Abstract
The fungal class I hydrophobin SC3 self-assembles into an amphipathic membrane at hydrophilic-hydrophobic interfaces such as the water-air and water-Teflon interface. During self-assembly, the water-soluble state of SC3 proceeds via the intermediate α-helical state to the stable end form called the β-sheet state. Self-assembly of the hydrophobin at the Teflon surface is arrested in the α-helical state. The β-sheet state can be induced at elevated temperature in the presence of detergent. The structural changes of SC3 were monitored by various mass spectrometry techniques. We show that the so-called second loop of SC3 (C39–S72) has a high affinity for Teflon. Binding of this part of SC3 to Teflon was accompanied by the formation of α-helical structure and resulted in low solvent accessibility. The solvent-protected region of the second loop extended upon conversion to the β-sheet state. In contrast, the C-terminal part of SC3 became more exposed to the solvent. The results indicate that the second loop of class I hydrophobins plays a pivotal role in self-assembly at the hydrophilic-hydrophobic interface. Of interest, this loop is much smaller in case of class II hydrophobins, which may explain the differences in their assembly.
INTRODUCTION
Class I hydrophobins are a family of small secreted fungal proteins. They have eight conserved cysteine residues and share similar hydropathy patterns. Yet, their amino acid sequences are diverse. Class I hydrophobins fulfill a wide variety of functions in fungal growth and development (Wessels, 1997; Wösten and Wessels, 1997; Wösten, 2001). For instance, they are involved in emergence of aerial structures such as fruiting bodies, make aerial structures hydrophobic, and attach fungal hyphae to hydrophobic surfaces. The class I hydrophobins self-assemble at hydrophilic/hydrophobic interfaces (e.g., those between water and air, water and oil, and water and a hydrophobic solid) into a highly stable amphipathic membrane. This membrane has an ultrastructure consisting of a mosaic of 10-nm-wide parallel rodlets (Wösten et al., 1993, 1994a,b,c). The rodlets have an amyloid-like nature. For instance, they bind the dyes Thioflavine T and Congo red (Wösten and de Vocht, 2000; Mackay et al., 2001; Butko et al., 2001). Due to their remarkable surface activity hydrophobins have potential industrial and medical applications (Wessels, 1997; Scholtmeijer et al., 2001). For instance, they can be used for surface patterning with nanometer accuracy, immobilization of antibodies in biosensors, and drug delivery using vesicles stabilized by hydrophobin.
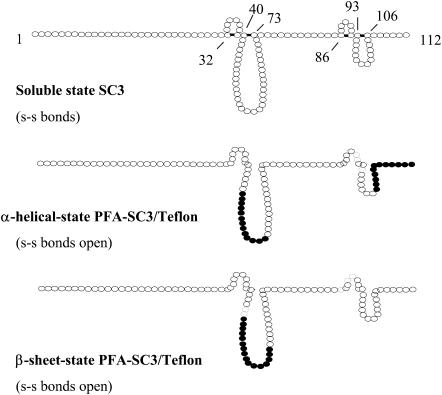
The SC3 hydrophobin of Schizophyllum commune is the best-studied hydrophobin. Its eight cysteine residues are assumed to form four disulfide bridges (de Vries et al., 1993), dividing the molecule into four loops of 6, 32, 5, and 12 amino acids (see Fig. 8). The first loop is preceded by an N-terminal sequence of 29 amino acids, whereas the third loop is preceded by 12 amino acids. The N-terminal region is modified with 16–22 O-linked mannose residues (de Vocht et al., 1998) that are exposed at the hydrophilic side of the membrane after self-assembly (Wösten et al., 1994b; de Vocht et al., 1998). The C-terminal region only has six amino acids. The disulfide bridges in SC3 prevent self-assembly in an aqueous environment (i.e., in the absence of a hydrophilic-hydrophobic interface) (de Vocht et al., 2000). When the cysteine residues were reduced and alkylated with iodoacetamide, SC3 readily aggregated in water. On the other hand, SC3 alkylated with iodoacetic acid, introducing eight negative charges, remained soluble. The completely unfolded protein, however, refolded at a Teflon surface into a structure similar to that observed for SC3 at this interface.
FIGURE 8.
Structural changes of SC3 during self-assembly are accompanied with changes in the solvent exposure of residues in the second loop and C-terminal part, as was determined by deuterium exchange experiments. The highly H/D exchange-protected regions are indicated as • (k > 0.1 min−1), whereas ○ indicates regions that are less exchange protected.
Self-assembly of SC3 is accompanied by conformational changes (de Vocht et al., 1998, 2002; Wang et al., 2002). At the water-air interface, soluble state SC3 proceeds via an intermediate, called the α-helical state, to the stable end form called the β-sheet state. Initially, this state has no clear ultrastructure (β-sheet I state) but after a few hours the typical rodlets have been formed (β-sheet II state). SC3 is arrested at the water-Teflon interface in the intermediate α-helical state (de Vocht et al., 1998). By treating with detergent at elevated temperature, the protein is induced to adopt the β-sheet end state.
The structure/function relationships of class I hydrophobins are poorly understood. The soluble-state structure of the class I hydrophobin EAS from Neurospora crassa has been studied by NMR. The protein was found to be largely unstructured, except for a small core region composed of three antiparallel β-strands, which are probably stabilized by the four disulfide bridges (Mackay et al., 2001). Folding of a derivative of SC3, called TrSC3 (Scholtmeijer et al., 2002), was studied by molecular dynamics simulations (MD) (Zangi et al., 2002). TrSC3 that lacks 25 N-terminal amino acids of SC3 and that is not glycosylated was found to be a largely unstructured globular protein in solution. It underwent a rapid disorder to order folding process at the water/hexane interface, resulting in an elongated planar structure with extensive β-sheet. The formation of β-sheet at the interface was a dynamic process starting with a two-stranded β-sheet element to which additional β-sheet strands were added from the fourth predicted loop.
Secondary structure predictions suggest that the region T52–S63 in the predicted second loop has a high tendency to form an amphipathic helix. The hydrophobic face of this helix was suggested to bind to hydrophobic solid surfaces (de Vocht et al., 1998). We here show that this part indeed strongly adsorbs to Teflon, which is accompanied by α-helix formation and a reduced accessibility to the solvent. On the other hand, the C-terminal part becomes exposed to the solvent during self-assembly.
