Abstract
DNA release from lipoplexes is an essential step during lipofection and is probably a result of charge neutralization by cellular anionic lipids. As a model system to test this possibility, fluorescence resonance energy transfer between DNA and lipid covalently labeled with Cy3 and BODIPY, respectively, was used to monitor the release of DNA from lipid surfaces induced by anionic liposomes. The separation of DNA from lipid measured this way was considerably slower and less complete than that estimated with noncovalently labeled DNA, and depends on the lipid composition of both lipoplexes and anionic liposomes. This result was confirmed by centrifugal separation of released DNA and lipid. X-ray diffraction revealed a clear correlation of the DNA release capacity of the anionic lipids with the interfacial curvature of the mesomorphic structures developed when the anionic and cationic liposomes were mixed. DNA release also correlated with the rate of fusion of anionic liposomes with lipoplexes. It is concluded that the tendency to fuse and the phase preference of the mixed lipid membranes are key factors for the rate and extent of DNA release. The approach presented emphasizes the importance of the lipid composition of both lipoplexes and target membranes and suggests optimal transfection may be obtained by tailoring lipoplex composition to the lipid composition of target cells.
INTRODUCTION
Cationic lipids are used for delivery of plasmid DNA vectors in both experimental biology and medicine. In recent years, encouraging results have been obtained in gene therapy of cystic fibrosis, cancer, and cardiovascular diseases (Caplen et al., 1995; Laitinen et al., 2000; Nabel et al., 1993; Nishikawa and Hashida, 2002; Woodle and Scaria, 2001). However, understanding of the DNA delivery process is still limited and the low efficiency of lipid-based transfection is a concern. The problem can be described in terms of specific biological barriers along the lipoplex delivery pathway (Bally et al., 1999; Wiethoff and Middaugh, 2003). In understanding the machinery of DNA delivery, the question of how DNA dissociates from the lipoplexes and is released into the cytoplasm is clearly important. It was demonstrated (Ashley et al., 1996; Szoka Jr. et al., 1996; Xu and Szoka Jr., 1996) that negatively charged liposomes initiate DNA release from lipoplexes as a result of membrane fusion and lipoplex charge neutralization by anionic lipids. A similar process of lipid exchange between lipoplexes and endosomal membranes could be responsible for DNA release into the cytoplasm during transfection (Nakanishi and Noguchi, 2001; Noguchi et al., 1998). A number of investigators observed release of DNA molecules from various lipoplexes after treatment with anionic lipids (Bhattacharya and Mandal, 1998; Harvie et al., 1998; Itaka et al., 2002; MacDonald et al., 1999a; Pantazatos and MacDonald, 2003; Sakurai et al., 2000; Zuhorn et al., 2002).
In this study, we focused on the mechanism of DNA release from lipoplexes by anionic lipids. Fluorescent probes were covalently attached to DNA and to lipid to evaluate DNA release with a FRET assay. This procedure indicated that the release of DNA is much slower than previously thought. Importantly, the DNA releasing activity of different classes of anionic lipids varies considerably. As to rationalize these differences, we performed x-ray diffraction experiments and established correlation of the DNA release capacity of the anionic lipids with the interfacial curvature of the mesomorphic structures developed when the anionic and cationic liposomes were mixed.
MATERIALS AND METHODS
Lipids
The triflate derivative of EDOPC was synthesized as previously described (MacDonald et al., 1999b,a; Rosenzweig et al., 2000) or purchased as the chloride salt from Avanti Polar Lipids (Alabaster, AL). DOPG Na-salt, DOPS Na-salt, DOPA Na-salt, cardiolipin (heart, Na-salt), PI (bovine liver, Na-salt), DOTAP, DOPE, liver total lipid extract, all from Avanti, and oleic acid from Sigma (Sigma-Aldrich, St. Louis, MO) were used without further purification. The phospholipids migrated as a single spot by thin-layer chromatography. Lipids were stored at −20°C in chloroform.
Liposome preparation
All lipids were dissolved in chloroform at concentrations of 10 mg/ml and stored at −20°C. The quality of lipids was tested by thin layer chromatography. The aliquots of lipids were placed into borosilicate glass tubes, dried with an argon stream, and kept under vacuum for at least 2 h to remove the remains of chloroform. After that, samples were hydrated with phosphate-buffered saline (PBS) buffer (pH 7.4), vortexed for 5 min, and stored at 4°C during a few next days.
Lipoplex formation and DNA release assays
FRET-assay
Plasmid DNA pCMVSport-β-Gal DNA (DNA β-Gal) was from Clontech (Palo Alto, CA) and propagated and purified by Bayou Biolabs (Harahan, LA). It was covalently labeled with Cy3 fluorophore using the Label IT labeling kit (Mirus, Madison, WI) according to the protocol supplied. The labeled DNA was purified on Microspin columns included in the kit, or by ethanol precipitation according to the suggested protocol. The number of labels per DNA molecule was measured as suggested by Mirus (www.genetransfer.com, FAQ Q26). In our experiments, one basepair per 80–100 was labeled. The lipid consisted of EDOPC with 1% DHPE headgroup labeled with BODIPY FL dye (Molecular Probes, Eugene, OR); 10 μl of labeled DNAβ-Gal (0.25 μg/ml) were added to 15 μl of 0.1mM EDOPC dissolved in 180 μl PBS buffer (100 mM phosphate and 150mM NaCl, pH 7.2) with constant stirring. The final charge ratio of lipoplexes was R(±) = 1.8. All fluorescence measurements were performed under continuous stirring at room temperature with an Alphascan fluorometer (Photon Technology International, Princeton, NJ) in 6 × 50 mm Durex borosilicate glass tubes. For excitation of BODIPY as a donor, we used its smaller maximum at 483 nm to minimize the direct excitation of acceptor Cy3 and measured the fluorescence of BODIPY at 516 nm. For DNA release, we added 15 μl (0.5 mM) of negatively charged liposomes, which corresponds to a fivefold excess of liposome negative charge over the positive charge of cationic lipid contained in the lipoplexes (unless indicated otherwise). All liposomes were prepared as described above and stored in PBS buffer at 4°C.
EtBr assay
For staining DNA, 0.75 μg EtBr (from stock solution of 0.25 mg/ml) was added to 5 μg DNA β-Gal dissolved in 200 μl PBS buffer. Labeled DNA was treated with 15 μl of EDOPC of 1 mM solution in PBS (DNA/lipid charge ratio = 1) and the fluorescence intensity changes were recorded at 610 nm (excitation 500 nm) under continuous stirring at room temperature. After the new level of fluorescence stabilized, the sample was treated with 15 μl of 4 mg/ml DOPS (∼5 times excess of anionic lipid over cationic lipid).
Membrane fusion
EDOPC cationic liposomes were prepared with 1% BODIPY FL-DHPE and 1% rhodamine-DPPE. The labeled lipids were added to a chloroform solution of EDOPC as in the initial step of liposome preparation (see above). The lipoplexes were prepared according to the procedure described above. Labeled lipoplexes were placed in the fluorometer and treated with a 5-time molar excess of unlabeled negative charged lipids at room temperature with constant stirring. The concentration of lipids, as well as the fluorescence measurement, corresponded to those in the previous paragraph. The level of fluorescence after complete mixing of lipids was measured in presence of 0.2% Triton X100 and this intensity was used for normalization of measurements.
