Abstract
Quasielastic neutron scattering measurements were performed in hydrated samples of ds-DNA and ss-DNA. The samples were hydrated in a high relative humidity atmosphere, and their final water content was 0.559 g H2O/g ds-DNA and 0.434 g H2O/g ss-DNA. The measurements were performed at 8 and 5.2 Å for the ds-DNA sample, and at 5.2 Å for the ss-DNA sample. The temperature was in both cases 298 K. Analysis of the obtained data indicates that in the ds-DNA sample we can distinguish two types of protons—those belonging to water molecules strongly attached to the ds-DNA surface and another fraction belonging to water that diffuses isotropically in a sphere of radius 2.8 Å, with a local diffusion coefficient of 2.2 × 10−5 cm2 s−1. For ss-DNA, on the other hand, no indication was found of motionally restricted or confined water. Further, the fraction of protons strongly attached to the ds-DNA surface corresponds to 0.16 g H2O/g ds-DNA, which equals the amount of water that is released by ds-DNA upon thermal denaturation, as studied by one of us (G.M.) by differential scanning calorimetry. This value also equals the difference between the critical hydration values of ds-DNA and ss-DNA, also determined by DSC. These results represent, thus, a completely independent measurement of water characteristics and behavior in ds- and ss-DNA at critical hydration values, and therefore substantiate the previous suggestions/conclusions of the results obtained by calorimetry.
INTRODUCTION
Water is essential for the stability and function of “the aperiodic crystal of heredity” (Stent, 1995)—the DNA double helix. Hydration plays a major role not only in the stability of the three-dimensional structure of double-stranded DNA (ds-DNA), but also on the assembly of the different forms (A-form, B-form, Z-form) of double helix and their conformational dynamics (Bloomfield et al., 2000; Saenger, 1984; Blackburn and Gait, 1990). Albiser et al. (2001) showed that the effective parameter that is relevant for the A-B transition of DNA is the average number of water molecules associated to a nucleotide pair, and further, that the number of water molecules necessary to induce the B-form depends on the base composition of DNA. Moreover, water molecules near some specific sequences of basepairs of DNA are important to the understanding of the activity of enzyme proteins and are part of the recognition process (Bloomfield et al., 2000; Blackburn and Gait, 1990). Thus, the water molecules that make up the hydration shell in the immediate vicinity of the surface of DNA are particularly relevant to the function of the double helix. An x-ray crystallographic investigation and solution nuclear magnetic resonance study on a model dodecamer B-DNA duplex showed that the minor groove is hydrated in an extensive and regular way, with a “spine” of first and second hydration shells. In contrast, the hydration of the major groove was found to be mainly confined to a monolayer of water molecules (Liepinsh et al., 1992). Water molecules at the surface of DNA are critical to its equilibrium structure, as well as to function and DNA-ligand recognition. Very recent measurements using time-resolved fluorescence have probed the dynamics of hydration, with femtosecond resolution, for both DNA dodecamer and calf thymus DNA in solution as well as with minor groove binding drugs (Pal et al., 2003a,b). Two timescales were observed for hydration water, one ∼1 ps (“bulk-like”) and another ∼10–12 ps (weakly bound type). The authors stress the importance of hydration water in structural integrity maintenance and recognition. It has also been pointed out in another work that the different hydration found for the different motifs for the tandem wobble G • U mismatches may reflect that hydration plays a role in their stability; and further, that their results imply that hydration of nucleic acids must be important in the genetic code recognition (Sundaralingam and Pan, 2002).
Despite the increasing knowledge and the considerable progress made in the x-ray analysis of short oligonucleotides (Egli et al., 1998), the complex surface of the double helix and its hydration are still the most poorly defined parts of the structures obtained by crystallographic methods, because they are topologically disordered or dynamically averaged and have conformational fluctuations (especially for the high molecular, semiflexible, highly charged DNA chains in aqueous solution). Hydration water is supposed to interact directly with phosphate groups, sugar oxygen atoms, and the polar groups of the bases. It has been shown that water molecules are mainly concentrated in six hydration sites per phosphate and that the positions and occupancies of these sites depend on the DNA conformation and on its nucleotide composition (Schneider et al., 1998). Hydration is also very important regarding protein-DNA interactions, because it has been suggested that hydration sites mark the binding sites at protein-DNA interfaces (Woda et al., 1998).
