Abstract
The active pool of internalized cholera toxin (CT) moves from the endosomes to the Golgi apparatus en route to the endoplasmic reticulum (ER). The catalytic CTA1 polypeptide is then translocated from the ER to the cytosol, possibly through the action of the ER-associated degradation (ERAD) pathway. Translocation was previously measured indirectly through the downstream effects of CT action. We have developed a direct biochemical assay for CTA1 translocation that is independent of toxin activity. Our assay is based upon the farnesylation of a CVIM motif-tagged CTA1 polypeptide (CTA1-CVIM) after it enters the cytosol. When expressed from a eukaryotic vector in transfected CHO cells, CTA1-CVIM was targeted to the ER, but was not secreted. Instead, it was translocated into the cytosol and degraded in a proteosome-dependent manner. Translocation occurred rapidly and was monitored by the appearance of farnesylated CTA1-CVIM in the detergent phase of cell extracts generated with Triton X-114. Detergent-phase partitioning of CTA1-CVIM resulted from the cytoplasmic addition of a 15-carbon fatty acid farnesyl moiety to the cysteine residue of the CVIM motif. Our use of the CTA1-CVIM translocation assay provided supporting evidence for the ERAD model of toxin translocation and generated new information on the timing of CTA1 translocation.
Infection with Vibrio cholerae can lead to life-threatening diarrheal disease. Diarrhea results from the osmotic movement of water that follows secretion of chloride ions into the intestinal lumen. These cellular events are triggered by elevated levels of intracellular cyclic AMP (cAMP) resulting from the activation of Gsα and its adenylate cyclase target. The causative agent for this process is cholera toxin (CT), an AB5-type protein toxin that constitutively activates Gsα by ADP ribosylation (reviewed in references 9 and 33).
CT consists of a single A subunit (CTA) and a homopentameric B subunit. Proteolytic nicking of CTA generates an A1 polypeptide with latent catalytic activity and an A2 polypeptide that interacts with the B pentamer and maintains the stability of the nicked holotoxin (9, 18, 28, 33). A KDEL motif at the C terminus of CTA2 also increases the efficiency of holotoxin targeting to the endoplasmic reticulum (ER) (16). The B pentamer binds to GM1 gangliosides that are clustered in glycosphingolipid-enriched regions of the eukaryotic plasma membrane, an event that leads to internalization of the ganglioside-bound enterotoxin within the endocytic system (1, 22, 25, 37). CT is then transferred to the trans-Golgi network, transported through the Golgi apparatus, and delivered to the ER, where the nicked CTA polypeptide is reduced and the CTA1 fragment is translocated to the cytoplasm (15, 16, 20, 23, 24, 30, 35). Whether translocation of the A2 subunit occurs is currently unknown, and the fate of B pentamers that enter the ER is also uncertain.
It is hypothesized that CTA1 translocation involves the quality control mechanism known as ER-associated degradation (ERAD) (8, 19). This system recognizes misfolded or misassembled proteins in the ER and exports them to the cytosol for ubiquitination and proteasomal degradation (reviewed in references 26 and 29). Hazes and Read (8) proposed that an exposed hydrophobic stretch within the A13 subdomain of CTA1 identifies the toxin as a misfolded protein and thereby triggers ERAD activity. After transfer to the cytosol, CTA1 is thought to escape proteasomal degradation because it has a paucity of the lysine residues that are required for ubiquitination. Although this is an attractive hypothesis, supporting evidence is limited, and few direct studies on CTA1 translocation have been reported.
One difficulty in examining CTA1 translocation is the lack of appropriate techniques. Most assays for translocation rely upon the downstream effects of toxin activity, such as morphological alterations to Y1 or CHO cells (7, 21), elevated intracellular cAMP levels (12), and chloride secretion from epithelial monolayers (2). All of these techniques measure translocation indirectly. They are not quantitative measures of translocation, they cannot be used with nonfunctional CT mutants, and they are dependent on multiple signaling events that occur after CTA1 translocation. A direct assay for translocation that is independent of toxin activity would thus greatly facilitate study of the translocation event.