Centrifugal separation of DNA and lipid
EDOPC liposomes contained 3% rhodamine-DPPE. Plasmid DNA was covalently labeled with Cy3 (see above). We performed two sets of experiments. In the first set, the lipoplexes were prepared by addition of labeled liposomes to unlabeled DNA. In the second set, unlabeled liposomes were added to DNA covalently labeled with Cy3. Lipoplexes were incubated at least for 1 h at room temperature, after which aliquots of lipoplexes with labeled lipid, or in a separate experiment, the lipoplexes with labeled DNA were treated with negatively charged liposomes and again incubated for 1 h. The charge ratio of compounds was the same as in the lipoplex formation and DNA release assay, except that the final concentration of DNA and lipids was ∼10 times higher. Centrifugation was performed in 6 × 50 mm borosilicate glass tubes for 1 h at 10,000 × g in a Sorvall HB-4 swinging bucket rotor. Fractions of 100 μl were removed from the top of tubes with MultiFlex Round Tips (Sorenson BioScience, Salt Lake City, UT). Each fraction was treated with 100 μl of 2% solution of Zwittergent 314 (Calbiochem, San Diego, CA) and fluorescence was measured at 588 nm, with excitation at 530 nm for samples with rhodamine and at 565 nm, with excitation at 520 nm, for samples with Cy3.
Electron microscopy
Thin-section electron microscopy
Thin-section electron microscopy was performed as described earlier (MacDonald et al., 1999a). For lipoplex preparation, we used unstained herring sperm DNA (Invitrogen, Carlsbad, CA) and lipids under the conditions described in previous paragraphs. The concentration of DNA was ∼1 mg/ml, with corresponding concentrations of lipids. After preparation, lipoplexes were incubated for 1 h and then treated with a fivefold excess of negatively charged liposomes and incubated for 1 h. All samples were then fixed with 1% osmium tetroxide in 100 mM cacodylate buffer (pH 7.0) overnight, sedimented at 8,000 rev/min in an Eppendorf centrifuge, washed with cacodylate buffer three times, and postfixed with 1% tannic acid in 100 mM cacodylate (pH 7.0) overnight. The precipitate of the fixed liposomes was covered with a thin layer of 1% agar prepared in 100 mM cacodylate to prevent loss of the sample. Next, the samples were dehydrated in series of alcohol and propylene oxide solutions, and embedded in Epon resin according to standard procedures. Sections of 50–70 nm thickness were cut on a MT6000-XL microtome (RMC, Tucson, AZ), stained with uranyl acetate and lead citrate, and observed with a JEM-100CX (JEOL, Peabody, MA) electron microscope.
Negative contrast electron microscopy
Samples were placed on Formvar-coated 400 mesh grids, blotted with filter paper, stained with 1% phosphotungstic acid for 1 min and blotted again. After drying for 15–30 min, grids were observed with a JEM-100CX (JEOL) electron microscope.
Small-angle x-ray diffraction
Separate cationic and anionic lipid dispersions were prepared. Lipid aliquots were transferred to vials where the bulk of the chloroform was removed with a stream of argon. The vial was then placed under high vacuum for at least 1 h/mg lipid to remove residual solvent. Next, 20 mM phosphate buffer, pH 7.2, was added. The lipid concentration of the dispersions was 5 wt %. The dispersions were equilibrated at room temperature for 2 days and intermittently vortex-mixed. The anionic dispersions were then added to the cationic liposomes at 1:1 cationic/anionic lipid molar ratio. The mixtures were vortex-mixed, filled into glass capillaries (d = 1.5 mm) (Charles Super Co., Natick, MA), allowed to settle, and then the supernatant was partially removed. The capillaries were flame-sealed and allowed to equilibrate for 5 days before data collection. X-ray measurements were performed at Argonne National Laboratory (Argonne, IL), Advanced Photon Source, BioCAT (beamline 18-ID) using 12 keV x rays, as previously described (Koynova and MacDonald, 2003a). Two-dimensional diffraction patterns were recorded using a high-sensitivity charge-coupled device detector. Sample-to-detector distance was ∼2 m. Silver behenate (DuPont, Wilmington, DE) was used as a calibrant. Measurements were at 37°C. Exposure times were 1 s. Diffraction intensity versus reciprocal space s-plots were obtained by radial integration of the two-dimensional patterns using the interactive data-evaluating program FIT2D (Hammersley et al., 1996). No products of lipid degradation were detected by thin layer chromatography after the experiments.
RESULTS
DNA release
The estimation of DNA release by EtBr and FRET assays revealed a striking difference in the processes of lipoplex formation and DNA release initiated by anionic lipids (Fig. 1 A). EtBr is a well-known fluorescent DNA label; when it intercalates between the basepairs of the DNA helix, its fluorescence at 610 nm (excitation at 500 nm) is greatly enhanced. Aqueous solutions of DNA stained with EtBr consequently have an initial high level of fluorescence. After addition of cationic lipid, the fluorescence of such solutions immediately decreases, suggesting the displacement of the intercalated dye from DNA. After addition of anionic lipid, most of the EtBr fluorescence reappears almost immediately. Varying the anionic lipid type did not affect the extent of fluorescence recovery, and a maximum recovery of ∼80–90% was achieved at twofold or more excess of anionic lipid charge over the cationic lipid charge. This phenomenon has been described by a number of authors (Ashley et al., 1996; Bhattacharya and Mandal, 1998; MacDonald et al., 1999a; Szoka Jr. et al., 1996; Xu and Szoka Jr., 1996). It was concluded that displacement of EtBr from DNA correlates with displacement of DNA itself from the cationic lipid complex.
FIGURE 1.
Fluorometry of lipoplexes formation and disruption by anionic lipids: (A) Time course of EDOPC-DNA lipoplexes formation and disruption. (□), EtBr assay; (○) FRET assay. (1), Moment of mixing DNA and lipid; and (2), addition of DOPS. All details of the experiments are presented in the lipoplex formation and DNA release assay section (see Methods). (B) FRET assay of DNA release from EDOPC-DNA lipoplexes after addition of negatively charged lipids. (−)Lipid/(+)lipid charge ratio was 5:1. (1), Oleic acid; (2), dioleoylphosphatidic acid; (3), dioleoylphosphatidylglycerol; (4), bovine heart cardiolipin; (5), dioleoylphosphatidylserine; and (6), bovine liver phosphatidylinositol. (B′) The same data as in B, but 5 min scale is shown. (C) FRET assay of DNA release from EDOPC-DNA lipoplexes initiated by liposomes containing: (1), OA; (2), mixture of DOPC + DOPE + DOPS + OA = 1:1:1:2 mol/mol; (3), mixture of DOPC + OA = 1:1; (4), mixture DOPC + OA = 2:1; and (5), liver total lipid extract. (D) FRET assay of DNA release from (DOTAP + DOPE)-DNA lipoplexes initiated by anionic lipids: (1), OA; (2), DOPS; (3), bovine liver PI; and (4), DOPA. Lipoplexes contained BODIPY-DPPE; DNA was covalently stained with Cy3. Measurements at 516 nm. Excitation at 483 nm.