In this respect neutron quasielastic scattering (NQES) is probably the most powerful technique that can be used for the study of the mechanism of biopolymers—water interactions in hydrated powders, proteins, and DNA fibers (see, e.g., Schreiner et al., 1988; Grimm and Rupprecht, 1989; Bellissent-Funel et al., 1992; Zanotti et al., 1997; Foucat et al., 2002). DNA hydration has also been studied by neutron diffraction, and chains of ordered water in DNA grooves were thereby identified (Shotton et al., 1997).
In this article results of high-resolution NQES experiments are reported for hydrated samples of DNA in the double-stranded (ds) and single-stranded (ss) states, at equal guanine-cytosine content. According to previous calorimetric data, the transformation “double-stranded helix”→“single-stranded chains” was found to be accompanied by “melting” of the structural part of the hydration shell of ds-DNA (Mrevlishvili et al., 2001, 2002). Moreover, formation of the duplex from mixing of their complementary single strands was found to be mostly accompanied by the uptake of structural water molecules (Mrevlishvili et al., 2002; Kankia and Marky, 1999). We wanted then to study ds- and ss-DNA, at low, limiting hydration levels by NQES, to see if we could characterize the type of water present in each case, as well as to find whether a difference could be found between ds- and ss-DNA as regarding water behavior and mobility.
MATERIALS AND METHODS
Materials
ds-DNA (deoxyribonucleic acid sodium salt, from calf thymus) and ss-DNA (deoxyribonucleic acid, single stranded, from calf thymus) were purchased from Sigma-Aldrich, reference numbers D-1501 and D-8899, respectively (St. Louis, MS). Potassium sulfate was obtained from Merck (Whitehouse Station, NJ) (p.A.). All water used was purified in a Milli-Q filtration system (Millipore, Billerica, MA). The indium wire was obtained from Goodfellow (London, UK) (99% pure).
Sample preparation
Samples of ds-DNA and ss-DNA were hydrated by exposing them to a high relative humidity atmosphere, from a saturated solution of potassium sulfate (relative humidity 98.5% at 5°C). The initial water content of the “dry” samples was determined by coulometry (Karl Fischer titration), using a Karl Fischer coulometer, model XX (Metrohm, Herisau, Switzerland). This initial water content was found to be 18.8% (w/w) for ds-DNA and 14.4% (w/w) for ss-DNA. The samples were thereafter maintained in an exicator under the high relative humidity atmosphere and were weighted daily, until they reached the desired water content, 0.559 g H2O/g ds-DNA (B-DNA) and 0.434 g H2O/g ss-DNA. This final water content of each sample was calculated taking into account the amount of water already present in the starting sample.
In both cases, after the desired water content was attained, each sample was placed in a rectangular aluminum plate, which was the container to be used in the scattering experiments, (45 × 75 mm and 0.5-mm thickness) and tightly closed with eight screws, by use of an indium wire.
Neutron-scattering experiments
Data collection
Neutron-scattering experiments were performed at Laboratoire Léon Brillouin (Saclay, France) using a time-of-flight (TOF) spectrometer (MIBEMOL). In a neutron-scattering experiment, one measures the exchange of momentum, Q, and the exchange of energy, ω, due to the interaction of the incident neutron beam with the nuclei of the sample. Momenta and energy conservation are expressed by:
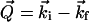 |
(1) |
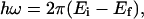 |
(2) |
where h is the Planck constant, k is the momentum and E is the energy, and the indexes i and f refer to the incident and scattered beam, respectively. Incoherent quasielastic neutron scattering takes advantage of the fact that cold neutrons have energy comparable to the energy barriers that hinder molecular orientation and diffusion and a wavelength comparable with interatomic distances. For our sample, the scattered intensity is almost totally due to hydrogen atoms. Because of the large incoherent neutron cross section of protons, the measured intensity is directly related to the time correlation function of individual atoms, which allows the study of single-particle diffusive motions of water molecules.
