in nonfunctional channels; residue replacements of Lys-27 (or Lys-28) in the toxins, or Tyr-400 or His-404 in the channel, decreased the binding affinities by 520- to 11,000-fold (Aiyar et al., 1996). Other residues involved in the interaction contacts listed in Tables 3 and 4 can also be used to explain the mutagenesis results. Residues Asp-386 and Gly-380 in Kv1.3, Arg-24 or Arg-25 (Asn-25 in Pi2), and Asn-30 or Asn-31 in the scorpion toxins also play important roles in the recognition of the potassium channel and the scorpion toxins. These are in accordance with the mutagenesis experiments of both potassium channel and scorpion toxins (Ellis et al., 2001; Fernandez et al., 1994; Goldstein et al., 1994; MacKinnon et al., 1988; Miller, 1995; Park and Miller, 1992; Peter et al., 2001; Thompson and Begenisich, 2000). The consistency between the 3D models and the mutation experiments demonstrates that the refined 3D models of toxin-Kv1.3 channel complexes are reliable.
CONCLUSIONS
The purpose of this study was to determine the binding structures of the scorpion toxins in complex with the pore of the Kv1.3 potassium channel. Based on the homology model of the Kv1.3 potassium channel, we have investigated the recognition between six scorpion toxins and the potassium channel by an approach combining the Brownian dynamics simulation, molecular dynamics simulation, and molecular mechanics (MM) refinement. Reasonable 3D models of the toxin-channel complexes have been obtained that can explain available mutagenesis results of toxin-channel binding. The docking process overcame some of the disadvantages of conventional BD, i.e., that it cannot easily consider the conformational flexibility of the associating proteins. We noticed that the conformation of a toxin indeed affected the simulation results. BD simulations predict that all the six scorpion toxins in this study use their β-sheets to associate with the extracellular entryway of the Kv1.3 channel, which is in line with the primary clues from the electrostatic interaction calculations and mutagenesis results. BD triplet analysis and MD/MM results show that residues Lys-27 or Lys-28, residues Arg-24 or Arg-25, and Asn-30 or Asn-31 in the separate six toxins, and residues Tyr-400, Asp-402, His-404, Asp-386 and Gly-380 in each subunit of the Kv1.3 potassium channel, are the key residues for the toxin-channel recognitions. Additionally, the electrostatic interaction energies between the toxins and Kv1.3 channel correlate well with the binding affinities (–logKds), R2 = 0.603, suggesting that the electrostatic interaction is a dominant component for toxin-channel binding specificity. Moreover, the simulation result demonstrates that the electrostatic interaction energies combined with the distribution frequencies from BD simulations might be used as criteria in ranking the binding configuration of a scorpion toxin to the Kv1.3 channel.
The present study illustrates the power of combination use of homology modeling, BD simulation, and MD/MM simulation methods for the structural characterization of macromolecular complexes. This strategy may be helpful when a high-resolution structure of a complex cannot be obtained by NMR or x-ray crystallography experiments even though the structures of the constituents are known. The consistency between the results of the BD/MD simulations and the experimental data indicated that our 3D model structures of the toxin-channel complex are reasonable and can be used in guiding the design of future biological studies, such as the rational design of the blocking agents of Kv1.3 channel and mutagenesis in both toxins and Kv1.3 channel.
Acknowledgments
We gratefully acknowledge financial support from the National Natural Science Foundation of China (Grants 20102007, 29725203, and 20072042), the State Key Program of Basic Research of China (Grant 2002CB512802), the 863 Hi-Tech Program of China (Grants 2002AA233011, 2002AA233061, 2001AA235051, and 2001AA235041), the Qi Ming Xing Foundation of Shanghai Ministry of Science and Technology (Grants 00QB14034 and 03QD14065), and the Key Program of New Drug Research and Development from the Chinese Academy of Sciences.