In this paper, we describe a direct and quantitative biochemical assay to measure CTA1 translocation. The procedure does not require an active toxin, but instead uses farnesylation as a marker for the ER-to-cytosol export of a CVIM-tagged CTA1 subunit (CTA1-CVIM). Farnesylation involves addition of a 15-carbon fatty acid moiety to the cysteine residue of a C-terminal CaaX motif (such as CVIM) of a target protein and can be detected by partitioning of the farnesylated protein into the detergent phase of a Triton X-114 solution (4, 6). Since farnesylation only occurs in the cytoplasm, any CTA1-CVIM receiving the modification must have been delivered to the cytosol. A related methodology was used to track the cytosolic appearance of a recombinant diphtheria toxin A polypeptide after cultured cells were exposed to extracellular diphtheria toxin (6). However, because only a minor portion of internalized CT reaches the ER (17, 22, 30), the cytosolic pool of CTA1 following exposure of cultured cells to extracellular CT was likely to be small and difficult to detect. We therefore used a eukaryotic expression vector for the production of recombinant CTA1-CVIM constructs and for delivery of the constructs to the ER of transfected CHO cells. Studies with a cell-free transcription-translation system had previously shown that the CTA1 leader sequence is sufficient for targeting newly synthesized CTA1 to the lumen of ER-derived microsomes (31), and a similar strategy was used to target the catalytic subunit of pertussis toxin to the ER of CHO-K1 and COS-1 cells in vivo (3, 36). Inactive variants of CTA1-CVIM were used in order to circumvent any possible complications that might arise from cAMP production by active forms of CTA1-CVIM. In the present study, we demonstrated that this strategy could be used to develop a direct biochemical assay for CTA1-CVIM translocation that is independent of CT toxicity, and we used the assay to investigate the timing of translocation and the role of ERAD in the translocation process.
MATERIALS AND METHODS
Materials.
Tissue culture reagents were purchased from Invitrogen (Carlsbad, Calif.), [35S]methionine was obtained from NEN Life Sciences (Boston, Mass.), endoglycosidase H (Endo H) was obtained from Roche (Indianapolis, Ind.), restriction enzymes were obtained from New England Biolabs (Beverly, Mass.) or Invitrogen, and other reagents were obtained from Sigma-Aldrich (St. Louis, Mo.). Immobilized anti-CTA antibody on beads was prepared by mixing 100 μl of the rabbit B9 antibody (11) and 100 mg of protein A-Sepharose (Sigma-Aldrich) overnight at 4°C in 1 ml of phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin.
Generation of CTA1 constructs.
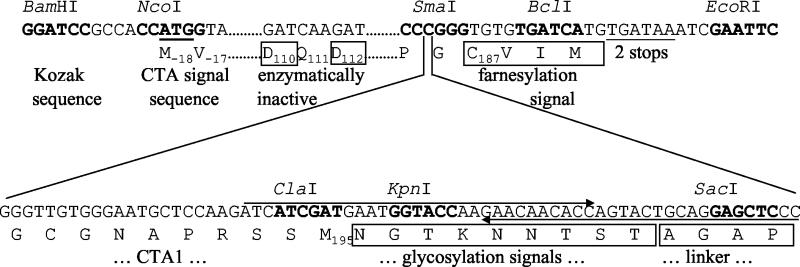
Standard DNA cloning procedures were performed with enzymes as described by the manufacturers. Silent mutations introducing a SmaI site into the codons encoding Pro185-Gly186 of CTA1 were made by oligonucleotide-directed mutagenesis in a wild-type ctxA1 gene cloned in a T7 expression vector. An SmaI-EcoRI-digested oligonucleotide linker encoding GCVIM followed by a stop codon was inserted in frame with ctxA1 to create pT7CTA1CVIM. Plasmid pMGJ6710 is a clone of a native ctxAB operon encoding an enzymatically inactive CTA variant with E110D and E112D substitutions (E110D+E112D) (11). The gene encoding the CTA1-CVIM variant was then subcloned in place of the ctxB gene of pMGJ6710, creating a tandem duplication of the inactive CTA-encoded variant followed by the active CTA1-CVIM-encoded construct. This construct was used to make clones producing the enzymatically inactive variants CTA1-CVIM and CTA1-Nglyc-CVIM (Fig. 1). First, the clone producing the enzymatically inactive CTA1-CVIM variant was made by digestion with BspEI (which cuts within both ctxA1 alleles, after the E110D+E112D- and before the CVIM-encoding sequences), followed by self-ligation. Second, the clone producing the enzymatically inactive CTA1-Nglyc-CVIM variant was made by digestion with ClaI (cutting after the E110D+E112D-encoding mutations) and SmaI (cutting prior to the CVIM-encoded tag) followed by ligation of a ClaI-cut SmaI linker encoding a glycosylation tag. This double-stranded linker was made by annealing the DNA primers shown in Fig. 1 and filling in the single-stranded extensions with DNA polymerase I Klenow fragment and deoxynucleoside triphosphates (dNTPs), followed by agarose gel-purification. A BamHI restriction site and a consensus Kozak sequence were then added to both constructs immediately preceding the start codon of the native ctxA signal sequence, by PCR amplification with CGGGATCCGCCACCATGGTAAAGATAATATTTGTG and the M13-20 vector-specific primer. These products were then cloned as BamHI-EcoRI fragments into the eukaryotic expression vector pcDNA3.1 (Invitrogen) to create the final constructs shown in Fig. 1. The first T of the Cys187 codon was mutated to A to generate an SVIM-encoding variant in which the SVIM sequence cannot serve as an acceptor motif for farnesylation. All manipulations were confirmed by DNA sequencing of the resulting clones.
