Abstract
This article presents a new formalism to perform a quantitative fluorescence analysis using the Stokes shift of AEDANS-labeled cysteine mutants of M13 major coat protein incorporated in lipid bilayers. This site-directed fluorescence spectroscopy approach enables us to obtain the topology of the bilayer-embedded transmembrane α-helix from the orientation and tilt angles, and relative bilayer location. Both in pure dioleoylphosphatidylcholine and dioleoylphosphatidylcholine/dioleoylphosphatidylglycerol (4:1 mol/mol) bilayers, which have a similar bilayer thickness, the tilt angle of the transmembrane helix of the coat protein turns out to be 23° ± 4. Upon decreasing the hydrophobic thickness on going from dieicosenoylphosphatidylcholine to dimyristoylphosphatidylcholine, the tilt angle and orientation angle of the transmembrane α-helix change. The protein responds to an increase of hydrophobic stress by increasing the tilt angle so as to keep much of its hydrophobic part inside the bilayer. At the same time, the transmembrane helix rotates at its long axis so as to optimize the hydrophobic and electrostatic interactions of the C-terminal phenylalanines and lysines, respectively. The increase of tilt angle cannot completely keep the hydrophobic protein section within the bilayer, but the C-terminal part remains anchored at the acyl-chain/glycerol backbone interface at the cost of the N-terminal section. In addition, our analysis results in the profile of the dielectric constant of the hydrophobic domain of the bilayer. For all phospholipid bilayers studied the profile has a concave shape, with a value of the dielectric constant of 4.0 in the center of the bilayer. The dielectric constant increases on approaching the headgroup region with a value of 12.4 at the acyl-chain/glycerol backbone interface for the various phosphatidylcholines with different chain lengths. For dioleoylphosphatidylcholine/dioleoylphosphatidylglycerol (4:1 mol/mol) bilayers the value of the dielectric constant at the acyl-chain/glycerol backbone interface is 18.6. In conclusion, the consistency of our analysis shows that the applied cysteine-scanning mutagenesis method with AEDANS labeling of a helical transmembrane protein in combination with a quantitative formalism offers a reliable description of the lipid bilayer topology of the protein and bilayer properties. This also indicates that the spacer link between the protein and AEDANS label is long enough to monitor the local polarity of the lipid environment and not that of the amino-acid residues of the protein, and short enough to have the topology of the protein imposing on the fluorescence properties of the AEDANS label.
INTRODUCTION
The determination of the structure and the embedding of membrane proteins is still a challenging question in membrane biophysics. As the more common techniques for structure determination, such as x-ray diffraction or solution nuclear magnetic resonance (NMR), encounter experimental difficulties, there is an ongoing search for alternative techniques and methodologies (Torres et al., 2003). A promising approach is the use of site-directed mutagenesis of the membrane protein in combination with fluorescence labeling. In the past years, we have explored this approach to study the membrane-bound state of the major coat protein of bacteriophage M13 in a range of applications, using the position of the fluorescence maximum of the labeled protein (Spruijt et al., 1996, 2000; Meijer et al., 2001a,b), as well as fluorescence resonance energy transfer (Fernandes et al., 2003, 2004; Nazarov et al., 2004).
In this study, we will focus again on the major coat protein of the filamentous bacteriophage M13. This protein has been used in the past in several reports as a model membrane protein, because it is a relatively small (50 amino-acid residues) integral membrane protein. These studies have resulted in structural models for both the detergent-solubilized and membrane-bound protein (for a recent review, see Stopar et al., 2003). Detailed information about the topology of the α-helical transmembrane domain was obtained from 13C MAS NMR experiments for the protein in dimyristoylphosphatidylcholine bilayers that clearly demonstrate the presence of a tilt angle of 20° ± 10 around amino-acid residues 29–31 (Glaubitz et al., 2000). Recently the three-dimensional structure of the membrane-bound coat protein of the fd bacteriophage, that closely resembles the M13 coat protein, was reported. In this work a tilt angle of 26° was found for the major part of the helical transmembrane section, whereas for the C-terminal part of the helix the tilt angle decreased to 16° (Marassi and Opella, 2003).
In this work our goal is to develop a new formalism to perform a quantitative fluorescence analysis of AEDANS-labeled cysteine mutants of the M13 major coat protein. This approach enables us to obtain the complete depth profile of the bilayer-embedded transmembrane α-helix including the orientation and tilt angles, in combination with a description of the polarity profile of the bilayer. In addition, we studied the effect of bilayer thickness on the orientation, tilt, and location of the transmembrane α-helical domain of the protein.
EXPERIMENTAL
Sample preparation
Site-specific cysteine mutants of M13 major coat protein were prepared, purified, and labeled with IAEDANS (Molecular Probes, Eugene, OR) as described previously (Spruijt et al., 2000). Labeled M13 coat protein mutants were reconstituted into phospholipid bilayers as reported earlier (Spruijt et al., 1989).
Dimyristoleoylphosphatidylcholine (14:1 PC), dipalmitoleoylphosphatidylcholine (16:1 PC), dioleoylphosphatidylcholine (DOPC, 18:1 PC), and dieicosenoylphosphatidylcholine (20:1 PC) were purchased from Avanti Polar Lipids (Alabaster, AL) and dioleoylphosphatidylglycerol (DOPG) was purchased from Sigma (St. Louis, MO). The various bilayer systems that were prepared consisted of: 1), DOPC and DOPG lipids in a 4:1 (mol/mol) ratio, denoted as DOPC/DOPG; 2), 100% 14:1 PC; 3), 100% 16:1 PC; 4), 100% 18:1 PC; and 5), 100% 20:1 PC.
Fluorescence measurements
AEDANS was excited using light of 340 nm and its emission was detected from 350 to 600 nm with a 2-nm bandpass in both the excitation and detection lightpaths on a Fluorolog 3.22 manufactured by Jobin Yvon-Spex (Edison, NJ). We will use the wavenumber of the fluorescence instead of the wavelength to characterize the fluorescence maximum. The spectral position of the fluorescence maximum νflu was taken from a six-termed polynomial fit to the top part of the emission spectrum. For this we used the program IGOR Pro 3.13 (Wavemetrics, Lake Oswego, OR). The Stokes shift Δν was taken as the difference between the spectral position of the (0,0) band in the fluorescence excitation spectrum νexc, which was found to be 27,322 cm−1 independent of label position, and νflu, as Δν = νexc − νflu. For the fluorescence studies highly diluted samples were prepared with a mutant protein concentration of ≈1 μM. The lipid/protein (L/P) ratio of the samples was ≈1500.
