Abstract
The interaction of Phormidium laminosum plastocyanin (PC) with P. laminosum cytochrome f (cyt f) was studied using Brownian dynamics (BD) simulations. Few complexes and a low rate of electron transfer were observed for wild-type PC. Increasing the positive electrostatic field on PC by the addition of a Zn2+ ion in the neighborhood of D44 and D45 on PC (as found in crystal structure of plastocyanin) increased the number of complexes formed and the calculated rates of electron transfer as did PC mutations D44A, D45A, E54A, and E57A. Mutations of charged residues on Phormidium PC and Phormidium cyt f were used to map binding sites on both proteins. In both the presence and absence of the Zn2+ ion, the following residues on PC interact with cyt f: D44, D45, K6, D79, R93, and K100 that lie in a patch just below H92 and Y88 and D10, E17, and E70 located on the upper portion of the PC molecule. In the absence of the Zn2+ ion, K6 and K35 on the top of the PC molecule also interact with cyt f. Cyt f residues involved in binding PC, in the absence of the Zn2+ ion, include E165, D187, and D188 that are located on the small domain of cyt f. The orientation of PC in the complexes was quite random in accordance with NMR results. In the presence of the Zn2+ ion, K53 and E54 in the lower patch of the PC molecule also interact with cyt f and PC interacts with E86, E95, and E123 on the large domain of cyt f. Also, the orientation of PC in the complexes was much more uniform than in the absence of the Zn2+ ion. The difference may be due to both the larger electrostatic field and the greater asymmetry of the charge distribution on PC observed in the presence of the Zn2+ ion. Hydrophobic interactions were also observed suggesting a model of cyt f-PC interactions in which electrostatic forces bring the two molecules together but hydrophobic interactions participate in stabilizing the final electron-transfer-active dock.
INTRODUCTION
Cytochrome f (cyt f) is a member of the cytochrome b6f complex. It accepts electrons from the Rieske FeS protein and passes them to plastocyanin (PC) (Hauska et al., 1983; Cramer et al., 1996). PC is a mobile electron carrier located in the lumen of the thylakoid where it accepts electrons from cyt f and donates them to P700 in Photosystem I (Freeman, 1981; Sykes, 1991; Gross, 1993, 1996; Redinbo et al., 1994; Sigfridsson, 1998; Hope, 2000).
Interactions between cyt f and PC have been intensely studied in higher plants and green algae. Studies using chemical modification (Takenaka and Takabe, 1984; Takabe et al., 1986; Anderson et al., 1987), cross-linking (Morand et al., 1989; Takabe and Ishikawa, 1989; Qin and Kostic, 1993), and mutagenesis (Lee et al., 1995; Kannt et al., 1996; Soriano et al., 1998; Gong et al., 2000) techniques showed that electrostatic forces bring the two molecules together. The specific electrostatic interactions occur between five positively charged residues on cyt f (Gray, 1992; Martinez et al., 1994, 1996) and a series of negatively charged residues surrounding Y83 on PC (Gross, 1996; Redinbo et al., 1993; Guss et al., 1992). NMR studies of complexes formed between spinach PC and turnip cyt f (Ubbink et al., 1998; Ejdeback et al., 2000) confirmed the role of electrostatic interactions and, in addition, showed that hydrophobic interactions between residues surrounding the heme on cyt f and those surrounding H87 on PC were also important for the formation of electron-transfer-active complexes.
In addition, Brownian dynamics (BD) simulations of the docking of higher plant (Pearson and Gross, 1998; Nelson et al., 1999; De Rienzo et al., 2001) and green algal (Gross and Pearson, 2003) PCs with cyt f have demonstrated the role of electrostatic interactions. The complexes formed also showed hydrophobic interactions (Pearson and Gross, 1998; Gross and Pearson, 2003). In conclusion, the experimental and computational results, taken together, lead to a model in which electrostatic forces bring higher plant and algal PCs to a hydrophobic dock on cyt f.