MATERIALS AND METHODS
Materials
Colloidal Teflon (average size of the beads is ∼150 nm) was kindly donated by DuPont de Nemours BV (The Netherlands). Endoproteinase Asp-N and pepsin were purchased from Roche Applied Science (Penzberg, Germany) and Boehringer Mannheim (Mannheim, Germany), respectively. Immobilized pepsin (cross-linked to 6% agarose, 2–3 mg of pepsin per ml of gel) was obtained from Pierce (Rockford, IL), whereas α-cyano-4-hydroxycinnamic acid (97%) and deuterium oxide (99.9 atom %D) were purchased from Sigma-Aldrich (Munich, Germany). Hydrogen peroxide (30%) and trifluoroacetic acid (TFA) were obtained from Merck (Darmstadt, Germany). All other chemicals and reagents used were of the highest grade commercially available.
Purification of SC3 and performic acid oxidation
SC3 was purified as described (Wang et al., 2002) and oxidized with performic acid (PFA) by adding 0.5 ml chilled PFA (50 μl 30% H2O2, 450 μl formic acid incubated for 2 h at room temperature) to 1 mg freeze-dried protein. After incubation for 2 h in the dark at 0°C, the oxidized protein was separated from the PFA by loading the reaction mixture onto a water equilibrated PD-10 desalting column. PFA-SC3 was collected by eluting the column with water. The oxidized hydrophobin was lyophilized and stored at room temperature until use.
SC3 and PFA-SC3 in the different conformational changes
Soluble-state SC3 (100 μg ml−1) was prepared by TFA treatment of lyophilized protein (Wösten et al., 1993), whereas soluble state PFA-SC3 was obtained by dissolving directly in water or buffer. To obtain the α-helical-state structure of SC3 and PFA-SC3, an amount of colloidal Teflon was added corresponding to a surface 10 times larger than needed to adsorb all protein. The amount of colloidal Teflon was calculated based on the fact that 1.5 mg SC3 covers 1 m2 of Teflon surface (Wösten et al., 1994a). To obtain the β-sheet-state structure, solutions containing Teflon-bound α-helical-state hydrophobin were heated at 65°C followed by the addition of Tween 80 to a final concentration of 0.1%. To separate bound SC3 or PFA-SC3 from hydrophobin occurring free in solution, Teflon spheres were centrifuged at 8000 g for 10 min and resuspended in water or buffer.
Asp-N and pepsin digestion of PFA-SC3
Asp-N (2 μg) dissolved in 30 μl of distilled water was gently mixed with 500 μl PFA-SC3 (2 mg ml−1) dissolved in 50 mM sodium phosphate, pH 7.5. PFA-SC3 was digested for 18 h at 37°C. The reaction was stopped by freezing the solution at −20°C. To desalt the digestion mixture, the ZipTipC18 (Millipore) procedure was used according to the instructions of the manufacturer. Purified samples in 50% acetronitrile/0.1% TFA were either subjected to MALDI-TOF analysis or frozen at −20°C.
Pepsin digestion was carried out at an enzyme to substrate ratio of 1:1. Before use, 0.5 ml immobilized pepsin slurry was washed twice with 1 ml of chilled 0.1% TFA by vortexing and subsequently centrifuging for 2 min at 7000 g. Lyophilized PFA-SC3 (0.5 mg) dissolved in 0.5 ml chilled 0.1% TFA was added to the immobilized pepsin. Digestion was carried out for 6 min on ice with occasional mixing. The immobilized pepsin was removed by centrifugation for 1 min at 14,000 g at 4°C.
Binding of peptides to colloidal Teflon
Asp-N (250 μl) digestion mixture was mixed with colloidal Teflon as described above. Teflon spheres were either centrifuged or not at 8000 g for 10 min, after which the pellet was resuspended in distilled water. For MALDI-TOF analysis, samples were lyophilized, extracted with TFA (Wösten et al., 1993), dried with a flow of nitrogen gas, and then dissolved in 10 μl of distilled water.
H/D exchange experiments with SC3 and PFA-SC3 in different conformational states
Soluble-state SC3 (10 mg ml−1) in 50 mM sodium phosphate, pH 6.8, was diluted 20-fold with 50 mM ammonium phosphate/D2O, pD 7.5. Samples of 50 μl were taken at different time points during incubation at room temperature and placed on ice. Chilled 5% TFA/D2O (4 μl) was added to quench the deuterium in-exchange (final pH ± 2). This was followed by the addition of 100 μl of immobilized pepsin slurry. Preparation of the slurry and pepsin digestion were as described above.
α-Helical-state and β-sheet-state PFA-SC3 were obtained as described above. After incubation for 10 min at room temperature, the colloidal Teflon was pelleted at 8000 g for 10 min and quickly suspended in 600 μl 50 mM ammonium phosphate/D2O, pD 7.5. The mixture was incubated at room temperature with occasional mixing. Samples were taken as described for soluble state hydrophobin, followed by the addition of 10 μl of 5 mg ml−1 pepsin solution in chilled 0.1% TFA.
Undeuterated protein (0% reference) was prepared by diluting the SC3 or PFA-SC3 solution 20-fold with 50 mM ammonium phosphate/H2O, pH 7.5. The completely deuterated protein (100% reference) was prepared by deuterating PFA-SC3 for 24 h at room temperature.
MALDI-TOF mass spectroscopy
Stainless steel or Teflon targets were used (Yuan and Desiderio, 2002). To prepare the Teflon target, 2 × 30 mm squares of Teflon tape (Aldrich) were flattened on the stainless-steel target and fixed in position with tape at each end. Mass spectroscopy with the stainless steel target was done as follows. Asp-N digestion mixture (10 μl) with or without ZipTipC18 desalting or 10 μl of TFA extracted peptides from colloidal Teflon were mixed with an equal volume of 10 mg ml−1 α-cyano-4-hydroxycinnamic acid in 50% acetronitrile, 0.1% TFA (v/v). Usually, the samples were diluted fivefold to achieve MS spectra with the highest quality. Aliquots of 1.5 μl were applied on the target and allowed to air dry. In case of the Teflon targets, 2 μl of digestion mixture was applied. After a few minutes at room temperature the fluid was removed and the target dried in the air. This was either followed or not with three washes with 2 μl of distilled water. Finally, 1.5 μl of MALDI matrix was added on top and allowed to dry (see above). MALDI-TOF mass spectra were recorded with a Micromass Tofspec E MALDI time-of-flight mass spectrometer (Waters, Milford, MA) operated in the reflectron mode in case of the stainless steel target and the linear mode in case of the Teflon target. Spectra were calibrated externally.
