Abstract
We employ 100-ns molecular dynamics simulations to study the influence of cholesterol on structural and dynamic properties of dipalmitoylphosphatidylcholine bilayers in the fluid phase. The effects of the cholesterol content on the bilayer structure are considered by varying the cholesterol concentration between 0 and 50%. We concentrate on the free area in the membrane and investigate quantities that are likely to be affected by changes in the free area and free volume properties. It is found that cholesterol has a strong impact on the free area properties of the bilayer. The changes in the amount of free area are shown to be intimately related to alterations in molecular packing, ordering of phospholipid tails, and behavior of compressibility moduli. Also the behavior of the lateral diffusion of both dipalmitoylphosphatidylcholine and cholesterol molecules with an increasing amount of cholesterol can in part be understood in terms of free area. Summarizing, our results highlight the central role of free area in comprehending the structural and dynamic properties of membranes containing cholesterol.
INTRODUCTION
Cholesterol is one of the most prominent molecular species in the plasma membranes of mammalian cells. It is a tremendously important molecule, a component essential for the very existence and multiplication of cells (Finegold, 1993; Ohvo-Rekilä et al., 2002, and references therein). It is abundant in the plasma membranes of higher organisms: depending on the exact lipid composition, the plasma membrane may contain the order of 20–50% cholesterol (Alberts et al., 1994).
Eukaryotic cells do not seem to be able to grow and differentiate properly without cholesterol. It has been firmly established that cholesterol modulates the physical properties of the plasma membrane (McMullen and McElhaney, 1996). A finite cholesterol content has been said to improve the characteristics of a simple phospholipid bilayer and allow for wider variations in the lipid composition of the membrane (Vist and Davis, 1990). Perhaps not surprisingly, cholesterol is one of the primary molecules in lipid rafts (Edidin, 2003; Silvius, 2003; Simons and Ikonen, 1997, and references therein), i.e., microdomains rich in cholesterol, sphingomyelin, and saturated phospholipids. Rafts have been thought to confine proteins involved in, e.g., signal transduction events, and hence act as platforms for adhesion and signaling. Consequently, one could well imagine that as cholesterol alters the properties of the bilayer, it might affect the functioning of the embedded proteins (Cantor, 1999; Yeagle, 1991).
The effects of cholesterol on the properties of phospholipid bilayers are diverse. In the physiologically relevant fluid phase, adding cholesterol to the bilayer leads to increased orientational order in the phospholipid tails (Chiu et al., 2002; Hofsäß et al., 2003; McMullen and McElhaney, 1996; Sankaram and Thompson, 1990b) and smaller average areas per molecule (Petrache et al., 1999). In other words, cholesterol modifies the packing of molecules in bilayers. Other important effects are changes in passive permeability of small solutes (Jedlovszky and Mezei, 2003; Xiang, 1999, and references therein) and suppressed lateral diffusion of phospholipids in bilayers with cholesterol (Almeida et al., 1992; Galla et al., 1979; Hofsäß et al., 2003; Polson et al., 2001; Vattulainen and Mouritsen, 2003). Both permeability and lateral diffusion, in turn, are strongly affected by the amount and distribution of free volume or area in a membrane, i.e., space not occupied by phospholipids, cholesterols, or water. Cholesterol thus seems to simultaneously influence packing, free area, diffusion, and permeability in lipid bilayers, and it is reasonable to expect that the changes in these properties are somehow coupled.
Although there is a wealth of information on the effects of cholesterol on lipid bilayers, the interplay of packing, free area, diffusion, and permeability has not yet been studied systematically. Experimental electron density profiles (McIntosh, 1978) and deuterium nuclear magnetic resonance (NMR) data (Sankaram and Thompson, 1990b) suggest that cholesterol should influence the packing inside membranes. Fluorescence recovery after photobleaching (FRAP) experiments, in turn, have been used to study the dependence of lateral diffusion coefficients on free area (Almeida et al., 1992). More information at the atomic level, however, is essential for gaining a detailed understanding of the effect of cholesterol on lipid bilayers. Such atomic-level information can be obtained from computer simulations. Molecular dynamics in particular provides a unique tool to investigate both the structure and dynamics of lipid membranes with a level of detail missing in any experimental technique. Until recently, however, systematic simulation studies have been limited by the extensive computational requirements.
In the present study, we investigate the cholesterol-induced changes in packing, free area, ordering, and lateral diffusion in phospholipid bilayers. Specifically, we study the presumptive interplay between these changes. To this end, we employ 100-ns molecular dynamics simulations on dipalmitoylphosphatidylcholine (DPPC)/cholesterol bilayers, with cholesterol concentrations ranging from 0 to 50 mol %. Although detailed multi-nanosecond simulation studies on the atomic level have emerged only very recently (Hofsäß et al., 2003; Scott, 2002; Tieleman et al., 1997), there exist large amounts of experimental studies for DPPC/cholesterol bilayers (McMullen and McElhaney, 1996; Sankaram and Thompson, 1990a,b; Vist and Davis, 1990, and references therein). These previous studies and the experimental results in particular offer us an excellent platform for comparison.
To further enhance the understanding of the effect of cholesterol on bilayers, we introduce a novel method for investigating the packing and free area in bilayers. The scope of this technique is very wide. It allows us to estimate how much space DPPC, cholesterol, and water molecules on average occupy in different regions of the bilayer. Consequently, it yields information on the amount and location of free space in the bilayer. As discussed below, this is related to various structural aspects such as the ordering of lipids in a membrane. Our method also provides valuable insight into dynamic properties. For example, our approach allows us to determine the area compressibility modulus across a membrane, and hence yields information on rate-limiting regions for lateral diffusion. In addition, as the method enables us to examine changes in free area with an increasing cholesterol content, we may estimate diffusion coefficients in terms of free area theories for lateral diffusion. The present approach can be applied to a wide range of different kinds of membrane systems, including one- and multicomponent bilayers, and bilayers with embedded solutes, probes, and proteins.
We find that cholesterol strongly affects the amount of space occupied by molecules in different parts of a phospholipid bilayer. The close-packed areas occupied by the tails of DPPC molecules can be explained by the ordering of the tails, and a simple relation (Petrache et al., 1999) can be used for quantifying the dependence of close-packed area on ordering. The amount and location of free space is significantly reduced by an increasing cholesterol content, and clearly reflect the total space occupied by DPPC and cholesterol molecules. The lateral diffusion coefficients, too, show a substantial decrease with an increasing cholesterol concentration. We find that so-called free area theories (Almeida et al., 1992; Cohen and Turnbull, 1959; Galla et al., 1979), which are essentially two-dimensional mean-field models, correctly predict this reduction, but are not applicable to quantitatively describing lateral diffusion in lipid bilayers.
MODEL AND SIMULATION DETAILS
We studied fully hydrated lipid bilayer systems consisting of 128 molecules, i.e., DPPCs and cholesterols, and 3655 water molecules. Since the main focus of this article is on studying the effects of cholesterol on phospholipid bilayers, we were interested in bilayers with varying amounts of cholesterol. To this end, we studied a pure DPPC bilayer and composite DPPC/cholesterol bilayers with six different cholesterol molar fractions: χ = 0%, 4.7%, 12.5%, 20.3%, 29.7%, and 50.0%.
The starting point was a united atom model for a fully hydrated pure DPPC bilayer that has been validated previously (Tieleman and Berendsen, 1996; Patra et al., 2003). The parameters for bonded and nonbonded interactions for DPPC molecules were taken from a study of a pure DPPC bilayer (Berger et al., 1997) available at http://moose.bio.ucalgary.ca/Downloads/lipid.itp. The partial charges are from the underlying model description (Tieleman and Berendsen, 1996) and can be found at http://moose.bio.ucalgary.ca/Downloads/dppc.itp. For water, the SPC model (Berendsen et al., 1981) was used. As our initial configuration for the pure DPPC bilayer we used the final structure of run E discussed in Tieleman and Berendsen (1996) and available at http://moose.bio.ucalgary.ca/Downloads/dppc128.pdb. The bilayer is aligned such that it lies in the x,y plane, i.e., the bilayer normal is parallel to the z axis.
The cholesterol force field and the initial shape of an individual cholesterol molecule were taken from http://www.gromacs.org/topologies/uploaded_molecules/cholesterol.tgz (Höltje et al., 2001). Cholesterols were introduced to the bilayer by choosing DPPC molecules from the pure phospholipid bilayer at random and replacing them by cholesterols. The same number of DPPC molecules was replaced in each of the two monolayers. In practice, the center of mass (CM) of a cholesterol molecule was moved to the CM position of the removed DPPC molecule. The main axis of inertia of each inserted cholesterol was parallel to the z axis, and each molecule was rotated by a random angle around the z axis.
The molecular dynamics (MD) simulations were performed at a temperature T = 323 K using the GROMACS (Lindahl et al., 2001) molecular simulation package. The time step for the simulations was chosen to be 2.0 fs. The lengths of all bonds were kept constant with the LINCS algorithm (Hess et al., 1997). Lennard-Jones interactions were cut off at 1.0 nm without shift or switch functions. Long-range electrostatic interactions were handled using the particle-mesh Ewald (Essman et al., 1995) method, which has been shown to be a reliable method to account for long-range interactions in lipid bilayer systems (Patra et al., 2003). The details of the implementation of particle-mesh Ewald have been discussed elsewhere (Patra et al., 2004).