In previous work (Meijer et al., 2000; Spruijt et al., 2000) a PerkinElmer LS5 system (PerkinElmer Life and Analytical Sciences, Monza, Italy) was used to record fluorescence spectra, whereas for the quantitative analysis in the present work fluorescence spectra were measured on an advanced Fluorolog 3.22 spectrometer, as described above. Small differences were found in the wavelength of maximum fluorescence between both spectrometers. These differences arise from 1), an insufficient calibration of the wavelength scale for the PerkinElmer LS5; 2), the fact that in contrast to those recorded with the LS5, fluorescence spectra recorded with the Fluorolog 3.22 are corrected for the wavelength-dependent sensitivity of the detection part; and 3) spectra of the mutants are digitally corrected for background signals by subtracting the spectrum of a wild-type-containing sample having approximately the same protein concentration and L/P ratio.
METHODOLOGY
Polarity probing in a bilayer
The aromatic part of the AEDANS label used in this study resembles that of the dansyl compounds, which are known as polarity probes (Lakowicz, 1999). This effect arises from the relatively strong polar character of the photoexcited charge-transfer state, which causes a polarity-dependent fluorescence spectrum. Therefore the wavelength of maximum fluorescence is red-shifted with increasing polarity of the surrounding medium, known as the solvent-relaxation effect (Lakowicz, 1999). For DANSAEP, a dansyl derivative similar to AEDANS, it was already shown (Ren et al., 1999) that the Stokes shift Δν is almost linear with solvent polarity, according to the solvatochromic analysis proposed by Lippert and Mataga (see Lakowicz, 1999). This is described by the empirical equation for the Stokes shift,
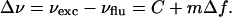 |
(1) |
In this equation C is a constant, and the so-called solvatochromic slope m is described by
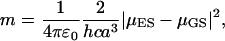 |
(2) |
and the orientational polarizability of the solvent is given by
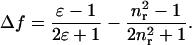 |
(3) |
In Eqs. 2 and 3, ɛ0 and ɛ are the dielectric constants of vacuum and solvent, respectively, and nr is the refractive index of the solvent. Parameter a is the radius of the Onsager cavity for the solute molecule, and h and c are Planck's constant and the velocity of light, respectively. The dipoles in the ground state and excited state are indicated by μGS and μES, respectively.
In a heterogeneous environment, such as a membrane, the relation between the Stokes shift Δν and solvent polarity is not straightforward. Recently we reported on the fluorescent properties of AEDANS-labeled cysteine mutants of the M13 major coat protein in lipid bilayers (Meijer et al., 2000; Spruijt et al., 1996, 2000). We found that after reconstitution in lipid bilayer systems the wavelength of maximum fluorescence is dependent on the position of the AEDANS label along the backbone of the coat protein. Assuming the refractive index nr to be constant in a bilayer, this indicates that the Stokes shift of the AEDANS label varies over the bilayer mainly as a result of a depth-dependent dielectric constant ɛ of the local environment of a transmembrane section of the protein. An effective dielectric constant for a bilayer system has already been introduced for the development of a total bilayer potential profile (Flewelling and Hubbell, 1986), taking into account a sigmoidal functional dependence of the dielectric constant on bilayer position. A sigmoidal shape of the polarity variation in phospholipid bilayers was used not long ago to fit isotropic hyperfine coupling constants obtained from ESR spectra of spin-labeled glycerophospholipids in phospholipid bilayers (Marsh, 2001). In this article we will introduce a bi-phase model using a similar functional dependence for the Stokes shift Δν of the AEDANS label given by
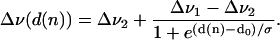 |
(4) |
In this equation Δν(d(n)) is the Stokes shift as a function of the distance d to the center of the bilayer; Δν1 and Δν2 are the limiting values of the Stokes shifts for the AEDANS label; d(n) is the distance of the AEDANS label to the bilayer center as a function of the amino-acid residue number of the mutant cysteine position n; d0 is the distance of maximum gradient with respect to the center of the bilayer; and σ is an exponential decay constant, which reflects the width of the transition region.
Because a phospholipid bilayer has more than one transition region over which its local solvent properties may change (i.e., hydrocarbon core to headgroup region, headgroup region to water phase), we will confine ourselves in this article to positions in, or close, to the hydrocarbon core. Therefore, our analysis is only roughly valid in the lipid bilayer domain that includes both the acyl-chain region with hydrophobic thickness dh (defined as the carbonyl-to-carbonyl distance; Ridder et al., 2002), and the glycerol ester regions on both sides of the bilayer. The interface at distance d = ½dh to the center of the bilayer is denoted as the acyl-chain/glycerol backbone interface.
Mathematical description of the bilayer topology of an α-helix
To relate the variation of the Stokes shift Δν for different positions of the AEDANS label along the backbone of the M13 major coat protein to a variation in bilayer depth, we assume the section of the protein located in the hydrophobic acyl-chain region to be α-helical. In Fig. 1 a we schematically represent an α-helical transmembrane part of the M13 major coat protein by a cylinder of radius R, being the distance of the AEDANS label to the protein helical axis (see Table 1 for a description of the parameters used in this article). To describe the various positions of the label attached to the amino-acid residues on the helix in Fig. 1 a, we define an axes system {x, y, z} relative to a reference position n0, such that the coordinates of n0 are (0, R, 0).
FIGURE 1.
Schematic representation of an α-helix before (a) and after positioning in a bilayer including a rotation and tilt (b).
TABLE 1.