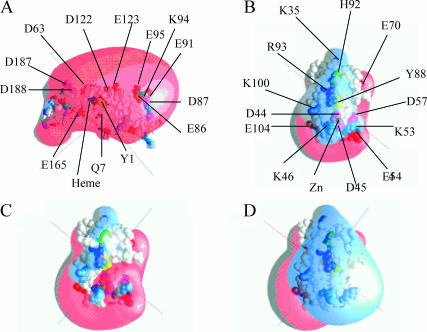
Recently, studies on complex formation and electron transfer have been extended to cyanobacteria. The cyanobacteria are of interest because they are the oldest and most primitive oxygen-evolving photosynthetic organisms. Also, the charge distribution of the surface of both cyt f and PC differs from that observed in higher plants and green algae. Like higher plant (Martinez et al., 1994, 1996) and green algal (Chi et al., 2000) cyt fs, the cyt f from Phormidium laminosum (Carrell et al., 1999) consists of two β-sheet domains and a transmembrane hydrophobic α-helical segment. The cyt f used for the structure determination is the truncated form lacking the transmembrane tail. The heme and Y1, the sixth ligand to the heme, are located on the large domain (Fig. 1 A). The heme is surrounded by a patch of hydrophobic amino acids, many of which are conserved in all cyt fs (Gray, 1992; Martinez et al., 1994, 1996). Cyt fs from higher plants (Martinez et al., 1994, 1996) and algae (Chi et al., 2000) have five highly conserved positively charged residues giving cyt f a positive electrostatic field (Pearson et al., 1996) that attracts negatively charged residues on PC. These residues are replaced by neutral or negatively charged amino acids (Gray, 1992) in cyt fs from cyanobacteria such as Phormidium that has a net charge of −13 (compared to −1 for Chlamydomonas cyt f) resulting in a very large negative electrostatic field (Fig. 1 A).
FIGURE 1.
Charged residues and electrostatic fields on Phormidium cyt f and PC. Heme (black); Y1 on cyt f and Y83 on PC (yellow); H87 on PC (green); Arg (dark blue); Lys (light blue); Glu (red); and Asp (magenta). Electrostatic field contours: +1 kT/e− (blue); and −1 kT/e (red). Electrostatic fields were calculated using GRASP (Nicholls and Honig, 1991). PC residues are numbered as in Bond et al. (1999). (A) WT cyt f; (B) WT PC plus Zn2+ (C) WT PC minus Zn2+; and (D) quadruple PC mutant (D44A + D45A + E54A + D57A) minus Zn2+.
Phormidium PC (Bond et al., 1999), like those from higher plants (Freeman, 1981; Guss et al., 1992; Gross, 1996) and green algae (Redinbo et al., 1993), is a β-sheet protein. The Cu atom is coordinated to four residues H39, C89, H92, and M97 in a distorted tetrahedral geometry. These residues correspond to H37, C84, H87, and M92, respectively, on higher plant and algal PCs (Gross, 1996). H92, which is located on the surface of PC molecule provides a possible electron transfer pathway between cyt f and the copper center and is surrounded by conserved hydrophobic residues in all PCs (Gross, 1996). The charge distribution on the surface of Phormidium PC is very different from that found in higher plants (Gross, 1996; Durell et al., 1990). Higher plant and algal PCs have a series of negatively charged residues (42–45, 51, and 59–61 in spinach PC) surrounding Y83. These negatively charged residues give rise to a large negatively charged electrostatic field (Durell et al., 1990) causing PC to be attracted to cyt f. In Phormidium PC, the only negatively charged residues remaining are D44 and D45 corresponding to D42 and D43 in higher plant PCs. The rest of the negatively charged residues surrounding Y83 in plants and algae are replaced by positive or neutral residues in Phormidium PC (Fig. 1 B).
Two types of studies have been carried out on the interaction of Phormidium cyt f with Phormidium PC: NMR studies of complex formation and electron transfer assays. NMR studies of complex formation showed that hydrophobic interactions were important for complex formation (Crowley et al., 2001). They also ruled out electrostatic interactions because complex formation was independent of ionic strength. On the other hand, complex formation between turnip cyt f and spinach PC (Ubbink et al., 1998; Ejdeback et al., 2000) showed both electrostatic interactions between positively charged residues on cyt f and negatively charged residues on PC and hydrophobic interactions between nonpolar residues surrounding the heme on cyt f and those surrounding H87 on PC. Interestingly, electrostatic interactions have been shown to play a part in the interaction of Prochlorothrix PC with Phormidium cyt f (Crowley et al., 2002).