Thermalized neutrons were used, with two different incident wavelengths, so as to obtain different resolutions. When the wavelength was 5.2 Å, a triangular resolution function with halfwidth at half-maximum (HWHM) of 64 μeV was obtained, whereas a value of HWHM of 17 μeV was obtained for the wavelength of 8 Å. The resolution function was in both cases Q independent. The aluminum plate (sample holder) was placed at an angle of 135° to the incident neutron beam. The scattered neutrons were detected at angles from 23.5° to 141.8°. The corresponding range of accessible values of the elastic momentum exchange Q was 0.346–1.254 Å−1 (8 Å) and 0.533–1.929 Å−1 (5.2 Å). Sampling times of 8 h were used for ds-DNA. For ss-DNA, the small amount of sample required a larger sampling time to improve the statistics, and therefore 16-h sampling times were used. The measurements were performed at 298 K. The resolution at these two wavelengths was determined by running the spectra of vanadium. The spectrum of the empty container was also recorded, and used to correct all experiments.
Data analysis
The TOF spectra were analyzed with programs, available at Laboratoire Léon Brillouin, that allow calibration of the detectors with the vanadium spectra, correction for the empty cell, transformation of the TOF spectra into energy spectra, and finally data grouping (GROUPE software) to improve signal/noise ratio. The data so treated was thereafter analyzed with the FITMIB software, from which the scattering laws S(Q,ω) could be obtained. The protons that appear as static within the spectrometer resolution give rise to an elastic contribution. In addition, mobile protons with similar dynamics appear in the spectra as Lorentzians, characterized by a HWHM. The scattering function, S(Q,ω), at each Q and ω can then be written as the sum of elastic and broadened terms:
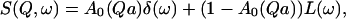 |
(3) |
where the first term is the elastic term, δ(ω), weighted by the elastic incoherent structure factor (EISF) Ao(Qa), and the second term is in principle an infinite sum of Lorentzians. In practice, due to the finite resolution of the instrument and to the experimental statistics, it can be approximated by a single Lorentzian having a HWHM, Γ1/2
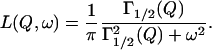 |
(4) |
The program FITMIB provides both the HWHM, Γ1/2 and the intensity of the Lorentzian, as well as the intensity of the elastic contribution.
From the experimentally obtained integrated intensities, the experimental EISF were calculated as
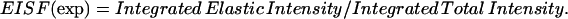 |
(5) |
This general formulation can be extended to extract more information from the obtained data. The goal is to characterize solvent dynamics, trying to discriminate between different types of solvent motion, and to quantify the respective fraction of protons. According to the model of Volino and Dianoux for the diffusion of a point particle inside a sphere of radius a (Volino and Dianoux, 1980) the EISF can be calculated according to:
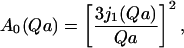 |
(6) |
where j1(Qa) is the first-order spherical Bessel function. This term represents the purely elastic component resulting from the motion confined within a sphere of radius a. Because we know that most protons in DNA do not rotate or diffuse (so they contribute to the elastic peak), and the water content of the samples is known, we can calculate the fraction of immobile protons in DNA, p, from the sample stoichiometry. Further, at this hydration level, we can also introduce in our model for data analysis the fraction of protons (from hydration water) strongly attached to the DNA surface, q. The previous model can then be extended to include these terms (Bellissent-Funel et al., 1992), as:
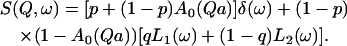 |
(7) |
After resolution broadening by a resolution function R(ω),
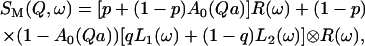 |
(8) |
where ⊗ means convolution in ω space. If the water molecules strongly attached to the DNA surface have a Lorentzian line narrower than the resolution function R(ω), then to a good approximation we can write
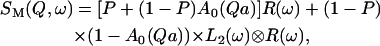 |
(9) |
where the parameter P was introduced to simplify the form of the equation, being defined as
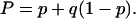 |
(10) |
The obtained scattering results were fitted to this model. The parameters Γ1/2 as a function of Q were extracted by use of the FITMIB program, and the experimental EISF values calculated (Eq. 5) from the integrated intensities also obtained from FITMIB. The calculation of the EISF values, coupled with the observation of the profile of the HWHM of the Lorentzian, allows the analysis of the geometry of the movements that give rise to the quasielastic broadening. The experimental values of EISF were used along with the ones that can be calculated from the model of Volino and Dianoux (1980) (Eq. 6) to obtain a and P by minimization (nonlinear minimum square fitting), as from Eq. 9 we can see that
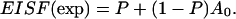 |
(11) |
After obtaining the value of P, the parameter q (fraction of protons (from hydration water) strongly attached to the DNA surface), could also be calculated, because p (fraction of immobile protons in DNA) was known from the sample stoichiometry (see Eq. 10 above).