Definition of the parameters used in the helix-membrane model
| Parameter | Unit | Description |
|---|---|---|
| n0 | Reference amino-acid residue position on helix | |
| n | Position of labeled amino-acid residue on helix | |
| hr | Å | Translation per amino-acid residue along the helix; this is 1.5 Å for a perfect α-helix |
| ϕr | ° | Rotation per amino-acid residue; this is 100° for a perfect α-helix |
| R | Å | Distance of the center of the chromophore moiety of AEDANS to the symmetry axis of the helix |
| α | ° | Helix orientation angle; rotation at the z axis of the axes system {x, y, z}; z is parallel to the normal to the bilayer |
| β | ° | Helix tilt angle; rotation at the x axis of the axes system {x, y, z} |
| hc | Å | Position of the center plane of the bilayer within the axes system {x, y, z} |
| d | Å | Distance of point n to the center plane of the bilayer along its normal |
| dh | Å | Hydrophobic thickness of the bilayer (two-times the lipid acyl-chain length) |
| nC | — | Position where the helix axis crosses the acyl-chain/glycerol backbone interface (given by dh) at the C-terminal protein part |
| nN | — | Position where the helix axis crosses the acyl-chain/glycerol backbone interface (given by dh) at the N-terminal protein part |
| νexc | cm−1 | Wavenumber of lowest energy excitation band (the (0,0) band) |
| νflu | cm−1 | Wavenumber of highest energy fluorescence band |
| Δν | cm−1 | Stokes shift; this is the difference νexc−νflu |
| Δf | — | Solvent polarity parameter (orientational polarizability) by Lippert and Mataga (see Lakowicz, 1999) |
| ɛ | — | Local dielectric constant |
| nr | — | Local refractive index |
To relate the position of another label at position n to this reference position, we have to take into account the translation hr and rotation ϕr per amino-acid residue of the helix. The coordinates of the label at amino-acid residue position n are then given by
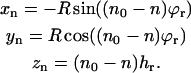 |
(5) |
To make the model more general, we introduce an orientation of the helix in the axes system {x, y, z} given by a rotation angle α at the symmetry axis of the helix (the z axis), together with a tilt angle β of the symmetry axis at the x axis (see Fig. 1 b). In this case the coordinates of the label at position n become
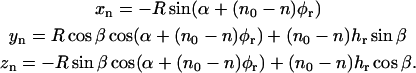 |
(6) |
Finally, we consider the location of the tilted helix in a bilayer. Assuming that the normal to the bilayer is parallel to the z axis in the axes system {x, y, z}, we define hc as the position of the center of the bilayer within the axes system {x, y, z}. After rotation, tilting, and translation of the helix, the distance of the AEDANS label at position n becomes d = |zn − hc|. By substituting zn from Eq. 6 we obtain a general expression for the distance d(n) of label position n at the tilted helix to the center of the bilayer,
 |
(7) |
Using the Levenberg-Marquardt nonlinear least-squares optimization in IGOR Pro 3.13, the experimental Δν(d(n)) data obtained for different amino-acid positions n were fitted to Eq. 4 with d(n) given by Eq. 7.
RESULTS AND DISCUSSION
AEDANS fluorescence of labeled mutants in lipid bilayers
In Fig. 2 the fluorescence spectra of coat protein mutants reconstituted in DOPC/DOPG with the AEDANS label at positions 22 and 46 (dotted and dashed curves, respectively) are clearly shifted to smaller wavenumbers (longer wavelengths) with respect to that of the mutant with the AEDANS label at position 34 (solid curve). For the latter mutant the AEDANS moiety is assumed to reside in the hydrophobic region of the lipid bilayer. The fluorescence spectra of the former two mutants are typical for AEDANS close to or within the phospholipid headgroup region, because the positions 22 and 46 are at the outer sides of the putative transmembrane domain of the protein (Spruijt et al., 2000). In all cases the overall lineshape of the fluorescence spectrum is independent of label position; however, the width of the AEDANS fluorescence band for mutants holding the AEDANS label in the hydrophobic domain of the bilayer is slightly larger than that for mutants holding the AEDANS label in the relatively polar domains of the bilayer (see also Fig. 2). According to Eq. 3 we can ascribe this difference in spectral width to increased sensitivity of the fluorescence properties toward polarity variations (caused by local mobility of the label) when the label is in the hydrophobic domain (ɛ relatively small) with respect to when the label is in a relatively polar domain (ɛ relatively large). Apart from what is described above, the fluorescence spectra do not show any broadening resulting from other conformational heterogeneities (e.g., coexistence of transmembrane and superficially bound proteins). There will be no effects coming from oligomerization because of the low L/P ratio of ∼1500 that is used.
FIGURE 2.
Normalized fluorescence spectra of different mutants, reconstituted in DOPC/DOPG bilayers with the AEDANS label attached at positions 22 (dashed curve), 46 (dotted curve), and 34 (solid curve) in the primary sequence.
Label position-dependent Stokes shift of AEDANS fluorescence
In Fig. 3 the Stokes shift Δν of the AEDANS fluorescence is shown as a function of the amino-acid residue number n for different-labeled M13 coat protein mutants in DOPC/DOPG lipid bilayers (Fig. 3 a) and pure 18:1 PC (Fig. 3 b), respectively. The minimum value of the Stokes shift (∼6000 cm−1) reflects a position in the center of the bilayer (n ≈ 35). The value of the Stokes shift of amino-acid residue position 49 in Fig. 3 a (7550 cm−1) is close to the value of 7520 cm−1 that was obtained for free AEDANS in an aqueous buffer solution without lipids.
FIGURE 3.
Stokes shift Δν of AEDANS-labeled M13 coat protein mutants as a function of the amino-acid residue number n at which the label is attached in DOPC/DOPG (a) and 18:1 PC (b). Computer fits are included represented by a solid line in the data range (n = 15–46). The dashed lines represent extrapolations of the fitted function.