FIG. 1.
Design and construction of CTA1-CVIM constructs. The BamHI-EcoRI fragments shown were cloned into pcDNA3.1. (Top) Relevant sequence of the CVIM-tagged CTA1 variant. Dots indicate sequence (data not shown) identical to the published sequence. Restriction sites are marked in boldface. Start and stop codons are underlined. Beneath the DNA sequence are relevant translations of the CTA sequence or tag. Subscript numbers denote signal sequence, active site, or the last-encoded residues of wild-type CTA1, respectively, relative to Asn(+1) of the mature CTA polypeptide. Residues different from wild-type CTA1 are boxed. Significant features are identified below the translation. (Bottom) DNA sequence and translation of the insert encoding the glycosylation tag, which replaces the third C of the SmaI site in the upper sequence. Long arrows denote synthetic DNA primers annealed together, extended and then digested with ClaI, to clone the glycosylation linker tag.
Cell culture and transfection.
CHO cells were transferred to six-well plates at 60 to 80% confluency the day before transfection. DNA was introduced into the cells by a 3-h incubation with 1 μg of DNA and 5 μl of Lipofectamine as per the manufacturer's instructions (Invitrogen), and experiments were conducted at ∼48 h posttransfection. Cells were maintained at 37°C and 5% CO2 under humidified conditions in Ham's F-12 medium supplemented with 10% fetal bovine serum.
Immunofluorescence.
Cells transferred to coverslips at ∼24 h posttransfection were fixed and permeabilized with a 5-min incubation in ice-cold acetone at ∼48 h posttransfection. The monoclonal anti-CTA antibody 35C2 (10) was then added for 1 h. After a second 30-min incubation with a FITC-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.), the intracellular distributions of CTA1-CVIM and CTA1-Nglyc-CVIM were visualized with a Nikon Eclipse TE200 microscope (Melville, N.Y.) and a ×63 objective.
Metabolic labeling and immunoprecipitation.
Transfected cell monolayers were washed twice with PBS, incubated in methionine-free medium for 1 h, and exposed to 150 μCi of [35S]methionine per ml for 1 h. After two additional PBS washes, the cells were either solubilized in 1 ml of lysis buffer (25 mM Tris [pH 7.4], 20 mM NaCl, 1% deoxycholic acid, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of pepstatin per ml, 1 μg of leupeptin per ml) for 20 min at 4°C or returned to serum-free medium containing an excess of cold methionine. Cell extracts were then collected after the stated chase intervals. When indicated, the chase medium was also collected. Triton-insoluble material was removed from the cell extracts by centrifugation, and anti-CTA-conjugated protein A Sepharose beads were added to the cleared supernatants for an overnight incubation on a 4°C rotator. The immunoprecipitated material was washed twice with NDET (1% NP-40, 0.4% deoxycholic acid, 5 mM EDTA, 10 mM Tris [pH 7.4], 150 mM NaCl) and once with water before resuspension in sample buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide) with PhosphorImager analysis (Bio-Rad, Hercules, Calif.) was subsequently used to visualize and quantitate the isolated samples. Background values were subtracted from the experimental values obtained for all samples before quantitation.
For experiments involving tunicamycin, transfected cells were washed twice with PBS and exposed to 1 ml of 10 μg of tunicamycin per ml for 3 h prior to metabolic labeling. This time frame included the 1-h preincubation in methionine-free medium. Cells were then pulse-labeled with 100 μCi of [35S]methionine per ml for 1 h in the continued presence of tunicamycin. The remaining steps of sample preparation were performed as described above.
For detergent-phase partitioning experiments, cell extracts were generated from a 20-min incubation at 4°C with 1 ml of 1% Triton X-114 containing protease inhibitors (1 mM PMSF, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml). Triton-insoluble material was removed by centrifugation, and the cleared supernatant was placed at 37°C for 15 min. Aqueous and detergent phases of the Triton X-114 extract were then separated with a short centrifugal spin. Water (0.9 ml) was added to the detergent phase, after which anti-CTA-conjugated protein A Sepharose beads were added to both aqueous and detergent-phase isolates for an overnight incubation on a 4°C rotator. The remaining steps were performed as described above. To quantitate the cytosolic pool of CTA1-CVIM at each time interval, the amount of farnesylated CTA1-CVIM in the detergent (i.e., cytoplasmic) phase was calculated as a percentage of the total CTA1-CVIM found in both aqueous and detergent-phase samples.
RESULTS
Design of CTA1-CVIM constructs.
