Abstract
Tolevamer, (GT160-246), is a sodium salt of styrene sulfonate polymer that is under development for the treatment of diarrhea caused by infection with Clostridium difficile. Pulsed ultrafiltration binding experiments in phosphate buffer containing 0.15 M Na+ provide per polymer chain dissociation constants of 133 nM and 8.7 μM for the binding of tolevamer to C. difficile toxins A and B, respectively. At 0.05 M Na+, the binding of toxin A to tolevamer is irreversible, whereas the dissociation constant to toxin B under these conditions is 120 nM. Binding constants obtained from fluorescence polarization data for toxin A binding to tolevamer at 0.15 M Na+ agree substantially with those obtained by pulsed ultrafiltration. The binding activity of tolevamer reported here correlates well with previously reported results for the inhibition of the biological activity of C. difficile toxins A and B. From the fluorescence polarization data, it is estimated that one toxin A molecule interacts with between 600 to 1000 monomer units on tolevamer at 0.15 M Na+. Thus, the data suggest a very large interaction surface between polymer and toxin A.
INTRODUCTION
Clostridium difficile infection is the major identified cause of antibiotic-associated diarrhea in hospitals. Under ordinary conditions, the presence of normal intestinal flora inhibits the growth of C. difficile. However, after antibiotic treatment, C. difficile can proliferate in the lower intestinal tract. C. difficile infection results in symptoms including profuse diarrhea and abdominal pain (Pothoulakis and LaMont, 1993; Kelly and LaMont, 1998). In severe cases, pseudomembranous colitis and toxic megacolon may occur (Pothoulakis and LaMont, 1993; Kelly and LaMont, 1998; Sheth and LaMont, 1998). C. difficile infection is typically treated with one of two antibiotics, metronidazole or vancomycin. Relapse of disease after such antibiotic treatment occurs in 5–20% of patients, most likely because such antibiotics continue to suppress not only C. difficile growth, but also the growth of normal competitive intestinal flora.
The symptoms of C. difficile infection are mediated by two high molecular mass protein toxins produced by this bacterium, toxins A and B. Toxin A is thought to play the primary role in antibiotic-associated diarrhea, though toxin B appears to be significant as well (Lyerly et al., 1988; Riegler et al., 1995; Limaye et al., 2000).
An attractive approach to the treatment of C. difficile infection would involve binding and neutralizing C. difficile toxins without disrupting the reestablishment of normal bacterial growth. Cholestyramine, a cationic resin that has been used clinically as a bile acid sequestrant, binds C. difficile toxins in vitro (Taylor and Bartlett, 1980), and has been tested in humans as a treatment for C. difficile colitis. However, the activity shown by this resin was modest, and it is not recommended for the treatment of severe colitis (Burbige and Milligan, 1975; George et al., 1980; Tedesco, 1982). In previous work, we have shown that modest doses of tolevamer, a high molecular mass nonantimicrobial polymer, neutralizes both toxin A and toxin B mediated inhibition of protein synthesis in Vero cells, and substantially decreases toxin A mediated fluid accumulation and permeability in a rat ileal loop model (Kurtz et al., 2001). Most significantly, tolevamer substantially reduces the mortality of C. difficile-infected hamsters (Kurtz et al., 2001). In the work reported here we demonstrate that tolevamer binds both toxins A and B with significant affinity, and that the binding affinities we have determined correlate well with the ability of tolevamer to neutralize the activities of these toxins. We demonstrate further that the binding of toxins A and B is not a general property of polyanions, since poly(2-acrylamido-2-methyl-1-propanesulfonate) (AMPS), a high molecular mass polyanion of similar charge density to tolevamer, does not bind either toxin to any measurable extent.
MATERIALS AND METHODS
Materials
Chemically, tolevamer is a high molecular mass sodium salt of polystyrene sulfonate, prepared by Genzyme Corporation (Cambridge, MA). For the fluorescence polarization measurements, fluorescein-labeled tolevamer was synthesized at Genzyme by copolymerization of styrene sulfonate with FITC-labeled 4-aminostyrene. As estimated by UV absorbance, the polymer was labeled to ∼1 mol %. AMPS was purchased from Aldrich (Milwaukee, WI) and purified by dialysis. The molecular masses of tolevamer and AMPS, measured by size-exclusion chromatography (SEC) with UV detection, were estimated to be 600 kDa and 200 kDa, respectively. 10x phosphate buffered saline buffer (PBS), pH = 7.0, was obtained from GIBCO (Carlsbad, CA), and diluted with Millipore (Billerica, MA) filtered water to give 50 mM sodium ion (low salt buffer) or 150 mM sodium ion (physiological buffer).