Previously we analyzed the variations in wavelength of maximum fluorescence λmax of the AEDANS fluorescence of M13 coat protein mutants as a function of label position n in a qualitative way (Spruijt et al., 1996, 2000; Meijer et al., 2001a,b). Here we use the Stokes shift Δν for which we have developed a quantitative formalism, as given by Eqs. 4 and 7. Therefore fitting of the experimental Stokes shifts in Fig. 3 to these equations provides information about the orientation angle α, tilt angle β, and relative position hc of the transmembrane part of the coat protein in the bilayer, as well as the polarity profile of the bilayer, given by the limiting values of the Stokes shift (Δν1 and Δν2) and the location (d0) and width of the transition region (σ). There are two basic restrictions in the present theory: 1), the assumption of a rigid perfect α-helix (3.6 residues and 5.4 Å translation along the helix per turn) in the transmembrane section of the protein, and 2), the description of the polarity profile of the membrane being limited to the hydrocarbon core and the glycerol ester part of the headgroup region. These restrictions imply that our analysis is valid only for a transmembrane α-helix within the apolar to moderately polar region of a lipid bilayer.
For an α-helix oriented perpendicular to the surface of the bilayer, one would expect a gradual decrease of Δν upon going from the relative polar headgroup region to the relatively apolar center of the bilayer. Consequently, again a gradual increase of Δν is expected after passing the center of the bilayer and approaching the opposite headgroup region. This tendency is reflected by the overall behavior of the plots of Δν(n) in Fig. 3. This shows that the protein spans the bilayer in both types of bilayers, similar to what it is doing in the other lipid systems studied (data not shown).
The Stokes shift in the plots in Fig. 3 shows oscillations in the range from n of ∼15 to ∼40 with a periodicity roughly in accordance with that of an α-helix. When the various mutants are dissolved in pure organic solvents the fluorescence properties of the AEDANS label are almost independent of residue position, showing that the AEDANS is probing the homogeneous solvent environment (R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga, unpublished). Therefore the presence of the oscillations in the Stokes shift indicates that the AEDANS label is probing the local polarity of the bilayer. The presence of these oscillations also indicates that the transmembrane α-helix is tilted with respect to the bilayer normal (R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga, unpublished). The oscillations are most pronounced for label positions within the hydrophobic region where the dielectric constant ɛ is relatively small, indicating that in this part of the bilayer the Stokes shift Δν(n) is relatively sensitive to changes in ɛ, as described by Eqs. 1 and 3. Apart from the tilt effect, the dependency of Δν on the polarity gradient over the bilayer will be determined by the length of the AEDANS-cysteine spacer, as will be discussed in the next paragraph.
Fitting the data to the α-helix model
To extract parameters such as orientation angle α and tilt angle β of the transmembrane helix of M13 coat protein in the various lipid bilayers, we fitted the Δν(n) data series to Eqs. 4 and 7. In Eq. 4 the limiting values Δν1 and Δν2 are expected to be independent of lipid chain length; however, Δν2 could be dependent on the chemical nature of the headgroup (i.e., zwitterionic or net-charged). Therefore, we fixed Δν1 at the measured value for free AEDANS label in hexane (5344 cm−1), expecting it to mimic the interior of a phospholipid bilayer beyond the influence of headgroups and water. The fitted value of Δν2 was found to be very similar (7020 ± 20 cm−1) for all pure PC bilayers used in this study and slightly different for mixed DOPC/DOPG bilayers (7189 cm−1).
Because the fit parameters R, β, and σ were found to be correlated in the data analysis, we decided to fix the AEDANS-to-helical axis distance R during the final fit procedures. From Table 2 it can be seen that changing of R results in slightly different values of both β and σ; however, it did not affect the quality of the fit (χ2 value; not in Table). To fit the DOPC/DOPG data we varied R between 6.5 Å, being the approximate distance of the center of mass of a tryptophan residue to the helical axis, and 9.5 Å, being the maximum effective distance as concluded from tryptophan-to-AEDANS energy transfer measurements (unpublished results). The high quality of the fits shown in Fig. 3 and of the fits to the data for 14:1, 16:1, and 20:1 PC (data not shown) justifies that we can approximate the transmembrane section of M13 coat protein by the α-helical model in Fig. 1 for positions n from 18 to 46. In Fig. 3 the variations between the Δν values for n = 15 to 18 are relatively small, and although the Δν values are not far from the theoretical curve, the indication of α-helical structure becomes less strong. The dotted curves outside the range 15 < n < 46 are extrapolations of the fit, showing that for positions in the amphipathic helix (n = 7 and 13) and for positions near the termini (e.g., n = 49), our bi-phase model is not valid.
TABLE 2.
Fit parameters characterizing the topology of the M13 major coat protein in mixed DOPC/DOPG bilayers
| R*/Å | α†/° | β†/° | hc†/Å | d0†/Å | σ† |
|---|---|---|---|---|---|
| 6.5 | −0.5 (±0.1) | 27 (±1) | −5.6 (±0.1) | 3.6 (±0.1) | 5.9 (±0.1) |
| 8.0 | −0.5 (±0.1) | 22 (±1) | −5.9 (±0.1) | 3.8 (±0.1) | 6.1 (±0.1) |
| 9.5 | −0.5 (±0.1) | 19 (±1) | −6.0 (±0.1) | 3.9 (±0.1) | 6.3 (±0.1) |
Fitting was performed using reference position n0 = 29 for n between positions 15 and 46 (except for position 43, see text). The limiting values Δν1 and Δν2 in Eq. 4 were fixed at the value for the AEDANS label in hexane (5344 cm−1) and the average value (7189 cm−1) of the individual fits, respectively.
Distance of AEDANS to backbone R was varied between 6.5, 8.0 and 9.5 Å for DOPC/DOPG (see text).
Errors follow from standard deviations produced by the fit program.
The sensitivity of the other fit parameters to a fixed input value of R is shown in Table 2. The fit parameters of the data for different hydrophobic thickness of the bilayer, using a fixed value of R = 8 Å, are collected in Table 3. This value of R is the average of the extreme values, as indicated above. The uncertainty in the value of R results in an error in the fitted tilt angle β of ±4°, whereas it has relatively small effects on the other parameters. The contribution of experimental errors (like wavelength accuracy of the spectrometer and reproducibility of the wavelength of maximum fluorescence) to the final error in the fit parameters is small compared to that following from the above-mentioned uncertainty in the value of R.
TABLE 3.