LC/MS analysis of peptic fragments of PFA-SC3
Aliquots of 20 μl were analyzed on a PE-Sciex API3000 LC-ESI triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA) using a C18 column (Xterra MS, 3.0 × 150 mm, Waters) at 200 μl min−1 flow rate and a gradient elution with 0.1% formic acid in water as solvent A and acetonitrile containing 0.1% formic acid as solvent B. After washing with 10% B for 5 min, the peptides were eluted with a linear gradient from 10 to 40% B in 35 min, followed by a gradient from 40 to 95% B in 10 min. The effluent from the column was split to reduce the flow rate before being introduced into the mass spectrometer. The eluted peptides showing significant intensities were selected and analyzed using MS/MS. Sequence information was obtained based on the product ions generated.
To determine the deuterium content in a peptide a faster HPLC elution profile was used. Samples (50 μl) frozen in liquid nitrogen were quickly thawed at 30°C and loaded with a 20-μl injection loop. Desalting at 10% B was done in 3 min, the linear gradient from 10 to 40% B in 5 min, and that of 40–95% B in 2 min. The HPLC injector and the column were submerged in an ice/water bath, and the transfer syringe was rinsed with D2O and precooled on ice before use.
Data analysis of H/D exchange data
To obtain an average molecular weight of a peptide, the data were processed by centroiding an isotopic distribution corresponding to the +1 charge state of each peptide. Corrections were made for deuterium back-exchange during analysis as reported (Zhang and Smith, 1993). With each set of samples, an undeuterated (0% reference) control and a completely deuterated (100% reference) control were also analyzed. Equation 1 was used to apply the correction, where D is the deuterium content of the peptide, and m, m0%, and m100% are the average molecular weights of the same peptide in the partially deuterated, the undeuterated, and the completely deuterated form, respectively, and N is the number of peptide amide hydrogens.
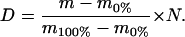 |
(1) |
Deuterium levels (D) were then plotted versus the exchange time (t) and fitted with first-order rate expressions with maximally three exponentials (Eq. 2), where N is the same as in Eq. 1, and ki is the hydrogen deuterium-exchange rate constant for each peptide linkage.
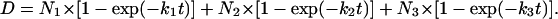 |
(2) |
Back-exchange during the experimental procedure was kept below 20%.
Circular dichroism spectroscopy
CD spectra of SC3 were recorded from 190 to 250 nm on an Aviv 62A DS CD spectrometer, using a 1-mm quartz cuvette. The temperature was kept at 4°C and the sample compartment was continuously flushed with nitrogen gas. The final spectra were obtained by averaging five scans, using a bandwidth of 1 nm, a step width of 1 nm, and a 5-s averaging per point. The spectra were then corrected for the background signal using a reference solution without the protein.
Peptide synthesis
The 16-amino-acid peptide (Ac-Ser-Ser-Ser-Pro-Val-Thr-Ala-Leu-Leu-Gly-Leu-Leu-Gly-Ile-Val-Leu-NH2) of SC3 was synthesized as follows. Individual N-acetyl peptide-amide derivatives were assembled on an automatic ABI 433A Peptide Synthesizer using the ABI FastMoc 0.25 mmol protocols (Fields et al., 1991), except that the coupling time was 45 min instead of 20 min. Fmoc-amino-acid derivatives, activated in situ using HBTU/HOBt and DiPEA in NMP, were used in the coupling steps. The peptides were deprotected and cleaved from the resin by treatment with TFA/TIS/H2O (95/2.5/2.5, v/v/v) at room temperature for 2 h. After this, the peptides were precipitated by MTBE/n-hexane (1/1, v/v) solvent mixture. Finally, the pellet was dissolved in MeCN/H2O (1/1, v/v) and lyophilized to obtain the crude peptides. The crude peptides were purified by preparative C18 reversed-phase high-performance liquid chromatography. The identity of the peptide was confirmed by MALDI-TOF MS. Similar to the treatment of SC3, the lyophilized peptide was first treated with pure TFA and dried with a flow of nitrogen gas. The dried material was then dissolved in water before being analyzed.
RESULTS
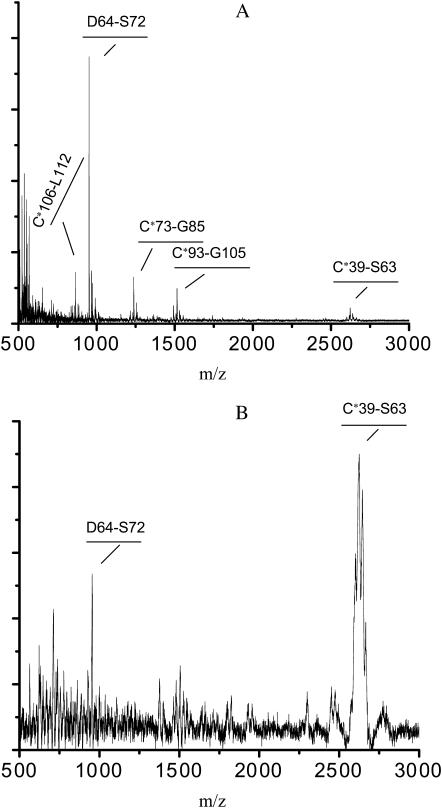
MALDI-TOF mass spectrometry analysis of Teflon-bound peptides after Asp-N digestion of performic-acid-oxidized SC3 (PFA-SC3)
Performic acid (PFA) converts cystine to cysteic acid with a yield of ∼90% (Sun and Smith, 1988; Chowdhury et al., 1995). SC3 treated with PFA (PFA-SC3) was found to be totally unstructured in solution; it did not assemble or aggregate (see next section). The eight cysteic acids and the aspartic acid in the second loop of PFA-SC3 served as target sites for Asp-N digestion. SDS-PAGE showed that digestion was complete after overnight incubation at 37°C (not shown). The resulting peptides were assigned to different parts of SC3 by MALDI-TOF analysis (Figs. 1 and 2, Table 1).
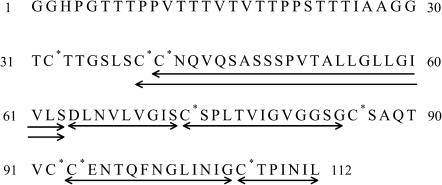
FIGURE 1.
Amino acid sequence of PFA-SC3. The asterisks indicate the cysteines oxidized upon PFA treatment. Arrows indicate the peptides generated by endoproteinase Asp-N digestion that could be assigned by MALDI-TOF analysis.
FIGURE 2.