Cotranslational polypeptide import into the ER lumen is triggered by an N-terminal stretch of hydrophobic residues that are preceded by at least one positively charged amino acid. A similar hydrophobic targeting motif will direct bacterial proteins to an sec-dependent secretory pathway. The eukaryotic translational machinery will thus recognize the bacterial consensus sequence for sec-dependent secretion as an ER targeting motif and will accordingly direct these exogenously expressed bacterial proteins to the ER lumen. Such a result was reported for in vitro-translated CTA1 (31) and for the catalytic subunit of pertussis toxin (3). We therefore placed our CTA1-CVIM constructs in pcDNA3.1, a eukaryotic expression vector. CTA1-CVIM consisted of the native CTA signal sequence, the sequence of mature CTA1 with inactivating glutamate-to-aspartate substitutions at residues 110 and 112, and a C-terminal CVIM farnesylation consensus sequence beginning at residue 187 of the mature CTA1 polypeptide. A second construct, CTA1-Nglyc-CVIM, contained three partially overlapping N-X-T/S consensus sequences for N glycosylation (27) just upstream of the CVIM motif (Fig. 1).
Expression of the CTA1-CVIM constructs.
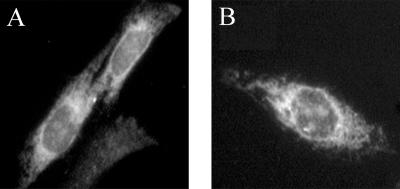
To determine if our constructs were properly targeted in cultured cells, we visualized the site of CTA1 expression with indirect immunofluorescence. Transiently transfected CHO cells were fixed and permeabilized by acetone treatment, incubated with a monoclonal anti-CTA antibody for 1 h, and incubated with an FITC-conjugated secondary antibody for 30 min. As shown in Fig. 2A and B, the CTA1-CVIM constructs produced a tubuloreticular staining pattern indicative of ER localization. Staining was most concentrated in the perinuclear region of the cell, but also extended throughout the cytosol in a tubularized network. Furthermore, labeling of the nuclear envelope was clearly visible. Such a distribution demonstrated that both CTA1-CVIM and CTA1-Nglyc-CVIM were properly targeted to the ER.
FIG. 2.
Immunofluorescent localization of the CTA1-CVIM constructs. CHO cells expressing either (A) CTA1-CVIM or (B) CTA1-Nglyc-CVIM were fixed and permeabilized with acetone at 48 h posttransfection, exposed to a monoclonal anti-CTA antibody for 1 h, and incubated with a FITC-conjugated goat anti-mouse IgG antibody for 30 min.
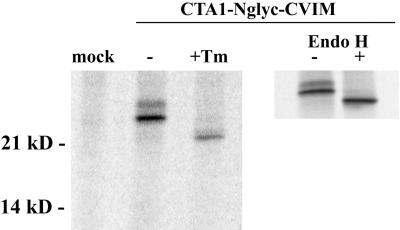
An ER staining pattern could be produced by proteins associated with either the lumenal or cytoplasmic face of the ER membrane, but only those proteins positioned within the ER lumen can serve as targets for modification with N-linked oligosaccharides. CTA1 targeting to the ER lumen was therefore confirmed by the glycosylation state of CTA1-Nglyc-CVIM (Fig. 3). Two glycosylated forms of CTA1-Nglyc-CVIM were detected in transfected cells after metabolic labeling and im-munoprecipitation. Pretreatment with tunicamycin, an inhibitor of N glycosylation, reduced the molecular masses of both bands to the predicted size of 22 kDa for unmodified CTA1-Nglyc-CVIM. Since the unmodified form of CTA1-Nglyc-CVIM was only observed after drug treatment, it appeared that the entire pool of transfected CTA1 was initially imported into the ER lumen. Targeting of transfected CTA1 to the ER was thus confirmed by both biochemical and morphological criteria.
FIG. 3.
Glycosylation of CTA1-Nglyc-CVIM. Mock-transfected cells or cells transfected with CTA1-Nglyc-CVIM were pulse-labeled for 1 h with 100 μCi of [35S]methionine per ml before solubilization in Triton X-100 lysis buffer. One sample was prepared by exposure to 10 μg of tunicamycin (Tm) per ml for a 3-h preincubation and continued coincubation during labeling. Anti-CTA immunoprecipitates from the cell extracts were visualized by SDS-PAGE and PhosphorImager scanning. In a separate experiment, the immunoprecipitated pool of CTA1 was divided into two equal aliquots and treated with (+) or without (−) 5 mU of Endo H for 1 h at 37°C before the addition of sample buffer and SDS-PAGE.