After an initial energy minimization, we needed to equilibrate the system to fill the small voids left by replacing DPPC molecules by somewhat smaller cholesterol molecules. The equilibration was commenced by 50 ps of NVT molecular dynamics with a Langevin thermostat using a coupling time of 0.1 ps, i.e., every 0.1 ps the velocities of all particles were completely randomized from a Maxwell distribution corresponding to the target temperature. This complete loss of memory after 0.1 ps reduces the amount of ballistic motion of atoms inside a void. The equilibration was continued by 500 ps of NpT molecular dynamics at a pressure of 1 bar with a Langevin thermostat and a Berendsen barostat (Berendsen et al., 1984). The time constant for the latter was set to 1 ps, and the height of the simulation box was allowed to vary separately from the cross-sectional area of the box.
Finally, for every cholesterol concentration, we performed 100 ns of MD in the NpT ensemble with a Berendsen thermostat and barostat (Berendsen et al., 1984). The barostat was the same as the one described above, and the thermostat was set to separately couple the DPPC, cholesterol, and water molecules to a heat bath with a coupling time of 0.1 ps. With such a setup, in the case of pure DPPC, the average dimensions of the simulation box are 6.5 nm × 6.5 nm × 6.5 nm. For 29.7% cholesterol the dimensions are 5.2 nm × 5.2 nm × 9.0 nm.
The six simulations took a total of ∼60,000 h of CPU time. For all systems up to and including the cholesterol molar fraction of 29.7%, a simulation time of 100 ns guarantees a good sampling of the phase space. The results for 50% cholesterol should be regarded with some caution, as the diffusion of the DPPC and cholesterol molecules is already quite slow; see Lateral Diffusion and Free Area, below. As mixing of DPPC and cholesterol molecules in this case is quite limited, the system probably bears traces of its initial configuration. This applies to all state-of-the-art simulation studies of phospholipid/cholesterol systems, and has been mentioned by other authors (Smondyrev and Berkowitz, 1999).
RESULTS AND DISCUSSION
Equilibration
One of the most important quantities describing lipid bilayers is the average area per molecule. The average area per molecule for a given configuration, A, is computed by dividing the size of the simulation box in the x,y plane, designated Atot, by N, the total number of molecules, i.e., DPPCs and cholesterols, in a monolayer (see Fig. 1). The average area per molecule can, among other things, be used for monitoring the equilibration of the membrane.
FIGURE 1.
Structural formulae of DPPC and cholesterol molecules. The molecular masses of DPPC and cholesterol are 734.1 and 386.7 amu.
Fig. 2 shows the temporal behavior of the area per molecule. It can be seen that after 20 ns the area per molecule has converged even for the highest cholesterol concentrations. It is, nevertheless, immediately obvious from the data that this type of MD simulation of bilayer systems should be at least of the order of tens of nanoseconds to reach equilibrium and surpass the longest characteristic timescales for area fluctuations. The first 20 ns of the total 100 ns were therefore considered as equilibration, and the last 80 ns were used for analysis.
FIGURE 2.
Temporal behavior of area per molecule. The curves correspond to (from top to bottom) cholesterol concentrations 0.0%, 4.7%, 12.5%, 20.3%, 29.7%, and 50.0%.
The data clearly show that the area per molecule decreases with the cholesterol content. Further, an increasing cholesterol concentration seems to suppress the fluctuations in the average area per molecule. The values of the average area per molecule (see also Fig. 6) are in excellent agreement with two recent simulation studies on the DPPC/cholesterol system (Chiu et al., 2002; Hofsäß et al., 2003). As for experimental results, we are only aware of an accurate measurement for the average area per molecule in the case of a pure DPPC bilayer (Nagle and Tristram-Nagle, 2000). In this case the average area per molecule was determined to be 0.64 nm2 at T = 323 K, in good agreement with (0.655 ± 0.005) nm2 obtained here. Measurements of the average area per molecule in DPPC/cholesterol monolayers (McConnell and Radhakrishnan, 2003) show trends similar to ours. The exact correspondence between average areas per molecule measured for bilayers and monolayers, however, is not evident (Nagle and Tristram-Nagle, 2000).
FIGURE 6.
Average area per molecule as function of cholesterol concentration. The inset shows the average areas for DPPC (•) and cholesterol (○), i.e., 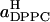 and
and 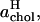 computed as in a recent simulation study by Hofsäß et al. (2003). The errors are smaller than the markers.
computed as in a recent simulation study by Hofsäß et al. (2003). The errors are smaller than the markers.
Ordering of acyl chains
Average areas per molecule are closely related to order parameters (Petrache et al., 1999), which are a measure of the orientational order of the phospholipid tails. Order parameters can be obtained from deuterium NMR experiments (Seelig and Seelig, 1974) or computer simulations (Tieleman et al., 1997). In united atom simulations such as ours, the orientational order can be characterized using tensors with elements Sαβ such that
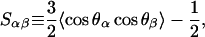 |
(1) |
where θα is the angle between the molecular α-axis and the bilayer normal (Tieleman et al., 1997). The molecular axes must be defined separately for each segment of an acyl chain: usually for the nth methylene group denoted as Cn, the z axis points in the Cn−1−Cn+1 direction, and Cn−1, Cn, and Cn+1 span the y,z plane. If the motion of the segments is assumed to be symmetric at the bilayer normal, the experimental deuterium order parameter SCD can be easily acquired, as
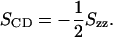 |
(2) |
As the two acyl chains sn-1 and sn-2 give rise to slightly different NMR quadrupole splittings (Seelig and Seelig, 1974), it is useful to compute the order parameters separately for both chains.
The order parameter profiles for the sn-1 and sn-2 chains are depicted in Fig. 3. The ordering effect of cholesterol is clearly visible: the order parameters grow significantly with an increasing cholesterol content. For pure DPPC and low cholesterol concentrations, the order parameter profiles show a plateau for small and intermediate values of n and decay near the center of the bilayer. When the cholesterol content increases, the plateau disappears, and there is a clear maximum at intermediate n. The ordering effect of cholesterol is most pronounced for n ∼ 6–10 and quite modest for segments near the phospholipid headgroups and bilayer center. This is due to the position of the cholesterol ring system in the bilayer along the bilayer normal (Smondyrev and Berkowitz, 1999): the largest ordering occurs for segments at roughly the same depth as the ring system. For instance, with 29.7% cholesterol, the order parameters for n ∼ 6–10 are increased roughly by a factor of 2.
FIGURE 3.
Order parameter profiles for (a) sn-1 and (b) sn-2 tails. The cholesterol concentrations are 0.0% (○), 4.7% (•), 12.5% (□), 20.3% (▪), 29.7% (⋄), and 50.0% (♦), and the index n increases toward the center of the bilayer.
Our results for the order parameters are in good agreement with other simulation studies (Smondyrev and Berkowitz, 1999; Chiu et al., 2002; Hofsäß et al., 2003). However, as most force fields yield qualitatively similar results, and various technical details may influence the detailed form of the order parameter profile (Patra et al., 2003), it is more interesting to make comparisons to experimental findings.
The results for the pure DPPC system are in good agreement with experiments (Brown et al., 1979; Douliez et al., 1995; Petrache et al., 2000). As for mixtures of DPPC and cholesterol, Sankaram and Thompson found that when 50% of the DPPC molecules were substituted by cholesterols in a pure DPPC bilayer at T = 325 K, the order parameter for intermediate n was increased by a factor of 2.65 (Sankaram and Thompson, 1990b). Similarly, when 30% of the dimyristoylphosphatidylcholines (DMPCs) were replaced by cholesterols in a pure DMPC bilayer at T = 308 K, the order parameter increased by a factor of 2. Vist and Davis, in turn, observed an increase by a factor of 2 when replacing 24% of the DPPC molecules by cholesterol at T = 323 K (Vist and Davis, 1990). Similar agreement is found when our results are compared to other experiments (Douliez et al., 1996; Kintanar et al., 1986). In all, our simulations agree well with experimental findings. The only detail which our, or any other, united-atom MD simulations cannot reproduce is the behavior of the experimental deuterium order parameter for sn-2 at n = 2 (Sankaram and Thompson, 1990b; Seelig and Seelig, 1975).
Electron density profiles
Additional information about the structure of the bilayer along the normal or z direction can be obtained by computing density profiles for the whole system, different molecular species, or certain atomic groups of interest. In simulations it is possible to calculate atom density, mass density, and electron density profiles. These give information on the distribution of atoms in the normal direction. Related information can be acquired from x-ray and neutron diffraction studies. Due to fluctuations, x-ray diffraction studies on fully hydrated bilayers in a fluid phase only yield total electron density profiles, whose maxima are associated with the electron dense phosphate groups (Nagle and Tristram-Nagle, 2000). The distance between the maxima allows one to estimate the distance between the headgroups in the opposite leaflets, but does not yield accurate predictions for the hydrocarbon thickness or the true phosphate-phosphate distance (Nagle et al., 1996). Additional information, most importantly about the average location of various atomic groups, can be gained from neutron diffraction studies either with selective deuteration or in combination with x-ray diffraction (Nagle and Tristram-Nagle, 2000).
Fig. 4 shows the total electron densities calculated for the different cholesterol concentrations. The density profiles have a characteristic shape reminiscent of x-ray diffraction studies, with maxima approximately corresponding to the location of the phosphate groups, and a minimum, a so-called methyl trough, in the bilayer center, where the terminal methyl groups reside. For pure DPPC and low cholesterol concentrations, the densities decrease monotonically from the maxima to the minimum in the bilayer center. This medium density region corresponds to the methylene groups in the DPPC tails. When more cholesterol is present, the headgroup-headgroup distance increases, i.e., the bilayer gets thicker, and the electron density in the bilayer center decreases slightly. In addition, the density in the tail region increases, and the density profile between the center and the headgroups is no longer monotonically decreasing. The elevation is due to the fact that the cholesterol ring structure, which resides in the phospholipid tail region, has a higher electron density than do phospholipid tails.