MALDI-TOF analysis of the Asp-N digestion mixture of PFA-SC3. Sample was concentrated and desalted by using ZipTipC18, and detected with reflectron mode (A). The digestion mixture was dried on a Teflon target, which was washed with water and then overlaid with matrix compound. MALDI-TOF analysis was performed in linear mode (B).
TABLE 1.
MALDI-TOF determination of the peptides generated by endoproteinase Asp-N
| Peptide | Theoretical mass (Da) | Measured m/z | Number of clustered sodium | Theoretical m/z | |
|---|---|---|---|---|---|
| 1 | C*39-S63 | 2555.3 | 2622.7 | 3 | 2622.3 |
| 2 | D64-S72 | 928.5 | 951.4 | 1 | 951.5 |
| 3 | C*73-G85 | 1193.6 | 1238.4 | 2 | 1238.6 |
| 4 | C*93-G105 | 1469.7 | 1514.4 | 2 | 1514.7 |
| 5 | C*106-L112 | 820.4 | 865.3 | 2 | 865.4 |
The asterisks indicate the oxidized cysteines upon PFA treatment.
All the peptide masses listed are monoisotopic masses.
Sodium adducts were identified by the isotope clusters that were separated by 22 mass units. A large part of the SC3 sequence was covered by the assigned peptides. The N-terminal glycosylated domain and the first and third loop were missing, probably due to poor ionization of the fragments.
Asp-N digested samples of PFA-SC3 were dried on Teflon targets for MALDI-TOF analysis. All the peptides shown in Figs. 1 and 2 A could be detected when the targets were not washed. However, after washing only the peptides D64–S72 (951 mol wt) and C39–S63 (2623 mol wt) were observed (Fig. 2 B). These peptides correspond to the second and first half of the second loop of SC3, respectively, indicating that this loop binds strongly to a hydrophobic surface.
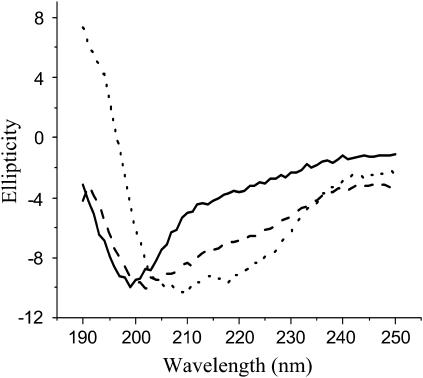
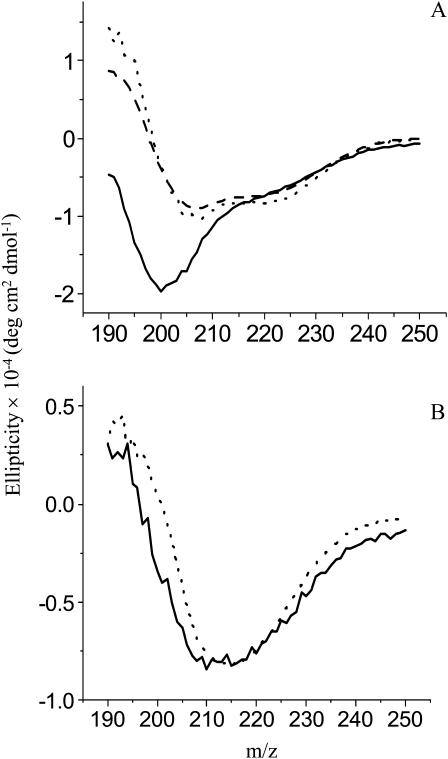
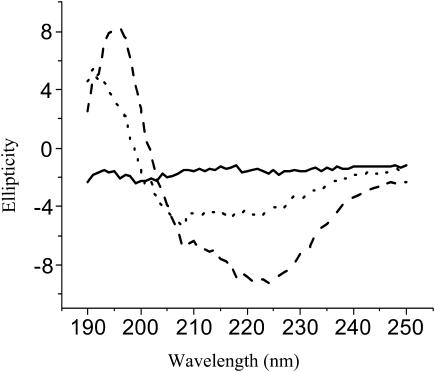
Colloidal Teflon was added to the digestion mixture of Asp-N-treated PFA-SC3. After incubation for 0.5 h, the beads were pelleted by centrifugation. The peptides contained in the supernatant, having a low binding affinity for Teflon, showed a typical CD spectrum for unstructured proteins/peptides with an ellipticity minimum at ∼200 nm (solid line in Fig. 3). The CD spectrum of peptides with high binding affinity for Teflon (i.e., peptides D64–S72 and C39–S63 as shown by mass spectroscopy) was determined by resuspending the pelleted Teflon beads in water. The CD spectrum was indicative for protein with α-helix (dotted line in Fig. 3). Heating this sample to 65°C did not affect the CD spectrum, but subsequent addition of detergent removed some of the peptides from the Teflon surface. The dissociated peptides showed a spectrum more typical for unstructured peptide (dashed line in Fig. 3). From these results it is concluded that the predicted second loop of SC3 binds strongly to a hydrophobic surface in an α-helical state. However, these peptides partly dissociate by treating with hot detergent.
FIGURE 3.
Normalized CD spectra of peptides of SC3 with low and high binding affinities to colloidal Teflon. Supernatant after removal of the Teflon-bound species by centrifugation (solid line). Peptides bound to Teflon beads before (dotted line) and after (dashed line) heating at 65°C in the presence of 0.1% Tween 80.
Probing the various states of SC3 by amide hydrogen/deuterium exchange
PFA-SC3, which has eight negative charges, did not assemble in solution. Its CD spectrum was typical for that of an unfolded peptide (solid line in Fig. 4 A). Upon binding to colloidal Teflon, PFA-SC3 adopted an α-helical-like secondary structure (dotted line in Fig. 4 A). The spectrum was similar to that of native SC3 adsorbed to Teflon (dashed line in Fig. 4 A). Both SC3 and PFA-SC3 bound to Teflon converted to the β-sheet state upon treatment with detergent at 65°C (Fig. 4 B). From these data it is concluded that PFA treatment does not affect structural changes of SC3.
FIGURE 4.
CD spectra of PFA-SC3 upon binding to a Teflon surface. (A) CD spectra of PFA-SC3 in solution (solid line) and after binding to colloidal Teflon (dotted line), SC3 bound to colloidal Teflon serving as a control (dashed line). (B) CD spectra of Teflon-bound PFA-SC3 (solid line) and SC3 (dashed line) heated at 65°C in the presence of 0.1% Tween 80.
