Abstract
Chlorosomes of green photosynthetic bacteria constitute the most efficient light harvesting complexes found in nature. In addition, the chlorosome is the only known photosynthetic system where the majority of pigments (BChl) is not organized in pigment-protein complexes but instead is assembled into aggregates. Because of the unusual organization, the chlorosome structure has not been resolved and only models, in which BChl pigments were organized into large rods, were proposed on the basis of freeze-fracture electron microscopy and spectroscopic constraints. We have obtained the first high-resolution images of chlorosomes from the green sulfur bacterium Chlorobium tepidum by cryoelectron microscopy. Cryoelectron microscopy images revealed dense striations ∼20 Å apart. X-ray scattering from chlorosomes exhibited a feature with the same ∼20 Å spacing. No evidence for the rod models was obtained. The observed spacing and tilt-series cryoelectron microscopy projections are compatible with a lamellar model, in which BChl molecules aggregate into semicrystalline lateral arrays. The diffraction data further indicate that arrays are built from BChl dimers. The arrays form undulating lamellae, which, in turn, are held together by interdigitated esterifying alcohol tails, carotenoids, and lipids. The lamellar model is consistent with earlier spectroscopic data and provides insight into chlorosome self-assembly.
INTRODUCTION
Photosynthesis is the ultimate source of energy for most current life forms, including humans. The first step in utilization of solar energy is photon capture by a light-harvesting system (antenna). Typically, the antennae are composed of pigment-protein complexes, in which the protein framework determines pigment orientations and optical properties, and ensures efficient flow of excitation energy to the photosynthetic reaction center. The only known exception is the chlorosome. Chlorosomes are large enclosures of BChl aggregates, which are organized by pigment-pigment rather than pigment-protein interactions, and are attached to the inner side of the cytoplasmic membrane of green photosynthetic bacteria (Blankenship et al., 1995; Frigaard et al., 2003). Two bacterial families, the green sulfur and the green nonsulfur bacteria, belong to this group and are only distantly related, but use chlorosomes as the main light harvesting system. The green sulfur bacteria are able to survive at the lowest light conditions of all known photosynthetic organisms (Overmann et al., 1992; Frigaard et al., 2003). In effect, the chlorosome is the most efficient antenna known. The efficiency is in part due to the large size of the chlorosome (ellipsoidal particle with typical dimensions 1500 × 500 × 200 Å, 1 Å = 0.1 nm) and the large number of pigment molecules inside. A typical chlorosome contains on the order of 105 BChl molecules (Montano et al., 2003) (BChl c, d, e, depending on the species) in the form of aggregates. The aggregation modulates the optical properties of the BChls and results in fast energy transfer rates (Prokhorenko et al., 2000; Psencik et al., 2003, and references therein), which are a prerequisite for the light-harvesting efficiency.
Chlorosomes were first reported in 1963 and their structure was subsequently characterized by electron microscopy (Cohen-Bazire et al., 1964). The electron micrographs of Cohen-Bazire and co-workers revealed 12–20 Å wide striations, arrayed more or less parallel to the long axis of the chlorosome, but no interpretation was given. Later, this observation was overshadowed by freeze-fracture electron microscopy study (Staehelin et al., 1978, 1980), in which micrographs of chlorosome interiors were interpreted in terms of rod-like elements with a diameter of 50 Å (nonsulfur bacteria) or 100 Å (sulfur bacteria). This interpretation was reinforced by freeze-fracture (Oelze and Golecki, 1995) and disruption (Wullink and van Bruggen, 1988) studies and became the basis for all subsequent chlorosome models. Perhaps because of the large size and unusual organization, no crystals of chlorosomes or aggregated BChls have been obtained, and the chlorosome remains the last known light-harvesting complex for which no high-resolution structural information is available.
Several models for the organization of BChl aggregates into rod-like elements were proposed (Holzwarth and Schaffner, 1994; Nozawa et al., 1994; Blankenship et al., 1995; van Rossum et al., 2001). These models can be classified into two principal groups based on the asymmetric repeating unit: 1), parallel-chain model with BChl monomer as the building block (Holzwarth and Schaffner, 1994; Balaban et al., 1995; Chiefari et al., 1995; van Rossum et al., 2001); and 2), antiparallel double-chain model with an antiparallel so-called “piggy-back” BChl dimer as a building block (Smith et al., 1986; Nozawa et al., 1994; Umetsu et al., 1999; Wang et al., 1999a; Umetsu et al., 2002). The terms parallel and antiparallel refer to the mutual orientation of the Qy transition dipoles. In all these models, short-range order was based on the results of NMR and optical spectroscopy (Smith et al., 1986; Hildebrandt et al., 1991; Holzwarth and Schaffner, 1994; Nozawa et al., 1994; Balaban et al., 1995; Chiefari et al., 1995; Umetsu et al., 1999, 2002; Wang et al., 1999a; Mizoguchi et al., 2000; van Rossum et al., 2001). Most notably, the long-range order followed from constraining the models to resemble the rod-like elements of Staehelin et al. (1978, 1980). Furthermore, none of these models has provided insight into chlorosome assembly.