Failure to modify all three of the potential CTA1-Nglyc-CVIM N-glycosylation sites in the ER was the most likely explanation for the immunoisolation of two glycosylated forms of CTA1-Nglyc-CVIM. It was possible, however, that the upper CTA1-Nglyc-CVIM band resulted from additional, Golgi apparatus-specific oligosaccharide modifications. An immunoprecipitated sample of CTA1-Nglyc-CVIM was treated with Endo H in order to exclude this possibility. Endo H cleaves the sugar residues from proteins that have received N-linked oligosaccharides in the ER, but it will not act upon N-linked oligosaccharides that have received further modifications in the medial Golgi apparatus (34). Both forms of CTA1-Nglyc-CVIM were sensitive to Endo H (Fig. 3), thus confirming that CTA1 did not travel from the ER to the Golgi apparatus. No larger forms of CTA1-Nglyc-CVIM that would result from Golgi apparatus-specific modifications were generated during a 4-h chase (data not shown). CTA1 was thus effectively retained in the ER before its degradation.
CTA1-CVIM degradation.
Although ERAD processing can involve a number of distinct pathways, the general mechanism of ERAD is the same for all substrates: degradation does not require lysosomal proteolysis or vesicular transport from the ER, but it does depend upon substrate export from the ER to the cytosol and proteosomal action in the cytosol (26, 29). We conducted a series of pulse-chase experiments to determine whether the turnover of CTA1 resembled an ERAD-mediated process. Cells transiently transfected with CTA1-CVIM were left untreated or were incubated with various drugs before and throughout the time course of the assay. These drugs—brefeldin A (BfA), chloroquine, and N-acetyl-Leu-Leu-Norleu-Al (ALLN)—were chosen because their mode of action would inhibit secretion, lysosomal proteolysis, or proteosomal degradation. BfA blocks vesicular transport from the ER and would therefore prevent secretion as well as the delivery of lysosomal substrates to their site of degradation (13). Chloroquine dissipates the lysosomal pH gradient and thus negates the activity of acid-dependent lysosomal proteases (32). ALLN, in contrast, inhibits proteosomal action and would therefore impact ERAD-mediated proteolysis (14). We can thus infer the route and mechanism of CTA1 turnover from the effects of these drugs on CTA1-CVIM degradation.
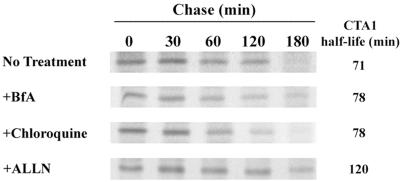
As shown in Fig. 4, a half-life of 71 min was calculated for CTA1-CVIM. Neither 5 μg of BfA per ml nor 100 μM chloroquine affected the degradation of CTA1-CVIM. Treatment with 100 μM NH4Cl, another inhibitor of lyosomal proteolysis, also failed to disrupt the turnover of CTA1-CVIM (data not shown). Incubation with 100 μM ALLN, however, extended the half-life of CTA1-CVIM from 71 min to 120 min. No detectable pool of CTA1-CVIM could be immunoprecipitated from the medium after 2 or 3 h of chase. Similar results were obtained with CTA1-Nglyc-CVIM, although the half-life of this construct was 144 min and the inhibitory effect of ALLN was more pronounced (data not shown). Collectively, our results supported the ERAD model of toxin translocation by demonstrating that the degradation of CTA1 was proteosome dependent and did not require lysosomal proteolysis or vesicular transport from the ER.
FIG. 4.
CTA1-CVIM degradation. Transfected cells were incubated with 150 μCi of [35S]methionine per ml for 1 h and chased in serum-free medium containing an excess of cold methionine. Exposure to 5 μg of BfA per ml, 100 μM chloroquine, or 100 μM ALLN was initiated 1 h prior to metabolic labeling and maintained throughout the experiment. Anti-CTA immunoprecipitates collected from cell extracts at the indicated time points were visualized by PhosphorImager scanning of SDS-PAGE gels. Sample quantitation from three to four independent experiments for each condition was used to calculate the half-life of CTA1.
CTA1-CVIM translocation.
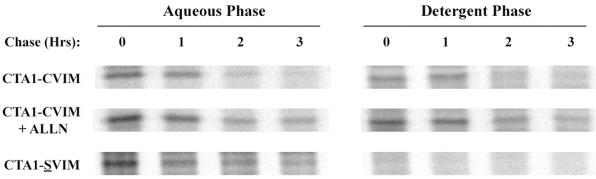
Having established that CTA1 degradation resembled an ERAD-mediated process, we next attempted to directly measure the translocation of CTA1-CVIM from the ER to the cytosol (Fig. 5). Pulse-chase experiments were conducted as described above, except cell extracts were generated with Triton X-114 instead of Triton X-100. A short centrifugal spin was then used to separate the aqueous and detergent phases of the Triton X-114 extracts. Both phases were subjected to anti-CTA immunoprecipitation, after which SDS-PAGE in conjunction with PhosphorImager analysis was used to visualize and quantitate the isolated material.
FIG. 5.