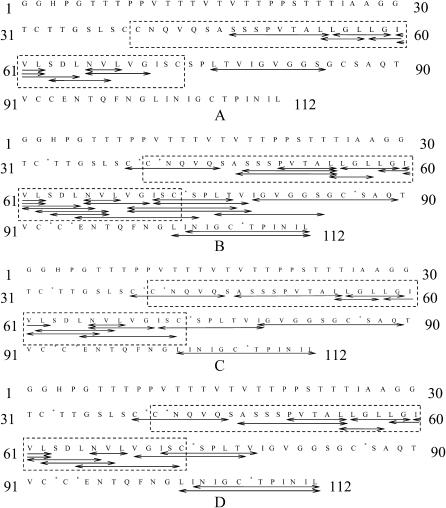
When soluble-state SC3 was digested with pepsin for 30 min at 0°C, only the fragments corresponding to the predicted second loop and the fragment L76–S84 could be detected by LC/MS (Fig. 5 A, Table 2). The rest of the molecule might be resistant to pepsin digestion and/or difficult to ionize (similar to soluble-state SC3 without digestion). LC/MS of soluble-state PFA-SC3 treated with pepsin and the following MS/MS analysis of the peptides resulted in a much higher sequence coverage (Fig. 5 B). In contrast, pepsin digestion of Teflon-bound PFA-SC3 (either in α-helical or β-sheet state) was less efficient than soluble-state SC3. This was especially the case for the predicted second loop (C39–C73), yielding fewer peptic peptides although the sequence coverage remained the same (compare Fig. 5, B–D). These results may be explained by shielding of pepsin cleavage sites when the protein is bound to a solid surface. Similarly, peptide I79–Q89 was missing when PFA-SC3 was bound to Teflon in the β-sheet. On the other hand, the amount of peptides L101–L112 and I102–L112, corresponding to the C-terminal end of SC3, largely increased when the protein was converted from α-helical state into β-sheet state. This indicates that the C-terminal tail is exposed to the solvent especially when the protein is in the β-sheet state.
FIGURE 5.
Peptides observed by LC/MS of pepsin digests of soluble state SC3 (A), soluble state PFA-SC3 (B), PFA-SC3 in α-helical state (C), and PFA-SC3 in β-sheet state (D). Arrows in the SC3 sequence indicate the assigned peptides. The dotted rectangles highlight the predicted second loop of SC3. The asterisks indicate the oxidized cysteines upon PFA treatment.
TABLE 2.
Distribution of rate constants (k, min−1) for H/D exchange at amide bonds in peptic peptides generated from soluble-state SC3
| Number of amide hydrogens*
|
|||||
|---|---|---|---|---|---|
| Peptide | Sequence | NHs | Fast (k > 1000) | Slow (100 > k > 1) | Very slow (0.1 > k) |
| S47-L54 | SSSPVTAL | 6 | 3.6 | 2.0 | 0.40 |
| L54-L57 | LLGL | 3 | 1.1 | 1.2 | 0.7 |
| L55-G59 | LGLLG | 4 | 1.6 | 1.7 | 0.7 |
| L58-L62 | LGIVL | 4 | 2.6 | 1.3 | 0.1 |
| I60-L62 | IVL | 2 | 1.1 | 0.9 | 0 |
| I60-L65 | IVLSDL | 5 | 4.6 | 0.3 | 0.1 |
| S63-V67 | SDLNV | 4 | 2.8 | 0.7 | 0.5 |
| N66-G70 | NVLVG | 4 | 1.1 | 2.1 | 0.8 |
| N66-L68 | NVL | 2 | 0 | 1.8 | 0.2 |
| L76-S84 | LTVIGVGGS | 8 | 7.4 | 0 | 0.6 |
Soluble-state SC3 and PFA-SC3, and Teflon-bound α-helical-state and β-sheet-state PFA-SC3 were exposed to D2O at neutral pH for periods ranging from 1 min to 48 h. Samples were cooled on ice and quickly digested by pepsin at pH 2. The peptic peptides were analyzed by LC/MS and isotopic distributions were determined. Although MS signals were less intense for the Teflon-bound samples, isotopic distributions were still distinguishable in most cases. The exchange of deuterium into peptide amide bonds results in an increase of the molecular mass (Fig. 6). All the peptides detectable in the H/D exchange experiments showed a single envelope of isotope peaks, indicating that they were structurally homogeneous in the various states. To compare the deuterium contents of the different peptides, an averaged mass was obtained by centroiding the envelope of isotopic peaks.
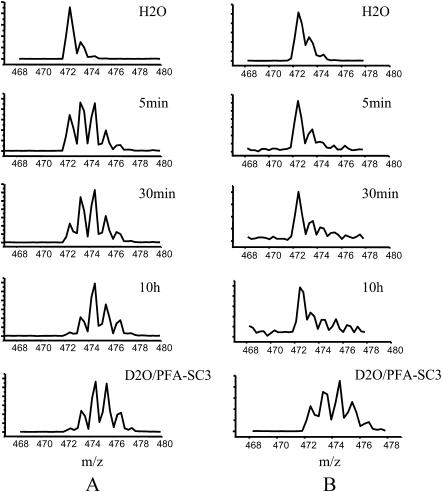
FIGURE 6.
Time-dependent deuterium exchange as detected by the shift of isotopic distributions for peptic fragment L55–G59. Deuterium in-exchange in soluble-state PFA-SC3 (A) and PFA-SC3 bound to a Teflon surface (α-helical state) (B). Complete exchange on PFA-SC3 (indicated as D2O/PFA-SC3) served as a control.