References
- Aiyar, J., J. P. Rizzi, G. A. Gutman, and K. G. Chandy. 1996. The signature sequence of voltage-gated potassium channels projects into the external vestibule. J. Biol. Chem. 271:31013–31016. [DOI] [PubMed] [Google Scholar]
- Alessandri-Haber, N., A. Lecoq, S. Gasparini, G. Grangier-Macmath, G. Jacquet, A. L. Harveyi, C. de Medeirosi, E. G. Rowani, M. Gola, A. Menez, and M. Crest. 1999. Mapping the functional anatomy of BgK on Kv1.1, Kv1.2, and Kv1.3. J. Biol. Chem. 274:35653–35661. [DOI] [PubMed] [Google Scholar]
- Beeton, C., J. Barbaria, P. Giraud, J. Devaux, A.-M. Benoliel, M. Gola, J. M. Sabatier, D. Bernard, M. Crest, and E. Beraud. 2001. Selective blocking of voltage-gated K+ channels improves experimental autoimmune encephalomyelitis and inhibits T cell activation. J. Immunol. 166:936–944. [DOI] [PubMed] [Google Scholar]
- Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comp. Phys. Commun. 91:43–56. [Google Scholar]
- Biggin, P. C., T. Roosild, and S. Choe. 2000. Potassium channel structure: domain by domain. Curr. Opin. Struct. Biol. 10:456–461. [DOI] [PubMed] [Google Scholar]
- Blanc, E., R. Romi-Lebrun, O. Bornet, T. Nakajima, and H. Darbon. 1998. Solution structure of two new toxins from the venom of the Chinese scorpion Buthus martensi Karsch blockers of potassium channels. Biochemistry. 37:12412–12418. [DOI] [PubMed] [Google Scholar]
- Blanc, E., J. M. Sabatier, R. Kharrat, S. Meunier, M. E. Ayeb, J. V. Rietschoten, and H. Darbon. 1997. Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels. Proteins. 29:321–333. [PubMed] [Google Scholar]
- Bontems, F., B. Gilquin, C. Roumestand, A. Menez, and F. Toma. 1992. Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications. Biochemistry. 31:7756–7764. [DOI] [PubMed] [Google Scholar]
- Cahalan, M. D., H. Wulf, and K. G. Chandy. 2001. Molecular properties and physiological roles of ion channels in the immune system. J. Clin. Immunol. 21:235–252. [DOI] [PubMed] [Google Scholar]
- Chandy, K. G., M. Cahalan, M. Pennington, R. S. Norton, H. Wulff, and G. A. Gutman. 2001. Potassium channels in T lymphocytes: toxins to therapeutic immunosuppressants. Toxicon. 39:1269–1276. [DOI] [PubMed] [Google Scholar]
- Chandy, K. G., T. E. Decoursey, M. D. Cahalan, C. McLaughlin, and S. Gupta. 1984. Voltage-gated potassium channels are required for human T lymphocyte activation. J. Exp. Med. 160:369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., J. H. Shen, J. M. Biggs, W. Fu, J. J. Wu, Y. M. Zhang, X. M. Luo, Z. W. Chi, R. Ji, H. Jiang, and K. Chen. 2002. Brownian dynamics simulations of the recognition of the scorpion toxin P05 with the small-conductance calcium activated potassium channels. J. Mol. Biol. 318:417–428. [DOI] [PubMed] [Google Scholar]
- Cui, M., J. H. Shen, J. M. Biggs, X. M. Luo, X. J. Tan, H. L. Jiang, K. Chen, and R. Ji. 2001. Brownian dynamics with hydrodynamic interactions. Biophys. J. 80:1659–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- Dauplais, M., B. Gilquin, L. Possani, G. Gurrola-Briones, C. Roumestand, and A. Menez. 1995. Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins. Biochemistry. 34:16563–16573. [DOI] [PubMed] [Google Scholar]
- Davis, M. E., J. D. Madura, B. A. Luty, and J. A. McCammon. 1991. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program. Comp. Phys. Commun. 62:187–197. [Google Scholar]
- DeCoursey, T. E., K. G. Chandy, S. Gupta, and M. D. Cahalan. 1984. Voltage-gated K+ channels in human T lymphocytes: a role in mitogenesis? Nature. 307:465–468. [DOI] [PubMed] [Google Scholar]
- Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Ellis, K. C., T. C. Tenenholz, H. Jerng, M. Hayhurst, C. S. Dudlak, W. F. Gilly, M. P. Blaustein, and D. J. Weber. 2001. Interaction of a toxin from the scorpion Tityus serrulatus with a cloned K+ channel from Squid (sqKv1A). Biochemistry. 40:5942–5953. [DOI] [PubMed] [Google Scholar]
- Ermak, D. L., and J. A. McCammon. 1978. Brownian dynamics with hydrodynamic interactions. J. Chem. Phys. 69:1352–1360. [Google Scholar]
- Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh ewald potential. J. Chem. Phys. 103:8577–8592. [Google Scholar]
- Fajloun, Z., G. Ferrat, E. Carlier, M. Fathallah, C. Lecomte, G. Sandoz, E. di Luccio, K. Mabrouk, C. Legros, H. Darbon, H. Rochat, J. M. Sabatier, and M. De Waard. 2000. Synthesis, 1H NMR structure, and activity of a three-disulfide-bridged maurotoxin analog designed to restore the consensus motif of scorpion toxins. J. Biol. Chem. 275:13605–13612. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gomez, J. D., G. R. Smith, and M. S. P. Sansom. 2002. Setting up and optimization of membrane protein simulations. Eur. Biophys. J. 31:217–227. [DOI] [PubMed] [Google Scholar]
- Fernandez, I., R. Romi, S. Szendeffy, M. F. Martin-Eauclaire, H. Rochat, J. Van Rietschoten, M. Pons, and E. Giralt. 1994. Kaliotoxin (1–37) shows structural differences with related potassium channel blockers. Biochemistry. 33:14256–14263. [DOI] [PubMed] [Google Scholar]
- Freudenthaler, G., M. Axmann, H. Schindler, B. Pragl, H.-G. Knaus, and G. J. Schütz. 2002. Ultrasensitive pharmacological characterisation of the voltage-gated potassium channel KV1.3 studied by single-molecule fluorescence microscopy. Histochem. Cell Biol. 117:197–202. [DOI] [PubMed] [Google Scholar]
- Fu, W., M. Cui, J. M. Briggs, X. Huang, B. Xiong, Y. Zhang, X. Luo, J. Shen, R. Ji, H. Jiang, and K. Chen. 2002. Brownian dynamics simulations of the recognition of the scorpion toxin maurotoxin with the voltage-gated potassium ion channels. Biophys. J. 83:2370–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Calvo, M., R. J. Leonard, J. Novick, S. P. Stevens, W. Schmalhofer, G. J. Kaczorowaki, and M. L. Garcia. 1993. Purification, characterization, and biosynthesis of margatoxin, a component of centruroides margaritatus venom that selectively inhibits voltage-dependent potassium channels. J. Biol. Chem. 268:18866–18874. [PubMed] [Google Scholar]
- Gilquin, B., J. Racape, A. Wrisch, V. Visan, A. Lecoq, S. Grissmer, A. Menez, and S. Gasparini. 2002. Structure of the BgK-Kv1.1 complex based on distance restraints identified by double mutant cycles. J. Biol. Chem. 277:37406–37413. [DOI] [PubMed] [Google Scholar]
- Goldstein, S. A., and T. J. Colatsky. 1996. Ion channels: too complex for rational drug design? Neuron. 16:913–919. [DOI] [PubMed] [Google Scholar]
- Goldstein, S. A., D. J. Pheasant, and C. Miller. 1994. The charybdotoxin receptor of a Shaker K+ channel: peptide and channel residues mediating molecular recognition. Neuron. 12:1377–1388. [DOI] [PubMed] [Google Scholar]
- Gross, A., and R. MacKinnon. 1996. Agitoxin footprinting the Shaker potassium channel pore. Neuron. 16:399–406. [DOI] [PubMed] [Google Scholar]
- Gulbis, J. M., M. Zhou, S. Mann, and R. MacKinnon. 2000. Structure of the cytoplasmic β-subunit-T1 assembly of voltage-dependent K+ channels. Science. 289:123–127. [DOI] [PubMed] [Google Scholar]
- Harvey, A. L., H. Vatanpour, E. G. Rowan, S. Pinkasfeld, C. Vita, A. Menez, and M.-F. Martin-Eauclaire. 1995. Structure-activity studies on scorpion toxins that block potassium channels. Toxicon. 33:425–436. [DOI] [PubMed] [Google Scholar]
- Hermans, J., H. J. C. Berendsen, W. F. van Gunsteren, and J. P. M. Postma. 1984. A consistent empirical potential for water-protein interactions. Biopolymers. 23:1513–1518. [Google Scholar]
- Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- Liu, H., Y. Li, X. Tan, M. Song, F. Cheng, S. Zheng, J. Shen, X. Luo, R. Ji, J. Yue, G. Y. Hu, H. L. Jiang, and K. Y. Chen. 2003. Structure-based discovery of potassium channel blockers from natural products: virtual screening and electrophysiological assay testing. Chem. Biol. 10:1103–1113. [DOI] [PubMed] [Google Scholar]
- Hopkins, W. F. 1998. Toxin and subunit specificity of blocking affinity of three peptide toxins for heteromultimeric, voltage-gated potassium channels expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 285:1051–1060. [PubMed] [Google Scholar]