In contrast, electron transfer assays tell a different story. Schlarb-Ridley et al. (2002) observed that mutation of charged residues on Phormidium PC had significant effects on the rate of electron transfer. Smaller effects were observed for mutants of cyt f (Hart et al., 2003).
In this article, we will use Brownian dynamics simulations to examine the effect of electrostatic interactions on complex formation between Phormidium cyt f and Phormidium PC. We will examine the effect of mutating charged residues on both cyt f and PC on the rates of interaction and will also determine the structure of the complexes formed. We will show that electrostatic interactions do play an important role in bringing the two molecules together to a hydrophobic dock. However, the structure of the complexes formed depends on the presence or absence of the Zn2+ ion found in the crystal structure of Phormidium PC (Bond et al., 1999).
BD simulations (Northrup et al., 1988; Gabdoulline and Wade, 1998) have been used to study the interaction between turnip cyt f with poplar (Pearson and Gross, 1998) or spinach (Nelson et al., 1999; De Rienzo et al., 2001) PC as well as the interaction of Chlamydomonas cyt f with both Chlamydomonas PC and Chlamydomonas cyt c6 (Gross and Pearson, 2003). A preliminary report on the interaction of Phormidium PC with Phormidium cyt f has previously appeared (Gross, 2001).
METHODS
Molecular structures
The protein structures were obtained from the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/; Berman et al., 2000). P. laminosum cyt f (structure 1CI3) was taken from Carrell et al. (1999), and P. laminosum PC was structure A (1BAW) from Bond et al. (1999). The Chlamydomonas cyt f and PC used was the second cyt f record within 1CFM (Chi et al., 2000) and 2PLT (Redinbo et al., 1993), respectively. The structures of Phormidium cyt f-Phormidium PC complexes (Crowley et al., 2001) were provided by M. Ubbink, Leiden University, The Netherlands.
Structures for mutant molecules were generated using the MacroDox program (Northrup et al., 1987a, 1993; Northrup, 1999). All mutant residues were kept in the same orientation as their wild-type counterparts (i.e., no energy minimization was performed on the mutants). Mutants were divided into different classes depending on their relative effects (see Tables 2–5).
TABLE 2.
The effect of mutation of charged residues on Phormidium PC in the absence of the Zn2+ ion on its interaction with Phormidium cyt f
| Ionic strength (mM)
|
|||||||
|---|---|---|---|---|---|---|---|
| 10
|
100
|
||||||
| Class* | Mutant† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† |
| WT | 20.4 ± 0.5 | 100 | 5.9 ± 0.3 | 8.5 ± 0.6 | 100 ± 10 | 2.5 ± 0.4 | |
| I | >200% WT complexes formed at 10 mM ionic strength | ||||||
| D44(42)A | 70.8 ± 0.9 | 403 | 20.2 ± 0.9 | 16.0 ± 0.7 | 188 ± 15 | 4.6 ± 0.5 | |
| D45(43)A | 65.1 ± 1.3 | 370 | 18.7 ± 1.0 | 15.0 ± 0.7 | 176 ± 15 | 4.4 ± 0.4 | |