C. difficile toxins were obtained from TECHLAB (Blacksburg, VA). The concentration of toxin A was 2 mg/ml and the concentration of toxin B varied between 0.2 to 0.44 mg/ml. The molecular masses of toxins A and B are 308 and 270 kDa, respectively.
Pulsed ultrafiltration methods
The pulsed ultrafiltration (PUF) cell used in this study followed the design of Woodbury and Venton (Chen et al., 1998; Woodbury and Venton, 1998,1999). The cell volume was 1 ml. The Millipore ultrafilter membranes used in the cell had a nominal molecular mass cutoff of 500 kDa. The cell was kept at a constant temperature of 25°C by immersing in a constant temperature water bath. A Waters 2690 Separation pump was used to control the sample injection and buffer flow rate (0.2 ml/min). A Waters 996 Photodiode Array Detector was used for detection at 280 nm and data were collected in digital format. Before the start of the experiment, toxin samples were stored at 5°C.
PUF experiments consisted of four steps, and took ∼6 h. Each new membrane was first flushed through with buffer for 2–3 h or until a stable baseline was achieved. Protein ligand was injected and monitored for 1 h in the absence of polymer. Then, polymer was injected and washed with buffer for ∼2 h. Finally, the same amount of protein ligand was again injected and monitored for 1 h to assess polymer-protein binding.
The mathematical analysis of the PUF method follows closely that described by Chen et al. (1998). Briefly, in the absence of ligand binding, the flow curve after the injection of a short pulse of ligand into the cell reflects the dilution of the ligand by the continuous flow of buffer through the system:
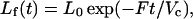 |
(1) |
where Lf(t) is the concentration of ligand exiting the cell, as monitored by UV absorption, F is the flow rate, in ml/min, t is the time in minutes, and Vc is the physical volume of the cell, in milliters. L0 = N0/Vc, where N0 is the total moles of ligand injected into the cell. For an infinitely narrow pulse, L0 would thus be the initial total concentration of ligand in the cell immediately after injection of the pulse of ligand. In our analysis, we determined Vc by fitting the blank flow curve to Eq. 1. The values thus obtained agreed to within 10% to the physical volume of the cell, as estimated by injecting liquid into the cell.
In the presence of polymer in the cell, Nb, the number of moles of bound ligand at any time is given by the equation of mass balance:
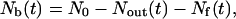 |
(2) |
where Nout(t) is the number of moles of ligand that have exited the cell, and Nf(t) is the number of moles of free ligand remaining in the cell. Nout(t) can be obtained by integrating from time t to ∞:
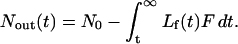 |
(3) |
Substituting this expression into (2) and simplifying, we obtain:
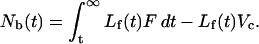 |
(4) |
Hence, we can obtain Nb(t) at any point in the flow curve by integrating over time, from some convenient starting point t. A better method, which we used in our analysis, is the constant concentration method (Chen et al., 1998). This method corrects for the effects of nonspecific binding, and does not require explicit knowledge of Vc. According to this method, the difference at constant concentration Lf(t) is taken between the area under the curve for the sample containing polymer and the blank sample containing no polymer. This difference defines Nb(t):
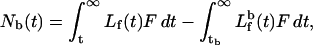 |
(5) |
where the first integral reflects integration of the sample from the time t at which the free concentration of ligand exiting the sample cell is equal to Lf, and the second integral reflects integration from the time tb, where the free concentration of ligand exiting the blank cell is equal to the same value Lf(t). Data processing was performed by fitting the free ligand flow curve to extract the flow cell volume, and calculating the free and bound ligand concentration from the flow curves. The fitting of the extracted bound-free ligand curve to obtain the binding parameters was performed using the program SigmaPlot (SPSS Inc.).
Fluorescence polarization methods
Fluorescence polarization provides another method for determining binding parameters (Jameson and Sawyer, 1995). Fluorescence polarization measurements were performed in a 96-well format (LJL Analyst) using an excitation filter centered at 485 nm, an emission filter centered at 530 nm, and a dichroic mirror with a short wavelength cutoff of 505 nm.