RESULTS
In the analysis of the results at 8 Å, only one Lorentzian was used in the fit. For 5.2 Å, two different models were tried—with one Lorentzian and with two Lorentzians. In both cases a narrow Lorentzian, whose parameters were Q dependent, was obtained. When two Lorentzians were used, a second Γ1/2 value was obtained, which was Q independent. Further, the Γ1/2 values for the narrow Lorentzian were identical, within error. It turned out, then, that for this analysis, either could be used. The results presented correspond to the fit obtained with two Lorentzians at 5.2 Å, because the EISF values obtained with this model fit perfectly with those obtained at 8 Å.
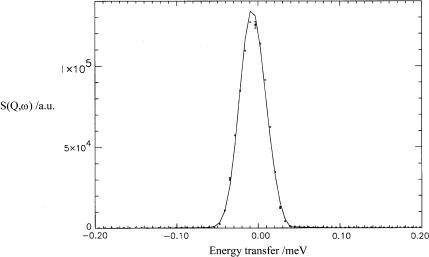
In Fig. 1 one example of obtained fits for ds-DNA is presented, at 8 Å, at a scattering angle θ = 23.5° (for 8 Å). These results are representative of the general goodness of fit obtained at the other scattering angles and wavelengths.
FIGURE 1.
Typical NQES spectrum as a function of energy transfer, for ds-DNA at 8 Å, T = 298 K, at a scattering angle θ = 23.5°. Circles are the experimental points and the solid line is the fit.
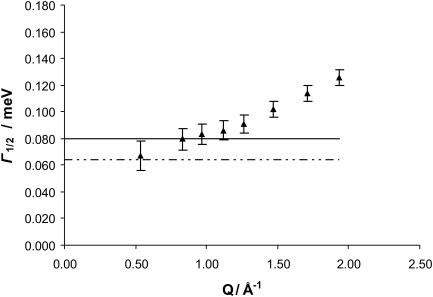
In Fig. 2, the obtained values of Γ1/2 for the narrow Lorentzian at 5.2 Å are represented as a function of Q. As can be seen in the figure, Γ1/2 presents, within error bars, a plateau value of 0.080 meV until 1.12 Å−1. This feature can be accounted for by the model of Volino and Dianoux referred to above as being due to confinements, and represented by diffusion within a sphere (Volino and Dianoux, 1980). This plateau value, observed between Q = 0 and Q = π/a, allows the calculation of a, the radius of the sphere, as
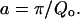 |
(12) |
FIGURE 2.
Γ1/2 as a function of Q for ds-DNA, at 5.2 Å. The solid triangles are the Γ1/2 values obtained from FITMIB for the thin Lorentzian at 5.2 Å. The error bars are the uncertainties provided by the FITMIB program. The solid line represents the plateau value of Γ1/2 = 0.080 meV and the dashed line the instrument resolution in this experimental condition (64 μeV).
The value obtained for the radius of the confinement sphere is a = 2.8 Å. The local diffusion coefficient can thereafter be calculated as
 |
(13) |
providing the value of D = 2.2 × 10−5 cm2 s−1 for this parameter.
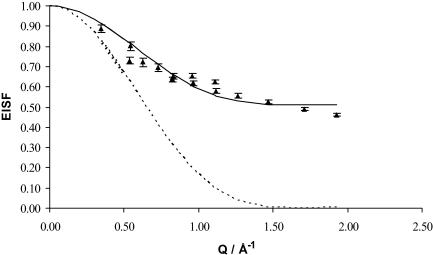
As explained in the data-analysis section, the experimental values of EISF (Eq. 5) along with the ones that can be calculated from the model of Volino and Dianoux (1980) (Eq. 6) were used to obtain a and P by minimization (nonlinear minimum square fitting) We did the fitting either by fixing a to the value of 2.8 Å obtained above and just fitting for the value of P, or by allowing both parameters to vary. The obtained results were in perfect agreement. The P-value obtained for ds-DNA was 0.51 and a = 2.8 Å.