In addition to BChl aggregates, chlorosomes contain BChl a, carotenoids, quinones, lipids, and proteins. Lipids presumably form an enveloping monolayer of the chlorosome and the coupling to the cytoplasmic membrane is achieved via a BChl a-containing protein baseplate (Blankenship et al., 1995; Frigaard et al., 2003). Proteins constitute a minor component and are thought to reside in the chlorosome baseplate and envelope (Blankenship et al., 1995; Frigaard et al., 2003).
Here we present the first EM images of intact Chlorobium tepidum chlorosomes embedded in vitreous ice. EM was complemented by solution SAXS and WAXS. Both the high-resolution EM images and SAXS revealed fine internal structure with spacing of ∼20 Å, which can be explained by a simple lamellar organization of pigment molecules. The data were clearly incompatible with the rod-like model. Based on these results we propose a new lamellar structure of chlorosomes, which yields a model of chlorosome assembly.
MATERIALS AND METHODS
Chlorosome preparation and characterization
The Chl. tepidum cultures (kind gift of Prof. G. Hauska, University of Regensburg) were grown for 3 days at 48°C in a modified Pfennig's medium (Wahlund et al., 1991) under constant illumination (60 W Tungsten lamp, 25 cm illumination distance). The culture was stored at 4°C (dark) and harvested by centrifugation (7000 RPM, Sorvall SLA-3000 rotor, 10 min, 4°C). The chlorosomes were isolated by a method of Gerola and Olson (1986) with modifications. Cell pellets from 250 ml culture were resuspended in 5 ml of 50 mM Tris buffer pH 8, containing 2M sodium isothiocyanide (NaSCN) and lysed by three passages through a French pressure cell at 20,000 psi. Cell debris was removed by centrifugation (10,000 RPM, Sorvall SS-34 rotor, 10 min, 4°C). The chlorosome-containing supernatant was loaded onto 10–40% sucrose density gradient and centrifuged for 24 h at 220,000 g, 5°C. The chlorosome-containing band was recovered, concentrated by centrifugation, and further purified by another round of density-gradient centrifugation. The density and the optical absorption spectrum of the resulting chlorosomes were measured to assure sample integrity. The absorption spectra were measured before and after each experiment to ensure that no degradation occurred during exposure to x rays or during the handling required for EM sample preparation.
Electron microscopy
Small drops (10 μl) of fresh chlorosome solution (OD ∼80 per cm at 748 nm) were dialyzed for 1 min using Millipore 0.025 μm pore membrane (Millipore, Billerica, MA), against 5 mM Tris, pH 8. Protein-A gold (50 Å, Dept. of Cell Biology, University of Utrecht) was added to chlorosome solutions before dialysis as fiducial markers for tilt experiments. Grid preparation and vitrification was accomplished by the guillotine method of Dubochet et al. (1988) in liquid-nitrogen cooled ethane. The vitrified samples were examined in a Tecnai F20 transmission electron microscope using an Oxford CT3500 cryo-holder (EM Unit, Institute of Biotechnology, University of Helsinki). Micrographs were recorded at 200 kV, 50,000× magnification, 0.8–3.2 μm underfocus, on Kodak S0163 film using low dose. Images free from astigmatism and drift were scanned at 7 μm step size (1.4 Å per pixel) using a Zeiss Photoscan TD scanner. The defocus of the scanned micrographs was calculated using the program CTFFIND3 (Grigorieff, 1998). Individual chlorosomes were boxed (1000 × 1000 pixels) from the original images and subregions containing the fine structure were selected (area of 125 × 125 pixels). The subregions were padded to 2048 × 2048 array with the average image intensity, and power spectra were calculated.
X-ray scattering
Samples for x-ray scattering were prepared by rapid dialysis against 1 mM Tris, pH 8, followed by controlled concentration under low vacuum to avoid complete drying and salt crystallization. The concentrated but fluid sample (OD ∼2000 per cm at 748 nm) was loaded into a steel-framed sample cell (thickness 1 mm) sealed with two 13 μm Capton windows. The measurements in the q-range of 0.1–2.5 Å−1 (q = 4 × π sin(ϑ/2)/λ, where ϑ is the scattering angle and λ is the wavelength) were made on the in-house x-ray scattering apparatus in Division of X-ray Physics, University of Helsinki. The quasi-monochromatic x rays (Cu Kα) scattered from the sample were detected with a two-dimensional proportional counter. The x-ray transmission of the samples was determined during the scattering experiments using a transparent beam stop. The q-scale was calibrated with a standard sample of silver behenate.
Measurements at very low-angles were conducted at the synchrotron beamline BW4 in HASYLAB, Hamburg, Germany (Gehrke, 1992). The energy of the x rays was 8.979 keV and two measurement distances of 4 m and 13 m were used. The q-ranges achieved with these two configurations were 0.011–0.10 Å−1 and 0.0028–0.033 Å−1, respectively. The beam intensity and sample absorption was monitored by two ionization chambers, placed before and after the sample, and by a PIN diode at the beamstop. A standard sample of rat tail collagen was used to calibrate the q-scale.