Toxin dilutions were made directly in the 96-well plates, for each constant concentration of fluorescein-labeled tolevamer (FL-tolevamer) monitored. Duplicate data points were obtained for each concentration of toxin and polymer. The variation in these duplicate points provided the reported estimates of the standard deviation. For each sample, the background fluorescence was subtracted using blanks containing the same concentration of toxin, but no added polymer. Background subtracted intensities of polarized light were monitored, and the polarization in mP was determined from the equation
 |
(6) |
where I‖ is the intensity of light parallel to the direction of the incident light, and I⊥ is the intensity of light perpendicular to the polarization of the incident light. Defined in this manner, the polarization in mP has a theoretical range from 0 for complete depolarization to 500 for a completely rigid system.
Upon binding to toxin, the polarization of FL-tolevamer increases as the rotational mobility of the fluorescein probe decreases. Using the method of isoparametric analysis (Chatelier and Sawyer, 1987; Winzor and Sawyer, 1995), we have determined binding curves for the interaction. In this method, the toxin concentration is varied at constant polymer concentration. Data are fitted to smooth curves, and from these curves, protein concentrations are evaluated at constant polarization, as illustrated below.
Assuming that constant polarization corresponds to constant binding density, and that constant binding density corresponds to constant free toxin concentration, then at constant polarization:
 |
(7) |
where Ltot is the total toxin concentration, Lf is the unbound toxin concentration, Ptot is the total polymer concentration, and r is the binding density (toxin bound per unit of polymer concentration). From this equation we see that a plot of Ltot versus Ptot at constant polarization (constant r) will give a plot with a slope of the binding density and an intercept of the free toxin concentration.
Binding analysis
Values of r versus Lf obtained from the flow data and from the isoparametric analysis are fitted to the model for binding to independent and identical sites:
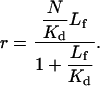 |
(8) |
In this equation we have defined N as the average number of toxin binding sites on each polymer molecule. Kd is the dissociation binding constant (on a per site basis), and the other parameters are as defined above. Note that the dissociation constant on a per molecule basis is equal to Kd/N. N/Kd thus defines the y-intercept on a Scatchard plot of r/Lf versus r.
RESULTS
Pulsed ultrafiltration
To assess the net electrostatic contribution to the binding free energy, binding measurements were performed at two different salt concentrations. Fig. 1, a and b, show the PUF curves for toxin A binding to tolevamer in low salt (0.05 M Na+) and physiological salt (0.15 M Na+) phosphate buffer, respectively. It can be discerned from Fig. 1 a that the area under the curve in the presence of polymer is less than the area in the absence of polymer, and hence that the binding of toxin A under these low salt conditions is irreversible. A quantitative comparison of the areas shows that toxin A leaving the cell is reduced by 28% when tolevamer is present in the cell. The amount of toxin A that remains in the cell under these low salt conditions corresponds to ∼2.5 toxin A molecules bound per every polymer molecule. In contrast, a quantitative comparison of the curves in Fig. 1 b shows that the areas are equal to within 2%. Hence, under physiological salt conditions, binding is reversible. Qualitatively, it can be seen from a comparison of Fig. 1, c and d, that the binding of toxin B to tolevamer is stronger at low salt than at physiological salt. A quantitative discussion of this point will be presented below.
FIGURE 1.
Flow profiles for toxins A and B at two salt concentrations. (a) 0.04 mg toxin A, and 0.015 mg tolevamer, in 0.05 M phosphate buffer. (b) 0.02 mg toxin A and 0.03 mg tolevamer, in 0.15 M phosphate buffer. (c) 0.031 mg toxin B and 0.06 mg tolevamer in 0.05 M phosphate buffer, and (d) 0.0176 mg toxin B and 1.0 mg tolevamer in 0.15 M phosphate buffer. In all cases, the dotted curve is for the toxin in the absence of polymeric drug, whereas the solid curve is for the toxin in the presence of the indicated amount of tolevamer.
For comparison, we examined the binding of toxins A and B to another high molecular mass sulfonated polyanion, AMPS. Fig. 2 demonstrates that AMPS has no effect on the flow behavior of either toxins A or B, even under low salt conditions. Hence, the binding of AMPS to these toxins is too weak to measure under the conditions of our experiments.