CTA1-CVIM translocation from the ER to the cytosol. Cells transfected with CTA1-CVIM or CTA1-SVIM were incubated with 150 μCi of [35S]methionine per ml for 1 h and chased in serum-free medium containing an excess of cold methionine. When indicated, exposure to 100 μM ALLN was initiated 1 h prior to metabolic labeling and was maintained throughout the experiment. Cell extracts were generated after the stated intervals by solubilization in Triton X-114. Aqueous (i.e., ER lumen) and detergent (i.e., cytosolic) phases of the cell extracts were separated by centrifugation following warming to 37°C, and both phases were subjected to anti-CTA immunoprecipitation. Isolated material was visualized by SDS-PAGE and PhosphorImager scanning.
We found that 26% of CTA1-CVIM had already passed from the ER to the cytosol by the end of a 1-h pulse label (n = 3). The level of CTA1-CVIM expression precluded use of a shorter labeling period to minimize the initial pool of cytosolic protein. In control cells, the cytosolic pool of CTA1-CVIM remained at 27% of pulse-labeled protein after 1 h of chase, but then fell to negligible amounts at 2 and 3 h of chase. Proteosomal inhibition with ALLN did not prevent CTA1 translocation, but did allow some amount of the protein to persist in the cytosol after 2 and 3 h of chase. Importantly, the detergent-phase partitioning of CTA1 was completely abolished when an inactivating cysteine-to-serine alteration was introduced into the CVIM farnesylation motif to generate CTA1-SVIM. Farnesylation in the cytosol, rather than an inherent physical property of CTA1, was therefore responsible for the detergent-phase partitioning of CTA1. Finally, an 85-min half-life for CTA1-SVIM was calculated from the data prepared for Fig. 5 (n = 2). Because CTA1-SVIM and CTA1-CVIM were degraded with similar kinetics, the farnesylation of CTA1-CVIM did not appear to significantly alter the rate of its turnover in the cytosol.
DISCUSSION
Most assays for the cytosolic translocation of CTA1 are indirect, depending upon prior delivery of holotoxin from the cell surface to the ER and subsequent toxic effects of the translocated CTA1 polypeptide. Such approaches do not allow direct quantitative assessment of CTA1 translocation and make it difficult to study the structural requirements for translocation. They cannot be used to study translocation of nontoxic CTA1 variants and have limited power for analyzing the kinetics and physiology of toxin translocation. We therefore sought to develop a translocation assay that is independent of both retrograde toxin transport to the ER and subsequent toxic activity in the cytosol. In this study, we characterized two CVIM-tagged CTA1 constructs and demonstrated that they could be used to monitor toxin trafficking, translocation, and degradation in living cells by direct biochemical methods.
Indirect immunofluorescence was used to confirm the ER localization of CTA1 in CHO cells after transient transfection with expression plasmids encoding CTA1-CVIM or CTA1-Nglyc-CVIM. Furthermore, the addition of N-linked oligosaccharides to CTA1-Nglyc-CVIM—a modification that only occurs in the ER lumen—demonstrated that the entire pool of transfected protein was initially inserted into the ER lumen. CTA1 targeting to the ER was thus established by both morphological and biochemical methods. CTA1-Nglyc-CVIM did not receive oligosaccharide modifications indicative of Golgi processing events, thereby confirming that CTA1 was efficiently retained in the ER before its translocation to the cytosol. These findings demonstrated that CTA1-Nglyc-CVIM was useful for monitoring the intracellular distribution and trafficking of CTA1. However, because the native A1 subunit lacks carbohydrate modifications, and because the rate-limiting step in ERAD often involves interactions between lectin-like ER chaperones and the carbohydrate residues of an ERAD substrate (26, 29), we used the CTA1-CVIM construct instead of the CTA1-Nglyc-CVIM construct to study the degradation and translocation of CTA1.
Pulse-chase experiments with transiently transfected cells were used to calculate a half-life of 71 min for CTA1-CVIM. A similar half-life of 85 min was calculated for CTA1-SVIM, thus indicating that farnesylation did not significantly affect the rate of CTA1-CVIM degradation. BfA, chloroquine, and ammonium chloride did not affect the turnover of CTA1-CVIM. This demonstrated that neither vesicular trafficking from the ER nor lysosomal proteolysis was required for the degradation of CTA1. The proteosome inhibitor ALLN, however, increased the half-life of CTA1-CVIM to 120 min and thereby demonstrated that CTA1 degradation is proteosome dependent. Finally, we could not detect a secreted pool of CTA1. The standard characteristics of ERAD were thus established for the processing of CTA1. Similar findings on the kinetics and mechanism of toxin degradation were recently reported for the catalytic polypeptide of the plant toxin ricin (5).