Fit parameters characterizing the topology of the M13 major coat protein in lipid bilayers of different hydrophobic thicknesses
| Lipid (dh*/Å) | α†/° | β‡/° | hc†/Å | d0†/Å | σ† | nN | nC |
|---|---|---|---|---|---|---|---|
| 20:1 PC (33.0) | −18 (±7) | 19 (±4) | −4.4 (±0.2) | 2.5 (±0.3) | 7.4 (±0.3) | 20 | 44 |
| 18:1 PC (29.5) | −27 (±5) | 23 (±4) | −4.8 (±0.2) | 2.5 (±0.3) | 5.4 (±0.3) | 22 | 43 |
| 16:1 PC (26.0) | −31 (±1) | 26 (±4) | −5.7 (±0.1) | 1.1 (±0.1) | 6.2 (±0.1) | 24 | 43 |
| 14:1 PC (22.5) | −36 (±1) | 33 (±4) | −6.4 (±0.1) | 1.2 (±0.1) | 5.6 (±0.1) | 25 | 43 |
The various parameters are described in Table 1. Fitting was performed using reference position n0 = 29 for n between positions 15 and 46 (except for position 43, see text). The limiting value Δν1 (see Eq. 4) was fixed at the value for the AEDANS label in hexane (5344 cm−1) for all PC bilayers, and Δν2 was fixed at 7020 cm−1 being the average of individual fit values. The distance of AEDANS to backbone R was fixed at 8.0 Å for all PC bilayers.
Hydrophobic thickness (Ridder et al., 2002).
Errors follow from standard deviations produced by the fit program.
Errors follow from uncertainty in R (6.5 Å ≤ R ≤ 9.5 Å).
Effect of lipid headgroup on the topology of the protein
Taking into account the uncertainty in the distance parameter R we may conclude that in DOPC/DOPG and 18:1 PC bilayers, with both having a hydrophobic thickness of 29.5 Å, the tilt angle β of the transmembrane helix of the M13 major coat protein is 23° ± 4 (see Tables 2 and 3). A comparison between this tilt angle and those reported in literature will be made after evaluation of the effect of hydrophobic thickness on the tilt angle in one of the next sections.
As can be seen in Tables 2 and 3, the helix orientation angle α is found to be different for DOPC/DOPG as compared to 18:1 PC: −0.5 and −27°, respectively. An orientation angle of 0.5° implies that in this lipid system our reference position (n = 29) is almost exactly facing the tilt. To visualize this situation, we have calculated the coordinates of positions at a distance of 5 Å to the helical axis for all amino-acid residues in the fitted region using the fit parameters in Tables 2 and 3. The distance of 5 Å is considered to be the average thickness of a model α-helix. A projection of the calculated helix in DOPC/DOPG is presented in Fig. 4, clearly showing that the protein section holding the three positively charged lysines (n = 40, 43, and 44) is able to interact with the negatively charged headgroups as reported earlier (Meijer et al., 2001b; Strandberg et al., 2002). In this configuration both phenylalanines (n = 42 and 45) are facing the hydrophobic interior, which is expected to be more favorable to aromatic residues (Stopar et al., 2003). The functional significance of this topology for the bacteriophage assembly process has been noted earlier (Meijer et al., 2000; Marassi and Opella, 2003).
FIGURE 4.
Schematic projection of the calculated helix in a DOPC/DOPG bilayer, showing the bilayer depth of various typical amino-acid residues, Trp-26 (W); Phe-42 and Phe-45 (F); Lys-40, Lys-43, and Lys-44 (K); and of the reference position n = 29 (#), with respect to the acyl-chain/glycerol backbone interfaces (dashed lines). For this lipid system the reference position is almost exactly facing the tilt. Positions are calculated for a model helix with a radius of 5 Å using the parameters in Table 2.
The different orientation angle in pure 18:1 PC as compared to DOPC/DOPG can be explained by the fact that the positively charged lysines probably anchor differently to the lipid headgroup region in the absence of net negatively charged PG lipids. In line with this observation, for DOPC/DOPG (Fig. 3 a) the relatively strong deviation of Δν(43) from the fit (although residing in the region that is described by a single bi-phase model) may be an effect of anchoring as well: replacing the positively charged Lys-43 by an AEDANS-labeled cysteine probably has an effect on the interactions at the headgroup region, thereby altering the location and orientation of the mutant protein. However, this demonstrates that care should be taken with mutations of highly functional amino-acid residues. For this reason, we decided to exclude the data with n = 43 from the fitting of the DOPC/DOPG data.
Effect of hydrophobic thickness on the topology of the protein
Since our analysis gives a full description of the topology of the transmembrane part of the M13 coat protein, given by the orientation angle α, tilt angle β, and location hc, it is interesting to follow these parameters under the condition that the bilayer thickness is changed, so that the protein is put under hydrophobic stress. The results are given in Table 3 and further illustrated in Figs. 5 and 6.
FIGURE 5.
Orientation angle α (□) and tilt angle β (○) for PC bilayers from Table 3 as a function of hydrophobic thickness.
FIGURE 6.
Distances of typical amino-acid residues to the bilayer center for isoleucine (32, top left) and to the acyl-chain/glycerol backbone interface for tryptophan (26, top right), phenylalanines (42 and 45, bottom left), and lysines (40, 43, and 44, bottom right) plotted as a function of the hydrophobic thickness for a model helix with a radius of 5 Å, using the parameters in Table 3.