In addition to the EtBr procedure, we also used a FRET assay to study the process of DNA-lipid lipoplex formation and subsequent DNA release after addition of anionic lipids. As shown in Fig. 1 A, the BODIPY fluorescence at 516 nm was considerably reduced after addition of Cy3-labeled DNA, as a result of resonance transfer of BODIPY energy to the acceptor Cy3. This kind of fluorescent energy resonance interaction appears when the distance between the two fluorophores is close to or less than the Förster distance of ∼31 Å (Wiethoff et al., 2002). In these experiments it corresponds to DNA adsorption to the surface of the lipid membranes. After solubilization of DNA-lipid complexes with nonionic detergent (Triton X-100), the fluorescence of Cy3 labeled DNA is restored while the fluorescence of BODIPY labeled lipid drops (not shown) because the distance between fluorophores significantly exceeds the Förster distance. As seen in Fig. 1 A, complex formation between DNA and lipid as revealed by FRET was considerably slower than that estimated by the EtBr assay. After addition of a negatively charged lipid, DOPG, BODIPY fluorescence partially recovered (Fig. 1 A) possibly due to a decrease of energy transfer as a consequence of the distance between fluorophores exceeding 31 Å. The increase of the distance could be a result of DNA displacement from the lipid surface and release into the surrounding solution. The release of DNA according to the FRET assay was much slower than that revealed by EtBr assay. The percent of the DNA released was also considerably smaller.
The initial rate and the maximum level of the DNA release depended considerably on both lipoplex and anionic liposome composition (Fig. 1, B–D). In some cases the process took a few hours and was not completed within the duration of our experiments (Fig. 1 D). With EDOPC (Fig. 1 B), the anionic lipids known to be prevalent in mammalian cells, PS, PI, and CL, were weak (in term of rate) DNA releasers whereas minor cellular lipids like OA and PA possessed strong DNA releasing activity. PG exhibited intermediate ability to release DNA. A somewhat different sequence in effectiveness of DNA release was found for the anionic lipids when lipoplexes were prepared from a DOTAP + DOPE mixture (Fig. 1 D). The rate of DNA release from these lipoplexes was slow for all anionic lipids except OA. The DNA releasing activity of DOPA was weaker than that of all other lipids tested. It is evident from these data that the releasing activity of a given anionic lipid depends on the nature of cationic component of the lipoplexes.
Mixtures of negatively charged lipids and the lipid extract of bovine liver revealed variable ability to release DNA from interaction with EDOPC (Fig. 1 C); however, in most cases investigated, the rate of DNA release estimated by FRET was lower than that indicated by EtBr assay (Bhattacharya and Mandal, 1998; Szoka Jr. et al., 1996; Xu and Szoka Jr., 1996). The cationic lipid/DNA ratio influenced both lipoplex formation (Fig. 2 A) and DNA release after addition of negatively charged lipid (Fig. 2 B). As an indicator of the lipoplex formation, we estimated the variation of BODIPY fluorescence intensity from its initial level (S) to the level of lipoplex formation (L) after addition of labeled DNA to lipid. We measured the restoration of the BODIPY fluorescence intensity (R-L) produced upon addition of anionic lipid as an estimate of the extent of DNA release. Here, the experiments with oleic acid are presented (Fig. 2 B) because this lipid released DNA faster and most reproducibly relative to other lipids. The (R-L) value obtained was divided to initial change of fluorescence (S-L) for normalization. Both processes were considerably intensified when the cationic lipid/DNA ratio was increased. In DNA release experiments (Fig. 2 B), the plateau was achieved at (±) ≥2.
FIGURE 2.
Formation of lipoplexes (A), and DNA release (B), depending on lipid/DNA ratio. DNA release was initiated by five times excess of OA. Lipoplexes were prepared from EDOPC + DNAβ-Gal. Lipid contained 1% DHPE-BODIPY, whereas plasmid DNA was covalently labeled with Cy3. It is shown (C) that during the process of lipoplex formation and DNA release, we can distinguish the initial level of DHPE-BODIPY fluorescence (S), the level of lipoplex fluorescence (L), and the level of fluorescence after DNA was released by negative charged lipid (R). Lipoplex formation was estimated from the decrease of DOPE-BODIPY fluorescence after addition of Cy3-labeled DNA (S-L). DNA release was estimated from the restoration of DOPE-BODIPY fluorescence after addition of OA (R-L). The latter value was divided to (S-L) for normalization.
Membrane fusion
The ability of anionic lipids to initiate DNA release could depend on the extent of membrane fusion (strictly, lipid mixing) between anionic liposomes and lipoplexes. To study such a fusion process, we used a FRET assay involving two fluorescent dyes, BODIPY-DHPE and Rh-DPPE, both incorporated into the cationic lipid EDOPC. This mixture of lipids was then used for the preparation of lipoplexes with plasmid DNA under standard conditions. As a result of energy transfer between donor BODIPY and acceptor rhodamine, the fluorescence of BODIPY at 516 nm was considerably reduced when excited at 473 nm. However, the BODIPY fluorescence was restored after fusion with anionic liposomes because of probe dilution and increased fluorophore separation. Fig. 3 shows the recovery of BODIPY fluorescence after addition of different anionic liposomes to the lipoplexes. The membrane fusion ability of the best DNA releaser, OA, was considerably better than that of all of the other lipids tested. The weak DNA releasers, PI and PS, exhibited the weakest fusion activity. PA exhibited intermediate activity that also correlated with the DNA releasing activity of this lipid.
FIGURE 3.
Fusion of EDOPC + DNA lipoplexes with anionic lipids: (▿), OA; (○), DOPA; (□), bovine liver PI; (▵), DOPS. (−)Lipid/(+)lipid ratio was 5:1. FRET assay at 516 nm, excitation at 473 nm for cationic liposomes labeled with BODIPY-DPPE and with Rh-DPPE.
Further, we estimated the initial rates of DNA release and membrane fusion by fitting our experimental data for the time evolution of the DNA release (Fig. 1 B) and membrane fusion (Fig. 3) to an asymptotic regression model: E = E∝(1 − exp(−V0t/E∝)), where E is the extent of i), DNA release, or ii), membrane fusion, as estimated from the fluorescence intensity increase in Figs. 1 B and 3, respectively; t is time, and E∝ and V0 are fitting parameters, E∝ characterizing the asymptote at t = ∝, and V0 is the initial rate of the process at t = 0 (Ratkowsky, 1983). The initial rates of membrane fusion and of DNA release obtained this way (not presented), which illustrate the relative releasing and fusion activities of the various classes of anionic lipids, arrange in the order: OA > PA > PG > CL > PS > PI. It should be emphasized that this sequence of activities pertains only to EDOPC-containing lipoplexes. Our experiments with DNA release from DOTAP containing lipoplexes revealed a different sequence of lipid activities (Fig. 1 D), although OA was the most active in both cases.