FIGURE 4.
Total electron density profiles as functions of distance z from bilayer center. The curves correspond to the various cholesterol concentrations as 0.0% (dash-dotted gray), 4.7% (solid black), 12.5% (solid gray), 20.3% (dashed black), 29.7% (dashed gray), and 50.0% (dash-dotted black).
To gain more insight into the structure of the bilayer, we can investigate the electron densities for DPPC molecules, cholesterols, water molecules, phospholipid tails, phosphate groups, and cholesterol rings, portrayed in Fig. 5. All density profiles are consistent with a thickening of the bilayer with an increasing amount of cholesterol: the molecules and their constituent atomic groups are pushed toward the water phase. Still, it is clear that for all cholesterol concentrations, DPPC molecules largely stay within a distance of 3 nm from the center, whereas cholesterols and DPPC tails can be found within ∼2 nm. We can conclude that cholesterol is located in the hydrophobic interior of the bilayer. The penetration of water into the bilayer becomes more difficult with increasing amounts of cholesterol: this reflects both the thickening of the bilayer and the increasing densities in the headgroup region. The lipid/water interface also seems to become steeper. The electron density of DPPC in the hydrophobic tail region decreases with the cholesterol content, which is compensated by an increasing cholesterol electron density. By comparing the electron densities for cholesterol and cholesterol ring systems, we can conclude that only the short acyl chain of cholesterol can approach the bilayer center.
FIGURE 5.
Electron density profiles for molecular species and atomic groups: (a) DPPC, (b) cholesterol, (c) water, (d) DPPC tails (atoms 15–31 and 34–50), (e) phosphate groups, and (f) cholesterol ring system. The curves correspond to the cholesterol concentrations as indicated in Fig. 4. For reasons of clarity, the densities for the system with 50% cholesterol are not shown.
Both the total electron density profile and the densities for molecular species and atomic groups can be compared to previous simulations. Here we will concentrate on simulations on DPPC with cholesterol at T = 323 K (Hofsäß et al., 2003; Smondyrev and Berkowitz, 1999; Tu et al., 1998). In all simulation studies, the peaks that indicate the location of headgroups for pure DPPC are located approximately at the same distance from the bilayer center. With 10–12.5% cholesterol, only minor changes in the total densities can be observed. Except in the case of Tu et al., increasing amounts of cholesterol lead to a larger bilayer thickness and a slightly decreased total density in the bilayer center. All studies show an increased density in the phospholipid tail region. By investigating the DPPC or water densities, one may also note that all studies clearly indicate that the lipid/water interface becomes more abrupt. Our findings for the distribution of phosphorus atoms agree well with those of Smondyrev and others: when the cholesterol content increases, the peaks are narrowed and shifted toward the water phase. In all, density profiles computed using slightly different force fields are, for the most part, consistent with each other.
Our results are also consistent with diffraction experiments on DPPC and DMPC bilayers. Nagle et al. (1996) have determined the structure of a fully hydrated pure DPPC bilayer in the liquid-disordered phase using x-ray diffraction. The form of the density profile from our simulations of pure DPPC closely resembles Nagle's electron density profile for pure DPPC at T = 323 K. The head-head distance obtained from Nagle's experiment and that determined from our density profiles also are in good agreement. As for the influence of cholesterol, McIntosh (1978) has published x-ray diffraction experiments on model membranes containing cholesterol and phospholipids with saturated tails containing 12–18 carbons. His DLPC/cholesterol systems in the fluid phase behave in a qualitatively similar way as do our DPPC/cholesterol bilayers. By comparing the electron densities from systems with different phospholipids and cholesterol to the densities from pure phospholipid bilayers, McIntosh also establishes the location of the cholesterol ring structure in the bilayer. Our studies support his view. In addition, there are more recent neutron diffraction studies of DMPC/cholesterol bilayers. The studies by Douliez et al. (1996) and Léonard et al. (2001) clearly show that substituting 30% of the phospholipids by cholesterol in a pure DMPC bilayer in the liquid-disordered phase increases the bilayer thickness. Léonard and co-workers have also investigated the location of cholesterol in the bilayer, and concluded that cholesterol is located well within the hydrophobic core. Although DPPC has longer hydrocarbon tails than DMPC, the cholesterol ring structure should be located in the same region of the bilayer (McIntosh, 1978). Our simulations indicate that cholesterol is indeed situated in the nonpolar region, as is the case in Douliez's and McIntosh's experiments.
Radial distribution functions
Together, the above results ascertain that our model correctly describes the behavior of the dimensions of the bilayer and the ordering of the nonpolar phospholipid tails as functions of the cholesterol content. Further, the structure of our DPPC/cholesterol bilayer in the normal direction is consistent with results from previous computations and experiments. This is very satisfactory, but in addition, we need to ensure that our bilayers truly are in the fluid state, i.e., that there is no translational long-range order. This can be ascertained by examining the radial distribution functions for, e.g., phosphorus and nitrogen atoms in the DPPC headgroups. For instance, the N–N radial distribution functions calculated in two dimensions for various cholesterol concentrations have large nearest-neighbor peaks at r ∼ 0.82 nm and show essentially no structure beyond r = 1.5 nm (data not shown). Additional calculations for other pairs of atoms and for the CM positions of the DPPC and cholesterol molecules lead to a similar conclusion, i.e., that there is no lateral long-range structure. Hence, we can be confident that our bilayers are either in the liquid-disordered or liquid-ordered phase, as they should. With this, we consider our model to be valid.
Estimating average areas per molecule in multicomponent bilayers
The average area per molecule, which is obtained by dividing the total area of the bilayer by the total number of molecules, is a well-defined concept in one-component lipid bilayers. It includes both area actually occupied by a lipid, the so-called close-packed area, and some free area. A similar quantity can be defined for multicomponent bilayers. It is a useful quantity when simulation results are compared to experiments. Its interpretation, however, is less clear: different lipids and sterols could occupy significantly different amounts of area. Hence, it would be desirable to be able to estimate the average area occupied by each molecular species present in the bilayer.
The average area per molecule 〈A〉 as a function of cholesterol concentration χ is portrayed in Fig. 6. As mentioned in Equilibration, above, it is evident that 〈A〉 decreases with the cholesterol content, and that the results agree well with previous simulation studies (Chiu et al., 2002; Hofsäß et al., 2003).
We would not, however, like Chiu et al. (2002), conclude that 〈A 〉 decreases linearly with χ and use this assumption to compute the average areas per phospholipid and cholesterol. It is not obvious, in the first place, that the average area per cholesterol or DPPC is independent of cholesterol content, as these authors seem to imply.
Another way to divide the total area between DPPC and cholesterol molecules has also been suggested (Hofsäß et al., 2003). By computing the total area and volume of the simulation box as functions of the cholesterol content and making a number of assumptions, one can arrive at estimates for the average areas occupied by DPPC and cholesterol molecules. In this case, an important assumption is that the average volume of a cholesterol molecule can be, for all concentrations, taken to be the volume occupied by a cholesterol molecule in a cholesterol crystal. Further, it is assumed that all space is occupied by DPPC, cholesterol, or water, i.e., that there is no free volume or area. The average areas per DPPC and cholesterol, 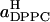 and
and 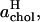 obtained along these lines from our data, are shown in the inset of Fig. 6. These closely resemble the corresponding results by Hofsäß et al.
obtained along these lines from our data, are shown in the inset of Fig. 6. These closely resemble the corresponding results by Hofsäß et al.
A yet further method of distributing the area among the molecular species in a bilayer is to apply Voronoi analysis in two dimensions (Jedlovszky et al., 2004; Patra et al., 2003; Shinoda and Okazaki, 1998). In Voronoi tessellation for a bilayer, the center of mass (CM) coordinates of the molecules comprising the bilayer are projected onto the x,y plane. An arbitrary point in this plane is considered to belong to a particular Voronoi cell, if it is closer to the CM position associated with that cell than to any other one. In this way one can calculate the total area associated with the CM positions of, e.g., the DPPC molecules and then scale this quantity by the number of DPPC molecules in a monolayer. The resulting average areas per DPPC and cholesterol, 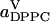 and
and 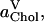 as functions of the cholesterol content, are depicted in Fig. 7.
as functions of the cholesterol content, are depicted in Fig. 7.
FIGURE 7.
Areas per DPPC (•) and cholesterol (○), i.e.,  and
and 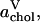 computed using Voronoi tessellation. The errors for DPPC are smaller than the markers.
computed using Voronoi tessellation. The errors for DPPC are smaller than the markers.
These values for the areas per DPPC and cholesterol differ markedly from those reported by Hofsäß et al. The values obtained using the formulae introduced by Hofsäß et al. are sensible and meaningful, if one is interested in studying quantities containing both close-packed and free area. The values obtained from the Voronoi analysis, on the other hand, do not appear to be quite as sensible: in the case of 29.7% cholesterol, the area of a DPPC molecule is already smaller than that of a DPPC molecule in a bilayer in the gel state (Nagle and Tristram-Nagle, 2000; Venable et al., 2000). The differences are in part due to the fact that the assumptions inherent to the respective methods lead to different ways of distributing the free area in the bilayer. This, however, does not fully explain the large differences and the peculiar values obtained using the Voronoi analysis at high cholesterol concentrations. A more important reason is that basic Voronoi analysis does not, in any way, allow one to take into account the close-packed sizes of the molecules. It may well be that the area (which from the point of view of the Voronoi analysis belongs to cholesterol), as a matter of fact would be covered by projected coordinates of atoms from a DPPC molecule. Concluding, although Voronoi analysis may be a useful tool for studying, e.g., phase separation or local effects, the values for molecular areas from such analysis have no quantitative meaning.