In Fig. 5 it can be seen that upon decreasing the hydrophobic thickness from 33.0 Å (20:1 PC) to 22.5 Å (14:1 PC) (Ridder et al., 2002) the tilt angle β increases from 19 to 33° and the orientation angle α increases from 18 to 36°. This indicates that the protein responds to hydrophobic stress by increasing the tilt angle to keep its hydrophobic part inside the bilayer. At the same time, the transmembrane helix rotates at its axis presumably to optimize the hydrophobic interactions of Phe-42 and Phe-45 and electrostatic interactions of Lys-40, Lys-43, and Lys-44 at the C-terminal region, similar to what has been observed in going from DOPC/DOPG to 18:1 PC. In view of these results our tilt angles can be compared to those reported in literature only for bilayer systems having comparable hydrophobic thickness. From a solid-state NMR study of the M13 major coat protein in oriented bilayers of fully saturated dimyristoyl phosphatidylcholine (DMPC) bilayers at 243 K (Glaubitz et al., 2000), a tilt angle of 20 ± 10° was obtained. At the given temperature DMPC bilayers are in the gel state, having a hydrophobic thickness of 31.5 Å (Dumas et al., 1999). Also from solid-state NMR studies a tilt angle of 26° is reported for the main part of the transmembrane section of the very similar fd bacteriophage coat protein in oriented bilayers of palmitoyl-oleoyl-phosphatidylcholine/palmitoyl-oleoyl-phophatidylglycerol (4:1), which have a hydrophobic thickness of 30 Å (Marassi and Opella, 2003). Considering the similarity between the hydrophobic thicknesses of the above-mentioned oriented bilayers (31.5 Å and 30 Å), and that of our DOPC bilayers (29.5 Å), we conclude that the tilt angles (20 ± 10°; 26°; and 23 ± 4°, respectively) are fairly in agreement.
In addition to a change in orientation angle α and tilt angle β, the relative bilayer location of the helix, as given by hc, changes upon varying the hydrophobic thickness. Together these parameters make up the major mechanism for the M13 major coat protein to adapt to hydrophobic mismatch conditions. To visualize the combined effect of these parameters, we calculated in Fig. 6 the positions of typical amino-acid residues of the transmembrane helix relative to the closest interface.
In Fig. 6 the positions (at 5 Å distance to the helical axis) of the phenylalanines (42 and 45) and lysines (40, 43, and 44) turn out to be less shifted with respect to the nearest interface than the tryptophan (26). Therefore it is evident that the section containing the lysines and phenylalanines at the C-terminal end of the helix is a stronger interfacial anchor than the tryptophan at the N-terminal end. In addition to this, the almost constant location of isoleucine at position 32 suggests that the hydrophobic section of the protein (from position 29 to 33), containing four valines and one isoleucine, functions as an anchor to the hydrophobic interior of the bilayer.
Another way of interpreting the parameters α, β, and hc of Table 3 is to calculate the section of the transmembrane α-helix that still fits into the hydrophobic domain of the bilayer. To analyze this effect, we define the amino-acid residue positions nC and nN as the positions where the helical axis crosses the acyl-chain/glycerol backbone interface (given by the hydrophobic thickness dh) at the C- and N-terminal protein parts, respectively. For these positions it can be evaluated that
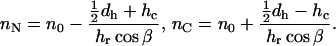 |
(8) |
The calculated amino-acid positions are compiled in Table 3 as well. In this table we can see that on decreasing the bilayer thickness, position 43 in the C-terminal protein domain remains at a constant position in the interface, whereas the N-terminal protein part sticks out more and more from the hydrophobic bilayer phase. For a relatively thick membrane (20:1 PC), position 20 is still within the hydrophobic core, whereas for a relatively thin membrane (14:1 PC), the length of the membrane-embedded α-helix is reduced by five amino-acid residues. Obviously, the increase of tilt angle β cannot completely keep the hydrophobic protein section within the hydrophobic core of the bilayer, but the C-terminal part remains anchored at the acyl-chain/glycerol backbone interface at the cost of the N-terminal section.
From our analysis it seems that there is not a direct role put aside for the hinge region and N-terminal protein domain in controlling the embedding of the bilayer of the transmembrane α-helix. This could be due to the fact that the hinge has a flexible connection to the amphipathic N-terminal domain (Stopar et al., 2003), so that it can easily adapt to environmental stress conditions. Interestingly Trp-26, which is at the N-terminal end of the transmembrane α-helix, does not seem to play a role in the membrane anchoring mechanism either. This observation is in agreement with conclusions from previous work (Meijer et al., 2001a).
Polarity profile of the bilayers
For describing the bilayer polarity, we have introduced a bi-phase model for the Stokes shift, as given by Eq. 4. The limiting values of the Stokes shift are given by the parameters Δν1 and Δν2 at the center of the bilayer and in the headgroup region, respectively. The shape of the profile is described by the parameters d0 (the value of d at the point of maximum gradient, corresponding to Δν = ½(Δν1 + Δν2)) and σ, an exponential decay constant, which gives rise to a broadening of the gradient.
For 18:1 PC and DOPC/DOPG the Stokes shift Δν is plotted in Fig. 7, using the parameters from Tables 2 and 3. It is surprising to see that the combination of relatively small values of d0 and large values of σ derived from the data analysis lead to an almost linear change of Δν within the hydrophobic core of the bilayer for all lipid systems investigated. On approaching the headgroup region, i.e., for d-values approaching dh, the Stokes shift starts to level off to Δν2. This leads to an insensitivity of the Stokes shift to locations within the headgroup region. This effect is observed in Fig. 3 as oscillations of Δν with decreasing amplitude for values of n < 20 and n > 43.
FIGURE 7.
Calculated Δν profiles (top) and ɛ(d) profiles (bottom) for bilayers of 18:1 PC (solid curves) and DOPC/DOPG (dotted curves). The ɛ(d) curves are calculated according to Eqs. 1–3 using nr = 1.5 (Salomon et al., 2000), a = 0.36 nm, Δμ = 5.63 D (Ren et al., 1999), and C = Δνhexane (Δf ≈ 0) = 5344 cm−1 (this article). Vertical lines represent locations of the acyl-chain/glycerol backbone interface following from the published hydrophobic thicknesses dh (Ridder et al., 2002) at d = ½dh.