In all experiments, the measured data were corrected for the detector response and averaged circularly to obtain one-dimensional scattering curves. The background scattering from the cuvette and solvent buffer were measured separately. The intensity I of scattering by the sample was obtained (in arbitrary units) by subtracting the weighed intensity Ib of the cuvette and the solvent background from the sample measurement Is:
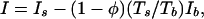 |
where Ts and Tb are the transmission coefficients of the sample and background, respectively, and φ is the estimated volume fraction of the chlorosomes. A volume fraction of 0.2 was estimated for the sample used in measurements with synchrotron radiation, whereas the sample used in the wide angle measurements with the conventional x-ray source was more concentrated, having a volume fraction of 0.3.
In addition, a wide-angle x-ray scattering measurement was made in-house (as described above) from completely dry, flake-like sample of chlorosomes. The scattering background corresponding to an empty sample holder was subtracted.
RESULTS
Electron microscopy
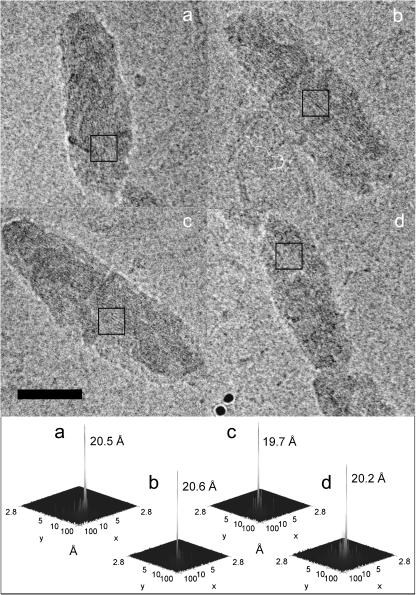
To retain high-resolution information, chlorosomes were embedded in vitreous ice and imaged without further treatment or staining. Fig. 1 shows typical projection images of chlorosomes obtained by EM. The chlorosomes appear as approximately elliptic objects with ruffled edges and exhibit dimensions 1400–1800 Å × 500 Å. The most remarkable feature, which is observed in nearly all chlorosomes, is a striation pattern formed by parallel dark and light stripes oriented close to parallel with the long axis of the chlorosome.
FIGURE 1.
EM analysis of chlorosomes. Image of four representative chlorosomes embedded in vitreous ice. The bottom panels show power spectra of the boxed areas and the corresponding striation spacings. Bar, 500 Å. The defocus value for panels a, b, and d was 2.5 μm and 2.55 μm for panel c.
The distance between centers of two neighboring dark (or light) striae is ∼20 Å. The distance was determined more precisely by calculating Fourier transforms of selected regions. A single intense band was usually observed for the chlorosome region showing pronounced striation (Fig. 1). The peak was always found on the axis perpendicular to the direction of the striation (long axis of the chlorosome). The distance of the peak from the center was found in a narrow range corresponding to spacing of 19.5–21.5 Å with a mean value of 20.5 Å.
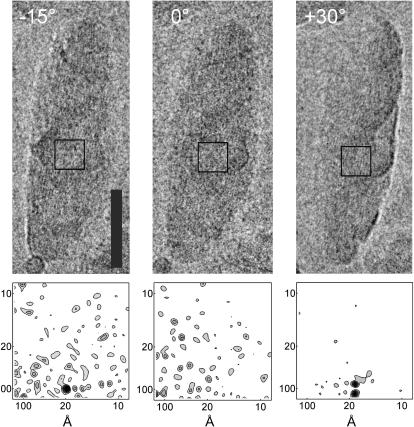
To further characterize the origin of the striation, tilt series were collected. Three tilt angles (−15, 0, and 30°) were selected to determine a plausible pigment packing. For example, hexagonally packed pigments would yield projections in which the spacing would strongly depend on the tilt angle. Striae with similar spacing were observed under different tilts, often within the same region of the chlorosome. An example is shown in Fig. 2 of striation observed under −15 and 30° tilts. In other areas, striation was observed under both 0° and 30° tilts (not shown), which made a hexagonal arrangement of pigments unlikely. A splitting of the diffraction peak in the power spectrum (Fig. 2, +30°, bottom right) indicates that two domains with the same spacing but slightly different orientation coexist within the projected volume.
FIGURE 2.
(Upper panels) Tilted series images (−15, 0, and 30°) of a representative chlorosome. Bar, 500 Å. The defocus value was 1.6 μm. The tilt axis is oriented almost parallel to the striae and intersects through the middle of the chlorosome. Lower panels show the power spectra computed from the boxed regions (shown as a square in each upper panel), which were selected to correspond to the same volume of the chlorosome throughout the tilt.