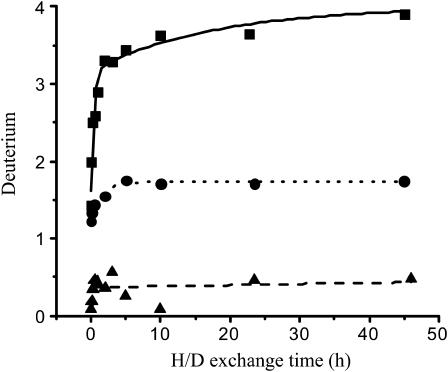
The deuterium levels in the peptic peptides were plotted against the exchange time and the data were fitted with a series of first-order exponential equations. The plot of peptide L55–G59 of SC3 is shown in Fig. 7 as an example. The kinetics of H/D exchange allows classification of exchangeable amide hydrogens in terms of rate constants (Tables 2–4). The second loop and the C73–C86 segment of soluble-state SC3, with the only peptides that could be observed by LC/MS, showed exclusively fast or intermediate exchanged residues. This shows that these regions were readily accessible from the solvent. On the other hand, highly H/D exchange-protected regions could be found in the Teflon-bound α-helical and β-sheet state of SC3, with a remarkable consistency for the first half of the predicted second loop (Fig. 8). The peptic peptides in this region, A46–L54, L54–L57, L55–G59, and L58–L62, showed almost complete protection from deuterium labeling in the Teflon-bound forms. Some other peptic peptides also contained some slowly exchanging residues. However, due to the difficulty of assignment of the residues, they are not indicated in the figure. Once PFA-SC3 was bound to a Teflon surface and converted into the α-helical state, at least 13 residues in the second loop became highly protected against deuterium exchange. The highly protected region in the predicted second loop of α-helical-state PFA-SC3 extended upon conversion into the β-sheet state, indicating that more residues in this region are involved in the structure formation. The peptide L101–L112, which corresponds to the last part of the fourth loop and the whole C-terminal tail, was detectable in both Teflon-bound states. In the α-helical state on Teflon, this peptide showed both intermediate and slow deuterium exchange (Table 3, Fig. 8). Interestingly, the deuterium exchange rates for the same peptide increased dramatically upon the formation of β-sheet-state structure (Table 4). This indicates that, in contrast to the α-helical state on Teflon, the C-terminal end of the molecule in the β-sheet state is largely exposed to the solvent.
FIGURE 7.
Exponential fitting of time-dependent deuterium exchange of peptic peptide L55–G59 of soluble state SC3 (solid line), Teflon-bound α-helical state SC3 (dotted line) and Teflon-bound β-sheet state SC3 (dashed line).
TABLE 3.
Distribution of rate constant (k, min−1) for H/D exchange at amide bonds in peptic peptides generated from Teflon-associated α-helical-state PFA-SC3
| Number of amide hydrogens*
|
|||||
|---|---|---|---|---|---|
| Peptide | Sequence | NHs | Fast (k > 1000) | Slow (100 > k > 1) | Very slow (0.1 > k) |
| C*39-Q44 | C*C*NQVQ | 5 | 1.5 | 2.1 | 1.4 |
| A46-L54 | ASSSPVTAL | 7 | 0.6 | 0.7 | 5.7 |
| L54-L57 | LLGL | 3 | 0 | 0.2 | 2.8 |
| L55-G59 | LGLLG | 4 | 1.3 | 0.3 | 2.4 |
| L58-L62 | LGIVL | 4 | 0.5 | 3.0 | 0.5 |
| V61-N66/L62-V67 | VLSDLN/LSDLNV | 5 | 0 | 3.5 | 1.5 |
| L65-S74 | LNVLVGISC*S | 9 | 3.7 | 0.3 | 5.0 |
| N66-G70 | NVLVG | 4 | 0.3 | 1.3 | 2.4 |
| N66-L68 | NVL | 2 | 0.4 | 1.0 | 0.6 |
| 171-179 | ISC*SPLTVI | 7 | 4.2 | 0.8 | 2.0 |
| 179-Q89 | IGVGGSGC*SAQ | 10 | 0.7 | 1.1 | 8.2 |
| L101-L112 | LINIGC*TPINIL | 10 | 0 | 3.6 | 6.4 |
The asterisks indicate the oxidized cysteines upon PFA treatment.
See Table 2.
TABLE 4.
Distribution of rate constant (k, min−1) for H/D exchange at amide bonds in peptic peptides generated from Teflon-associated β-sheet-state PFA-SC3
| Number of amide hydrogens*
|
|||||
|---|---|---|---|---|---|
| Peptide | Sequence | NHs | Fast (k > 1000) | Slow (100 > k > 1) | Very slow (0.1 > k) |
| C*39-Q44 | C*C*NQVQ | 5 | 0.7 | 1.4 | 2.9 |
| A46-L54 | ASSSPVTAL | 7 | 0.1 | 0.7 | 6.2 |
| P50-L54 | PVTAL | 3 | 0.3 | 0.5 | 2.2 |
| L54-L57 | LLGL | 3 | 0 | 0 | 3.0 |
| L55-G59 | LGLLG | 4 | 0 | 0 | 4.0 |
| L58-L62 | LGIVL | 4 | 1.2 | 06 | 2.2 |
| I60-L62 | IVL | 2 | 0 | 0.3 | 1.7 |
| V61-N66/L62-V67 | VLSDLN/LSDLNV | 5 | 0 | 4.6 | 0.4 |
| L65-S74 | LNVLVGISC*S | 9 | 1.4 | 3.4 | 4.2 |
| N66-L68 | NVL | 2 | 0 | 1.1 | 0.9 |
| V69-T77 | VGISC*SPLT | 7 | 0 | 2.4 | 4.6 |
| 171-V78 | ISC*SPLTV | 6 | 0 | 4.2 | 1.8 |
| L101-L112 | LINIGC*TPINIL | 10 | 5.5 | 4.4 | 0.1 |
| I102-L112 | INIGC*TPINIL | 9 | 4.7 | 4.0 | 0.3 |
The asterisks indicate the oxidized cysteines upon PFA treatment.
See Table 2.
Chemical synthesis of a part of the predicted second loop and its α-helical-state-like structure on the Teflon surface
The 16 amino acid peptide, corresponding to the highly H/D exchange-protected region (S47–L62) in the predicted second loop of SC3 in the Teflon-bound forms (Fig. 8), was synthesized. The peptide was highly water insoluble and at the maximum concentration in solution, the peptide yielded only a very low CD signal (solid line in Fig. 9). However, upon addition of 10% (v/v) colloidal Teflon to the peptide solution, α-helix structure was formed and the CD signal was highly enhanced (dotted line in Fig. 9). In fact, the spectrum of the peptide was similar to that of α-helical-state SC3. When the Teflon beads were spun down and resuspended in the same volume of buffer, the intensity and the shape of the spectrum did not change (data not shown). Thus, the change in conformation was caused by binding of the peptide to the Teflon surface. Treating the peptide bound to Teflon with hot detergent (e.g., 0.1% Tween 80, 65°C) affected neither the binding of the peptide nor its CD spectrum (data not shown).
FIGURE 9.
CD spectra of the chemically synthesized SC3 peptide (S47–L62) after dissolving in 10 mM sodium phosphate, pH 7 (solid line) and after subsequent binding to colloidal Teflon (dotted line) or vortexing (dashed line).