Slicing membranes
We are now confronted by fundamental questions relevant to both one- and multicomponent bilayer systems. How can we find estimates for the average close-packed cross-sectional areas for the molecular species present in a one-component or composite bilayer? Further, how can we estimate the average amount of free area in a membrane?
Our approach to answer these questions bears a certain resemblance to tomography. Related grid approaches have been used in other applications (see, e.g., Kandt et al., 2004, and references therein). We map each configuration on a number of rectangular three-dimensional grids as follows. If a grid point lies within the van der Waals radius of an atom belonging to a DPPC molecule, this point is considered occupied, and otherwise empty, on a grid keeping account of DPPC molecules. Grid points within van der Waals radiae of atoms belonging to cholesterol, in turn, will be occupied on a grid characterizing the cholesterol molecules. Finally, a grid for water molecules is constructed analogously. In the x,y plane the grids have 100 × 100 elements. Because the system size fluctuates weakly, the size of an element will vary slightly from configuration to configuration. In the z direction, on the other hand, the size of the elements has been fixed to 0.1 nm, and we only consider grid points within 3 nm from the bilayer center.
The grids can be used to view given slices of the bilayers: they show cross sections of DPPC, cholesterol, and water molecules, as well as patches of free area. Pictures of slices can be illustrative as such, and Fig. 8 contains a selection of such slices for the case of 20.3% cholesterol. From Figs. 5 and 8 a we can conclude that there are quite large amounts of free area in the bilayer center, and that cholesterol tails from a given monolayer extend to the opposite monolayer. Fig. 8 b portrays the region where DPPC tails and cholesterol ring structures should, according to Fig. 5, dominate. DPPC tails can be recognized as circular red structures, and the green formations are cross sections of cholesterol ring structures. Fig. 8 c is a cross section of the bilayer at a distance z ∼ 1.7 nm from the bilayer center. Some cholesterol is still present in this slice, and there are also small amounts of water. The amount of free area is significantly smaller than in the bilayer center. Fig. 8 d finally shows a cross section at z ∼ 2 nm: there are DPPC headgroups, substantial amounts of water, and very little cholesterol.
FIGURE 8.
Cross sections of bilayer with 20.3% cholesterol at 100 ns. DPPC grid elements have been colored red, cholesterol is green, and water blue. The remaining area, i.e., the free area, is white. The panels correspond to slices at different distances z from the bilayer center: (a) bilayer center, (b) z ∼ 1 nm, (c) z ∼ 1.7 nm, (d) z ∼ 2 nm.
From the grids constructed for DPPC, cholesterol, and water, we can compute total area profiles for the various molecular species, that is, average total areas occupied by the molecules as functions of the distance from the bilayer center. In addition, we can calculate free area profiles, i.e., the amount of free area as a function of the distance from the bilayer center. In practice, this is achieved by traversing the grids slice by slice and augmenting the various area profiles. If a grid element in a certain slice at a distance z from the bilayer center is occupied in, say, the DPPC grid, but not in the cholesterol or water grids, we increment the total area of DPPC in that slice, ADPPC(z), by an area corresponding to a grid element. If, on the other hand, a grid point at a distance z from the center is occupied by neither DPPC nor cholesterol, nor water, the total free area Afree(z) in the slice in question is incremented. In the end we average over the total area profiles constructed separately for each configuration. This procedure leads to a definition of free area which is similar in nature to the concept of empty free volume introduced by Marrink et al. (1996) to characterize a pure DPPC bilayer. Fig. 9 exemplifies the computation of the various area profiles for a bilayer with 20.3% cholesterol.
FIGURE 9.
Area profiles for bilayer with 20.3% cholesterol scaled by total bilayer area: DPPC area profile 〈ADPPC(z)〉/〈Atot〉 (solid black); cholesterol area profile 〈Achol(z)〉/〈Atot〉 (solid gray); water area profile 〈Awater(z)〉/〈Atot〉 (dashed black); and free area profile 〈Afree(z)〉/〈Atot〉 (dashed gray). The errors of the scaled areas are of the order of a few percent.
Close-packed areas for DPPC and cholesterol
To gain understanding of the effect of cholesterol on the properties of phospholipid bilayers, we first concentrate on the behavior of the cross-sectional area profiles for DPPC and cholesterol. Hence, we need to know both the total areas of DPPC and cholesterol and the average numbers of respective molecules as functions of the distance from the bilayer center. The total areas occupied by DPPC and cholesterol molecules, denoted by 〈ADPPC(z)〉 and 〈Achol(z)〉, for the different cholesterol concentrations are computed in the manner described above.
To find the average numbers of DPPC and cholesterol molecules in each slice, we locate the maximum and minimum z coordinates of each molecule with respect to the bilayer center, taking into account the finite size of the constituent atoms. The molecule is considered to be present in all the slices between these points. By averaging over all molecules of a certain species and over all configurations, we arrive at the average numbers of DPPC molecules and cholesterols as functions of the distance from the bilayer center, denoted by 〈NDPPC(z)〉 and 〈NChol(z)〉, shown in Fig. 10. Perhaps the most notable feature in Fig. 10 is that all curves peak in the bilayer center. This is due to so-called interdigitation: a substantial part of both DPPC and cholesterol molecules extend to the opposite monolayer. On both sides of the peak, there are broad plateaus, which reflect the amount of molecules of a certain species in a monolayer. Eventually, at ∼3 nm from the bilayer center for DPPC and 2 nm for cholesterol, the curves decay to zero.
FIGURE 10.
Numbers of (a) DPPC and (b) cholesterol molecules as functions of distance from bilayer center. The curves correspond to the cholesterol concentrations as indicated in Fig. 4. The errors are of the order of <1%.
There seem to be two effects that together contribute to the thickening of the bilayer, both visible in Fig. 10. First, the DPPC molecules are extended. Cholesterol molecules, on the other hand, are not significantly elongated. These observations are quite plausible, as the presence of cholesterol leads to a smaller amount of gauche defects in the acyl chains of the DPPC molecules (Hofsäß et al., 2003). Further, the tilt of the DPPC molecules with respect to the bilayer normal decreases (Róg and Pasenkiewicz-Gierula, 2001). Cholesterol, on the other hand, with its rigid ring structure and short tail, does not undergo such significant extension.
The second effect partially responsible for the thickening is that with a larger cholesterol content χ, a smaller amount of DPPC and cholesterol molecules extend to the opposite monolayer. Further, the ones that protrude do not penetrate quite as deep into the opposite leaflet as they do at low cholesterol concentrations. In a pure DPPC bilayer, 53% of the DPPC molecules protrude to the opposite monolayer, whereas at 29.7% cholesterol the corresponding figure is 40%. The effect is stronger for cholesterol: at 4.7% and 29.7% concentrations, respectively, 41% and 17% of the molecules extend to the opposite bilayer. As the cholesterol hydroxyl is thought to be anchored to the DPPC headgroup via direct hydrogen bonding or through water bridges (Chiu et al., 2002; Pasenkiewicz-Gierula et al., 2000), this effect may be coupled to the elongation of the DPPC molecules.
Equipped with the total areas occupied by the molecular species together with the average numbers of these molecules as functions of distance from the bilayer center, we can now compute the average cross-sectional areas for DPPC and cholesterol across a membrane, aDPPC(z) ≡ 〈ADPPC(z)〉/〈NDPPC(z)〉 and achol(z) ≡ 〈Achol(z)〉/〈Nchol(z)〉, shown in Fig. 11.
FIGURE 11.
Cross-sectional close-packed areas for (a) DPPC and (b) cholesterol molecules as functions of distance from bilayer center. The curves correspond to the cholesterol concentrations as indicated in Fig. 4. The errors are of the order of a few percent. In the water phase, the relative errors for achol are somewhat larger.
It should not come as a surprise that the cross-sectional close-packed area occupied by a DPPC or cholesterol molecule is not constant along the bilayer normal. In the case of DPPC, there are significant changes in the form of aDPPC(z) when the cholesterol content is increased. A maximum located at ∼1 nm from the bilayer center for a pure DPPC bilayer becomes at intermediate cholesterol concentrations a plateau at 0.5–1.5 nm from the center, and finally with 29.7% cholesterol in the bilayer develops into two small maxima at 0.5 nm and 2 nm with a shallow minimum in between.
These changes in aDPPC(z) are in part due to the behavior of the phospholipid tails: significant changes occur in regions where the tail densities are high and where there are few or no headgroups. This can be deduced by comparing the electron densities for DPPC molecules and DPPC tails in Fig. 5, a and d. This allows us to partially interpret the behavior of aDPPC(z) from the point of view of ordering. The most substantial ordering effect with large amounts of cholesterol present in the bilayer occurs for carbons in the middle of the tail; see Fig. 3. Close to the headgroups and in the bilayer center the ordering effects of cholesterol are more modest. As increased order correlates with a decreasing area occupied by the tails, one expects that with an increased cholesterol content the cross-sectional area per DPPC approximately at a distance 1 nm from the bilayer should decrease. Our findings are consistent with this picture.