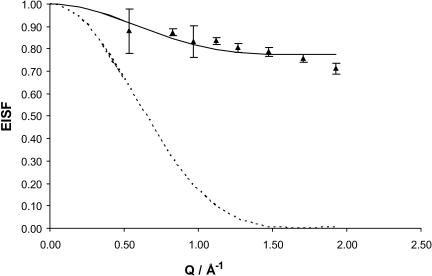
The experimental values of EISF, at 8 and 5.2 Å, as well as the calculated EISF according to Eq. 6 (using the parameters a and P just obtained) are plotted in Fig. 3, as a function of Q.
FIGURE 3.
EISF for ds-DNA at 5.2 and 8 Å as a function of Q. The solid line represents the fit of the experimental EISF according to Eq. 11, with a = 2.8 Å and P = 0.510. The triangles are the experimental EISF (Eq. 5). The error bars are the calculated uncertainties of the experimental EISF values. The dotted line represents the calculated Ao values (Eq. 6), also for a = 2.8 Å.
Finally the value of q was obtained according to Eq. 10—from the sample stoichiometry, the value of p was calculated to be p = 0.32. This value, along with P = 0.51 referred to above leads to the value q = 0.28.
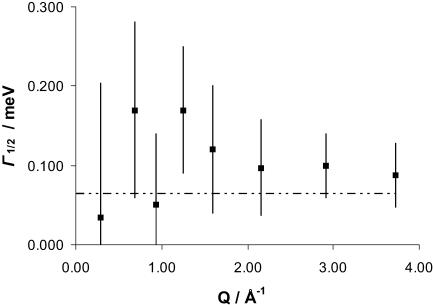
The same initial data treatment with FITMIB was applied to the results obtained at 5.2 Å for ss-DNA, and the Γ1/2 values as a function of Q as well as the integrated intensities were obtained. In Fig. 4 we can see the plot of Γ1/2 as a function of Q and in Fig. 5 the EISF(exp) values are represented also as a function of Q. Indeed, the error bars are much larger for ss-DNA, as the amount of substance in the sample was reduced, and although we used double sampling time (16 h), we have rather poor statistics. Nevertheless, we can say that no momentum dependence is observed for the obtained Γ1/2 values. The same is apparent from the EISF values plotted on Fig. 5. Even so, we did the minimization of the EISF to obtain the P-value (Eq. 11), considering a = 2.8 Å, just for the sake of comparison with the ds-DNA results. The value obtained was P = 0.78, which equals, within uncertainty, the p-value calculated from the sample stoichiometry, p = 0.76.
FIGURE 4.
Γ1/2 as a function of Q for ss-DNA at 5.2 Å. The solid squares are the Γ1/2 values obtained from FITMIB for the Lorentzian at 5.2 Å. The error bars are the uncertainties provided by the FITMIB program. The dashed line represents the instrument resolution in these conditions (64 μeV).
FIGURE 5.
EISF as a function of Q for ss-DNA at 5.2 Å. The solid line represents the fit of the experimental EISF according to Eq. 11, with a = 2.8 Å and P = 0.775. The triangles are the experimental EISF (Eq. 5). The error bars are the calculated uncertainties of the experimental EISF values. The dotted line represents the calculated Ao values (Eq. 6), also for a = 2.8 Å.
DISCUSSION
The momentum transfer dependence of the EISF values, as well as the broadening of the quasielastic peak and its Q dependence are consistent with the presence of local motion, or confinement, in the samples of ds-DNA studied here. The value obtained for the radius of the confinement sphere (a = 2.8 Å) is in good agreement with similar ones obtained for proteins and DNA at low or limiting hydrations: a = 3 Å was obtained for C-phycocyanin (100% hydration) at 333 K (Bellissent-Funel et al., 1992); a = (1.7 ± 0.1) Å was obtained for parvalbumin (0.65 g D2O/g protein) at 298 K (Zanotti et al., 1999); a = (3.6 ± 0.4) Å for hen egg-white lyzozyme crystals (∼30% (v/v) of solvent, which corresponds to ∼0.38 g H2O/g protein) at 300 K (Bon et al., 2002); a = 3 Å for DNA fibers (9.5 mol H2O/mol bp, A-DNA) (Schreiner et al., 1988).