By vortexing an aqueous solution of the peptide for 5 min a large structural change of the peptide was observed. This was accompanied with an increase in intensity of the CD signal (Fig. 9; from solid line to dashed line), indicating that the content of secondary structure elements might have largely increased upon such a treatment. However, the spectrum was different from that of β-sheet-state SC3 as formed at the water-air interface upon vortexing. The latter spectrum has a maximal ellipicity at ∼215 nm.
DISCUSSION
Class I and class II hydrophobins were distinguished on the basis of their solubility characteristics and their hydropathy patterns (Wessels, 1994). Class II hydrophobins also self-assemble at hydrophilic-hydrophobic interfaces but their membranes are less stable (see Wösten and de Vocht, 2000). One of the main differences in the primary structure is that class I hydrophobins have a much larger second loop (17–39 residues) compared to that of class II (11 residues) (Wösten and de Vocht, 2000). Part of the second loop (T52–S63) of the class I hydrophobin SC3 was suggested to have the propensity to form an α-helix structure with hydrophobic residues at one face and hydrophilic or small residues at the other face of the helix. This amphipathic α-helix was proposed to form an anchor that binds the hydrophobin strongly to hydrophobic surfaces (de Vocht et al., 1998). The function of this part would agree with the observation that class II hydrophobins, with a smaller second loop, bind much less to a hydrophobic surface (A. Askolin and H. A. B. Wösten, unpublished data). We here show that the second loop of the SC3 hydrophobin indeed has a strong affinity for Teflon and that it forms α-helix structure upon binding. This was accompanied by a strong decrease in solvent accessibility.
The results presented in this study are based on mass spectroscopy of peptides resulting from Asp-N and pepsin digestion combined with binding experiments to Teflon and deuterium exchange. Only few peptides of soluble state SC3 could be detected by mass spectroscopy, explained by poor proteolytic digestion and/or poor ionization during mass spectroscopy. Therefore, the protein was oxidized with performic acid (PFA-SC3). In contrast to soluble state SC3, PFA-SC3 was completely unstructured in solution. However, PFA-SC3 adopted a similar secondary structure as SC3 (i.e., the intermediate α-helical state) when bound to colloidal Teflon; a stable β-sheet state could also be induced by treating with hot detergent. From these results it was concluded that PFA-SC3 could be used instead of native SC3.
When the Asp-N digestion mixture of PFA-SC3 was incubated with Teflon only peptides contained in the predicted second loop of SC3 were shown to bind. The CD spectrum of bound peptides was indicative for α-helical structure. In agreement, a chemically synthesized peptide corresponding to a large part of the second loop (Ser-47–Leu-62) bound strongly to Teflon, in an α-helix conformation. The result indicates that amino acid residues S47–L62 of SC3 are at least partially responsible for the strong binding of SC3 to the Teflon surface and for the α-helical-state structure of SC3 on this hydrophobic solid. This conclusion was strengthened by H/D exchange experiments that were combined with LC/ESI-MS analysis. All the deuterium exchanged peptic peptides of SC3 showed a single envelope of isotope peaks. Such a single envelope is expected when the sample is structurally homogeneous and when the exchange is uncorrelated with the folding-unfolding process of protein molecules. Bimodal isotope patterns are expected otherwise (Smith et al., 1997; Raza et al., 2000). Although the assignment of individual residues turned out to be difficult due to the lack of enough overlapping peptic peptides, it is clear that most residues in the first half of the predicted second loop underwent very slow H/D exchange once the molecule became structured on a hydrophobic surface (Fig. 8). In contrast, most detectable peptic peptides in soluble-state SC3, such as L58–L62, I60–L62, I60–L65, S63–V67, and L76–S84 (see Table 2), which covered a part of the predicted second loop and the segment behind it (C73–C86) underwent fast H/D exchange, indicating that at least these parts of the molecule lacked a tight core structure. The 13 residues (from A46 to G59) of the predicted second loop undergoing very slow H/D exchange when SC3 adopted α-helical state structure on Teflon extended to 16 residues (from A46 to L62) when the structure was converted to β-sheet state (see Tables 3 and 4). Assuming that the number of protected residues is related to the content of secondary structure elements, these results agree with a change in the SC3 structure, from disordered, to ordered, to highly ordered during self-assembly.
Peptides responsible for the tight binding of β-sheet-state SC3 to Teflon could not be identified. They dissociated from the Teflon during treatment with hot detergent. This contrasts with intact SC3, which tightly binds to Teflon. This apparent discrepancy may be explained with results from a molecular dynamics simulation of SC3 assembling at the water/hexane interface (Zangi et al., 2002). This simulation predicted a long-range β-sheet network throughout the whole protein rather than a single structural element adopting this structure. Moreover, we have previously shown that conversion of SC3 from the α-helix to β-sheet structure on Teflon is accompanied by a major increase in protein-protein interaction as reflected in increased fluorescence resonance energy transfer (Wang et al., 2002). The results indicated that during the conversion SC3 molecules moved toward one another. Although there are obviously enough interactions between the individual SC3 molecules and the Teflon surface to maintain or restore the surface-bound nature of SC3 during these association processes, this is not the case with the isolated second loop peptide.
H/D exchange data showed that the C-terminal tail of SC3 is probably exposed to the solvent when SC3 assembles on a hydrophobic surface and forms a β-sheet-state structure. Previously, it was shown that the N-terminal glycosylation part is also exposed to the solvent after self-assembly whereas the N-terminal end is more buried and probably closer to the Teflon surface (Wösten et al., 1994b; de Vocht et al., 1998; Wang et al., 2002). Gross properties of the hydrophobin were not affected when the N-terminal part was genetically engineered. However, the biophysical properties of the hydrophilic side of the assemblage did change (Scholtmeijer et al., 2002). In fact, a RGD peptide could be exposed at the hydrophilic side after assembly in this way. Similar experiments could be done with the C-terminal part. This should reveal whether self-assembly and surface properties of the hydrophilic part will be affected. Of interest, the C-terminal part of class I hydrophobins is less variable in length compared to that of the N-terminus (i.e., 2–13 amino acids following the last cysteine residue versus 26–85 residues preceding the first cysteine residue) (see Wösten and de Vocht, 2000). This suggests that the C-terminal part of the hydrophobin is perhaps more important for self-assembly than the N-terminal part.