FIGURE 2.
Flow profiles for toxins A and B in the presence of AMPS, in 0.05 M phosphate buffer. (a) 0.02 mg toxin A, and 0.03 mg AMPS. (b) 0.044 mg toxin B and 0.06 mg AMPS. In both cases, the dotted curve is for the toxin in the absence of AMPS, whereas the solid curve is for the toxin in the presence of the indicated amount of AMPS.
Binding curves extracted from the data of Fig. 1 are shown in Fig. 3 for toxins A and B, at 0.15 M Na+, and for toxin B at 0.05 M Na+. These data are plotted as r versus Lf. For the data taken at 0.15 M Na+, the low range of binding densities covered does not allow us to obtain an estimate of the N, the total number of toxin binding sites on each polymer. From a linear fit of the data we obtain Kd/N = 133 nM and 8.7 μM for toxins A and B, respectively. From the data for toxin B at 0.05 M Na+, shown in Fig. 3 c, we are able to fit the data to Eq. 8, and from this fitting we obtain Kd/N = 120.9 ± 0.4 nM, and N = 0.203 ± 0.0004. The small N obtained for toxin B at 0.05 M NaCl is notable, and suggests that under these low salt conditions a single molecule of toxin B can bind ∼5 molecules of polymeric drug.
FIGURE 3.
Binding curves extracted from the data of Fig. 1, plotted as r versus Lf. (a) Binding curve for 0.02 mg toxin A, and 0.03 mg tolevamer, in 0.15 PBS. The solid curve is a fitting of the data to a single site binding model with Kd = 133 nM. (b) Binding curve for 0.0176 mg toxin B, and 1.0 mg tolevamer, in 0.15 PBS. The solid curve is a fitting of the data to a single site binding model with Kd = 8.7 μM. (c) Binding curve for 0.031 mg toxin B, and 0.06 mg tolevamer, in 0.05 PBS. The solid curve is a fitting of the data to Eq. 8 with Kd/N = 121 ± 0.4 nM, and N = 0.203 ± 0.0004.
If a net number m of sodium ions are released during the binding process, then the observed association constants will increase according to Anderson and Record (1990)
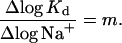 |
(9) |
Using this equation, and equilibrium constants determined at 0.05 M Na+ and at 0.15 M Na+, we estimate that m = 3.9 sodium ions are released during the association process for toxin B. This small net number of sodium ions released suggests that ionic interactions do not dominate the binding free energy.
Fluorescence polarization
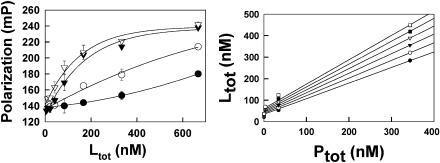
Polarization data for toxin A binding to FL-tolevamer are shown in Fig. 4 a. The relatively low polarization values that are observed for tolevamer are not surprising, and reflect the high internal flexibility of this linear chain molecule. By curve-fitting the data we can obtain plots of Ltot versus Ptot at constant polarization, as shown in Fig. 4 b. In obtaining these plots, we used polarization values over the range of 150–160 mP. By using this polarization range we were able to use well-defined values of Ltot at each value of Ptot. Linear fits of Ltot versus Ptot allowed us to determine Lf and r from the intercepts and slopes, respectively. These values of Lf and r are plotted in Fig. 5, and fitted to Eq. 8 to give Kd/N = 36 ± 4 nM, and N = 3.6 ± 0.2 for the binding of toxin A to Fl-tolevamer at 0.15 M Na+.
FIGURE 4.
(a) Raw polarization data as a function of toxin A concentration determined at different drug concentrations. On going from the upper to the lower curve, the concentration of FL-tolevamer is equal to 344 nM (▿), 34.4 nM (▾), 3.44 nM(○) and 0.34 nM (•), on a per molecule basis, assuming a polymer molecular mass of 600 kDa. (b) Derived data of the total toxin concentration versus total concentration of tolevamer, at constant polarization. The lines represent best least squares linear fits to constant polarizations of 150 mP (•), 152 mP (○), 154 mP (▾), 156 mP (▿), 158 mP (▪) and 160 mP (□).