Regarding the water diffusion, our value D = 2.2 × 10−5 cm2 s−1 is similar to the value for bulk water at this temperature, D = 2.3 × 10−5 cm2 s−1 (Teixeira et al., 1985). It is also similar to the value D = 2.6 × 10−5 cm2 s−1 obtained for fully hydrated C-phycocyanin at 300 K (Bellissent-Funel et al., 1992) and to the range of values estimated for ds-DNA with water contents between 0.5 and 2.5 g H2O/g ds-DNA, 1.4–4.2 × 10−5 cm2 s−1 (Beta et al., 2003), and larger than the values D = 0.68 × 10−5 cm2 s−1 obtained for parvalbumin with 0.65 g D2O/g protein, at 298 K (Zanotti et al., 1999), D = 0.39 × 10−5 cm2 s−1 for hen egg-white lyzozyme crystals with 0.38 g H2O/g protein (Bon et al., 2002) and D = 0.5 × 10−5 cm2 s−1 for DNA fibers. Regarding this last value, we should note that they have used a lower hydration regime (9.5 mol H2O/mol bp whereas we have 20 mol H2O/mol bp), which reflects in a lower limiting Γ1/2 value (20 μeV) and should correspond to a lower diffusion coefficient. If we except the value obtained for parvalbumin, which is very low taking into account the water content of the sample, the other results, including ours, are not surprising, as it is known that only with degrees of hydration of ∼0.6 (w/w) the properties of interfacial water become close to bulk solvent properties (Goldanskii and Krupanskii., 1995).
The use of a refined model, introducing the parameters p and q, allowed us to distinguish between two types of water present in ds-DNA. Our results indicate that a fraction q = 0.28 of the water protons are strongly attached to the ds-DNA surface, whereas the remaining water has a motion that can be interpreted within the model diffusive motion confined within a sphere of radius 2.8 Å. Very interestingly, a recent study of water at DNA surfaces by femtosecond-resolved fluorescence (Pal et al., 2003b) also showed the presence of two types of water, that the authors call a “bimodal hydration behavior”, with two distinct time constants: ∼1 ps (that they classify as bulk type, labile water, that represents 60% of the total water) and 20 ps (classified by the authors as weakly bound, ordered water, representing 40% of total water). Not only have they observed the same behavior as we find here, namely of two types of water with different dynamics, but also our percentages are close to theirs; our value for the percentage of protons strongly attached to the ds-DNA surface (q = 0.28) is ∼30% and corresponds to their fraction of 40% of “weakly bound, ordered water.”
In the samples of ss-DNA, on the contrary, we could not find any indication of confinement, from the momentum transfer dependences of either the EISF or Γ1/2. This shows that the water present, although contributing to the quasielastic peak, does not present the typical behavior of confined water.
It is very enlightening to compare these results with the ones obtained recently by one of us (G.M.) in a differential scanning calorimetry study of low hydration samples of ds-DNA and ss-DNA (Mrevlishvili at al., 2001,2002). In these studies, the authors use the concept of “unfreezable water”, that is, the amount of water present in the DNA sample that does not undergo the liquid-to-solid phase transition when the sample is cooled down in a DSC experiment. This amount was found to be 0.55 g H2O/g DNA for ds-DNA and 0.40 g H2O/g DNA for ss-DNA. We chose our water contents for the scattering experiments accordingly. In the interpretation of the obtained calorimetric data, the transformation “double-stranded helix”→“single-stranded chains” is suggested to be accompanied by dehydration of ds-DNA, which amounts to 0.15 g H2O/g DNA. Further, this value is equal to the one obtained in the first study (Mrevlishvili et al., 2001) for the hydration change upon thermal denaturation of ds-DNA. This was interpreted as suggesting that the “double helix” → “single-stranded” thermal transition is accompanied by disruption of the ordered part of the water present in the hydration shell of the double helix. Now, if we consider the result obtained in this study by neutron scattering, we did obtain a value for q, the fraction of strongly attached protons to the ds-DNA surface, of 0.28. This value corresponds to 0.16 g H2O/g DNA. As referred to above, in the case of ss-DNA no evidence was found of strongly attached protons, or better said, we could not distinguish different types of protons in the ss-DNA sample. This shows that we have obtained, by a completely independent measurement, a confirmation of the previous results obtained by calorimetry.