X-ray scattering
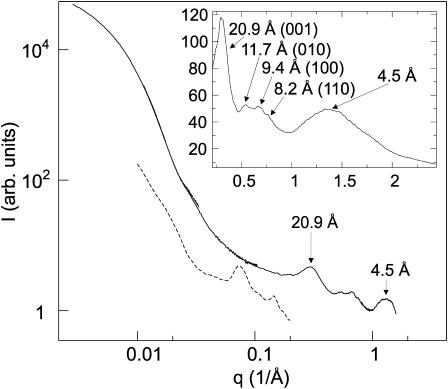
To determine the internal arrangement of the pigments, e.g., size and spacing of the putative rods, x-ray scattering from the chlorosome solution was obtained (Fig. 3). The initial portion of the scattering curve (q < 0.05) reflects the ellipsoidal shape of chlorosomes. Notably, no peak was observed in the range of q values 0.06–0.15 Å−1, which would be expected for hexagonally arranged rod-like elements with diameter of 50 to 100 Å (cf. the solid and dashed lines in Fig. 3). Moreover, the scattering pattern does not exhibit features typical for a dilute solution of rods (Gandini et al., 2003). Instead, the scattering curve contains a prominent scattering peak at q = 0.30 Å−1 which corresponds to a spacing of 20.9 Å. Additional maxima are observed at 0.54, 0.67, and 0.76 Å−1 (Bragg distances 11.7, 9.4, and 8.2 Å, respectively). The four observed diffraction maxima could not yield a unique lattice for pigment arrangement. However, a preliminary assignment, which is compatible with both EM and diffraction is a monoclinic lattice (a = 9.6 Å, b = 12.0 Å, c = 20.9 Å, and γ = 77.5°, Fig. 3, inset). Additionally, a broad feature at wide angles with a maximum at 1.4 Å−1 (spacing 4.5 Å) was observed (Fig. 3), which reflects short-range order, e.g., stacking of chlorin rings and intramolecular electron density correlation.
FIGURE 3.
X-ray scattering curves measured from a concentrated chlorosome solution combined from four individual measurements. The dashed curve shows scattering calculated from an analytical model containing 1000 Å long hollow cylinders of inner radius of 40 Å and outer radius of 50 Å in a 5 × 2 hexagonal lattice with a lattice constant of 100 Å (Prokhorenko et al., 2000), plus an exponentially decaying background. (Inset) X-ray scattering from a dry chlorosome sample. The positions of the diffraction maxima and corresponding monoclinic lattice indices are indicated by arrows.
The measurement of the dry chlorosome sample (Fig. 3, inset) contains the same structural features as the hydrated sample, demonstrating that water does not play a significant role in the internal structure of the chlorosome. The absence of sharp diffraction maxima in the wide angles of both the hydrated and the nonhydrated samples indicates that there is no rigid long-range order in the pigment packing.
DISCUSSION
Cryoelectron imaging and x-ray scattering reveal internal structure of the chlorosome
An important question is whether the striation observed in the EM images originates from the projection of BChl c aggregates inside the chlorosome or from the surface features (the baseplate or envelope). The latter possibility is unlikely, however, given the following arguments. The EM image is a projection of the whole chlorosome, in which the surface, namely the envelope and the baseplate, provides less contrast compared to the internal volume. The same argument holds for scattering power in SAXS. Furthermore, the ∼20 Å striation is seen in the same area under tilt angles of −15° and 30° (i.e., 45° difference) which would be impossible for a surface feature with the same spacing (Fig. 2). Finally, the strongly interacting and ordered chlorin rings are likely to contribute to the observed diffraction pattern. Thus, we conclude that the ∼20 Å spacing arises from the volume of the chlorosome rather than from its surface and reflects the organization of the BChl aggregates. Interestingly, a similar 12–20 Å striation was reported by Cohen-Bazire et al. (1964) for chlorosomes in sectioned cells and was observed within the chlorosome interior, remote from the baseplate.
Chlorosome pigments cannot be arranged in rod-like elements
Both experimental methods (SAXS and EM) show that the dominant spacing in the chlorosome is ∼20 Å. This value is clearly incompatible with the spacing of 86.6 Å expected for a hexagonal lattice of rods with 100 Å diameter (86.6 Å = 100 Å/cos(30°)) as proposed in all current models. Notably, absence of maxima between ∼40–100 Å in the measured SAXS (Fig. 3) and EM makes the presence of 50–100 Å diameter rods in any arrangement unlikely.
In principle, the SAXS data could be explained by pigments arranged in rods (20 or 24.1 Å diameter), which are organized on an oblique or hexagonal, two-dimensional, lattice, respectively. The latter possibility is not compatible with EM results. Attempts to build plausible molecular models with rods of 20 Å diameter, which would satisfy spectroscopic results and hydrogen bonding patterns, were unsuccessful due to the following reason: close packing required interdigitation of the rods and led to unrealistically high densities (>1.9 g/cm3).