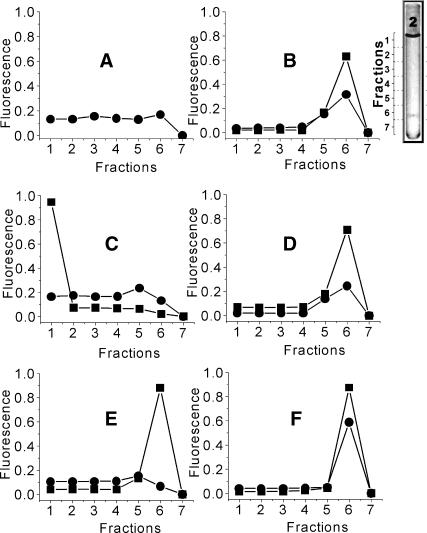
Centrifugation
Centrifugal analyses of lipoplexes treated with anionic lipids provided additional insight into the interaction between DNA and cationic lipids. Our experiments were based on the large difference between sedimentation parameters of DNA, lipid, and lipoplexes. A cushion of 30% sucrose was placed at the bottom of the centrifugation tube (Fig. 4 inset, Fraction 7) and the rest of the tube was filled with lipoplex suspension (Fig. 4 inset, Fractions 1–6). Free DNA did not sediment, but remained distributed throughout the volume of the tube (Fig. 4 A, Fractions 1–6). In contrast, lipoplexes sedimented to the surface of the sucrose cushion (Fig. 4 B), so that Fraction 6 contained both DNA and lipid. A similar location of DNA and lipid in Fraction 6 was observed in the samples containing lipoplexes that had been treated with DOPS or with cardiolipin (Fig. 4, D and F), i.e., those lipids that expressed weak DNA releasing activity according to the FRET assay. In contrast, in samples treated with OA and DOPA (Fig. 4, C and E), the distribution of DNA was similar to that of the control sample with free DNA (Fig. 4 A), and the lipid was found at the top of the tube (Fraction 1 in Fig. 4 C) in the case of OA, or above the sucrose cushion (Fraction 6; Fig. 4 E) in the case of DOPA. These results thus also correlate with the differences between OA and DOPA in releasing DNA from EDOPC-DNA lipoplexes as revealed by FRET assay.
FIGURE 4.
Centrifugal separation of lipoplexes treated with negatively charged lipids. Two sets of experiments are presented: (•), experiments with samples containing DNA covalently labeled with Cy3. Measurements at 565nm, excitation at 520 nm. (▪), experiments with samples containing lipid labeled with Rh-DPPE, measurements at 588nm, excitation at 530nm. (Inset) Illustration of centrifugation tube used for separation DNA-lipid complexes. Fraction 7 has 30% sucrose. Fractions 1–6 were filled with the suspension to be tested (not a density gradient). As is seen, this sample has a layer of stained material in Fraction 6 and is typical of the lipoplex separation that occurred after 1 h centrifugation at 10,000 × g. (A) The control sample of free DNA; (B) the control sample of EDOPC-DNA lipoplexes; (C) the lipoplexes treated with OA; (D) the lipoplexes treated with DOPS; (E) the lipoplexes treated with DOPA; and (F) the lipoplexes treated with bovine heart CL.
X-ray diffraction
In an effort to rationalize the differences in the efficiency of the anionic lipid classes in releasing DNA from the EDOPC lipoplexes, we studied the phase organization of the lipid aggregates formed upon mixing aqueous dispersions of EDOPC with dispersions of the same selection of anionic lipids described above. We were particularly interested in whether the mesomorphic organization of the positive-negative lipid mixtures might correlate with the DNA release pattern described above. The results are presented in Fig. 5 and summarized in Table 1.
FIGURE 5.
Small-angle x-ray diffraction patterns of mixtures of anionic lipids with EDOPC (1:1 molar ratio) mixed from previously prepared aqueous dispersions; patterns are arranged in ascending order of increasing polar/apolar interfacial curvature.
TABLE 1.
Liquid crystalline phases formed upon mixing of EDOPC dispersion with various anionic lipid aqueous dispersions
| Anionic lipid mixed with EDOPC liposomes | Phase identification | Structural parameters (Å) |
|---|---|---|
| Oleic acid | Micellar cubic Fd3m | 153.6 |
| + bicontinuous cubic Pn3m (traces) | 79.8, 73.7 | |
| DOPA | Hexagonal HII | 76.3 |
| + bicontinuous cubic Im3m (traces) | 209.3 | |
| + bicontinuous cubic Pn3m (traces) | 163.3 | |
| Liver - total lipid extract | Bicontinuous cubic Pn3m | 155 |
| DOPG | Lamellar | 63 |
| + bicontinuous cubic Ia3d | 257.7 | |
| Cardiolipin (heart) | Lamellar | 50.0 |
| + bicontinuous Im3m (traces) | 132 | |
| DOPS | Uncorrelated lamellae/sponges | |
| PI (liver) | Lamellar | 55.6 |
On the diffraction pattern of the mixture of EDOPC with oleic acid at least 15 diffraction maxima were visible. Twelve of those have reciprocal spacings fitting the ratio: √3:√8:√11:√12:√16:√19:√24:√27:√32:√35:√40:√44, which includes the full set of the initial 10 reflections characteristic for the cubic Fd3m phase (cubic aspect No. 15) (Kasper and Lonsdale, 1985), with a structural parameter a = 153.6 Å. The reciprocal spacing S versus √(h2 + k2 + l2) graph (h, k, and l are the Miller indices (Kasper and Lonsdale, 1985)) for these reflections fitted linear function at R2 = 0.9999 (not shown). That structure dominates the diffraction pattern. Since oleic acid has been reported to form inverse micelles in excess water (Seddon et al., 1990, 2000; Luzzati et al., 1992; Nakano et al., 2002), it is logical to suppose that the observed Fd3m micellar cubic phase is of inverted type. Traces of coexisting Pn3m phase could be suggested based on the rest of the observed reflections.
The diffraction pattern of the mixture of EDOPC with DOPA is dominated by diffraction peaks at 66.1 Å, 38.2 Å, and 33.0 Å, indexing as the (10), (11), and (20) reflections of a hexagonal phase with a 76.3 Å structural unit. Additionally, five minor reflections at low angles could be suggestive for traces of coexisting bicontinuous cubic Pn3m (a = 163.3 Å) and Im3m (a = 209.3 Å) phases.
The diffraction pattern of the EDOPC/liver extract mixture displays 14 diffraction peaks, with reciprocal spacings fitting the ratio: √2:√3:√4:√6:√8:√9:√10:√11:√12:√14:√16:√17:√18:√19, which clearly index as the full set of the initial 14 reflections of a single cubic Pn3m phase (cubic aspect No. 4) (Kasper and Lonsdale, 1985), with a structural node of a = 154.8 Å. The reciprocal spacing S versus √(h2 + k2+ l2) graph for these reflections fitted linear function at R2 = 0.9999.
The pattern of the EDOPC/DOPG mixture is dominated by a lamellar phase, with a lamellar repeat period of 63 Å. An additional 10 reflections with spacings fitting the ratio: √6:√8:√20:√22:√24:√26:√38:√40:√42:√46 (fit at R2 = 0.9996) suggest the presence of a coexisting Ia3d cubic phase (cubic aspect No. 12), with a = 259.5 Å (the √14 and √16 reflections are possibly hidden by the strong lamellar first-order peak).