This is not to say, however, that there would exist a simple way of mapping aDPPC(z) with order parameter profiles (see also Close-Packed Areas from Ordering of Acyl Chains, below). The maximum that develops at z ∼ 2 nm, e.g., is a result of contributions from glycerol, phosphate, and choline groups. From separate cross-sectional area profiles for the two tails on one hand and the glycerol, phosphate, and choline groups on the other hand (data not shown), we found that when the cholesterol concentration increases, the cross-sectional area occupied by the tail portion of a DPPC molecule decreases as a consequence of ordering, whereas the area occupied by the glycerol, phosphate, and choline groups seems to be increasing (data not shown). The increase is probably related to changes in the orientation of these groups. Concluding, the maximum at z ∼ 2 nm at intermediate and high cholesterol concentrations is related to the interplay of the decreasing tail contribution with a plateau centered at z ∼ 1 nm and the increasing head contribution that peaks at z ∼ 2 nm.
In the case of cholesterol the cross-sectional close-packed area of a molecule is changed only weakly when the cholesterol concentration χ is increased. The slight decrease with an increasing χ can be explained by the tilt of the cholesterol molecules. At high concentrations, almost all cholesterols are oriented nearly parallel to the bilayer normal (data not shown). At low concentrations, on the other hand, the distribution of the angle between the bilayer normal and the ring structure becomes more broad and flat, i.e., the molecules are more tilted with respect to the bilayer normal. Hence the cross sections appear larger at low concentrations.
The general form of achol(z) is compatible with our idea of the structure of the cholesterol molecule: narrow in the bilayer center where the small cholesterol tails reside and broad where the ring structure is located. It also reflects the thickening of the bilayer, as the maxima associated with the ring structures are pushed toward the water phase when more cholesterol is present. This picture, overall, supports the common belief that the average area per cholesterol in a phospholipid bilayer is largely unaltered by the amount of cholesterol in the bilayer.
Our results for achol(z) can be compared to the outcome of an old experiment (Rothman and Engelman, 1972), where a model of cholesterol made of plastic was immersed in a tube filled with water. This experiment resulted in a steric profile for cholesterol, i.e., a profile of the cross-sectional area occupied by cholesterol. This steric profile and our achol(z), especially at high cholesterol concentrations, bear a surprisingly good resemblance to each other. The steric profile measured by Rothman and Engelman displays a plateau where the cholesterol rings are located, with cross-sectional areas of the order of 0.25 nm2. In the region where the cholesterol tail is located, they report a small maximum: here the cross-sectional areas are of the order of 0.15 nm2.
It is clearly difficult to describe the close-packed area of a DPPC or cholesterol molecule by a single number. Of course, we could attempt to define the close-packed area of, e.g., a DPPC molecule in a given DPPC/cholesterol bilayer as the maximum of the relevant aDPPC(z) profile, but this would not give accurate information about the packing of DPPC and cholesterol molecules in a composite bilayer. Despite this, we may note that the maximum values are useful at least when assessing the plausibility of the close-packed area profiles for DPPC and cholesterol molecules.
In the case of DPPC the maxima assume values between 0.36 nm2 and 0.42 nm2. These values can be compared to the average area per molecule in a pure DPPC bilayer in the gel state, where the contribution of the free area to the total area assigned to a phospholipid molecule is expected to be rather minor. Experiments have yielded an area per molecule of ∼0.48 nm2 (Nagle and Tristram-Nagle, 2000), and MD simulations suggest that 〈A〉 = 0.46 nm2 (Venable et al., 2000). An exact comparison is not meaningful, since DPPC/cholesterol mixtures, especially with high cholesterol concentrations, have structures quite different from a pure DPPC bilayer in the gel state. However, the comparison shows that the magnitude of the close-packed areas for DPPC molecules is rational.
In a similar fashion, the maxima of the achol(z) profiles can be compared to values extracted from experiments on cholesterol crystals. The maxima found in this study decrease monotonically from 0.33 nm2 to 0.29 nm2 when the cholesterol concentration changes from 4.7% to 29.7%. In a cholesterol crystal, the area per cholesterol, which in this case contains both occupied and free area, has been reported to be 0.38 nm2 (Craven, 1979; Chiu et al., 2002; Hofsäß et al., 2003; and references therein).
Free area
We now turn our attention to the behavior of free area profiles for bilayers with different amounts of cholesterol. In Fig. 12, we show the average amount of free area per molecule, i.e., afree ≡ 〈Afree〉/N, where N is the total number of molecules—both phospholipids and cholesterol—in a monolayer. The figure clearly shows that the amount of free area per molecule decreases in all regions of the bilayer, i.e., for all values of z, with an increasing cholesterol content. Compared to the case of pure DPPC, 4.7% cholesterol in the bilayer leads to a free area per molecule reduced by ∼7% in all regions of the bilayer. With 12.5%, 20.3%, and 29.7% cholesterol in the bilayer, the free area per molecule is decreased by 20%, 35%, and 45%. One may note that the behavior of the total area of the bilayer cannot be explained by the reduced free area only. The occupied area, i.e., the area taken up by DPPC, cholesterol, or water molecules, also decreases with more cholesterol. For instance, when 29.7% of the DPPC molecules are substituted by cholesterol, the amount of occupied area decreases by ∼30%. The behavior of the total free and occupied volumes in a bilayer with an increasing cholesterol concentration will be discussed in detail elsewhere.
FIGURE 12.
Free areas per molecule as functions of distance from bilayer center for different cholesterol concentrations. The curves correspond to the cholesterol concentrations as indicated in Fig. 4.
Fig. 12 also demonstrates that an increasing cholesterol content in a bilayer implies that the form of the free area profile is altered. Nevertheless, the different curves corresponding to the various cholesterol concentrations have certain features in common: the free area profiles all have a maximum in the bilayer center, and there is more free area per molecule in the water phase than in the tail and headgroup regions. For pure DPPC and low cholesterol concentrations, we observe a minimum of free area per molecule at z ∼ 1.7 nm. For large cholesterol concentrations the minimum is still present, but due to the thickening of the bilayer it is pushed toward larger z: e.g., for 29.7% it can be found at z ∼ 2 nm. This minimum can, for all cholesterol concentrations, be associated with a peak in the density profile of DPPC molecules located slightly behind the headgroups in a region where tails, glycerol, phosphate, and choline groups are present. The density of water in this region is already substantial, whereas there is very little cholesterol. When the cholesterol concentration is increased, also another flat, plateau-like minimum starts to develop between the bilayer center and the minimum associated with the maximum in the DPPC density, i.e., at z ∼1–2 nm. The plateau is almost constant through the tail and headgroup regions. It has counterparts in the area profiles of DPPC and cholesterol: the cross-sectional DPPC area displays here a flat minimum and the cholesterol area a broad maximum (see Fig. 11). We can thus conclude that the changes in the form of the free area profile are intimately related to modifications in the packing of the molecules in the bilayer.
It is evident that the free area profiles are related to the relocation and diffusion of solutes inside membranes. MD simulations suggest that solutes such as ubiquinone (Söderhäll and Laaksonen, 2001) and benzene (Bassolino-Klimas et al., 1993) are preferentially located in the hydrophobic core region of a membrane. Also, it is known that certain nonpolar probe molecules, e.g., diphenylhexatriene, prefer the bilayer center to the lipid/water interface (Lentz, 1993). These observations are in accord with our suggestion that the free area is largest in the bilayer center.
There are two other simulation studies where quantities similar in nature to our free area profile have been calculated for DPPC or DPPC/cholesterol bilayers. Marrink et al. (1996) have calculated a so-called empty free volume profile for pure DPPC. This should give essentially the same information about the amount of average free area in a given cross section of the bilayer as does our free area profile for pure DPPC. Our profile does indeed show the same general features as Marrink's: a maximum in the bilayer center and minima near the headgroup region. Tu et al. (1998) have also looked at the influence of 12.5% cholesterol on a so-called empty free volume fraction, which is equivalent to our total free area scaled by the total area of the bilayer. If we compare such scaled free areas (data not shown) to Tu's data, we see that the scaled profiles have many features in common. One difference is that the bilayer thickening is not visible in Tu's results, whereas it can be clearly distinguished from ours. The thickening has also been verified experimentally. Further, there are some differences in the detailed form of the profiles in the bilayer interior, i.e., the location of the minima are slightly different. As Tu et al. point out, the differences are probably due to different computational models.
Lateral diffusion and free area
We have seen that an increasing cholesterol concentration reduces the amount of free area per molecule in the bilayer and simultaneously alters the packing of the molecules. On the other hand, it is well known from experiments that lateral diffusion of both DPPC and cholesterol molecules is affected by changes in the cholesterol content (Almeida et al., 1992; Filippov et al., 2003a; König et al., 1992). It is reasonable to expect that these properties of the bilayer and the observed modifications in them with the cholesterol concentration are related. Free volume theory is a simple but appealing model for explaining such dependencies.
Free volume theory was originally developed for describing the transport properties of glass-forming fluids (Cohen and Turnbull, 1959; Macedo and Litovitz, 1965; Turnbull and Cohen, 1961, 1970). It was subsequently adapted to modeling two-dimensional diffusion (Galla et al., 1979; MacCarthy and Kozak, 1982; Vaz et al., 1985; Almeida et al., 1992) and is usually in this context dubbed free area theory. Free area theory, a two-dimensional mean-field model for diffusion, can be used to at least qualitatively describe lateral self-diffusion in lipid bilayers (Almeida et al., 1992). According to free area theory, lateral diffusion of a lipid or sterol in a bilayer is restricted by the occurrence of a free area greater than some critical area adjacent to the diffusing molecule. A diffusing molecule spends a comparatively long time—of the order of tens of nanoseconds (Tieleman et al., 1997; Vattulainen and Mouritsen, 2003)—in a cage formed by its neighbors, and then, given a large enough activation energy and an adjacent free area, jumps.