Lamellar model
As all of our results indicated that the rod model was not feasible, we searched for other plausible arrangements. EM images (Fig. 1) suggested that pigments could be arranged in parallel planes, i.e., lamellae parallel to the long axis of the chlorosome. Indeed, the most prominent SAXS feature at q = 0.30 Å−1 (Fig. 3) is consistent with the lamellar model with ∼20 Å spacing between planes.
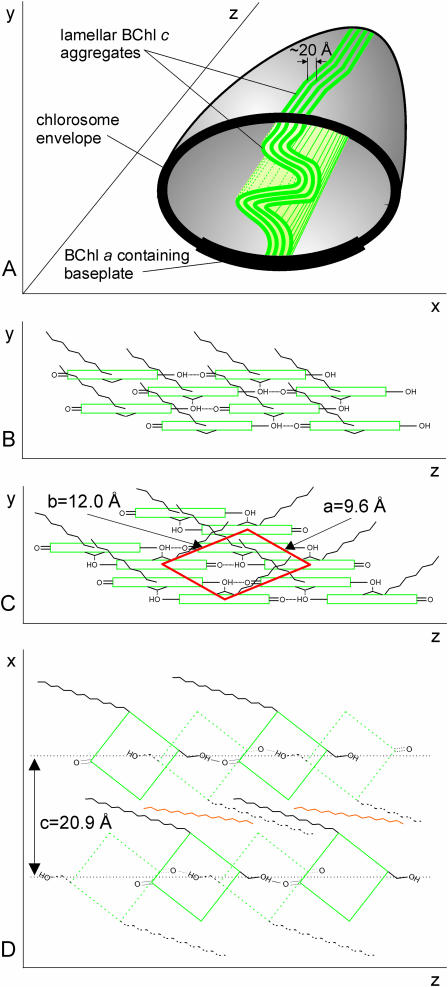
The EM tilt series gave further information about the organization of lamellae within the chlorosome. If the lamellae maintained planarity throughout the chlorosome, the striation should disappear as the chlorosome is tilted. However, this was not the case and the striae were observed under different tilts often from the same region of the chlorosome (Fig. 2). Thus, the lamellae seem to undulate in the direction perpendicular to the long axis of the chlorosome. The width of the 20.9 Å SAXS peak also indicates that the order persists only over 60–80 Å distance and suggests considerable disorder. Fig. 4 A summarizes the lamellar model of the chlorosome structure.
FIGURE 4.
Schematic model of the BChl aggregates in the chlorosome. (A) Arrangement of lamellae inside the chlorosome. Each undulated plane (thick green line) extends in the long axis of the chlorosome (z), and through the height of the chlorosome (y). Planes are arranged into lamellae throughout the chlorosome (x). Only a few lamellae are depicted for clarity. Model of one plane of BChl aggregate with parallel (B) or antiparallel (C) pigment configuration. Coordinate system as in panel A. The monoclinic unit cell projection and lattice constants are shown in red. (D) Top view of chlorin (green) planes (antiparallel configuration) associated via interdigitated esterifying alcohol tails (black). An underlying layer of BChl molecules is shown dotted whereas carotenoids (orange) are interspersed between the alcohol tails.
The lamellar model is consistent with structural and spectroscopic results
It remained to be seen whether the lamellar arrangement of pigments could be achieved using the well-established short-range interaction patterns of BChl molecules. Two possible configurations of BChl molecules have been suggested for aggregates on the basis of NMR experiments: the parallel-chain model (Balaban et al., 1995) (Fig. 4 B) and the antiparallel-chain model (Nozawa et al., 1994) (Fig. 4 C). The interactions satisfied by these models are coordination of Mg atoms by the C31-OH of the stacked neighbor and a hydrogen bond between the C31-OH and C131 = O of the neighbor in the same chain (Fig. 4, B and C).
We propose that the linear chains as described above can be arranged into planar structures instead of rods via Mg coordination and/or chlorin ring stacking (Fig. 4). In this arrangement the esterifying alcohol tails stick out of the plane (Fig. 4, B and C) (Holzwarth and Schaffner, 1994; Nozawa et al., 1994) as hydrophobic surfaces for lamellar assembly (Fig. 4 D).
The monoclinic lattice that we propose based on x-ray scattering could provide clues about the pigment arrangement within the planes. The lattice dimensions are most compatible with a dimer in the unit cell as shown in Fig. 4 C. Indeed, the antiparallel dimer is stable in solution (Smith et al., 1986; Wang et al., 1999a,b; Umetsu et al., 2002) and has been suggested as the building block of larger aggregates (Brune et al., 1988; Nozawa et al., 1994; Umetsu et al., 1999; Wang et al., 1999a; Mizoguchi et al., 2000). Assuming the dimer model and the proposed lattice arrangement (Fig. 4 C) the stacking distance between chlorin rings is 3.3 Å whereas the Mg-Mg distance between BChls along the z axis is 16.9 Å. An alternative lattice orientation with the short diagonal along the z axis gives the stacking distance as 4.2 Å, whereas the Mg-Mg distance is 13.6 Å. The broad WAXS feature (q-range 1–2.5 Å−1 in Fig. 3, inset), notably the absence of a prominent peak corresponding to a fixed stacking distance between chlorin rings, suggests considerable disorder within the planes. Because of the disorder, the above distances represent limiting cases and the actual values will fluctuate within the range of 3.3–4.2 Å and 13.6–16.9 Å, respectively. These estimates compare favorably with the range of values obtained for the antiparallel dimer model from NMR constraints (stacking distance 3.2–3.4 Å (Wang et al., 1999a), Mg-Mg distance between BChls along the z axis 15.25–15.5 Å (Nozawa et al., 1994; Mizoguchi et al., 2000)). The proposed model yields a density of ∼1.15 g/ml (without carotenoids) or 1.21 (with 6% w/w carotenoids, Borrego et al., 1999), which is similar to the measured density of chlorosomes (1.16 ± 0.05 g/ml). However, the monoclinic lattice constitutes only the first approximation and delineation of detailed molecular arrangements within the planes will have to await further experiments.