The EDOPC/cardiolipin diffraction pattern is strongly dominated by a lamellar phase (d = 50 Å), with only small traces of Im3m cubic phase (seven diffraction peaks with spacings fitting the ratio: √2:√4:√6:√10:√12:√14:√16, with the missing √8 peak probably hidden by the lamellar first-order reflection), with structural parameter a = 132.8 Å. The mixture of EDOPC with DOPS consists of a broad halo centered at ∼46 Å, which possibly indicates uncorrelated lamellae or sponges. The mixture of EDOPC with liver PI is organized in the lamellar phase with a lamellar repeat spacing d = 55.6 Å.
Electron microscopy
Electron microscopy of EDOPC-DNA lipoplexes (Fig. 6 A) revealed that multilamellar structures were most typical, as described earlier (MacDonald et al., 1999b; Tarahovsky et al., 2002). Such structures represent lipid bilayers separated by oriented molecules of DNA (Koltover et al., 1998; Raedler et al., 1997). After addition of negatively charged liposomes to the preformed lipoplexes, the structural organization of membranes changes dramatically. For example, in the presence of negative charged lipids, various examples of destruction of initial multilamellar structure appear. After 30 min treatment by DOPS, most lamellar structures disaggregate and some example of cubic (or sponge) structure appears (Fig. 6 B). Numerous small unilamellar vesicles of ∼100–200 nm are also formed (Fig. 6 B′, arrows). The structural organization of lipoplexes treated by DOPA is different. Lamellar structure is not readily evident (Fig. 6 C). The sample contained a large amount of unordered—possible micellar—material (Fig. 6 C′).
FIGURE 6.
Thin-section electron microscopy of EDOPC + DNA lipoplexes (A) after treatment by DOPS liposomes (B and B′) or DOPA liposomes (C and C′). B′ represents a larger magnification of the selected area in B. The leaf in B indicates lamellar structures. The asterisk in B indicates examples of cubic (or sponge) structures. The arrows in B′ indicate small vesicles released from the surface of lipoplex. The periodicity of A is ∼65 Å. Magnification of B and C and C′ is given by the bar representing 1 μm.
DISCUSSION
The transfection of cells by lipoplexes is a very complex process with a number of different stages. Several publications have called attention to specific barriers for transfection that may be rate-limiting (Bally et al., 1999; Wiethoff and Middaugh, 2003; Zabner et al., 1995). One factor that is likely to be important in the efficiency of transfection is the dissociation of DNA from cationic lipids and its release into the cytoplasm or, perhaps, the nucleus. An earlier suggestion, that anionic lipids are factors responsible for DNA release, was based on experiments in vitro showing that anionic liposomes are able to effectively interact with lipoplexes, neutralize the positive charge of cationic lipids, and promote DNA release (Ashley et al., 1996; MacDonald et al., 1999a; Szoka Jr. et al., 1996; Xu and Szoka Jr., 1996; Zelphati and Szoka, 1996a). Szoka and collaborators reported that different types of anionic lipids were equally effective in DNA release and a twofold excess of anionic lipid was sufficient to release of 80–90% of the lipoplex DNA (Xu and Szoka Jr., 1996; Zelphati and Szoka, 1996b).
To estimate the time course of lipoplex formation and the ability of different anionic lipids to release DNA, we used a FRET assay involving fluorescent probes covalently attached to DNA and to lipid. By this measure, both the time course of interaction between DNA and cationic lipid during lipoplex formation and the time course of lipoplex dissociation after addition of anionic lipids to lipoplexes was considerably slower than that reported earlier by investigators utilizing fluorescent intercalating dyes, EtBr or To-PRO-1 (Bhattacharya and Mandal, 1998; Harvie et al., 1998; Sakurai et al., 2000; Szoka Jr. et al., 1996; Xu and Szoka Jr., 1996). Furthermore, according to the FRET assay, the kinetics of DNA release depended on the lipid composition of both the cationic and anionic liposomes.
The difference between the two assays may be related to the fact that those based on intercalator fluorescence may not directly relate to DNA behavior, since the fluorescence of intercalators depends upon the DNA environment, which, in turn, may not necessary correlate exactly with lipoplex formation or disruption. On the other hand, other approaches allowed monitoring of DNA release from lipoplexes in presence of anionic liposomes, one of which is based on accessibility of DNA molecule to enzymatic digestion (Xu and Szoka Jr., 1996; Zuhorn and Hoekstra, 2002). This approach presupposes that only free DNA is available to DNase, whereas interaction with cationic lipids protects DNA from enzyme action (Zhang et al., 1997). However, when assays are based on the enzymatic digestion of DNA, information about the time course of DNA release is limited (Harvie et al., 1998). Furthermore, a direct correlation between the EtBr assay and enzymatic digestion was not necessarily observed; in some cases, cationic lipids protected DNA from DNase 1 degradation but did not preclude its staining with EtBr (Crook et al., 1996; Zhang et al., 1997). Electron microscopy observations (Fig. 6) revealed a major reorganization of the initial multilamellar structure of lipoplexes subsequent to their interaction with anionic lipids. Although data on DOPA or oleic acid were not presented because of space considerations, these lipids generally expressed a greater ability to destroy the initial lamellar structure of the lipoplexes and produce unordered micellar structures than did lipids DOPS or DOPG with a smaller capacity to release DNA. During this process, the DNA was released from membrane surfaces, but was retained inside lipid structures, where it could become available for interaction with dyes or enzymes.
We found that the DNA-releasing activity of different anionic lipids was very different, and we could distinguish strong and weak DNA releasers, although such differentiation was not entirely independent of the cationic lipid. For example, we found that DOPA was a good DNA releaser in the case of EDOPC-containing lipoplexes, but a bad releaser in the case of the DOTAP-DOPE mixture. Thus, the characteristics of anionic lipids in terms of efficiency of DNA release may only be meaningful in the context of specific lipoplexes.
Centrifugal separation of DNA and lipid is one of the most direct approaches for demonstrating the integrity of lipoplexes; however, this procedure can provide little information about the dynamic aspects of the process. Nevertheless, there was a good correlation between the data obtained by FRET assay and those obtained by centrifugation (compare Figs. 1 B and 4). Both approaches revealed that OA and DOPA are able to release a considerable proportion of DNA from EDOPC-DNA lipoplexes, whereas DOPS and bovine liver PI expressed weak DNA releasing activity under similar conditions (Fig. 4).