More specifically, free area theory predicts that the lateral diffusion coefficient of a lipid or sterol diffusing in a bilayer depends on the free area and the packing properties as follows (Almeida et al., 1992):
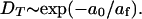 |
(3) |
Here a0 is an estimate for the average cross-sectional area for a DPPC or cholesterol molecule and af is a measure for the average amount of free area per molecule in the bilayer.
To examine the validity of Eq. 3 we compute the lateral diffusion coefficients for DPPC and cholesterol molecules at different cholesterol concentrations. The lateral tracer diffusion coefficients can be computed using the Einstein relation
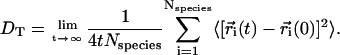 |
(4) |
Here 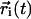 is the CM position of molecule i at time t and the sum is over all molecules of a given species. The lateral diffusion coefficients have been calculated by following the position of each molecule in the upper (lower) monolayer with respect to the CM position of the corresponding upper (lower) monolayer. Thus, should there be any drift, the motion of the CM of each monolayer has been taken into account.
is the CM position of molecule i at time t and the sum is over all molecules of a given species. The lateral diffusion coefficients have been calculated by following the position of each molecule in the upper (lower) monolayer with respect to the CM position of the corresponding upper (lower) monolayer. Thus, should there be any drift, the motion of the CM of each monolayer has been taken into account.
Results for lateral diffusion coefficients are shown in Fig. 13. The lateral diffusion coefficients for both DPPC and cholesterol decrease monotonically with an increasing cholesterol content. This reduction is qualitatively consistent with experiments (Almeida et al., 1992; Filippov et al., 2003a; König et al., 1992). Quantitative comparisons should preferably be made to experimental techniques that probe lateral diffusion of individual molecules at timescales comparable to those reached in MD simulations. Fluorescence correlation spectroscopy measurements should hence give us a good reference. In fluorescence correlation spectroscopy measurements for DLPC/cholesterol systems, Korlach et al. (1999) found that when the cholesterol concentration was increased from 0% to 60%, DT for DLPC was reduced by a factor of 10. Even though the acyl chains of DLPC molecules are shorter than those of DPPC molecules, our findings are in reasonable accord with Korlach's experiments.
FIGURE 13.
Lateral diffusion coefficients of DPPC (•) and cholesterol (○) molecules as functions of cholesterol concentration.
Let us now consider the implications of our results to free area theory for lateral diffusion. In free area theory, the critical area a0 is essentially a number describing the close-packed cross-sectional molecular area of the diffusant. In the same spirit, the average free area per molecule af should be characterized by a single number. We have, however, seen that the free areas per molecule and the areas per DPPC and cholesterol molecules are functions of the distance from the bilayer center. Hence, it seems that a two-dimensional mean-field model might be a too simplistic means of describing lateral diffusion.
In our opinion, one should at least not expect free area theory to yield quantitative results. It might, however, give qualitative predictions about trends in cases where, e.g., the cholesterol content in a bilayer is increased. With this in mind, let us assume that the cholesterol concentration in a DPPC/cholesterol bilayer rises from 4.7% to 29.7%. If we now use the largest possible values for the fractions a0/af, free area theory will give us upper bounds for the reduction of the lateral diffusion coefficients. The lateral diffusion coefficient for DPPC should, according to free area theory, now be reduced by a factor of 3 at most, and DT for cholesterol should decrease by a factor of 2. As a matter of fact, the lateral diffusion coefficients for both DPPC and cholesterol computed from the simulation data are reduced much more strongly; see Fig. 13. We can conclude that Eq. 3 tends to underestimate the changes in the values of the lateral diffusion coefficients.
Even though the discrepancies in the predictions of Eq. 3 and the computed lateral diffusion coefficients do exist, we cannot immediately declare free area theory incomplete. There is a detail that has been overlooked in our discussion so far, and the significance of this detail will now be considered. To jump to an adjacent empty site, a diffusing molecule needs energy to overcome an activation barrier. In free area theory this is accounted for by letting the lateral diffusion coefficient be proportional to a Boltzmann factor exp(−Ea/kBT), where Ea is the activation barrier. As a growing cholesterol concentration increases the ordering of the DPPC tails and therefore reduces the area per molecule, it seems reasonable to expect that Ea should increase with the cholesterol content. Experimental results (Almeida et al., 1992) do support this idea but are partly contradictory. This is, however, probably due to the fitting procedure used (Almeida et al., 1992). In a more recent study, Filippov et al. (2003b) used NMR to study the lateral diffusion in palmitoyloleoylphosphocholine/cholesterol and dioleoylphosphocholine/cholesterol bilayers over a cholesterol concentration range of ∼0–45 mol %. At small χ, they found the apparent (Arrhenius) diffusion barrier to be approximately constant, whereas for large χ the diffusion barrier increased markedly. Hence, the neglect of the energy term might in our case lead to slight underestimates for the reduction of the lateral diffusion coefficients.
Summarizing, we have found that free area theory correctly predicts the reduction of the lateral diffusion coefficients with an increasing cholesterol concentration. At the same time it seems unnecessary to aim for a quantitative description with such a simple framework. Instead of being based on mean-field arguments, a full theoretical description of lateral diffusion should account for local free volume fluctuations in the vicinity of diffusing molecules. Atomic-scale MD studies in this direction should be feasible in the near future.
Area compressibility modulus
Lateral diffusion is clearly influenced by the average amount of free area in the bilayer. However, not only the average free area, but also fluctuations in the amount of free area should play a role here. Recall that free area theory states that a diffusion jump is not possible unless there is a large enough free area next to the diffusant (Almeida et al., 1992). Large enough free areas are a result of fluctuations, and hence we would expect diffusion to depend on the magnitude of the fluctuations: decreasing fluctuations and slowed lateral diffusion should be coupled. For similar reasons, it is likely that permeation of molecules across membranes can at least partially be explained by area fluctuations in membranes.
We may quantify area fluctuations in different regions of the membrane as follows. The starting point is the average occupied area 〈Aocc(z)〉, i.e., the area which is not free but occupied by DPPC, cholesterol, or water molecules. The occupied area obviously varies with the distance from the bilayer center z. Based on the definition of compressibility modulus given in Feller and Pastor (1999) and Hofsäß et al. (2003), we now define an area compressibility modulus for the occupied area as
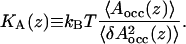 |
(5) |
Here kB is the Boltzmann constant and 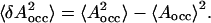 The area compressibility modulus is a measure of the fluctuations in the occupied area: a high compressibility modulus indicates small fluctuations and a low compressibility modulus, correspondingly, large fluctuations. Hence, the area compressibility modulus should be related to the permeation of small solutes, as well as to the lateral diffusion of lipids and sterols.
The area compressibility modulus is a measure of the fluctuations in the occupied area: a high compressibility modulus indicates small fluctuations and a low compressibility modulus, correspondingly, large fluctuations. Hence, the area compressibility modulus should be related to the permeation of small solutes, as well as to the lateral diffusion of lipids and sterols.
Fig. 14 shows the area compressibility modulus profiles computed for systems with different amounts of cholesterol. Before focusing on the behavior of KA(z), let us stress that these quantities are sensitive to force fields and various computational details and can only be used for discussing qualitative trends with an increasing cholesterol content.
FIGURE 14.
Area compressibility moduli as functions of distance from bilayer center. The curves correspond to the cholesterol concentrations as indicated in Fig. 4. The errors are between 10 and 20%.
Regardless of the cholesterol content, all compressibility modulus profiles have a minimum in the bilayer center. Moreover, the values are identical in the center. The situation in the water phase makes sense: we expect that the moduli, irrespective of the cholesterol content, should be approximately similar. The interesting regions are the tail and head ones. The compressibility modulus profiles show two maxima between the bilayer center and the water phase, the first at ∼1 nm from the bilayer center and the second at 1.7–2.0 nm, depending on the cholesterol concentration. Between these we observe a local minimum. For pure DPPC and at low cholesterol concentrations there is a very flat, plateau-like maximum centered at 1 nm. With more cholesterol, the maximum grows considerably. Returning to Fig. 5 f, we note that the position of the growing maximum coincides with the location of the cholesterol ring structure. Therefore we can conclude that the cholesterol steroid rings strongly reduce the area fluctuations in the bilayer. From the point of view of free area theory, the region with the ordered DPPC tails and cholesterol rings seems to be the rate-limiting region for lateral diffusion of lipids and sterols.
The local minimum between the two maxima moves from a distance 1.5 nm from the bilayer center to 1.9 nm from the center. This means that the minimum is located in a part of the bilayer containing mostly glycerol groups, some of the uppermost tail methylene groups, and to a lesser degree, phosphate and choline groups (data not shown). The densities of cholesterol backbone are quite small here, whereas the electron densities of the cholesterol hydroxyl groups peak (data not shown). Very few cholesterol rings, and hence tails with less order than at 1 nm, and possibly also the interface between hydrophobic and hydrophilic parts, lead to slightly larger area fluctuations here. This could have consequences for the permeation of small molecules. Knowing that there are larger area fluctuations in this region than elsewhere, does not, however, tell us how permeation is affected. Jedlovszky et al. (2004), e.g., found in a recent simulation study of DMPC/cholesterol that the region with the cholesterol hydroxyl groups is indeed important from the point of view of permeation (see also Jedlovszky and Mezei, 2003). The effect on the actual rate of the permeation process, nevertheless, must depend on the properties of the permeant molecule.