CTA1 was rapidly exported from the ER lumen to the cytosol. A quarter of the radiolabeled toxin had already entered the cytosol by the end of 1 h of pulse labeling, but this did not produce a long-lived cytosolic pool of CTA1. In fact, only a minimal amount of CTA1 remained in the cytosol after 2 h of chase. This indicated that CTA1 processing involved both efficient export from the ER and efficient degradation in the cytosol. Export and degradation of CTA1 during intoxication of susceptible target cells by CT holotoxin may be even more rapid than our findings indicate, because our transfection system could potentially saturate a rate-limiting mechanism for the processing of CTA1. Proteolysis of CTA1 occurred despite the paucity of lysine residues in CTA1, thereby suggesting a ubiquitin-independent mechanism for toxin degradation. The combination of inefficient CT trafficking to the ER (17, 22, 30) and rapid CTA1 turnover in the cytosol implies that very little CTA1 would be found in the cytosol at any given time. These findings highlight the potency of CTA1 and reinforce the importance of the prolonged activation of adenylate cyclase by ADP-ribosylated Gsα for the sustained toxic effects of small CT doses on the small intestine (12, 33).
ERAD substrates are degraded in a proteosome-dependent manner after or during extraction from the ER, but this general process can involve a number of separate pathways: branches of ERAD include ubiquitin-dependent degradation, ubiquitin-independent degradation, ubiquitin-dependent extraction from the ER, ubiquitin-independent extraction from the ER, proteosome-dependent extraction from the ER, and proteosome-independent extraction from the ER (26, 29). Our work suggests that ERAD processing of CTA1 involves proteosome-independent extraction from the ER, because treatment with ALLN did not prevent the cytosolic appearance of CTA1-CVIM. Others have reported that CTA1 translocation is also a ubiquitin-independent process (31). This proteosome- and ubiquitin-independent route of translocation may provide CTA1 with the opportunity to temporarily evade the efficient proteosomal degradation that usually accompanies ERAD-mediated protein translocation to the cytosol.
Most proteins inserted into the ER are transported to distal sites in the secretory pathway. Only misfolded proteins, misassembled proteins, aberrantly glycosylated proteins, or proteins with an ER retention-retrieval motif are held in the ER (26). As such, our results support the hypothesis that a structural feature of CTA1, such as a hydrophobic region in the CTA13 subdomain, identifies the toxin as a misfolded protein and promotes its ERAD-mediated passage into the cytosol. Future work with CVIM-tagged CTA1 constructs should allow us to directly explore this possibility and further elucidate the mechanism of CTA1 translocation.
Acknowledgments
This work was supported in part by an NIH National Research Service Award (AI10394) to K. Teter, an NIH Short-Term Training in Health Professional Schools award (2T35DK07496) to R. L. Allyn, and an NIH research grant (AI31940) to R. K. Holmes.
Editor: J. D. Clements
REFERENCES
- 1.Badizadegan, K., B. L. Dickinson, H. E. Wheeler, R. S. Blumberg, R. K. Holmes, and W. I. Lencer. 2000. Heterogeneity of detergent-insoluble membranes from human intestine containing caveolin-1 and ganglioside G(M1). Am. J. Physiol. Gastrointest. Liver Physiol. 278:G895-G904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, K. E. 1993. Positive and negative regulation of chloride secretion in T84 cells. Am. J. Physiol. 265:C859-C868. [DOI] [PubMed] [Google Scholar]
- 3.Castro, M. G., U. McNamara, and N. H. Carbonetti. 2001. Expression, activity and cytotoxicity of pertussis toxin S1 subunit in transfected mammalian cells. Cell. Microbiol. 3:45-54. [DOI] [PubMed] [Google Scholar]
- 4.Clarke, S. 1992. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 61:355-386. [DOI] [PubMed] [Google Scholar]
- 5.Di Cola, A., L. Frigerio, J. M. Lord, A. Ceriotti, and L. M. Roberts. 2001. Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98:14726-14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falnes, P. O., A. Wiedlocha, A. Rapak, and S. Olsnes. 1995. Farnesylation of CaaX-tagged diphtheria toxin A-fragment as a measure of transfer to the cytosol. Biochemistry 34:11152-11159. [DOI] [PubMed] [Google Scholar]
- 7.Guerrant, R. L., L. L. Brunton, T. C. Schnaitman, L. I. Rebhun, and A. G. Gilman. 1974. Cyclic adenosine monophosphate and alteration of Chinese hamster ovary cell morphology: a rapid, sensitive in vitro assay for the enterotoxins of Vibrio cholerae and Escherichia coli. Infect. Immun. 10:320-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazes, B., and R. J. Read. 1997. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36:11051-11054. [DOI] [PubMed] [Google Scholar]
- 9.Hirst, T. R. 1999. Cholera toxin and Escherichia coli heat-labile enterotoxin, p. 104-129. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins. Academic Press, San Diego, Calif.