Aqueous dispersions of polar lipids are known for their ability to form a large number of polymorphic and mesomorphic phases. Specifically, a variety of mesomorphic phases are known to appear upon hydration of anionic and cationic lipid mixtures that have been prepared from solutions in organic solvent. For example, mixtures of ethylphosphatidylcholine with different anionic lipids tend to produce not only lamellar but also different cubic and inverted hexagonal structures (Koynova and MacDonald, 2003b; Lewis et al., 2001; Tarahovsky et al., 2000). Such data provide information about the equilibrium (or quasiequilibrium) phase preferences of those mixtures, and the resultant phases could be rather different if the two lipids are mixed as preformed aqueous dispersions—a protocol much closer to the transfection scenario where kinetic barriers could substantially restrain the equilibration. The phase preferences of the anionic-cationic lipid mixtures might well be expected to influence DNA release efficiency. Indeed, the x-ray diffraction experiments revealed an unambiguous correlation between the releasing capacities of the aqueous dispersions of anionic lipids and the mesomorphic structures they form when mixed with EDOPC. The anionic lipids that were more efficient in releasing DNA (e.g., OA, DOPA) formed nonlamellar phases when mixed with preformed EDOPC liposomes. Conversely, the anionic lipids for which only inefficient release of DNA was observed (e.g., PI, DOPS) formed mostly lamellar phases. DOPG, which ranked intermediate as a DNA releaser, displayed a mixture of lamellar and nonlamellar phases in the mixed dispersion with EDOPC.
Moreover, the correlation could be extended: the capacity of anionic liposomes to release DNA corresponds to the position of the fluid phases they form when mixed with EDOPC within what is considered as the “natural” lyotropic phase sequence. This “natural” sequence in which the various possible fluid phases occur in a hypothetical lipid/water phase diagram (i.e., phase transitions are driven by water content) is arranged by the average mean curvature of the polar-nonpolar interface (see, e.g., (Charvolin, 1985; Scriven, 1977; Seddon, 1990; Winsor, 1968)). Another variable (aside from hydration), which is known to bring about the same “natural” phase sequence, is temperature (e.g., (Koynova and Tenchov, 2001; Seddon and Templer, 1995)). In our experiments, the same sequence of fluid phases ranked by their interfacial curvature was produced by cationic/anionic lipid mixtures as was found for the DNA release from lipoplexes. Indeed, the lamellar phase, for which the curvature is zero, was formed in the presence of PI, which was quite ineffective in DNA release. DOPA, an effective DNA releaser, formed an inverted hexagonal phase, characterized by higher interfacial curvature. The bilayer cubic phases—Pn3m, Ia3d, and Im3m—exhibit zero mean curvature, but they are still characterized with nonzero Gaussian curvature—and such cubic phases were formed by the anionic lipids that ranked intermediate with respect to their DNA release capacity. The micellar cubic phase exhibits virtually the highest interfacial curvature—and oleic acid is the most efficient in DNA release. Thus, the anionic lipids forming highly curved phases are the better DNA-releasing agents. To recapitulate, the ranking of anionic lipid dispersions with respect to their DNA-releasing efficiency: OA > DOPA > total liver extract > DOPG > cardiolipin > DOPS > PI (Fig. 1, B and B′) matches the interfacial curvature ranking of the mesophases formed when these dispersions are mixed with EDOPC dispersions: micellar cubic > hexagonal > bilayer cubic > uncorrelated lamellae/sponges > lamellar stacks (Fig. 5).
Of additional interest is the view that the mechanisms of lamellar-nonlamellar phase transformations in lipids are closely related to the mechanisms of fusion of lipid bilayers, in particular, that similar intermediates are implicated in both processes (Ellens et al., 1989; Siegel and Epand, 1997). A number of publications have suggested that membrane fusion could be an important factor in DNA translocation across the cell membrane during virus or bacterophage infection (Poranen et al., 2002; Tarahovsky et al., 1991) as well as during artificial transfection (Borovjagin et al., 1987; Chesnoy and Huang, 2000; Hafez et al., 2001; Hafez and Cullis, 2001; Koltover et al., 1998; MacDonald et al., 1999a; Noguchi et al., 1998; Zuhorn and Hoekstra, 2002). These suggestions are now credibly supported by our findings that the rate of anionic lipid fusion with EDOPC-DNA lipoplexes correlates with the DNA releasing activity of the corresponding lipids (Fig. 3). It thus seems likely that those membrane events that precede the formation of nonbilayer structures, such as destabilization of the bilayer and formation of interbilayer contacts, could well promote those processes of membrane fusion and lipid mixing that presumably lead to charge neutralization and DNA release from lipoplexes. Thus, lipids with a preference to form nonlamellar phases are thought to promote membrane fusion as a result of their ability to impart negative intrinsic curvature to the lipid aggregates. We suppose that the lipid properties per se are critical, not exposed surface area. Given the correlation we observed between DNA-releasing capacity and intrinsic curvature, we conclude that membrane fusion might be suspected to be a key step in DNA release and, as indicated by our fusion experiments, that lipids efficient in promoting membrane fusion are also efficient in releasing DNA from lipoplexes.
The ultrastructural analyses revealed considerable reorganization of the highly ordered multilamellar structure of EDOPC-DNA lipoplexes upon treatment with anionic liposomes. Subsequently, sponge or cubic structures, or unilamellar vesicles, appeared (Fig. 6). Structural reorganization of membranes could be responsible for DNA release, although it seems unlikely that all DNA could be released into the external aqueous volume (Fig. 7). Indeed, data from other laboratories indicates that after lipoplex reorganization by anionic lipid treatment, DNA molecules remain inside of internal aqueous volume of lipoplexes, but become accessible to enzymatic treatment and labeling (Bhattacharya and Mandal, 1998; Harvie et al., 1998; Itaka et al., 2002; MacDonald et al., 1999a; Pantazatos and MacDonald, 2003; Sakurai et al., 2000; Zuhorn et al., 2002). Such findings may lead to an overestimation of DNA release, and we have concluded that, even after the cationic lipid charge has been neutralized and the electrostatic interactions with DNA have been eliminated, a considerable amount of DNA may remain trapped inside the resultant mesophases.
FIGURE 7.
Schematic presentation of presumptive structural changes in multilamellar lipoplexes (A) after interaction with anionic liposomes (white on B and C). According to the scheme presented, some anionic lipids could initiate formation of sponge or more ordered bicontinuous cubic strictures (B), where diffusion of large linear molecules like DNA is restricted, whereas more compact molecules could easily diffuse through numerous pores. Other anionic lipids initiate disintegration of multilamellar structure of lipoplexes and formation of monolamellar liposomes within which some proportion of the original DNA could be trapped in the internal volume of vesicles (C). During the processes of membrane reorganization, DNA could become available to intercalating fluorescent probes, or perhaps even some nucleases.
Acknowledgments
We thank Jaime Stearns and David Yuan for synthesis of lipids and Ed Pozharsky for consultations related to fluorescent techniques.
This work was supported by National Institutes of Health grant GM52329. Synchrotron x-ray measurements were performed at the Biophysics Collaborative Access Team (BioCAT) Synchrotron Research Center of the Advanced Photon Source, Argonne National Laboratory. BioCAT is an NIH-supported Research Center, through grant RR08630. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research under contract No. W-31-102-Eng-38. We are grateful to Elena Kondrashkina and Boris Tenchov for the assistance throughout the synchrotron experiments.
Rumiana Koynova is also an associate member of the Institute of Biophysics, Bulgarian Academy of Sciences.
Abbreviations used: EDOPC, dioleoylethylphosphatidylcholine; DOPG, dioleoylphosphatidylglycerol; DOPS, dioleoylphosphatidylserine; DOPA, dioleoylphosphatidic acid; PI, phosphatidylinositol; DOTAP, dioleoyltrimethylammoniumpropane; DOPE, dioleoylphosphatidylethanolamine; FRET, fluorescence resonance energy transfer; EtBr, ethidium bromide; PG, phosphatidylglycerol; PS, phosphatidylserine; PA, phosphatidic acid; OA, oleic acid.