- Jiang, Y.-X., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002a. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang, Y.-X., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002b. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Shen, J., X. Xu, F. Cheng, H. Liu, X. Luo, J. Shen, K. Chen, W. Zhao, X. Shen, and H. Jiang. 2003. Virtual screening on natural products for discovering active compounds and target information. Curr. Med. Chem. 10:2327–2342. [DOI] [PubMed] [Google Scholar]
- Johnson, B. A., S. P. Stevens, and J. M. Williamson. 1994. Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry. 33:15061–15070. [DOI] [PubMed] [Google Scholar]
- Kaczorowski, G. J., and M. L. Garcia. 1999. Pharmacology of voltage-gated and calcium-activated potassium channels. Curr. Opin. Chem. Biol. 3:448–458. [DOI] [PubMed] [Google Scholar]
- Krezel, A. M., C. Kasibhatla, P. Hidalgo, R. Mackinnon, and G. Wagner. 1995. Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry. Protein Sci. 4:1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R. S., and M. D. Cahalan. 1988. Subset-specific expression of potassium channels in developing murine T lymphocytes. Science. 239:771–775. [DOI] [PubMed] [Google Scholar]
- Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- Lu, Z., A. M. Klem, and Y. Ramu. 2001. Ion conduction pore is conserved among potassium channels. Nature. 413:809–813. [DOI] [PubMed] [Google Scholar]
- MacKerell, A. D., Jr., D. Bashford, M. Bellott, R. L. Dunbrack, Jr., J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher III, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- MacKinnon, R. 1991. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 350:232–235. [DOI] [PubMed] [Google Scholar]
- MacKinnon, R., P. H. Reinhart, and M. M. White. 1988. Charybdotoxin block of Shaker K+ channels suggests that different types of K+ channels share common structural features. Neuron. 1:997–1001. [DOI] [PubMed] [Google Scholar]
- Madura, J. D., J. M. Briggs, R. C. Wade, M. E. Davis, B. A. Luty, A. Ilin, J. Antosciewicz, M. K. Gilson, B. Bagheri, L. R. Scott, and J. A. MacCammon. 1995. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program. Comp. Phys. Commun. 91:57–95. [Google Scholar]
- Matthew, J. B. 1985. Electrostatic effects in proteins. Annu. Rev. Biophys. Chem. 14:387–417. [DOI] [PubMed] [Google Scholar]
- Matthew, J. B., and F. R. Gurd. 1986. Calculation of electrostatic interactions in proteins. Methods Enzymol. 130:413–436. [DOI] [PubMed] [Google Scholar]
- Miller, C. 1995. The Charybdotoxin family of K+ channel-blocking peptides. Neuron. 15:5–10. [DOI] [PubMed] [Google Scholar]
- Mourre, C., M. N. Chernova, M.-F. Martin-Eauclaire, R. Bessone, G. Jacquet, M. Gola, S. L. Alper, and M. Crest. 1999. Distribution in rat brain of binding sites of kaliotoxin, a blocker of Kv1.1 and Kv1.3 α-subunits. J. Pharmacol. Exp. Ther. 291:943–952. [PubMed] [Google Scholar]
- Mullmann, T. J., K. T. Spence, N. E. Schroeder, V. Fremont, E. P. Christian, and K. M. Giangiacomo. 2001. Insights into α-K toxin specificity for K+ channels revealed through mutations in noxiustoxin. Biochemistry. 40:10987–10997. [DOI] [PubMed] [Google Scholar]
- Naranjo, D., and C. Miller. 1996. A strongly interacting pair of residues on the contact surface of charybdotoxin and a Shaker K+ channel. Neuron. 16:123–130. [DOI] [PubMed] [Google Scholar]
- Neria, E., S. Fischer, and M. Karplus. 1996. Simulation of activation free energies in molecular systems. J. Chem. Phys. 105:1902–1921. [Google Scholar]
- Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–296. [DOI] [PubMed] [Google Scholar]
- Northrup, S. H., J. O. Boles, and J. C. L. Reynolds. 1987. Electrostatic effects in the Brownian dynamics of association and orientation of heme protein. J. Phys. Chem. 91:5991–5998. [Google Scholar]
- Northrup, S. H., T. Laughner, and G. Stevenson. 1999. MacroDox Macromolecular Simulation Program. Tennessee Technological University, Cookeville, TN.
