Single molecule experiments have added a new dimension to the exploration of complex biochemical behavior (Bustamante et al., 2003). By observing reactions at their singular constituent level, new windows into microscopic behavior were opened, that heretofore were unavailable with standard bulk techniques due to their inherent ensemble averaging. Elegant works include the characterization of various molecular motors such as kinesin (Visscher et al., 1999), RNA polymerase (Wang et al., 1998), DNA polymerase (Maier et al., 2000; Wuite et al., 2000), and F1-ATPase (Yasuda et al., 1998). Other works concentrated on the observation of structural effects induced by proteins on DNA such as formation of loops (Finzi and Gelles, 1995; Lia et al., 2003) and nucleosomes' disruption (Brower-Toland et al., 2002). During the last few years our lab has utilized single molecule elasticity measurements to characterize the sequence nonspecific interaction of three nucleoid-associated proteins IHF, H-NS, and HU with DNA (Ali et al., 2001; Amit et al., 2003; B. Schnurr, R. Amit, A. B. Oppenheim, and J. Stavans, unpublished). We gained new insight into both the microscopic interaction mechanism and large-scale elastic effects induced by the binding of many such proteins on a single molecule of λ-DNA, with important implications for nucleoid architecture and function.
In a recent letter (Dame and Wuite, 2003) question the validity of single molecule methodology as compared with more standard bulk techniques by alleging a purported “single molecule effect.” Specifically, they bring to question results obtained in our study on the interaction of H-NS with DNA (Amit et al., 2003). In our work, we provided evidence for H-NS polymerization on DNA, resulting in a complex of higher bending rigidity than bare DNA. H-NS polymerization is consistent with its functions in silencing (Falconi et al., 1998), and its response to changing ionic conditions (Williams and Rimsky, 1997) and temperature (McLeod and Johnson, 2001). In addition, polymerization has been observed directly by electron microscopy (Tupper et al., 1994). However, the H-NS polymerization mechanism appears inconsistent with 1), previous notions that implicate H-NS in the compaction of the nucleoid (Spassky et al., 1984) and 2), with recent experiments by Dame et al. (2000, 2001) and Schneider et al. (2001) in which H-NS bridging between distal DNA segments has been observed.
Dame and Wuite asserted that single molecule experiments miss certain regimes of behavior due to the large excess of protein over binding sites on DNA (Dame and Wuite, 2003). Note that standard bulk DNA binding assays are normally carried out in solution conditions that contain a 100–1000-fold excess of ligand to substrate (Liu-Johnson and Gartenberg, 1986). For the case of the DNA-H-NS complexes observed in our study, Dame and Wuite claimed that the large excess of H-NS over DNA places our system in saturating or near-saturating conditions, thereby precluding any observation of H-NS-induced compaction.
In this Letter we claim that within the range of change of protein concentrations used in our experiments, we span the full behavior between that of bare DNA to that which is characteristic of near-saturation conditions. We plot in Fig. 1 the degree of saturation ν as a function of total protein concentration Ptot, for parameters relevant to our experiment (KD ∼ 1 μM and total DNA Dtot = 10−11 M). Previous measurements of the dissociation constant of H-NS to nonspecific sites lie within the range 0.5–1.0 μM (Rimsky et al., 2001; Umanski et al., 2002). This curve has been computed using the same equation for the fractional occupancy ν employed by Dame and Wuite to compute the curves of Fig. 2 in their Letter, and corresponds to the height of the plateau in their curves as Ptot is varied for fixed KD and Dtot:
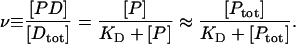 |
FIGURE 1.
Fractional occupancy ν as function of total protein concentration Ptot, for KD = 10−6 M and a total DNA concentration of 10−11 M.
The near equality applies very well under the condition [Ptot]≫[Dtot], which is true in our experiments. Fig. 1 shows that ν changes from negligible values to values near 1, as Ptot changes from 10−7 to 10−5 M. Note that the two-decade range of Ptot brackets KD. Consistently with this, our experimental results indeed exhibit a smooth variation in DNA end-to-end extension within the same range of Ptot, and not an abrupt change to near-saturation behavior. Our results together with those of others suggest that H-NS may have pleiotropic effects in vivo. H-NS can both bridge between distal DNA segments and polymerize along double-stranded DNA tracts.