| D10(8)A | 63.0 ± 1.3 | 358 | 7.6 ± 0.5 | 14.1 ± 0.7 | 165 ± 14 | 4.1 ± 0.4 | |
| E70(68)A | 50.9 ± 0.3 | 289 | 6.2 ± 0.4 | – | – | – | |
| D57(55)A | 50.0 ± 1.1 | 284 | 14.6 ± 0.9 | 12.4 ± 0.6 | 146 ± 13 | 2.6 ± 0.5 | |
| E17A | 39.1 ± 1.2 | 221 | 4.8 ± 0.3 | 12.4 ± 0.6 | 146 ± 12 | 3.6 ± 1.4 | |
| II | 130–200% WT complex formation at 10 mM ionic strength | ||||||
| E54(52)A | 30.9 ± 1.1 | 176 | 3.8 ± 0.3 | 9.3 ± 0.6 | 110 ± 12 | 0.8 ± 0.2 | |
| E1(−3)A | 29.4 ± 0.5 | 167 | 8.9 ± 0.8 | – | – | – | |
| E73(71)A | 29.4 ± 0.8 | 167 | 3.7 ± 0.3 | – | – | – | |
| E104(99)A | 28.1 ± 1.0 | 160 | 3.5 ± 0.3 | – | – | – | |
| III | 100–133% WT complex formation at 10 mM ionic strength | ||||||
| D79(77)A | 23.4 ± 1.0 | 133 | 2.9 ± 0.3 | 9.7 ± 0.6 | 114 ± 11 | 2.7 ± 0.4 | |
| D27(25)A | 22.5 ± 0.7 | 128 | 2.8 ± 0.4 | – | – | – | |
| IV | 40–100% WT complexes formed at 10 mM ionic strength | ||||||
| K58(56)A | 10.1 ± 0.7 | 58 | 1.3 ± 0.3 | 8.0 ± 0.5 | 94 ± 9 | 2.3 ± 0.4 | |
| K53(51)A | 9.9 ± 0.8 | 56 | 2.6 ± 0.5 | 5.8 ± 0.6 | 68 ± 8 | 1.7 ± 0.3 | |
| K30(28)A | 8.5 ± 0.6 | 48 | 1.1 ± 0.3 | – | – | – | |
| V | <40% WT complexes formed at 10 mM ionic strength | ||||||
| K100(95)A | 5.4 ± 0.3 | 31 | 0.8 ± 0.2 | 6.1 ± 0.7 | 76 ± 0.8 | 1.9 ± 0.4 | |
| K46(44)A | 4.6 ± 0.8 | 26 | 1.8 ± 0.4 | 5.5 ± 0.6 | 65 ± 9 | 1.6 ± 0.3 | |
| K6(4)A | 3.9 ± 0.4 | 22 | 0.6 ± 0.2 | 6.4 ± 0.6 | 76 ± 8 | 1.9 ± 0.4 | |
| R93(88)A | 1.3 ± 0.3 | 7 | 0.8 ± 0.3 | 3.0 ± 0.5 | 35 ± 7 | 0.9 ± 0.3 | |
| K35(33)A | 1.1 ± 0.4 | 6 | 0.2 ± 0.2 | 4.1 ± 0.6 | 48 ± 7 | 1.2 ± 0.3 | |
The number of complexes formed in the absence of the electrostatic field (2.2 ± 0.2 complexes per 1000 trajectories) was subtracted from the total number of complexes formed to obtain the number of complexes formed due to electrostatic forces. The mutants were divided into classes based on their effects on complex formation. Class I, significant stimulation of complex formation (i.e., the number of complexes formed was >200% WT at 10 mM ionic strength; class II, 133–200% WT at 10 mM; class III, the number of complexes formed was between 100 and 133% WT at 10 mM; class IV, the number of complexes formed was between 40 and 100% WT at 10 mM; class V, the number of complexes formed was <40% WT at 10 mM ionic strength. The rates of electron transfer were calculated as described in the Methods section and for Table 1. The rate of electron transfer found in the absence of the electrostatic field (0.6 ± 0.2 × 102 M−1 s−1) was subtracted from the observed rates to obtain the rate of electron transfer due to electrostatic forces alone. Five sets of 5000 and 10,000 trajectories were carried out at 10 and 100 mM ionic strength, respectively.
The mutants were made as described in the Methods section. The number of complexes formed and the rates of electron transfer were calculated as described for Table 1 and in the Methods section.
TABLE 3.