Thus, our results are compatible with an antiparallel dimer arrangement similar to those based on NMR. The antiparallel chain model also nicely explains results of Stark spectroscopy (Frese et al., 1997), which demonstrated a lack of permanent dipole moment difference between the ground- and excited states in chlorosomes (note that individual BChl c possesses such dipole moment difference). In the antiparallel dimer the dipole moments compensate each other, which is not the case for the parallel arrangement.
The EM and SAXS data indicate that the aggregates are significantly disordered by undulation. The disorder and undulation in the direction perpendicular to the z axis can be rationalized within the context of the antiparallel model. In this model the interfaces between the dimer chains are not stabilized by hydrogen bonding (Fig. 4 C) leading to weaker interactions than within the dimer and providing preferable interface for line defects and bending.
Despite the disorder, the lamellar packing produces an alteration of the electron-dense, tightly packed, chlorin layers with the less dense, lipid-like, interdigitated esterifying alcohol interfaces (Fig. 4 D). Such alteration provides enough contrast to be observed by EM and gives rise to the SAXS pattern. However, the striation would be hard to detect in freeze-fracture electron micrographs and the rod-like elements observed by Staehelin et al. (1978, 1980) most likely originated from the cleavage along the undulated (oblique) planes within the lamellae.
EM projection and tilt series show that the lamellae are preferentially ordered in the direction of the long chlorosome axis (z) whereas undulation and disorder is prevalent in the perpendicular directions (Fig. 4 A). Because the transition dipole moments of the BChl c molecules in the model are nearly parallel with the z axis of the chlorosome (Fig. 4 D) the observed strong order along this axis will produce strong linear dichroism (Blankenship et al., 1995; Frese et al., 1997). The disorder observed in SAXS and expected to originate from undulations along the y axis may well explain the broad NMR resonances previously observed and attributed to two forms of BChl c aggregates in the bilayer rod model (van Rossum et al., 2001).
Chlorosome assembly
The lamellar organization presented here provides clues about how self-assembly of the chlorosome may occur. The BChl a molecules and proteins of the crystalline baseplate form a regular lattice and may serve as the nucleation site for lamellar assembly. The rest of the assembly process proceeds via propagation of the planes utilizing the nonspecific hydrophobic interactions of the tails while maintaining order via the specific interactions of the chlorin rings. Arrangement of BChl molecules, in which the alcohol tails extend from both sides of the plane, would permit interdigitation of the hydrophobic tails and facilitate assembly of the planes into the lamellae. The antiparallel configuration yields automatically such arrangement of the tails (Nozawa et al., 1994) (Fig. 4 C) although a parallel configuration with similar properties can be envisioned (not shown).
The model also suggests that hydrophobic carotenoids may occupy the lipid-like space between the planes interacting with the esterifying alcohol chains (Fig. 4 D). The volume fraction of carotenoids within this layer may then modulate its thickness and the lamellar spacing. Indeed, preliminary SAXS and EM examination of Chl. phaeobacteroides chlorosomes, which possess a larger amount of carotenoids compared to Chl. tepidum, yielded a spacing of 28 Å (unpublished data) as opposed to ∼20 Å obtained for Chl. tepidum in this study. This tendency fits well with the results of x-ray diffraction experiments previously reported for BChl c aggregates in nonpolar solvents (without carotenoids) where the largest observed spacing was ∼18 Å (Umetsu et al., 1999). Similar interactions are expected between esterifying alcohol chains in the outer part of the chlorosome and hydrophobic tails of monogalactosyl diglyceride, which is thought to form the chlorosome envelope.
CONCLUSIONS
EM and x-ray scattering data rule out the existence of previously proposed rod-like aggregates with 50–100 Å diameter in chlorosomes. A new lamellar model of pigment organization is proposed. The model is consistent with current and previously published structural and spectroscopic constraints and provides insight into the assembly of the chlorosome.
Acknowledgments
The authors express their gratitude to Prof. G. Hauska (University of Regensburg) for providing the Chl. tepidum strain and to Dr. F. Vacha (University of South Bohemia) for growing the bacteria. HASYLAB is thanked for providing beam time.