- Northrup, S. H., K. A. Thomasson, C. M. Miller, P. D. Barker, L. D. Eltis, J. G. Guillemette, S. C. Inglis, and A. G. Mauk. 1993. Effects of charged amino acid mutations on the bimolecular kinetics of reduction of yeast iso-1-ferricytochrome c by bovine ferrocytochrome b5. Biochemistry. 32:6613–6623. [DOI] [PubMed] [Google Scholar]
- Park, C. S., and C. Miller. 1992. Interaction of charybdotoxin with permeant ions inside the pore of a K+ channel. Neuron. 9:307–313. [DOI] [PubMed] [Google Scholar]
- Pearlman, D. A., D. A. Case, J. W. Caldwell, W. S. Ross, T. E. Cheatham, I. S. DeBolt, D. Ferguson, G. Seibel, and P. Kollman. 1995. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91:1–41. [Google Scholar]
- Peter, J. M., Z. Varga, P. R. Hajdu, J. Gaspar, S. Damjanovich, E. Horjales, and L. D. Possani. 2001. Effects of toxins Pi2 and Pi3 on human T lymphocyte Kv1.3 channels: the role of Glu-7 and Lys24. J. Membrane Biol. 179:13–25. [DOI] [PubMed] [Google Scholar]
- Romi, R., M. Crest, M. Golag, F. Sampieri, G. Jacquets, H. Zerroukfl, P. Mansuelle, O. Sorokine, M. V. Dorsselaer, H. Rochat, M. F. M. Eauclair, and J. V. Rietschoten. 1993. Synthesis and characterization of kaliotoxin. J. Biol. Chem. 268:26302–26309. [PubMed] [Google Scholar]
- Tenenholz, T. C., R. S. Rogowski, J. H. Collins, M. P. Blaustein, and D. J. Weber. 1997. Solution structure for Pandinus toxin K-alpha (PiTX-K alpha), a selective blocker of A-type potassium channels. Biochemistry. 36:2763–2771. [DOI] [PubMed] [Google Scholar]
- Terlau, H., and W. Stühmer. 1998. Structure and function of voltage-gated ion channels. Naturwissenschaften. 85:437–444. [DOI] [PubMed] [Google Scholar]
- Thompson, J., and T. Begenisich. 2000. Electrostatic interaction between charybdotoxin and a tetrameric mutant of Shaker K+ channels. Biophys. J. 78:2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman, D. P., and H. J. C. Berendsen. 1998. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 74:2786–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman, D. P., H. J. C. Berendsen, and M. S. P. Sansom. 1999. An alamethicin channel in a bilayer: molecular dynamics simulations. Biophys. J. 76:1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hunenberger, P. Kruger, A. E. Mark, W. R. P. Scott, and I. G. Tironi. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Hochschulverlag AG an der ETH Zurich, Zurich, Switzerland.
- Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134. [DOI] [PubMed] [Google Scholar]
- Wickenden, A. D. 2002. K+ channels as therapeutic drug targets. Pharmacol. Ther. 94:157–182. [DOI] [PubMed] [Google Scholar]
- Wrisch, A., and S. Grissmer. 2000. Structural differences of bacterial and mammalian K+ channels. J. Biol. Chem. 275:39345–39353. [DOI] [PubMed] [Google Scholar]
- Zhou, M., J. H. Morais-Cabral, S. Mann, and R. MacKinnon. 2001a. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 411:657–661. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. Mackinnon. 2001b. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]