The effect of mutation of charged residues on Phormidium PC in the presence of the Zn2+ ion on its interaction with Phormidium cyt f
| Ionic strength (mM)
|
|||||||
|---|---|---|---|---|---|---|---|
| 10
|
100
|
||||||
| Class* | Mutant† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† |
| WT | 227 ± 11 | 100 | 56 ± 3 | 39.2 ± 0.8 | 100 ± 3 | 11.6 ± 0.8 | |
| II | 130–200% WT complex formation at 10 mM ionic strength | ||||||
| D45(43)A‡ | 361 ± 22 | 160§ | 83 ± 3 | 76.3 ± 1.2 | 191 ± 5 | 21.3 ± 0.7 | |
| D57(55)A | 335 ± 16 | 149 | 78 ± 3 | 56.8 ± 1.3 | 143 ± 5 | 16.1 ± 1.0 | |
| D44(42)A | 321 ± 7 | 143 | 78 ± 3 | 71.8 ± 0.9 | 180 ± 4 | 20.0 ± 1.0 | |
| E54(52)A | 317 ± 15 | 141 | 78 ± 3 | 50.0 ± 2.8 | 127 ± 8 | 14.0 ± 0.9 | |
| D10(8)A | 314 ± 8 | 140 | 74 ± 3 | 47.3 ± 3.6 | 120 ± 9 | 13.5 ± 1.0 | |
| E17(15)A | 310 ± 7 | 138 | 73 ± 3 | 46.0 ± 0.3 | 117 ± 3 | 13.1 ± 0.7 | |
| E70(68)A | 303 ± 5 | 135 | 72 ± 3 | – | – | – | |
| III | 100–133% WT complex formation at 10 mM ionic strength | ||||||
| E73(71)A | 278 ± 4 | 124 | 67 ± 3 | – | – | – | |
| D27(25)A | 252 ± 8 | 111 | 61 ± 3 | – | – | – | |
| E104(99)A | 246 ± 12 | 108 | 60 ± 4 | – | – | – | |
| E1(−3)A | 244 + 11 | 108 | 61 ± 4 | – | – | – | |
| D79(77)A | 241 ± 12 | 107 | 59 ± 3 | 41.8 ± 1.3 | 107 ± 4 | 11.8 ± 0.9 | |
| IV | 40–100% WT complex formation at 10 mM ionic strength | ||||||
| K30(28)A | 169 ± 8 | 75 ± | 44 ± 3 | 37.2 ± 1.3 | 95 ± 4 | 10.7 ± 0.7 | |
| K58(56)A | 162 ± 6 | 72 ± | 42 ± 3 | 34.3 ± 1.3 | 88 ± 4 | 9.8 ± 0.9 | |
| K6(4)A | 151 ± 12 | 67 | 39 ± 3 | 35.0 ± 1.1 | 90 ± 3 | 10.0 ± 0.8 | |
| K35(33)A | 141 ± 10 | 63 | 37 ± 3 | 31.8 ± 0.8 | 82 ± 3 | 9.2 ± 0.9 | |
| K100(95)A | 121 ± 5 | 54 | 32 ± 3 | – | – | – | |
| K53(51)A | 114 ± 7 | 51 | 30 ± 2 | 25.3 ± 0.7 | 66 ± 2 | 7.2 ± 0.6 | |
| V | <40% WT at 10 mM ionic strength | ||||||
| K46(44)A | 71 ± 11 | 32 | 20 ± 2 | 20.7 ± 1.0 | 54 ± 3 | 6.0 ± 0.6 | |
| R93(88)A | 55 ± 7 | 25 | 15 ± 3 | 13.4 ± 0.4 | 37 ± 1 | 3.9 ± 0.5 | |
| R93(88)Q | 51 ± 10 | 23 | 14 ± 2 | 14.9 ± 0.5 | 40 ± 2 | 4.3 ± 0.5 | |
The number of complexes formed in the absence of the electrostatic field (2.2/1000 trajectories) was subtracted from the total number of complexes formed to determine the electrostatic contribution. The mutants were divided into classes based on their effects on complex formation. Class I, significant stimulation of complex formation (i.e., the number of complexes formed was >200% WT at 10 mM ionic strength; class II, 130–200% WT at 10 mM; class III, the number of complexes formed was between 100 and 133% WT at 10 mM; class IV, the number of complexes formed was between 40 and 100% WT at 10 mM; and class V, the number of complexes formed was <40% WT at 10 mM ionic strength. Five sets each of 1000 and 5000 trajectories were carried out at 10 and 100 mM, respectively.
The mutants were made as described in the Methods section. Rates were determined as described in the Methods section and for Table 1. The rate obtained in the absence of the electrostatic field (0.6 × 108 M−1 s−1) was subtracted from the total rate to determine the electrostatic contribution.
The numbers in the parentheses refer to the consensus sequence of Pearson (2000).
Standard deviations averaged 5% of the values shown.