This study was supported by the Academy of Finland (projects 206926, R.T.; 208661, S.J.B.; 205138, J.P.; and 172622, R.E.S.; and the Finnish Centre of Excellence Program 2000-2005, R.T and S.J.B.) and by the Czech Science Foundation and Czech Ministry of Education, Youth and Sports (contracts 206/02/0942, LN00A141 to J.P.). T.P.I. is supported by the National Graduate School in Informational and Structural Biology.
Abbreviations used: BChl, Bacteriochlorophyll; Chl., chlorobium; EM, cryoelectron microscopy; OD, optical density (cm−1); SAXS, small angle x-ray scattering; WAXS, wide angle x-ray scattering.
References
- Balaban, T. S., A. R. Holzwarth, K. Schaffner, G. J. Boender, and H. J. de Groot. 1995. CP-MAS 13C-NMR dipolar correlation spectroscopy of 13C-enriched chlorosomes and isolated bacteriochlorophyll c aggregates of Chlorobium tepidum: The self organization of pigments is the main structural feature of chlorosomes. Biochemistry. 34:15259–15266. [DOI] [PubMed] [Google Scholar]
- Blankenship, R. E., J. M. Olson, and M. Miller. 1995. Antenna complexes from green photosynthetic bacteria. In Anoxygenic Photosynthetic Bacteria. R. E. Blankenship, M. T. Madigan, and C. E. Bauer, editors. Kluwer Academic Publisher, Dordrecht, The Netherlands. 399–435.
- Borrego, C. M., P. G. Gerola, M. Miller, and R. P. Cox. 1999. Light intensity effects on pigment composition and organisation in the green sulfur bacterium Chlorobium tepidum. Photosynth. Res. 59:159–166. [Google Scholar]
- Brune, D. C., G. H. King, and R. E. Blankenship. 1988. Interactions between bacteriochlorophyll c molecules in oligomers and in chlorosomes of green photosynthetic bacteria. In Photosynthetic Light-Harvesting Systems. H. Scheer,and S. Schneider, editors. Walter de Gruyter, Berlin. 141–151.
- Chiefari, J., K. Griebenow, F. Fages, N. Griebenow, T. S. Balaban, A. R. Holzwarth, and K. Schaffner. 1995. Models for the pigment organization in the chlorosomes of photosynthetic bacteria: Diastereoselective control of in vivo bacteriochlorophyll cs aggregation. J. Phys. Chem. 99:1357–1365. [Google Scholar]
- Cohen-Bazire, G., N. Pfennig, and R. Kunisawa. 1964. The fine structure of green bacteria. J. Cell Biol. 22:207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129–228. [DOI] [PubMed] [Google Scholar]
- Frese, R., U. Oberheide, I. H. M. van Stokkum, R. van Grondelle, M. Foidl, J. Oelze, and H. van Amerongen. 1997. The organization of bacteriochlorophyll c in chlorosomes from Chloroflexus aurantiacus and the structural role of carotenoids and protein—an absorption, linear dichroism, circular dichroism and Stark spectroscopy study. Photosynth. Res. 54:115–126. [Google Scholar]
- Frigaard, N. U., A. G. M. Chew, H. Li, J. A. Maresca, and D. A. Bryant. 2003. Chlorobium tepidum: insights into the structure, physiology, and metabolism of a green sulfur bacterium derived from a complete genome sequence. Photosynth. Res. 78:93–117. [DOI] [PubMed] [Google Scholar]
- Gandini, S. C. M., E. L. Gelamo, R. Itri, and M. Tabak. 2003. Small angle x-ray scattering study of meso-tetrakis (4-Sulfonatophenyl) porphyrin in aqueous solution: a self-aggregation model. Biophys. J. 85:1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke, R. 1992. An ultrasmall angle scattering instrument for the doris-III bypass. Rev. Sci. Instrum. 63:455–458. [Google Scholar]
- Gerola, P. D., and J. M. Olson. 1986. A new bacteriochlorophyll a-protein complex associated with the chlorosomes of green sulfur bacteria. Biochim. Biophys. Acta. 848:69–76. [DOI] [PubMed] [Google Scholar]
- Grigorieff, N. 1998. Three-dimensional structure of bovine NADH: Ubiquinone oxidoreductase (Complex I) at 22 angstrom in ice. J. Mol. Biol. 277:1033–1046. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, P., K. Griebenow, A. R. Holzwarth, and K. Schaffner. 1991. Resonance Raman spectroscopic evidence for the identity of the bacteriochlorophyll c organisation in protein-free and protein-containing chlorosomes from Chloroflexus aurantiacus. Z. Naturforsch. 46c:228–232. [Google Scholar]
- Holzwarth, A. R., and K. Schaffner. 1994. On the structure of bacteriochlorophyll molecular aggregates in the chlorosomes of green bacteria. A molecular modelling study. Photosynth. Res. 41:225–233. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., K. Hara, H. Nagae, and Y. Koyama. 2000. Structural transformation among the aggregate forms of bacteriochlorophyll c as determined by electronic-absorption and NMR spectroscopies: Dependence on the stereoisomeric configuration and on the bulkiness of the 8-C side chain. Photochem. Photobiol. 71:596–609. [DOI] [PubMed] [Google Scholar]
- Montano, G. A., B. P. Bowen, J. T. LaBelle, N. W. Woodbury, V. B. Pizziconi, and R. Blankenship. 2003. Characterization of Chlorobium tepidum chloromes: A calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys. J. 85:2560–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, T., K. Ohtomo, M. Suzuki, H. Nakagawa, Y. Shikama, H. Konami, and Z. Y. Wang. 1994. Structures of chlorosomes and aggregated BChl c in Chlorobium tepidum from solid state high resolution CP/MAS 13C NMR. Photosynth. Res. 41:211–233. [DOI] [PubMed] [Google Scholar]
- Oelze, J., and J. R. Golecki. 1995. Membranes and chlorosomes of green bacteria: structure, composition, and development. In Anoxygenic Photosynthetic Bacteria. R. E. Blankenship, M. T. Madigan, and C. E. Bauer, editors. Kluwer Academic Publishers, Dordrecht, The Netherlands. 259–278.