References
- Ashley, G. W., M. M. Shida, R. Qiu, M. K. Lahiri, P. C. Levisay, R. D. Jones, K. A. Baker, and R. C. MacDonald. 1996. Phosphatidylcholinium compounds: a new class of cationic phospholipids with transfection activity and unusual physical properties. Biophys. J.. 70:88 (Abstr.). [Google Scholar]
- Bally, M. B., P. Harvie, F. M. Wong, S. Kong, E. K. Wasan, and D. L. Reimer. 1999. Biological barriers to cellular delivery of lipid-based DNA carriers. Adv. Drug Deliv. Rev. 38:291–315. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, S., and S. S. Mandal. 1998. Evidence of interlipidic ion-pairing in anion-induced DNA release from cationic amphiphile-DNA complexes. Mechanistic implications in transfection. Biochemistry. 37:7764–7777. [DOI] [PubMed] [Google Scholar]
- Borovjagin, V. L., A. G. Sabelnikov, Y. S. Tarahovsky, and I. A. Vasilenko. 1987. Polymorphic behavior of gram-negative bacteria membranes. J. Membr. Biol. 100:229–242. [Google Scholar]
- Caplen, N. J., E. W., F. W. Alton, P. G. Middleton, J. R. Dorin, B. J. Stevenson, X. Gao, S. R. Durham, P. K. Jeffery, M. E. Hodson, C. Coutelle, L. Huang, D. J. Porteous, R. Williamson, and D. M. Geddes. 1995. Liposome-mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Nat. Med. 1:39–46. [DOI] [PubMed] [Google Scholar]
- Charvolin, J. 1985. Crystals of interfaces: the cubic phases of amphiphile/water systems. J. Phys. (Paris) Colloq. 46:C173–190. [Google Scholar]
- Chesnoy, S., and L. Huang. 2000. Structure and function of lipid-DNA complexes for gene delivery. Annu. Rev. Biophys. Biomolec. Struct. 29:27–47. [DOI] [PubMed] [Google Scholar]
- Crook, K., G. McLachlan, B. J. Stevenson, and D. J. Porteous. 1996. Plasmid DNA molecules complexed with cationic liposomes are protected from degradation by nucleases and shearing by aerosolisation. Gene Ther. 3:834–839. [PubMed] [Google Scholar]
- Ellens, H., D. P. Siegel, D. Alford, P. L. Yeagle, L. Boni, L. J. Lis, P. J. Quinn, and J. Bentz. 1989. Membrane-fusion and inverted phases. Biochemistry. 28:3692–3703. [DOI] [PubMed] [Google Scholar]
- Hafez, I. M., and P. R. Cullis. 2001. Roles of lipid polymorphism in intracellular delivery. Adv. Drug Deliv. Rev. 47:139–148. [DOI] [PubMed] [Google Scholar]
- Hafez, I. M., N. Maurer, and P. R. Cullis. 2001. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8:1188–1196. [DOI] [PubMed] [Google Scholar]
- Hammersley, A. P., S. O. Svensson, M. Hanfland, A. N. Fitch, and D. Hausermann. 1996. Two-dimensional detector software: from real detector to idealised image or two-theta scan. High Pressure Res. 14:235–248. [Google Scholar]
- Harvie, P., F. M. Wong, and M. B. Bally. 1998. Characterization of lipid DNA interactions. I. Destabilization of bound lipids and DNA dissociation. Biophys. J. 75:1040–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaka, K., A. Harada, K. Nakamura, H. Kawaguchi, and K. Kataoka. 2002. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromolecules. 3:841–845. [DOI] [PubMed] [Google Scholar]
- Kasper, J. S., and K. Lonsdale. 1985. International Tables for X-ray Crystallography. Riedel Publishing, Dordrecht, The Netherlands.
- Koltover, I., T. Salditt, J. O. Raedler, and C. R. Safinya. 1998. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 281:78–81. [DOI] [PubMed] [Google Scholar]
- Koynova, R., and R. C. MacDonald. 2003a. Cationic O-ethylphosphatidylcholines and their lipoplexes: phase behavior aspects, structural organization and morphology. Biochim. Biophys. Acta. 1613:39–48. [DOI] [PubMed] [Google Scholar]
- Koynova, R., and R. C. MacDonald. 2003b. Mixtures of cationic lipid O-ethylphosphatidylcholine with membrane lipids and DNA: Phase diagrams. Biophys. J. 85:2449–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova, R., and B. Tenchov. 2001. Interactions of surfactants and fatty acids with lipids. Curr. Opin. Colloid Interface Sci. 6:277–286. [Google Scholar]
- Laitinen, M., J. Hartikainen, M. O. Hiltunen, J. Eranen, M. Kiviniemi, O. Narvanen, K. Makinen, H. Manninen, M. Syvanne, J. F. Martin, M. Laakso, and S. Yla-Herttuala. 2000. Catheter-mediated vascular endothelial growth factor gene transfer to human coronary arteries after angioplasty. Hum. Gene Ther. 11:263–270. [DOI] [PubMed] [Google Scholar]
- Lewis, R. N., I. Winter, M. Kriechbaum, K. Lohner, and R. N. McElhaney. 2001. Studies of the structure and organization of cationic lipid bilayer membranes: calorimetric, spectroscopic, and x-ray diffraction studies of linear saturated P-O-ethyl phosphatidylcholines. Biophys. J. 80:1329–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzati, V., R. Vargas, A. Gulik, P. Mariani, J. M. Seddon, and E. Rivas. 1992. Lipid polymorphism: A correction. The structure of the cubic phase of extinction symbol Fd– consists of two types of disjoined reverse micelles embedded in a three-dimensional hydrocarbon matrix. Biochemistry. 31:279–285. [DOI] [PubMed] [Google Scholar]
- MacDonald, R. C., G. W. Ashley, M. M. Shida, V. A. Rakhmanova, Y. S. Tarahovsky, D. P. Pantazatos, M. T. Kennedy, E. V. Pozharski, K. A. Baker, R. D. Jones, H. S. Rosenzweig, K. L. Choi, R. Z. Qiu, and T. J. McIntosh. 1999a. Physical and biological properties of cationic triesters of phosphatidylcholine. Biophys. J. 77:2612–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, R. C., V. A. Rakhmanova, K. L. Choi, H. S. Rosenzweig, and M. K. Lahiri. 1999b. O-ethylphosphatidylcholine: a metabolizable cationic phospholipid which is a serum-compatible DNA transfection agent. J. Pharm. Sci. 88:896–904. [DOI] [PubMed] [Google Scholar]
- Nabel, G. J., E. G. Nabel, Z. Y. Yang, B. A. Fox, G. E. Plautz, X. Gao, L. Huang, S. Shu, D. Gordon, and A. E. Chang. 1993. Direct gene transfer with DNA-liposome complexes in melanoma: expression, biologic activity, and lack of toxicity in humans. Proc. Natl. Acad. Sci. USA. 90:11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, M., and A. Noguchi. 2001. Confocal and probe microscopy to study gene transfection mediated by cationic liposomes with a cationic cholesterol derivative. Adv. Drug Deliv. Rev. 52:197–207. [DOI] [PubMed] [Google Scholar]
- Nakano, M., T. Teshiqawara, A. Sugite, W. Leesajakul, A. Taniguchi, T. Kamo, H. Matsuoka, and T. Handa. 2002. Dispersions of liquid crystalline phases of the monoolein/oleic acid/Pluronic F127 systems. Langmuir. 18:9283–9288. [Google Scholar]
- Nishikawa, M., and M. Hashida. 2002. Nonviral approaches satisfying various requirements for effective in vivo gene therapy. Biol. Pharm. Bull. 25:275–283. [DOI] [PubMed] [Google Scholar]
- Noguchi, A., T. Furuno, C. Kawaura, and M. Nakanishi. 1998. Membrane fusion plays an important role in gene transfection mediated by cationic liposomes. FEBS Lett. 433:169–173. [DOI] [PubMed] [Google Scholar]
- Pantazatos, S. P., and R. C. MacDonald. 2003. Real-time observation of lipoplex formation and interaction with anionic bilayer vesicles. J. Membr. Biol. 191:99–112. [DOI] [PubMed] [Google Scholar]
- Poranen, M. M., R. Daugelavicius, and D. H. Bamford. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56:521–538. [DOI] [PubMed] [Google Scholar]
- Raedler, J. O., I. Koltover, T. Salditt, and C. R. Safinya. 1997. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science. 275:810–814. [DOI] [PubMed] [Google Scholar]
- Ratkowsky, D. A. 1983. Nonlinear Regression Modeling: A Unified Practical Approach. Marcel Dekker, New York.