CONCLUSION
At this state of knowledge, we can say that in ds-DNA at this limiting hydration value (B-DNA) we can distinguish two types of water: one that presents the behavior of confined water (∼70%), within a sphere of radius 2.8 Å, with local diffusion coefficient D = 2.2 × 10−5 cm2 s−1, that is, slightly smaller than bulk water (probably water inside the grooves), and another type of water, attached to the ds-DNA surface (∼30%). This second type of water could be inside or outside the grooves. This water is lost upon ds-DNA thermal denaturation, as opposite to the first one, that is kept even after thermal denaturation. The fact that we found this second type of water to be absent in ss-DNA, and that the numerical values obtained here for the difference do agree with the ones obtained by calorimetry, strengthens the interpretation put forward before, i.e., that the thermal transition between ds-DNA and ss-DNA is accompanied by disruption of the ordered water fraction in the inner hydration of the double helix, and stresses the importance of hydration effects on the maintenance of the double-helix structure.
Acknowledgments
We thank Laboratoire Léon Brillouin for awarding beam time for the neutron-scattering experiments. M.B. thanks Mário Pinto from INEGI, Porto, for making aluminum sample holders to be used in these experiments. We gratefully acknowledge Rémi Kahn for help with the measurements at MIBEMOL, as well as with the programs for data handling.
M.B. and V.C. also thank, for the economical support at Laboratoire Léon Brillouin, the European Union through the Access to Research Infrastructures action of the Human Potential Program Access to Research Infrastructures. We acknowledge Fundação para a Ciência e Tecnologia for financial support to Centro de Investigação em Química (Universidad do Porto), Unidade de Investigação 81.
G. Mrevlishvili's present address is Dept. of Physics, Tbilisi State University, 380028 Republic of Georgia.
References
- Albiser, G., A. Lamiri, and S. Premilat. 2001. The A-B transition: temperature and base composition effects on hydration of DNA. Int. J. Biol. Macromol. 28:199–203. [DOI] [PubMed] [Google Scholar]
- Bellissent-Funel, M.-C., J. Teixeira, K. F. Bradley, and S. H. Chen. 1992. Dynamics of hydration water in protein. J. Phys. I. 2:995–1001. [Google Scholar]
- Beta, I. A., I. Michalarias, R. C. Ford, J. C. Li, and M.-C. Bellissent-Funel. 2003. Quasi-elastic neutron scattering study of hydrated DNA. Chem. Phys. 292:451–454. [Google Scholar]
- Blackburn, G. M., and M. J. Gait. 1990. Nucleic Acids in Chemistry and Biology. Oxford University Press, Oxford, UK.
- Bloomfield, V. A., D. M. Crothers, and I. Tinoco, Jr. 2000. Nucleic Acids. University Science Books, Sausalito, CA.
- Bon, C., A. J. Dianoux, M. Ferrand, and M. S. Lehmann. 2002. A model for water motion in crystals of lyzozyme based on an incoherent quasielastic neutron scaettering study. Biophys. J. 83:1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, M., V. Tereshko, M. Teplova, G. Minasov, A. Joachimiak, R. Sanishvili, C. M. Weeks, R. Miller, M. Maier, H. An, P. Dan Cook, and M. Manoharan. 1998. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers. 48:234–252. [DOI] [PubMed] [Google Scholar]
- Foucat, L., J.-P. Renou, C. Tengroth, S. Janssen, and H. D. Middendorf. 2002. Dynamics of proteins at low temperatures: fibrous vs. globular. Appl. Phys. A. 74:S1290–S1292. [Google Scholar]
- Goldanskii, V. I., and Y. F. Krupanskii. 1995. Protein Dynamics in Protein-Solvent Interactions. R. B. Gregory, editor. Marcel Dekker, New York/Basel, Switzerland/Hong Kong.