FIGURE 5.
Binding curve derived from isoparametric analysis of fluorescence binding data for tolevamer binding to toxin A over the range of 150–160 mP. The solid line is a best fit to a single site model with Kd/N = 36 ± 4 nM, and N = 3.6 ± 0.2.
DISCUSSION
Binding of toxins A and B provides a quantitative explanation of the biological activity of tolevamer
Under conditions of a large excess of polymer over toxin, as are anticipated for patients under treatment with this polymer, the free concentration of polymer should nearly equal the total concentration of polymer, and the fraction fb of bound toxin will be given by
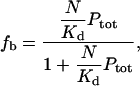 |
(10) |
where Ptot is the total concentration of polymer. Note that fb does not depend on the total concentration of toxin, provided that the polymer concentration is in excess.
Using Eq. 10 as a starting point, we suggest that the binding parameters that we have determined provide a quantitative explanation for the effect of tolevamer on toxin-mediated inhibition of protein synthesis (Kurtz et al., 2001). In previous work with Vero cell monolayers we have shown that, at a concentration of 5 mg/ml, tolevamer completely abolished the inhibition of protein synthesis by 5 ng/ml toxin A (Kurtz et al., 2001). Note first that 5 ng/ml corresponds to a toxin A concentration of 17 pM. 5 mg/ml tolevamer gives a polymer concentration of 8.3 μM. Hence, under the conditions of these experiments, the polymer is in substantial excess over the toxin, and Eq. 10 applies. If we substitute the value of Kd/N = 133 nM for toxin A that we have obtained from the PUF experiment, we determine fb = 0.984. Therefore, 98% of all toxin A should be bound by tolevamer under the conditions of these experiments. Hence, under the conditions of these experiments, the free concentration of toxin A should be 0.016 (5 ng/ml) = 0.08 ng/ml, which, according to the curve shown in Fig. 1 A of Kurtz et al. (2001), is well below the threshold for observable biological response.
Kurtz et al. (2001), previously reported that toxin B completely inhibited protein synthesis in Vero cell monolayers at a concentration of 1.25 ng/ml. In contrast, in the presence of 5 mg/ml polymer, it required 5 ng/ml of toxin B to completely inhibit protein synthesis. Substituting Kd/N = 8.7 μM, we calculate that ∼50% of all toxin B present under these conditions should be free. Hence, at 5 ng/ml toxin B, in the presence of 5 mg/ml polymer, the free toxin concentration should be 2.5 ng/ml. From the results reported by Kurtz et al. (2001), 2.5 ng/ml of free toxin B should completely inhibit protein synthesis. Hence, the binding data agree with the protein synthesis inhibition data in that the polymer diminishes, but does not eliminate the effect of toxin B over the concentration ranges tested in these studies (Kurtz et al, 2001). Our findings suggest that tolevamer may exert somewhat greater activity on the inhibition of protein synthesis than would be anticipated based on the binding data, although the modest discrepancy is perhaps not remarkable given the very different experimental parameters.
The ability of tolevamer to neutralize the enterotoxic activity of Toxin A in a rat ileal loop assay (Kurtz et al., 2001) is likewise in quantitative agreement with the binding results reported here. In the ileal loop assay, tolevamer was mixed with toxin A and injected into rat ileal loops. Fluid accumulation and permeability were monitored over a 4-h time period. In these experiments, a dose of 5 mg of tolevamer abolished both the excess fluid accumulation and the increase in intestinal permeability that were mediated by 5 μg of toxin A. Since the average volume of a rat ileal loop is ∼0.5–1 ml, the concentration of tolevamer in these experiments was between 5–10 mg/ml. Under these conditions, the PUF binding analysis would predict that the fraction of bound toxin should vary from 98.5% to 99.2%.
The binding data reported here provide a reasonable physical chemical model to support our optimism that tolevamer may ultimately prove effective as a drug for the treatment of antibiotic-associated diarrhea. Based on clinical titers, toxin A in the stool of patients with C. difficile-associated diarrhea has been estimated as generally <1 μg/ml (McFarland et al., 1991). If the volume of liquid in the patient's gut is estimated at ∼1 L, then the concentration of polymer should be ∼5 mg/ml. If we take the polymer molecular mass as 600 kDa, and the toxin molecular mass as 300 kDa, then we can calculate the effective concentration of polymer in the gut to be 8.3 μM (assuming a theoretical dose of 5 g/day), and the concentration of toxin A to be <3.3 nM. The large excess of polymer over toxin assures Eq. 10 applies. Under these conditions, as per our previous calculations, 98% of all toxin A in the gut should be bound. For 5 mg/ml tolevamer, as per our previous calculations, we can estimate that ∼50% of all toxin B present in the gut should be bound. We note however, that our calculations do not take into account the unknown effect of other gut contents on the binding of toxins A and B to tolevamer.