TABLE 4.
The effect of mutants of Phormidium cytochrome f on their interaction with Phormidium PC in the presence of the Zn2+ ion
| Class* | Mutant† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† |
|---|---|---|---|---|
| Q7E | 271 ± 3 | 121 | 66 ± 3 | |
| WT | 225 ± 5 | 100 | 57 ± 3 | |
| I | Complex formation ≥ 89% WT | |||
| E14A | 222 ± 5 | 99 | 56 ± 4 | |
| D42A | 211 ± 6 | 94 | 54 ± 3 | |
| D79A | 205 ± 5 | 91 | 53 ± 3 | |
| D199A | 198 ± 3 | 88 | 62 ± 3 | |
| D109A | 191 ± 4 | 85 | 49 ± 3 | |
| D108A | 181 ± 4 | 81 | 47 ± 3 | |
| II | Complex formation between 59 and 77% WT | |||
| E198A | 173 ± 2 | 77 | 45 ± 3 | |
| Q7R | 172 ± 5 | 76 | 45 ± 3 | |
| D63A | 168 ± 6 | 75 | 44 ± 3 | |
| E217A | 165 ± 3 | 73 | 43 ± 3 | |
| E125A | 161 ± 2 | 72 | 42 ± 3 | |
| E91A | 161 ± 6 | 72 | 42 ± 3 | |
| E92A | 161 ± 5 | 72 | 42 ± 3 | |
| E212A | 153 ± 6 | 68 | 41 ± 3 | |
| E34A | 152 ± 6 | 68 | 40 ± 3 | |
| D205A | 144 ± 4 | 64 | 38 ± 3 | |
| D187A | 139 ± 5 | 62 | 37 ± 3 | |
| E96A | 138 ± 6 | 61 | 37 ± 3 | |
| E192A | 132 ± 4 | 59 | 35 ± 3 | |
| III | Complex formation between 50 and 59% WT | |||
| E165A | 120 ± 1 | 53 | 32 ± 3 | |
| D188A | 119 ± 4 | 53 | 32 ± 3 | |
| D87A | 116 ± 3 | 52 | 32 ± 3 | |
| D122A | 112 ± 3 | 50 | 31 ± 3 | |
| IV | Complex formation between 44 and 48% WT | |||
| E95A | 105 ± 3 | 47 | 29 ± 3 | |
| E123A | 101 ± 2 | 45 | 28 ± 2 | |
| E86A | 100 ± 3 | 45 | 27 ± 3 | |
Class I: complex formation was >80% WT; class II: 59–77% WT; class III: 50–59% WT; and class IV: 44–48% WT. The effects on the rates of electron transfer were identical to those on complex formation.
The mutants were made as described in the Methods section. The number of complexes formed and the rates of electron transfer were calculated as described for Table 1 and in the Methods section.
TABLE 5.
The effect of mutants of Phormidium cytochrome f on their interaction with Phormidium PC in the absence of Zn2+
| Class* | Mutant† | Complexes formed per 1000 trajectories† | % WT | 10−8 × k2 (M−1 s−1)† |
|---|---|---|---|---|
| Q7E | 23.3 ± 0.6 | 114 | 6.7 ± 0.7 | |
| WT | 20.4 ± 0.5 | 100 | 5.9 ± 0.3 | |
| II | Complex formation between 59 and 77% WT | |||
| E92A | 14.7 ± 0.7 | 72 | 4.3 ± 0.4 | |
| E217A | 13.0 ± 0.5 | 63 | 3.8 ± 0.3 | |
| D199A | 12.6 ± 0.3 | 62 | 3.6 ± 0.3 | |
| Q7R | 12.6 ± 0.2 | 62 | 3.7 ± 0.3 | |
| III | Complex formation between 50 and 59% WT | |||
| E192A | 11.6 ± 0.3 | 57 | 3.4 ± 0.4 | |
| D42A | 11.6 ± 0.6 | 57 | 3.4 ± 0.3 | |
| D205A | 11.4 ± 0.3 | 56 | 3.4 ± 0.4 | |
| D87A | 11.0 ± 0.4 | 54 | 3.2 ± 0.4 | |
| E79A | 11.0 ± 0.2 | 54 | 3.2 ± 0.3 | |
| D109A | 10.6 ± 0.5 | 52 | 3.1 ± 0.3 | |
| E95A | 10.6 ± 0.2 | 52 | 3.1 ± 0.3 | |
| E14A | 10.5 ± 0.4 | 51 | 3.1 ± 0.4 | |
| IV | Complex formation between 44 and 48% WT | |||
| D122A | 9.8 ± 0.5 | 48 | 2.9 ± 0.3 | |
| E91A | 9.6 ± 0.4 | 47 | 2.6 ± 0.3 | |
| E123A | 9.5 ± 0.2 | 46 | 2.7 ± 0.3 | |
| D63A | 9.1 ± 0.2 | 45 | 2.7 ± 0.2 | |
| E86A | 9.0 ± 0.4 | 44 | 2.7 ± 0.3 | |
| V | Complex formation <40% WT | |||
| D187A | 7.2 ± 0.3 | 35 | 2.1 ± 0.3 | |
| E165A | 6.8 ± 0.3 | 33 | 2.0 ± 0.3 | |
| D188A | 6.5 ± 0.2 | 32 | 2.0 ± 0.3 | |