- 10.Holmes, R. K., and E. M. Twiddy. 1983. Characterization of monoclonal antibodies that react with unique and cross-reacting determinants of cholera enterotoxin and its subunits. Infect. Immun. 42:914-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jobling, M. G., and R. K. Holmes. 2001. Biological and biochemical characterization of variant A subunits of cholera toxin constructed by site-directed mutagenesis. J. Bacteriol. 183:4024-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassis, S., J. Hagmann, P. H. Fishman, P. P. Chang, and J. Moss. 1982. Mechanism of action of cholera toxin on intact cells. Generation of A1 peptide and activation of adenylate cyclase. J. Biol. Chem. 257:12148-12152. [PubMed] [Google Scholar]
- 13.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 15.Lencer, W. I. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal toxins. V. cholerae: invasion of the intestinal epithelial barrier by a stably folded protein toxin. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G781-G786. [DOI] [PubMed] [Google Scholar]
- 16.Lencer, W. I., C. Constable, S. Moe, M. G. Jobling, H. M. Webb, S. Ruston, J. L. Madara, T. R. Hirst, and R. K. Holmes. 1995. Targeting of cholera toxin and Escherichia coli heat labile toxin in polarized epithelia: role of COOH-terminal KDEL. J. Cell Biol. 131:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lencer, W. I., C. Delp, M. R. Neutra, and J. L. Madara. 1992. Mechanism of cholera toxin action on a polarized human intestinal epithelial cell line: role of vesicular traffic. J. Cell Biol. 117:1197-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lencer, W. I., T. R. Hirst, and R. K. Holmes. 1999. Membrane traffic and the cellular uptake of cholera toxin. Biochim. Biophys. Acta 1450:177-190. [DOI] [PubMed] [Google Scholar]
- 19.Lord, J. M., and L. M. Roberts. 1998. Toxin entry: retrograde transport through the secretory pathway. J. Cell Biol. 140:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majoul, I. V., P. I. Bastiaens, and H. D. Soling. 1996. Transport of an external Lys-Asp-Glu-Leu (KDEL) protein from the plasma membrane to the endoplasmic reticulum: studies with cholera toxin in Vero cells. J. Cell Biol. 133:777-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maneval, D. R., R. R. Colwell, S. W. J. Grays, and S. T. Donta. 1981. A tissue culture method for the detection of bacterial enterotoxins. J. Tissue Cult. Methods 6:85-90. [Google Scholar]
- 22.Montesano, R., J. Roth, A. Robert, and L. Orci. 1982. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature 296:651-653. [DOI] [PubMed] [Google Scholar]
- 23.Orlandi, P. A. 1997. Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J. Biol. Chem. 272:4591-4599. [PubMed] [Google Scholar]
- 24.Orlandi, P. A., P. K. Curran, and P. H. Fishman. 1993. Brefeldin A blocks the response of cultured cells to cholera toxin. Implications for intracellular trafficking in toxin action. J. Biol. Chem. 268:12010-12016. [PubMed] [Google Scholar]
- 25.Orlandi, P. A., and P. H. Fishman. 1998. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlmutter, D. H. 1999. Misfolded proteins in the endoplasmic reticulum. Lab. Investig. 79:623-638. [PubMed] [Google Scholar]
- 27.Rapak, A., P. O. Falnes, and S. Olsnes. 1997. Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc. Natl. Acad. Sci. USA 94:3783-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodighiero, C., A. T. Aman, M. J. Kenny, J. Moss, W. I. Lencer, and T. R. Hirst. 1999. Structural basis for the differential toxicity of cholera toxin and Escherichia coli heat-labile enterotoxin. Construction of hybrid toxins identifies the A2-domain as the determinant of differential toxicity. J. Biol. Chem. 274:3962-3969. [DOI] [PubMed] [Google Scholar]
- 29.Romisch, K. 1999. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112:4185-4191. [DOI] [PubMed] [Google Scholar]
- 30.Sandvig, K., O. Garred, and B. van Deurs. 1996. Thapsigargin-induced transport of cholera toxin to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 93:12339-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz, A., H. Herrgen, A. Winkeler, and V. Herzog. 2000. Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 148:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seglen, P. O. 1983. Inhibitors of lysosomal function. Methods Enzymol. 96:737-764. [DOI] [PubMed] [Google Scholar]
- 33.Spangler, B. D. 1992. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol. Rev. 56:622-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trimble, R. B., and F. Maley. 1984. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal. Biochem. 141:515-522. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, B., C. Rodighiero, W. I. Lencer, and T. A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937-948. [DOI] [PubMed] [Google Scholar]
- 36.Veithen, A., D. Raze, and C. Locht. 2000. Intracellular trafficking and membrane translocation of pertussis toxin into host cells. Int. J. Med. Microbiol. 290:409-413. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, A. A., M. G. Jobling, S. Wimer-Mackin, M. Ferguson-Maltzman, J. L. Madara, R. K. Holmes, and W. I. Lencer. 1998. Ganglioside structure dictates signal transduction by cholera toxin and association with caveolae-like membrane domains in polarized epithelia. J. Cell Biol. 141:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]