The correlations that we find between binding and biological activity for tolevamer contrast sharply with our findings for the control polyanion AMPS. AMPS has been tested alongside tolevamer and has failed to demonstrate C. difficile toxin A or toxin B neutralization in cellular models of toxin activity (unpublished data and Kurtz et al., 2001). In the hamster model of C. difficile colitis, AMPS also showed no measurable activity in preventing toxin-mediated colitis and mortality. Both of these observations are consistent with the inability of AMPS to bind toxins A or B in either of our binding assays.
The binding of tolevamer to C. difficile toxins is not purely electrostatic in origin
Only four sodium ions are thermodynamically released upon binding of toxin B to tolevamer. This small number of sodium ions suggests that the nonspecific, electrostatic interaction between tolevamer and the toxins, although significant at lower salt concentrations, does not dominate the binding thermodynamics under physiological conditions. This conclusion is further supported by the observation that neither toxin A nor toxin B binds to any measurable extent to AMPS, despite the fact that the overall charge density on this polyanion is comparable to that on tolevamer. This difference between AMPS and tolevamer cannot be explained by the higher molecular mass of the tolevamer sample (600 kDa) compared to the AMPS sample (200 kDa), since based on our fluorescence polarization results, the AMPS sample should be long enough to contain at least one strong toxin-binding site.
Multiple contacts stabilize the polymer-toxin interaction
The data that we have obtained at higher binding densities suggests that tolevamer interacts with toxins A and B through multiple weak contacts. Thus, analysis of the fluorescence polarization data for the binding of FL-tolevamer to toxin A suggests that a single 600 kDa polymer can bind ∼3–4 toxin molecules. Since the monomer molecular mass of the polymer is 206 Da, this implies that a single toxin molecule interacts with ∼800 monomer units on the polymer. The extended length of an individual monomer is ∼1.8 Å, implying that a single toxin molecule interacts with a linear region of the polymer of ∼1400 Å. Assuming a roughly spherical shape and a specific volume of 0.7 cm3/g (van Holde, 1985), for a protein of molecular mass 300 kDa, we can estimate a radius of ∼44 Å, and a circumference of ∼300 Å. As illustrated in the cartoon shown in Fig. 6, if such a toxin were to interact with an extended length of a polymer in the range of 1000–2000 Å, the polymer would need to wrap around the toxin on average ∼4–5 times, and could bind a maximum of ∼3–4 toxin molecules. Though we were unable to determine N for the interaction of toxin B with tolevamer at 0.15 M Na+, the binding curves shown in Fig. 3 c indicate that, at least under low salt conditions, toxin B also interacts with a large number of individual monomer units on tolevamer.
FIGURE 6.
A schematic illustration of how multiple contacts stabilize the interaction of individual toxin molecules with a linear polymer of tolevamer. Based on our calculations, 800 monomer units of tolevamer, or a linear stretch of 1400 Å, bind one molecule of toxin A. Since there are ∼3,000 monomer units for a polymer of an average molecular mass of 600 kDa, this implies that, as illustrated, each polymer molecule can bind, on average, 3–4 toxin molecules.
We hypothesize that the interaction of C. difficile toxins with such large stretches of tolevamer is likely to seriously impair the ability of these toxins to recognize and bind to cell surfaces. The binding of tolevamer may also exert its effects by inhibiting endocytosis of toxins into the cytoplasm, or by interfering with the glucosyltransferase activity of RhoA and related GTPases. As intriguing as these hypotheses are, resolving the biological effects of tolevamer binding is beyond the scope of the current investigation.
Acknowledgments
The authors thank Professors C. Woodbury and D. Stimson for helpful discussions, and for help with the design of the pulsed ultrafiltration cell.
Qiuwei Xu's present address is Merck and Company, WP44-I130, PO Box 4, West Point, PA 19486.