It has been noticed that in an environment where hydrogen bonding can occur, Stokes shifts can be larger than allowed according to the Lippert-Mataga theory (see Lakowicz, 1999). Such an effect may take place in the headgroup region, where the number of water molecules is expected to increase when approaching the headgroup/water interface (White and Wimley, 1998). Such specific solvent interactions could be the origin of the significantly increased Δν values for n = 7 and 13 (see Fig. 3) as compared to the extrapolated fitted curve. This also indicates that these positions in the N-terminal region are located close to the headgroup/water interface. It should be noted that this effect does not play a role in the results presented in Tables 2 and 3, since we here confine ourselves to the bilayer interior. On the other hand, because Δν is invariable to any increase of ɛ in the headgroup region in the direction of the bulk water phase (according to Eqs. 1–3), another explanation for the deviating values of Δν(7) and Δν(13) could be that, within the headgroup region, a gradient in refractive index nr exists. The similarity between the refractive index of the compounds 9-octadecene (nr = 1.447), glycerol tripropanoate (nr = 1.432), and glycerol 1-methyl ether (nr = 1.442) (which, because of their structure, may mimic the regions holding the lipid tails, the lipid backbones, and the PG headgroup glycerols, respectively), suggests that nr can be considered to be constant within most of the bilayer. However, on approaching the headgroup/water interface there could be a gradual decrease of nr from 1.4–1.5 for the bilayer to 1.33 for the water phase. A decrease of nr will result in increased Stokes-shift values (e.g., Δν(7) and Δν(13)) compared to theoretical values according to our bi-phase model. The argument of a gradient in nr over the headgroup/water interface is supported by the fact that only in changing nr and ɛ into values of bulk water (nr = 1.33 and ɛ = 80) and leaving the other parameters given in Fig. 7 unaltered, is the observed Stokes shift for the AEDANS label at the expected water-exposed C-terminus (Δν(49) = 7550 cm−1 for DOPC/DOPG) close to the theoretical Stokes shift (Δνwater = 7547 cm−1).
By using Eqs. 1–3, it is possible to calculate the profile of the dielectric constant ɛ from that of Δν within the apolar part of the phospholipid bilayer, i.e., where our bi-phase model is valid. For this we make use of the fact that for hexane, with ɛ = 1.89 and nr = 1.372, according to Eq. 3, Δf ≈ 0. Then in Eq. 1 C = Δνhexane = 5344 cm−1. Also we make use of averages of published values for dansyl compounds similar to AEDANS, giving for the change in dipole upon excitation Δμ = μGS − μES = 5.63 D and for the Onsager radius a = 0.36 nm (Ren et al., 1999). A refractive index of 1.4 to 1.45 is often used for bilayers in theoretical calculations (Koppaka and Axelsen, 2001; Binder, 2003; Fernandes et al., 2003). However, a value of 1.5 has recently been obtained experimentally (Salomon et al., 2000). Here we will use the latter value to convert Δν to ɛ. The result of this conversion is shown in Fig. 7 as well.
Interestingly, whereas the Δν profiles are almost linear with a convex shape near the acyl-chain/glycerol backbone interface, the profile of ɛ has a concave shape. This shape of the ɛ-profile is in agreement with what one would intuitively expect on the basis of the charge density profile of a phosphatidylcholine bilayer (White and Wimley, 1998): a relatively low polarity in the center of the hydrophobic bilayer part (given by ɛc), and a sigmoidal transition over the acyl-chain/glycerol backbone interface (given by ɛi) to a higher polarity in the polar headgroup region. There is only a small difference in the Δν profile for bilayers of 18:1 PC as compared to DOPC/DOPG. For the ɛ-profile a small difference shows up at the acyl-chain/glycerol backbone interface, the ɛ-values being larger for the DOPC/DOPG system, as would be expected based on the higher polarity of its headgroup region. It should be noted that the plots in Fig. 7 are limited to the distance region covering the hydrophobic core of the bilayer, which only reflects the first phase of the sigmoidal transition.
Values for ɛ at specific positions in the bilayer are collected in Table 4. For all PC systems studied, the value of the dielectric constant in the center of the bilayer (ɛc) is fairly constant, giving an average value of 4.0. A similar value (3.7) is found in the DOPC/DOPG system, indicating that for the lipid systems studied, the physical properties of the headgroup region and bilayer thickness do not strongly affect the dielectric constant in the bilayer center. These values are in agreement with values of 2–3 that are often reported in the literature (Elston et al., 1998; White and Wimley, 1998; Bechor and Ben-Tal, 2001). The average value of the dielectric constant at the acyl-chain/glycerol backbone interface (ɛi) turns out to be 12.4 for the PC systems and 18.6 for the DOPC/DOPG system. These values are in agreement with reported values of 10 and 30 for ester group region and headgroup/water interface, respectively (Petrov, 2001).
TABLE 4.
Calculated values of the dielectric constant ɛ at the center of the bilayer (ɛc at d = 0) and at the acyl-chain/glycerol backbone interface (ɛi at d = ½dh) for various lipid systems of different headgroup and chain lengths using the parameter values given in Fig. 7
| Lipid | ɛc (±0.1)* | ɛi (±0.5)* |
|---|---|---|
| 20:1 PC | 3.9 | 12.0 |
| 18:1 PC | 3.7 | 14.0 |
| 16:1 PC | 4.2 | 11.7 |
| 14:1 PC | 4.1 | 11.8 |
| Average | 4.0 | 12.4 |
| DOPC/DOPG | 3.7 | 18.6 |
Errors follow from uncertainty in R (6.5 Å ≤ R ≤ 9.5 Å).
CONCLUSIONS
In this article we have shown that site-directed fluorescence labeling of a helical transmembrane protein in combination with a quantitative formalism provides detailed information about the protein topology: the relative position of the protein in the bilayer as well as its tilt and orientation can be obtained and studied under various conditions. In addition, our analysis offers a detailed picture of the dielectric constant profile of the hydrophobic domain of the bilayer. The tilt angle of the transmembrane α-helix of the M13 coat protein increases with decreasing hydrophobic thickness of the bilayer, showing a mechanism for the protein to accommodate to mismatch situations. The consistency of the fits to our data indicates that the applied cysteine-scanning mutagenesis method with AEDANS labeling in combination with a reasonably large library of mutants offers a reliable description of the protein as well as the bilayer. Although the mutagenesis in combination with chemical modification by labeling will have effects on the protein-lipid interaction, in most cases the consequences are probably small and do not affect the overall conclusions about the protein topology. Also the spacer link between the protein and AEDANS label is long enough to monitor the local polarity of the lipid environment and not that of the amino-acid residues of the protein, and short enough to have the topology of the protein imposing on the fluorescence properties of the AEDANS label. In conclusion, site-directed fluorescence labeling offers a powerful tool to determine the topology of proteins in model membranes. The wealth of structure information that comes out, although of a low resolution, will enable geometry constraints to future molecular modeling and molecular dynamics studies.