- Overmann, J., H. Cypionka, and N. Pfennig. 1992. An extremely low-light-adapted phototrophic sulfur bacterium from the Black sea. Limnol. Oceanog. 37:150–155. [Google Scholar]
- Prokhorenko, V. I., D. B. Steensgaard, and A. R. Holzwarth. 2000. Exciton dynamics in the chlorosomal antennae of the green bacteria Chloroflexus aurantiacus and Chlorobium tepidum. Biophys. J. 79:2105–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psencik, J., Y. Z. Ma, J. B. Arellano, J. Hala, and T. Gillbro. 2003. Excitation energy transfer dynamics and excited-state structure in chlorosomes of Chlorobium phaeobacteroides. Biophys. J. 84:1161–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K. M., F. W. Bobe, D. A. Goff, and R. J. Abraham. 1986. NMR spectra of porphyrins. 28. Detailed solution structure of bacteriochlorophyllide d dimer. J. Am. Chem. Soc. 108:1111–1120. [Google Scholar]
- Staehelin, L. A., J. R. Golecki, R. C. Fuller, and G. Drews. 1978. Visualization of the supramolecular architecture of chlorosome (Chlorobium type vesicles) in freeze-fractured cells of Chloroflexus aurantiacus. Arch. Microbiol. 119:269–277. [Google Scholar]
- Staehelin, L. A., J. R. Golecki, and G. Drews. 1980. Supramolecular organization of chlorosome (Chlorobium vesicles) and of their membrane attachment site in Chlorobium limicola. Biochim. Biophys. Acta. 589:30–45. [DOI] [PubMed] [Google Scholar]
- Umetsu, M., Z. Y. Wang, J. Zhang, T. Ishii, K. Uehara, Y. Inoko, M. Kobayashi, and T. Nozawa. 1999. How the formation process influences the structure of BChl c aggregates. Photosynth. Res. 60:229–239. [Google Scholar]
- Umetsu, M., R. Seki, Z. Y. Wang, I. Kumagai, and T. Nozawa. 2002. Circular and magnetic circular dichroism studies of bacteriochlorophyll c aggregates: T-shaped and antiparallel dimers. J. Phys. Chem. B. 106:3987–3995. [Google Scholar]
- van Rossum, B. J., D. B. Steensgaard, F. M. Mulder, G. J. Boender, K. Schaffner, A. R. Holzwarth, and H. M. de Groot. 2001. A refined model of the chlorosomal antennae of the green bacterium Chlorobium tepidum from proton chemical shift constraints obtained with high-field 2-D and 3-D MAS NMR dipolar correlation spectroscopy. Biochemistry. 40:1587–1595. [DOI] [PubMed] [Google Scholar]
- Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from new-zealand hot-springs, chlorobium-tepidum sp-nov. Arch. Microbiol. 156:81–90. [Google Scholar]
- Wang, Z. Y., M. Umetsu, M. Kobayashi, and T. Nozawa. 1999a. Complete assignment of H1 NMR spectra and structural analysis of intact bacteriochlorophyll c dimer in solution. J. Phys. Chem. B. 103:3742–3753. [Google Scholar]
- Wang, Z. Y., M. Umetsu, M. Kobayashi, and T. Nozawa. 1999b. C-13- and N-15-NMR studies on the intact bacteriochlorophyll c dimers in solutions. J. Am. Chem. Soc. 121:9363–9369. [Google Scholar]
- Wullink, W., and E. F. J. van Bruggen. 1988. Structural studies on chlorosomes from Prosthecochlorisaestuarii. In Green Photosynthetic Bacteria. J. M. Olson, J. G. Ormerod, J. Amesz, E. Stackebrandt, and H. G. Trüper, editors. Plenum Press, New York. 3–14.