Close-packed areas from ordering of acyl chains
Let us return to the average area per DPPC and investigate whether anything can be said about the close-packed area of a DPPC molecule based on the chain order parameters. Traditionally, the use of deuterium NMR experiments to determine the average area per DPPC has resulted in a wide variety of values (Nagle and Tristram-Nagle, 2000). This is not so much due to the underlying results for the order parameters as due to the interpretation of the results. Petrache et al. (1999) have rather recently suggested a way of relating the deuterium order parameter to the average chain travel distance along the bilayer normal:
 |
(6) |
Here 〈Dn〉 is the average chain travel distance along the bilayer normal for segment n, DM the maximum travel per methylene for all-trans chains oriented perpendicularly to the bilayer, and 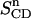 the deuterium order parameter for segment n. By assuming that
the deuterium order parameter for segment n. By assuming that  where
where 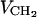 is the volume per methylene group (this is only true if
is the volume per methylene group (this is only true if 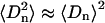 ; see Petrache et al., 1999) and recalling Eq. 2, we may write
; see Petrache et al., 1999) and recalling Eq. 2, we may write
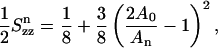 |
(7) |
where A0 is the area occupied by a fully ordered phospholipid molecule. By examination of Figs. 3 and 11 a, we can conclude that it is unrealistic to expect that Eq. 7 should allow one to extract the detailed form of aDPPC(z) from Szz. Nevertheless, Eq. 7 might be useful in predicting the average areas per DPPC molecule in the tail region, e.g., at a distance 1 nm from the bilayer center, where the headgroup density is negligible for all cholesterol concentrations.
To find the values of the order parameters at 1 nm from the bilayer center, we use electron density profiles calculated separately for each methylene group in the hydrocarbon tails (data not shown) to determine which segment is located at a distance 1 nm from the center for each cholesterol concentration separately. The order parameters at 1 nm are then calculated as averages over the segments whose electron density profiles peak at the close vicinity of 1 nm and over the sn-1 and sn-2 tails. The close-packed areas for DPPC at 1 nm from the center, in turn, can be easily obtained from the aDPPC(z) profiles. The resulting values and a fit to Eq. 7 are shown in Fig. 15. The fit is astonishingly good, given that Eq. 7 has been developed for a pure phospholipid bilayer and is based on a rather simple model. The best fit is obtained with A0 ≈ 0.28 nm2. A0 should in this case be interpreted as the area occupied by the fully ordered sn-1 and sn-2 tails. Hence, the agreement with Fig. 11 a is surprisingly good. Yet one should not pay too much attention to the exact numerical value here, as it probably depends on the details of the force field.
FIGURE 15.
Order parameters versus close-packed areas for DPPC at 1 nm from bilayer center. The markers represent values computed from the simulations and the solid line is a fit to these data based on Eq. 7.
Hofsäß et al. (2003) have used Eq. 7 in a slightly different setting. As order parameters they have used averages over the order parameter profiles from segment 3 to segment 8, and the average areas per DPPC they used contain some free area. They, too, find that Eq. 7 gives a very good fit to their data. However, we expect that order parameters are related to close-packed cross-sectional areas for DPPC chains rather than to average areas per DPPC containing an arbitrary amount of free area.
SUMMARY AND CONCLUSIONS
We have performed 100-ns molecular dynamics simulations at T = 323 K on a pure DPPC bilayer and composite DPPC/cholesterol bilayers with 4.7%, 12.5%, 20.3%, 29.7%, and 50.0% cholesterol. The main focus has been on the packing of molecules, free area in different parts of the bilayer, and lateral diffusion of DPPC and cholesterol molecules. Especially the interplay between these properties has been considered.
To investigate the packing and free area properties, we have introduced a novel method for estimating the average space occupied by DPPC, cholesterol, and water molecules, along with the average amount of free space, in different regions of the bilayer. Using this method we have computed the average cross-sectional areas for DPPC and cholesterol, as well as the total free area, as functions of the distance from the bilayer center. The method should be generally applicable for all kinds of pure and composite bilayers. Moreover, it could be used for investigating bilayers with integral proteins and in such a way finding out how the bilayer structure is changed in the vicinity of embedded proteins.
Inspection of the cross-sectional close-packed area profiles for DPPC and cholesterol, i.e., the close-packed areas as functions of the distance from the bilayer center, has shown that cholesterol alters the packing of molecules and reduces the amount of occupied space. These phenomena have been quite generally explained in terms of the form of the cholesterol molecule and the ordering effect of cholesterol on parts of the phospholipid tails.
Cholesterol has also been found to significantly reduce the average amount of free space in all regions of the bilayer. We have further discovered that the form of the free area profiles, i.e., the average amount of free area as a function of the distance from the bilayer center, is altered. These changes seem to reflect the ones observed in the close-packed area profiles for DPPC and cholesterol. We therefore conclude that the packing and free area properties are strongly coupled.
Also lateral diffusion of DPPC and cholesterol molecules has been found to be strongly reduced with an increasing cholesterol content. Further, the changes in the packing properties and the average amount of free area seem to be reflected in the behavior of the lateral diffusion coefficients for DPPC and cholesterol molecules. We have, however, learned that even though so-called free area theories correctly predict the suppressed lateral diffusion with reduced free area, the dependence cannot be quantitatively described by mean-field models such as free area theory. Not only are the average free areas or volumes relevant for diffusion, but also the size distribution, shape, and local fluctuations of the free volumes in the bilayer are important. It would hence be interesting to see how cholesterol influences the size distribution of free volumes in the bilayer.
Acknowledgments
We thank the Finnish IT Center for Science and the HorseShoe (DCSC) supercluster computing facility at the University of Southern Denmark for computer resources. We are grateful to Ole G. Mouritsen, Peter Lindqvist, and Perttu Niemelä for fruitful discussions.
This work has, in part, been supported by the Academy of Finland through its Center of Excellence Program (E. F. and I. V.), the National Graduate School in Materials Physics (E. F.), the Academy of Finland Grant Nos. 54113, 00119 (M. K.), 80246 (I. V.), and 80851 (M. H.), and the Jenny and Antti Wihuri Foundation (M. H.). M. P. acknowledges support through Marie Curie fellowship No. HPMF-CT-2002-01794.
References
- Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular Biology of the Cell, 3rd Ed. Garland Publishing, New York.
- Almeida, P. F. F., W. L. C. Vaz, and T. E. Thompson. 1992. Lateral diffusion in the liquid phases of dimyristoylphosphatidylcholine/cholesterol lipid bilayers: a free volume analysis. Biochemistry. 31:6739–6747. [DOI] [PubMed] [Google Scholar]
- Bassolino-Klimas, D., H. E. Alper, and T. R. Stouch. 1993. Solute diffusion in lipid bilayer membranes: an atomic level study by molecular dynamics simulation. Biochemistry. 32:12624–12637. [DOI] [PubMed] [Google Scholar]
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. B. Pullman, editor. Reidel, Dordrecht, The Netherlands.
- Berger, O., O. Edholm, and F. Jahnig. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M. F., J. Seelig, and U. Häberlen. 1979. Structural dynamics in phospholipid bilayers from deuterium spin-lattice relaxation time measurements. J. Chem. Phys. 70:5045–5053. [Google Scholar]
- Cantor, R. S. 1999. Lipid composition and the lateral pressure profile in lipid bilayers. Biophys. J. 76:2625–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, S. W., E. Jacobsson, R. J. Mashl, and H. L. Scott. 2002. Cholesterol-induced modifications in lipid bilayers: a simulation study. Biophys. J. 83:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, M. H., and D. Turnbull. 1959. Molecular transport in liquids and glasses. J. Chem. Phys. 31:1164–1169. [Google Scholar]
- Craven, B. M. 1979. Pseudosymmetry in cholesterol monohydrate. Acta Crystallogr. B35:1123–1128. [Google Scholar]
- Douliez, J.-P., A. Léonard, and E. J. Dufourc. 1995. Restatement of order parameters in biomembranes: calculation of C–C bond order parameters from C–D quadrupolar splittings. Biophys. J. 68:1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez, J.-P., A. Léonard, and E. J. Dufourc. 1996. Conformational order of DMPC sn-1 versus sn-2 chains and membrane thickness: an approach to molecular protrusion by solid state 2H-NMR and neutron diffraction. J. Phys. Chem. 100:18450–18457. [Google Scholar]
- Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- Essman, U., L. Perera, M. L. Berkowitz, H. L. T. Darden, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8592. [Google Scholar]
- Feller, S. E., and R. W. Pastor. 1999. Constant surface tension simulations of lipid bilayers: the sensitivity of surface areas and compressibilities. J. Chem. Phys. 111:1281–1287. [Google Scholar]
- Filippov, A., G. Orädd, and G. Lindblom. 2003a. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys. J. 84:3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov, A., G. Orädd, and G. Lindblom. 2003b. Influence of cholesterol and water content on phospholipid lateral diffusion in bilayers. Langmuir. 19:6397–6400. [Google Scholar]
- Finegold, L. editor, 1993. Cholesterol in Membrane Models. CRC Press, Boca Raton, FL.