William Braunlin's present address is Transgenomic, Inc., 10 Corporate Place South, Piscataway, NJ 08854.
References
- Anderson, C. F., and M. T. Record. 1990. Ion distributions around DNA and other cylindrical polyions: theoretical descriptions and physical implications. Annu. Rev. Biophys. Biophys. Chem. 19:423–465. [DOI] [PubMed] [Google Scholar]
- Burbige, E. J., and F. D. Milligan. 1975. Pseudomembranous colitis. Association with antibiotics and therapy with cholestyramine. JAMA. 231:1157–1158. [DOI] [PubMed] [Google Scholar]
- Chatelier, R. C., and W. H. Sawyer. 1987. Isoparametric analysis of binding and partitioning processes. J. Biochem. Biophys. Methods. 15:49–61. [DOI] [PubMed] [Google Scholar]
- Chen, C. J., S. Chen, C. P. Woodbury, and D. L. Venton. 1998. Pulsed ultrafiltration characterization of binding. Anal. Biochem. 261:164–182. [DOI] [PubMed] [Google Scholar]
- George, W. L., R. D. Rolfe, and S. M. Finegold. 1980. Treatment and prevention of antimicrobial agent-induced colitis and diarrhea. Gastroenterology. 79:366–372. [PubMed] [Google Scholar]
- Jameson, D. M., and W. H. Sawyer. 1995. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 246:283–300. [DOI] [PubMed] [Google Scholar]
- Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49:375–390. [DOI] [PubMed] [Google Scholar]
- Kurtz, C. B., E. P. Cannon, A. Brezzani, M. Pitruzzello, C. Dinardo, E. Rinard, D. W. Acheson, R. Fitzpatrick, P. Kelly, K. Shackett, A. T. Papoulis, P. J. Goddard, R. H. Barker, G. P. Palace, and J. D. Klinger. 2001. TOLEVAMER, a toxin binding polymer for treatment of Clostridium difficile colitis. Antimicrob. Agents Chemother. 45:2340–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye, A. P., D. K. Turgeon, B. T. Cookson, and T. R. Fritsche. 2000. Pseudomembranous colitis caused by a toxin A(−) B(+) strain of Clostridium difficile. J. Clin. Microbiol. 38:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland, L. V., G. W. Elmer, W. E. Stamm, and M. E. Mulligan. 1991. Correlation of immunoblot type, enterotoxin production, and cytotoxin production with clinical manifestations of Clostridium difficile infection in a cohort of hospitalized patients. Infect. Immun. 59:2456–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothoulakis, C., and J. T. LaMont. 1993. Clostridium difficile colitis and diarrhea. Gastroenterol. Clin. North Am. 22:623–637. [PubMed] [Google Scholar]
- Riegler, M., R. Sedivy, C. Pothoulakis, G. Hamilton, J. Zacherl, G. Bischof, E. Cosentini, W. Feil, R. Schiessel, and J. T. LaMont. 1995. Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J. Clin. Invest. 95:2004–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, S. G., and J. T. LaMont. 1998. Toxic megacolon. Lancet. 351:509–513. [DOI] [PubMed] [Google Scholar]
- Taylor, N. S., and J. G. Bartlett. 1980. Binding of Clostridium difficile cytotoxin and vancomycin by anion-exchange resins. J. Infect. Dis. 141:92–97. [DOI] [PubMed] [Google Scholar]
- Tedesco, F. J. 1982. Treatment of recurrent antibiotic-associated pseudomembranous colitis. Am. J. Gastroenterol. 77:220–221. [PubMed] [Google Scholar]
- van Holde, K. E. 1985. Physical Biochemistry. Prentice Hall, Englewood Cliffs, NJ.
- Winzor, D. J., and W. H. Sawyer. 1995. Quantitative Characterization of Ligand Binding. New York, Wiley-Liss.
- Woodbury, C. P., and D. L. Venton. 1998. Pulsed ultrafiltration: A new method for screening and measuring ligand binding. Am. Lab. 30:16–19. [Google Scholar]
- Woodbury, C. P., and D. L. Venton. 1999. Methods of screening combinatorial libraries using immobilized or restrained receptors. J. Chromatogr. B Biomed. Sci. Appl. 725:113–137. [DOI] [PubMed] [Google Scholar]