- Grimm, H., and A. Rupprecht. 1989. Hydration structure in natural DNA observed by thermal neutron scattering. Eur. Biophys. J. 17:173–186. [DOI] [PubMed] [Google Scholar]
- Kankia, B., and L. Marky. 1999. DNA, RNA, and DNA/RNA oligomer duplex: a comparative study of their stability, heat, hydration and Mg binding properties. J. Phys. Chem. B. 103:8759–8767. [Google Scholar]
- Liepinsh, E., G. Otting, and K. Wuthrich. 1992. NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Res. 20:6549–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrevlishvili, G. M., A. P. S. M. C. Carvalho, M. A. V. Ribeiro da Silva, T. D. Mdzinarashvili, G. Z. Razmadze, and T. O. Tarielashvili. 2001. The role of bound water on the energetics of DNA duplex melting. J. Therm. Anal. Cal. 66:133–144. [Google Scholar]
- Mrevlishvili, G. M., A. P. S. M. C. Carvalho, and M. A. V. Ribeiro da Silva. 2002. Low-temperature DSC study of the hydration of ss-DNA and ds-DNA and the role of hydrogen-bonded network to the duplex transition thermodynamics. Thermochim. Acta. 394:73–82. [Google Scholar]
- Pal, S. K., L. Zhao, T. Xia, and A. H. Zewail. 2003a. Site- and sequence selective ultrafast hydration of DNA. Proc. Natl. Acad. Sci. USA. 100:13746–13751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S. K., L. Zhao, and A. H. Zewail. 2003b. Water at DNA surfaces: ultrafast dynamics in minor groove recognition. Proc. Natl. Acad. Sci. USA. 100:8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. 1984. Principles of Nucleic Acid Structure. Springer-Verlag, New York/Berlin/Hidelberg/Tokyo.
- Schneider, B., K. Patel, and H. M. Berman. 1998. Hydration of the phosphate group in double-helical DNA. Biophys. J. 75:2422–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner, I. J., M. M. Pintar, A. J. Dianoux, P. Volino, and A. Rupprecht. 1988. Hydration of NaDNA by neutron quasi-elastic scattering. Biophys. J. 53:119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotton, M. W., L. H. Pope, T. Forsyth, P. Langan, R. C. Denny, U. Giesen, M.-T. Dauvergne, and W. Fuller. 1997. A high-angle neutron fibre diffraction study of the hydration of deuterated A-DNA. Biophys. Chem. 69:85–96. [DOI] [PubMed] [Google Scholar]
- Stent, G. S. 1995. The aperiodic crystal of heredity. Ann. N. Y. Acad. Sci. 758:25–31. [DOI] [PubMed] [Google Scholar]
- Sundaralingam, M., and B. Pan. 2002. Hydrogen and hydration of DNA and RNA nucleotides. Biophys. Chem. 95:273–282. [DOI] [PubMed] [Google Scholar]
- Teixeira, J., M.-C. Bellissent-Funel, S. H. Chen, and A. J. Dianoux. 1985. Experimental determination of the nature of diffusive motions of water molecules at low temperatures. Phys. Rev. A. 31:1913–1916. [DOI] [PubMed] [Google Scholar]
- Volino, F., and A. J. Dianoux. 1980. Neutron incoherent scattering law for diffusion in a potential of spherical symmetry: general formalism and application to diffusion within a sphere. Mol. Phys. 41:271–279. [Google Scholar]
- Woda, J., B. Schneider, K. Patel, K. Mistry, and H. M. Berman. 1998. An analysis of the relationship between hydration and protein-DNA interactions. Biophys. J. 75:2170–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti, J.-M., M.-C. Bellissent-Funel, and J. Parello. 1997. Dynamics of a globular protein as studied by neutron scattering and solid-state NMR. Phys. B. 234–236:228–230.
- Zanotti, J.-M., M.-C. Bellissent-Funel, and J. Parello. 1999. Hydration coupled dynamics in proteins studied by neutron scattering and NMR: the case of the typical EF-hand calcium-binding parvalbumin. Biophys. J. 76:2390–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]