- Galla, H.-J., W. Hartmann, U. Theilen, and E. Sackmann. 1979. On two-dimensional passive random walk in lipid bilayers and fluid pathways in biomembranes. J. Membr. Biol. 48:215–236. [DOI] [PubMed] [Google Scholar]
- Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- Höltje, M., T. Förster, B. Brandt, T. Engels, W. von Rybinski, and H.-D. Höltje. 2001. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 1511:156–167. [DOI] [PubMed] [Google Scholar]
- Hofsäß, C., E. Lindahl, and O. Edholm. 2003. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 84:2192–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlovszky, P., N. N. Medvedev, and M. Mezei. 2004. Effect of cholesterol on the properties of phospholipid membranes. III. Local lateral structure. J. Phys. Chem. B. 108:465–472. [DOI] [PubMed] [Google Scholar]
- Jedlovszky, P., and M. Mezei. 2003. Effect of cholesterol on the properties of phospholipid membranes. II. Free energy profile of small molecules. J. Phys. Chem. B. 107:5322–5332. [Google Scholar]
- Kandt, C., J. Schlitter, and K. Gerwert. 2004. Dynamics of water molecules in the bacteriorhodopsin trimer in explicit lipid/water environment. Biophys. J. 86:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintanar, A., A. C. Kunwar, and E. Oldfield. 1986. Deuterium nuclear magnetic resonance spectroscopic study of the fluorescent probe diphenylhexatriene in model membrane systems. Biochemistry. 25:6517–6524. [DOI] [PubMed] [Google Scholar]
- König, S., W. Pfeiffer, T. Bayerl, D. Richter, and E. Sackmann. 1992. Molecular dynamics of lipid bilayers studied by incoherent quasi-elastic neutron scattering. J. Phys. II (Fr.). 2:1589–1615. [Google Scholar]
- Korlach, J., P. Schwille, W. W. Webb, and G. W. Feigenson. 1999. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 96:8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz, B. R. 1993. Use of fluorescent probes to monitor molecular order and motions within liposome bilayers. Chem. Phys. Lipids. 64:99–116. [DOI] [PubMed] [Google Scholar]
- Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- Léonard, A., C. Escrive, M. Laguerre, E. Pebay-Peyroula, W. Néri, T. Pott, J. Katsaras, and E. J. Dufourc. 2001. Location of cholesterol in DMPC membranes: a comparative study by neutron diffraction and molecular mechanics simulation. Langmuir. 17:2019–2030. [Google Scholar]
- MacCarthy, J. E., and J. K. Kozak. 1982. Lateral diffusion in fluid systems. J. Chem. Phys. 77:2214–2216. [Google Scholar]
- Macedo, P. B., and T. A. Litovitz. 1965. On the relative roles of free volume and activation energy in the viscosity of liquids. J. Chem. Phys. 42:245–256. [Google Scholar]
- Marrink, S. J., R. M. Sok, and H. J. C. Berendsen. 1996. Free volume properties of simulated lipid membrane. J. Chem. Phys. 104:9090–9099. [Google Scholar]
- McConnell, H. M., and A. Radhakrishnan. 2003. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta. 1610:159–173. [DOI] [PubMed] [Google Scholar]
- McIntosh, T. J. 1978. The effect of cholesterol on the structure of phosphatidylcholine bilayers. Biochim. Biophys. Acta. 513:43–58. [DOI] [PubMed] [Google Scholar]
- McMullen, D. P. W., and R. N. McElhaney. 1996. Physical studies of cholesterol-phospholipid interactions. Curr. Opin. Colloid Interface Sci. 1:83–90. [Google Scholar]
- Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle, J. F., R. Zhang, S. Tristam-Nagle, W. Sun, H. I. Petrache, and R. M. Suter. 1996. X-ray structure determination of fully hydrated Lα phase dipalmitoylphosphatidylcholine bilayers. Biophys. J. 70:1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohvo-Rekilä, H., B. Ramstedt, P. Leppimäki, and J. P. Slotte. 2002. Cholesterol interactions with phospholipids in membranes. Prog. Lipid Res. 41:66–97. [DOI] [PubMed] [Google Scholar]
- Pasenkiewicz-Gierula, M., T. Róg, K. Kitamura, and A. Kusumi. 2000. Cholesterol effects on the phosphatidylcholine bilayer polar region: a molecular dynamics study. Biophys. J. 78:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra, M., M. Karttunen, M. T. Hyvönen, E. Falck, P. Lindqvist, and I. Vattulainen. 2003. Molecular dynamics simulations of lipid bilayers: major artifacts due to truncating electrostatic interactions. Biophys. J. 84:3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra, M., M. Karttunen, M. T. Hyvönen, E. Falck, and I. Vattulainen. 2004. Lipid bilayers driven to a wrong lane in molecular dynamics simulations by truncation of long-range electrostatic interactions. J. Phys. Chem. B. 108:4485–4494. [Google Scholar]
- Petrache, H. I., S. W. Dodd, and M. F. Brown. 2000. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys. J. 79:3172–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache, H. I., K. Tu, and J. F. Nagle. 1999. Analysis of simulated NMR order parameters for lipid bilayer structure determination. Biophys. J. 76:2479–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, J. M., I. Vattulainen, H. Zhu, and M. J. Zuckermann. 2001. Simulation study of lateral diffusion in lipid-sterol bilayer mixtures. Eur. Phys. J. E. 5:485–497. [Google Scholar]
- Róg, T., and M. Pasenkiewicz-Gierula. 2001. Cholesterol effects on the phosphatidylcholine bilayer nonpolar region: a molecular dynamics simulation study. Biophys. J. 81:2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J. E., and D. M. Engelman. 1972. Molecular mechanism for the interaction of phospholipid with cholesterol. Nat. New Biol. 237:42–44. [DOI] [PubMed] [Google Scholar]
- Sankaram, M. B., and T. E. Thompson. 1990a. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 29:10670–10675. [DOI] [PubMed] [Google Scholar]
- Sankaram, M. B., and T. E. Thompson. 1990b. Modulation of phospholipid acyl chain order by cholesterol: a solid-state 2H nuclear magnetic resonance study. Biochemistry. 29:10676–10684. [DOI] [PubMed] [Google Scholar]
- Scott, H. L. 2002. Modeling the lipid component of membranes. Curr. Opin. Struct. Biol. 12:495–502. [DOI] [PubMed] [Google Scholar]
- Seelig, A., and J. Seelig. 1974. Dynamic structure of fatty acyl chains in a phospholipid bilayer measured by deuterium magnetic resonance. Biochemistry. 13:4839–4845. [DOI] [PubMed] [Google Scholar]
- Seelig, A., and J. Seelig. 1975. Bilayers of dipalmitoyl-3-sn-phosphatidylcholine: conformational differences between the fatty acid chains. Biochim. Biophys. Acta. 406:1–5. [DOI] [PubMed] [Google Scholar]
- Shinoda, W., and S. Okazaki. 1998. A Voronoi analysis of lipid area fluctuation in a bilayer. J. Chem. Phys. 109:1517–1521. [Google Scholar]
- Silvius, J. R. 2003. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta. 1610:174–183. [DOI] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Söderhäll, J. A., and A. Laaksonen. 2001. Molecular dynamics simulations of ubiquinone inside a lipid bilayer. J. Phys. Chem. B. 105:9308–9315. [Google Scholar]
- Smondyrev, A. M., and M. L. Berkowitz. 1999. Structure of dipalmitoylphosphatidylcholine/cholesterol bilayer at low and high cholesterol concentrations: molecular dynamics simulation. Biophys. J. 77:2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman, D. P., and H. J. C. Berendsen. 1996. Molecular dynamics simulations of a fully hydrated dipalmitoylphosphatidylcholine bilayer with different macroscopic boundary conditions and parameters. J. Chem. Phys. 105:4871–4880. [Google Scholar]
- Tieleman, D. P., S. J. Marrink, and H. J. C. Berendsen. 1997. A computer perspective of membranes: molecular dynamics studies of lipid bilayer systems. Biochim. Biophys. Acta. 1331:235–270. [DOI] [PubMed] [Google Scholar]
- Tu, K., M. L. Klein, and D. J. Tobias. 1998. Constant-pressure molecular dynamics investigation of cholesterol effects in a dipalmitoylphosphatidylcholine bilayer. Biophys. J. 75:2147–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, D., and M. H. Cohen. 1961. Free-volume model of the amorphous phase: glass transition. J. Chem. Phys. 34:120–125. [Google Scholar]
- Turnbull, D., and M. H. Cohen. 1970. On the free-volume model of the liquid-glass transition. J. Chem. Phys. 52:3038–3041. [Google Scholar]
- Vattulainen, I., and O. G. Mouritsen. 2003. Diffusion in membranes. In Diffusion in Condensed Matter. J. Kärger, D. Heitjans, and R. Haberland, editors. Springer, New York.
- Vaz, W. L. C., R. M. Clegg, and D. Hallmann. 1985. Translational diffusion of lipids in liquid crystalline phase phosphatidylcholine multibilayers: a comparison of experiment with theory. Biochemistry. 24:781–786. [DOI] [PubMed] [Google Scholar]
- Venable, R. M., B. R. Brooks, and R. W. Pastor. 2000. Molecular dynamics simulations of gel (Lβi) phase lipid bilayers in constant pressure and constant surface area ensembles. J. Chem. Phys. 112:4822–4832. [Google Scholar]
- Vist, M. R., and J. H. Davis. 1990. Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry. 29:451–464. [DOI] [PubMed] [Google Scholar]
- Xiang, T.-X. 1999. Translational diffusion in lipid bilayers: dynamic free-volume theory and molecular dynamics simulation. J. Phys. Chem. B. 103:385–394. [Google Scholar]
- Yeagle, P. L. 1991. Modulation of membrane function by cholesterol. Biochimie. 73:1303–1310. [DOI] [PubMed] [Google Scholar]

