Abbreviations used: MD, molecular dynamics; CD, circular dichroism; TFA, trifluoroacetic acid; IAA, iodoacetic acid; PFA, performic acid; MALDI-TOF, matrix-assisted laser desorption ionization time of flight; ESI-MS, electrospray ionization-mass spectrometry; H/D exchange, hydrogen/deuterium exchange; HBTU, 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate; DiPEA, N,N-diisopropylethylamine; NMP, 1-methyl-2-pyrrolidinone; TIS, triisopropylsilane; MeCN, acetonitrile.
References
- Butko, P., J. P. Buford, J. S. Goodwin, P. A. Stroud, C. L. McCormick, and G. C. Cannon. 2001. Spectroscopic evidence for amyloid-like interfacial self-assembly of hydrophobin SC3. Biochem. Biophys. Res. Commun. 280:212–215. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S. K., J. Eshrghi, H. Wolfe, D. Forde, A. G. Hlavac, and D. Johnston. 1995. Mass spectrometric identification of amino acid transformations during oxidation of peptides and proteins: modifications of methionine and tyrosine. Anal. Chem. 67:390–398. [DOI] [PubMed] [Google Scholar]
- de Vocht, M. L., I. Reviakine, W.-P. Ulrich, W. Bergsma-Schutter, H. A. B. Wösten, H. Vogel, A. Brisson, J. G. H. Wessels, and G. T. Robillard. 2002. Self-assembly of the hydrophobin SC3 proceeds via two structural intermediates. Protein Sci. 11:1199–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vocht, M. L., I. R. Reviakine, H. A. B. Wösten, A. Brisson, J. G. H. Wessels, and G. T. Robillard. 2000. Structural and functional role of the disulfide bridges in the hydrophobin SC3. J. Biol. Chem. 275:28428–28432. [DOI] [PubMed] [Google Scholar]
- de Vocht, M. L., K. Scholtmeijer, E. W. van der Vegte, O. M. H. de Vries, N. Sonveaux, H. A. B. Wösten, J. M. Ruysschaert, G. Hadziioannou, J. G. H. Wessels, and G. T. Robillard. 1998. Structural characterization of the hydrophobin SC3, as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys. J. 74:2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, O. M. H., M. P. Fekkes, H. A. B. Wösten, and J. G. H. Wessels. 1993. Insoluble hydrophobin complexes in the walls of Schizophyllum commune and other filamentous fungi. Arch. Microbiol. 159:330–335. [Google Scholar]
- Fields, C. G., D. H. Lloyd, R. L. Macdonald, K. M. Otteson, and R. L. Noble. 1991. HBTU activation for automated Fmoc solid-phase peptide synthesis. Pept. Res. 4:95–101. [PubMed] [Google Scholar]
- Mackay, J. P., J. M. Matthews, R. D. Winefield, L. G. Mackay, R. G. Haverkamp, and M. D. Templeton. 2001. The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure. 9:83–91. [DOI] [PubMed] [Google Scholar]
- Raza, A. S., K. D. Dharmasiri, and D. L. Smith. 2000. Identification of non-covalent structure in apocytochrome c by hydrogen exchange and mass spectrometry. J. Mass Spectrom. 35:612–617. [DOI] [PubMed] [Google Scholar]
- Scholtmeijer, K., M. I. Janssen, B. Gerssen, M. L. De Vocht, B. M. van Leeuwen, T. G. van Kooten, H. A. Wösten, and J. G. Wessels. 2002. Surface modifications created by using engineered hydrophobins. Appl. Environ. Microbiol. 68:1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtmeijer, K., J. G. H. Wessels, and H. A. B. Wösten. 2001. Fungal hydrophobins in medical and technical applications. Appl. Microbiol. Biotechnol. 56:1–8. [DOI] [PubMed] [Google Scholar]
- Smith, D. L., Y. Deng, and Z. Zhang. 1997. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J. Mass Spectrom. 32:135–146. [DOI] [PubMed] [Google Scholar]
- Sun, Y., and D. L. Smith. 1988. Identification of disulfide-containing peptides by performic acid oxidation and mass spectrometry. Anal. Biochem. 172:130–138. [DOI] [PubMed] [Google Scholar]
- Wang, X., M. L. de Vocht, J. de Jonge, B. Poolman, and G. T. Robillard. 2002. Structural changes and molecular interactions of hydrophobin SC3 in solution and on a hydrophobic surface. Protein Sci. 11:1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, J. G. H. 1994. Developmental regulation of fungal cell wall formation. Annu. Rev. Phytopathol. 32:413–437. [Google Scholar]
- Wessels, J. G. H. 1997. Hydrophobins: proteins that change the nature of a fungal surface. Adv. Microb. Physiol. 38:1–45. [DOI] [PubMed] [Google Scholar]
- Wösten, H. A. B., O. M. H. de Vries, and J. G. H. Wessels. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell. 5:1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H. A. B., F. H. J. Schuren, and J. G. H. Wessels. 1994a. Interfacial self-assembly of a hydrophobin into an amphipathic membrane mediates fungal attachment to hydrophobic surfaces. EMBO J. 13:5848–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H. A. B., O. M. H. de Vries, H. C. van der Mei, H. J. Busscher, and J. G. H. Wessels. 1994b. Atomic composition of the hydrophobic and hydrophilic sides of self-assembled SC3p hydrophobin. J. Bacteriol. 176:7085–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H. A. B., S. A. Asgeirsdottir, J. H. Krook, J. H. H. Drenth, and J. G. H. Wessels. 1994c. The SC3p hydrophobin self-assembles at the surface of aerial hyphae as a protein membrane constituting the hydrophobic rodlet layer. Eur. J. Cell Biol. 63:122–129. [PubMed] [Google Scholar]
- Wösten, H. A. B., and J. G. H. Wessels. 1997. Hydrophobins, from molecular structure to multiple functions in fungal development. Mycoscience. 38:363–374. [Google Scholar]
- Wösten, H. A. B., and M. L. de Vocht. 2000. Hydrophobins, the fungal coat unraveled. Biochim. Biophys. Acta. 1469:79–86. [DOI] [PubMed] [Google Scholar]
- Wösten, H. A. B. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625–646. [DOI] [PubMed] [Google Scholar]
- Yuan, X., and D. M. Desiderio. 2002. Protein identification with Teflon as matrix-assisted laser desorption/ionization sample support. J. Mass Spectrom. 37:512–524. [DOI] [PubMed] [Google Scholar]
- Zangi, R., M. L. de Vocht, G. T. Robillard, and A. E. Mark. 2002. Molecular dynamics study of the folding of hydrophobin SC3 at a hydrophilic/hydrophobic interface. Biophys. J. 83:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and D. L. Smith. 1993. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]