Single molecule methodology has proven itself over the last decade to be extremely beneficial for the elucidation of complex biochemical behavior providing startling insights, consistent with bulk biochemical experiments. In this respect, the exploration of the architecture of both the bacterial nucleoid and eukaryotic chromatin has taken a major step forward due to the introduction of single molecule biophysical techniques. Specifically, the characterization of elastic characteristics induced by sequence-nonspecific interactions of proteins with DNA at the single molecule level has proved to be a tool that sheds important new light on structural and organizational problems that heretofore have gone largely unexplored. The wealth of new information is indeed providing a comprehensive new understanding to key, albeit old, problems.
References
- Ali, B. M. J., R. Amit, I. Braslavsky, A. Oppenheim, O. Gileadi, and J. Stavans. 2001. Compaction of single DNA molecules induced by integration host factor (IHF). Proc. Natl. Acad. Sci. USA. 98:10658–10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit, R., A. B. Oppenheim, and J. Stavans. 2003. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 84:2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland, B. D., C. L. Smith, R. C. Yeh, J. T. Lis, C. L. Peterson, and M. D. Wang. 2002. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 99:1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C., Z. Bryant, and S. B. Smith. 2003. Ten years of tension: single-molecule DNA mechanics. Nature. 421:423–427. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., and G. J. L. Wuite. 2003. On the role of H-NS in the organization of bacterial chromatin: from bulk to single molecules and back. Biophys. J. 85:4146–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2000. H-NS mediated compaction of DNA visualized by atomic force microscopy. Nucleic Acids Res. 28:3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie. 83:231–234. [DOI] [PubMed] [Google Scholar]
- Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility pf virF promotor to transcriptional repressor H-NS. EMBO J. 17:7033–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi, L., and J. Gelles. 1995. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 267:378–380. [DOI] [PubMed] [Google Scholar]
- Lia, G., D. Bensimon, V. Croquette, J. F. Allemand, D. Dunlap, D. E. Lewis, S. Adhya, L. Finzi, and J. Gelles. 2003. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Proc. Natl. Acad. Sci. USA. 100:11373–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Johnson, H. N., and M. R. Gartenberg. 1986. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 47:995–1005. [DOI] [PubMed] [Google Scholar]
- Maier, B., D. Bensimon, and V. Croquette. 2000. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl. Acad. Sci. USA. 97:12002–12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, S. M., and R. C. Johnson. 2001. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 4:152–159. [DOI] [PubMed] [Google Scholar]
- Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311–1323. [DOI] [PubMed] [Google Scholar]
- Schneider, R., R. Lurz, G. Luder, C. Tolksdorf, A. Travers, and G. Muskhelishvili. 2001. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 29:5107–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky, A., S. Rimsky, H. Garreau, and H. Buc. 1984. H1a, an E. coli DNA-binding protein which accumulates stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12:5321–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupper, A. E., T. A. Owens-Hughes, D. W. Usserly, D. S. Santos, D. J. P. Ferguson, J. M. Sidebotham, J. C. D. Hinton, and C. F. Higgins. 1994. The chromatin associated protein alters DNA topology in vitro. EMBO J. 13:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology. 148:2735–2744. [DOI] [PubMed] [Google Scholar]
- Visscher, K., M. J. Schnitzer, and S. M. Block. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189. [DOI] [PubMed] [Google Scholar]
- Wang, H. Y., T. Elston, A. Mogilner, and G. Oster. 1998. Force generation in RNA polymerase. Biophys. J. 74:1186–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175–185. [DOI] [PubMed] [Google Scholar]
- Wuite, G. J. L., S. B. Smith, M. Young, D. Keller, and C. Bustamante. 2000. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 404:103–106. [DOI] [PubMed] [Google Scholar]
- Yasuda, R., H. Noji, K. Kinosita, Jr., and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]