- Rosenzweig, H. S., V. A. Rakhmanova, T. J. McIntosh, and R. C. MacDonald. 2000. O-Alkyl dioleoylphosphatidylcholinium compounds: the effect of varying alkyl chain length on their physical properties and in vitro DNA transfection activity. Bioconjug. Chem. 11:306–313. [DOI] [PubMed] [Google Scholar]
- Sakurai, F., R. Inoue, Y. Nishino, A. Okuda, O. Matsumoto, T. Taga, F. Yamashita, Y. Takakura, and M. Hashida. 2000. Effect of DNA/liposome mixing ratio on the physicochemical characteristics, cellular uptake and intracellular trafficking of plasmid DNA/cationic liposome complexes and subsequent gene expression. J. Control Release. 66:255–269. [DOI] [PubMed] [Google Scholar]
- Scriven, L. E. 1977. Equilibrium bicontinuous structures. In Micellization, Solubilization, and Microemulsions. K. L. Mittal, editor. Plenum, New York. 877–93.
- Seddon, J. M. 1990. Structure of the inverted hexagonal (Hii) phase, and non-lamellar phase-transitions of lipids. Biochim. Biophys. Acta. 1031:1–69. [DOI] [PubMed] [Google Scholar]
- Seddon, J. M., and R. H. Templer. 1995. Polymorphism of lipid-water systems. In Handbook of Biological Physics. R. Lipowsky and E. Sackmann, editors. Elsevier Science, 97–160.
- Seddon, J. M., E. A. Bartle, and J. Mingins. 1990. Inverse cubic liquid-crystalline phases of phospholipids and related lyotropic systems. J. Phys. Condens. Matter. 2:SA285–SA290. [Google Scholar]
- Seddon, J. M., J. Robins, T. Gulik-Krzywicki, and H. Delacroix. 2000. Inverse micellar phases of phospholipids and glycolipids. Phys. Chem. Chem. Phys. 2:4485–4493. [Google Scholar]
- Siegel, D. P., and R. M. Epand. 1997. The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys. J. 73:3089–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoka, F. C., Jr., Y. Xu, and O. Zelphati. 1996. How are nucleic acids released in cell from cationic lipid-nucleic acid complexes. J. Liposome Res. 6:567–587. [Google Scholar]
- Tarahovsky, Y. S., A. L. Arsenault, R. C. MacDonald, T. J. McIntosh, and R. M. Epand. 2000. Electrostatic control of phospholipid polymorphism. Biophys. J. 79:3193–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarahovsky, Y. S., A. A. Khusainov, A. A. Deev, and Y. V. Kim. 1991. Membrane-fusion during infection of Escherichia coli cells by phage-T4. FEBS Lett. 289:18–22. [DOI] [PubMed] [Google Scholar]
- Tarahovsky, Y. S., V. A. Rakhmanova, R. E. Epand, and R. C. MacDonald. 2002. Thermal stabilization of DNA by cationic lipids. Biophys. J. 82:264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoff, C. M., M. L. Gill, G. S. Koe, J. G. Koe, and C. R. Middaugh. 2002. The structural organization of cationic lipid-DNA complexes. J. Biol. Chem. 277:44980–44987. [DOI] [PubMed] [Google Scholar]
- Wiethoff, C. M., and C. R. Middaugh. 2003. Barriers to nonviral gene delivery. J. Pharm. Sci. 92:203–217. [DOI] [PubMed] [Google Scholar]
- Winsor, P. A. 1968. Binary and multicomponent solutions of amphiphilic compounds. Solubilization and the formation, structure, and theoretical significance of liquid crystalline solutions. Chem. Rev. 68:1–40. [Google Scholar]
- Woodle, M. C., and P. Scaria. 2001. Cationic liposomes and nucleic acids. Curr. Opin. Colloid Interface Sci. 6:78–84. [Google Scholar]
- Xu, Y., and F. C. Szoka Jr. 1996. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 35:5616–5623. [DOI] [PubMed] [Google Scholar]
- Zabner, J., A. J. Fasbender, T. Moninger, K. A. Poellinger, and M. J. Welsh. 1995. Cellular and molecular barriers to gene-transfer by a cationic lipid. J. Biol. Chem. 270:18997–19007. [DOI] [PubMed] [Google Scholar]
- Zelphati, O., and F. C. Szoka. 1996a. Liposomes as a carrier for intracellular delivery of antisense oligonucleotides: a real or magic bullet? J. Control Release. 41:99–119. [Google Scholar]
- Zelphati, O., and F. C. Szoka. 1996b. Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl. Acad. Sci. USA. 93:11493–11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. P., D. L. Reimer, G. Zhang, P. H. Lee, and M. B. Bally. 1997. Self-assembling DNA-lipid particles for gene transfer. Pharm. Res. 14:190–196. [DOI] [PubMed] [Google Scholar]
- Zuhorn, I. S., and D. Hoekstra. 2002. On the mechanism of cationic amphiphile-mediated transfection. To fuse or not to fuse: Is that the question? J. Membr. Biol. 189:167–179. [DOI] [PubMed] [Google Scholar]
- Zuhorn, I. S., W. H. Visser, U. Bakowsky, J. B. Engberts, and D. Hoekstra. 2002. Interference of serum with lipoplex-cell interaction: modulation of intracellular processing. Biochim. Biophys. Acta. 1560:25–36. [DOI] [PubMed] [Google Scholar]