References
- Bechor, D., and N. Ben-Tal. 2001. Implicit solvent model studies of the interactions of the influenza hemagglutin fusion peptide with lipid bilayers. Biophys. J. 80:643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, H. 2003. The molecular architecture of lipid membranes—new insights from hydration-tuning infrared linear dichroism spectroscopy. Applied Spectrosc. Rev. 38:15–69. [Google Scholar]
- Dumas, F., M. C. Lebrun, and J. F. Tocanne. 1999. Is the protein/lipid hydrophobic matching principle relevant to membrane organization and functions? FEBS Lett. 458:271–277. [DOI] [PubMed] [Google Scholar]
- Elston, T., H. Wang, and G. Oster. 1998. Energy transduction in ATP synthase. Lett. Nature. 391:510–513. [DOI] [PubMed] [Google Scholar]
- Fernandes, F., L. M. S. Loura, R. B. M. Koehorst, R. B. Spruijt, M. A. Hemminga, and M. Prieto. 2004. Quantification of protein-lipid selectivity using FRET. Application to the M13 major coat protein. Biophys. J. 87:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, F., L. M. S. Loura, M. Prieto, R. B. M. Koehorst, R. B. Spruijt, and M. A. Hemminga. 2003. Dependence of M13 major coat protein oligomerization and lateral segregation on bilayer composition. Biophys. J. 85:2430–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flewelling, R. F., and W. L. Hubbell. 1986. The membrane dipole potential in a total membrane potential model. Biophys. J. 49:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz, C., G. Grobner, and A. Watts. 2000. Structural and orientational information of the membrane embedded M13 coat protein by C-13-MAS NMR spectroscopy. Biochim. Biophys. Acta. 1463:151–161. [DOI] [PubMed] [Google Scholar]
- Koppaka, V., and P. H. Axelsen. 2001. Evanescent electric field amplitudes in thin lipid films for internal reflection infrared spectroscopy. Langmuir. 17:6309–6316. [Google Scholar]
- Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy, 2nd Ed. Kluwer Academic/Plenum Publishers, New York.
- Marassi, F. M., and S. Opella. 2003. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 12:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, D. 2001. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 89:7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, A. B., R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga. 2000. Membrane assembly of the bacteriophage Pf3 major coat protein. Biochemistry. 39:6157–6163. [DOI] [PubMed] [Google Scholar]
- Meijer, A. B., R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga. 2001a. Configurations of the N-terminal amphipathic domain of the membrane-bound M13 major coat protein. Biochemistry. 40:5081–5086. [DOI] [PubMed] [Google Scholar]
- Meijer, A. B., R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga. 2001b. Membrane-anchoring interactions of M13 major coat protein. Biochemistry. 40:8815–8820. [DOI] [PubMed] [Google Scholar]
- Nazarov, P. V., V. V. Apanasovich, V. M. Lutkovski, M. M. Yatskou, R. B. M. Koehorst, and M. A. Hemminga. 2004. Artificial neural network modification of simulation-based fitting: application to a protein-lipid system. J. Chem. Inf. Comput. Sci. 44:568–574. [DOI] [PubMed] [Google Scholar]
- Petrov, A. G. 2001. Flexoelectricity of model and living membranes. Biochim. Biophys. Acta. 1561:1–25. [DOI] [PubMed] [Google Scholar]
- Ren, B., F. Gao, Z. Tong, and Y. Yan. 1999. Solvent polarity scale on the fluorescence spectra of a dansyl monomer copolymerizable in aqueous media. Chem. Phys. Lett. 307:55–61. [Google Scholar]
- Ridder, A., W. van de Hoef, J. Stam, A. Kuhn, B. de Kruijff, and J. A. Killian. 2002. Importance of hydrophobic matching for spontaneous insertion of a single-spanning membrane protein. Biochemistry. 41:4946–4952. [DOI] [PubMed] [Google Scholar]
- Salomon, Z., G. Lindblom, L. Rilfors, K. Linde, and G. Tollin. 2000. Interaction of phosphatidylserine synthase from E. coli with lipid bilayers: coupled plasmon-waveguide resonance spectroscopy studies. Biophys. J. 78:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt, R. B., A. B. Meijer, C. J. A. M. Wolfs, M. A. Hemminga. 2000. Localization and rearrangement modulation of the N-terminal arm of the membrane-bound major coat protein of bacteriophage M13. Biochim. Biophys. Acta. 1508:311–323. [DOI] [PubMed] [Google Scholar]
- Spruijt, R. B., C. J. A. M. Wolfs, and M. A. Hemminga. 1989. Aggregation-related conformational change of the membrane-associated coat protein of bacteriophage M13. Biochemistry. 28:9158–9165. [DOI] [PubMed] [Google Scholar]
- Spruijt, R. B., C. J. A. M. Wolfs, J. W. G. Verver, and M. A. Hemminga. 1996. Accessibility and environment probing using cysteine residues introduced along the putative transmembrane domain of the major coat protein of bacteriophage M13. Biochemistry. 35:10383–10391. [DOI] [PubMed] [Google Scholar]
- Stopar, D., R. B. Spruijt, C. J. A. M. Wolfs, and M. A. Hemminga. 2003. Protein-lipid interactions of bacteriophage M13 major coat protein. Biochim. Biophys. Acta. 1611:5–15. [DOI] [PubMed] [Google Scholar]
- Strandberg, E., S. Morein, D. T. S. Rijkers, R. M. J. Liskamp, P. C. A. van der Wel, and J. A. Killian. 2002. Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 41:7190–7198. [DOI] [PubMed] [Google Scholar]
- Torres, J., T. J. Stevens, and M. Samso. 2003. Membrane proteins: the “Wild West” of structural biology. Trends Biochem. Sci. 28:137–144. [DOI] [PubMed] [Google Scholar]
- White, S. H., and W. C. Wimley. 1998. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1376:339–352. [DOI] [PubMed] [Google Scholar]