Class II: the number of complexes was between 59 and 77% WT; class III: 50–59% WT; class IV: 44–48% WT; and class V: ≤35% WT. Note that the same class divisions were used for Tables 4 and 5 for ease of comparison.
The mutants were made as described in the Methods section. The number of complexes formed and the rates of electron transfer were calculated as described for Table 1 and in the Methods section.
Molecular representations
All molecular representations were made using the program GRASP (Nicholls and Honig, 1991). Electrostatic fields for the molecular representation only were also calculated using GRASP. The internal and external dielectric constants of the proteins were 4 and 78, respectively. The ionic strength was 10 mM, and the pH was 7.0.
Brownian dynamics simulations
BD simulations were carried out using the program MacroDox v. 3.2.1 (Northrup et al., 1987a,b, 1988, 1993; Northrup, 1999; http://gemini.tntech.edu/s;/index.html) as described by Pearson and Gross (1998), Pearson (2000), and Gross and Pearson (2003).
The equation of motion used by the BD algorithm is the Ermak-McCammon equation (Ermak and McCammon, 1978):
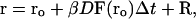 |
(1) |
where ro and r are the initial and final distances between the center of mass of the mobile molecule (molecule 2; in our case PC) and the center of mass of the target molecule (molecule 1, cyt f; note that Molecule 1 does rotate but the center of mass does not move. In contrast, molecule 2 both rotates and moves), before and after a time step Δt; β = (kT)−1; F(ro) is the external force on the molecule at ro, D is the relative diffusion coefficient of the two molecules, and R is a random (Brownian) vector with the following properties (McCammon and Harvey, 1987; Harvey, 1989):
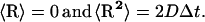 |
(2) |
Δt should be sufficiently small so that there is a minimal change in F (i.e., F(r) ∼ F(ro)). A similar equation can be derived for the torque (Northrup, 1996).
A typical BD experiment for 10 mM ionic strength at pH 7 consists of five sets of 1000 trajectories each (for a total of 5000 trajectories) unless otherwise stated. Each set of 1000 trajectories required from 15 min to 1 h on a Silicon Graphics O2 workstation (SGI, Mountain View, CA). For each trajectory, the center of mass of molecule 2 (PC) is positioned on the surface of a sphere 87 Å in radius (see Gross and Pearson, 2003, for details) from the center of mass of molecule 1 (cyt f). For each individual trajectory, the position and orientation of molecule 2 on the surface of the sphere is determined by the MacroDox program using a random number seed. Molecule 2 is subjected to a force F(r) and moves accordingly after which F(r) and R are recalculated. A trajectory is concluded when the mobile molecule leaves a sphere of 200-Å radius from the center of mass of molecule 1. The relative translational diffusion coefficient was 0.025 Å2/ps for cyt f-PC simulations. Both the fixed (cyt f) and diffusing (PC) molecules rotate with a rotational diffusion coefficient, calculated by MacroDox, of 0.32 × 10−4 ps−1. Exclusion volumes are determined as described by Northrup et al. (1993). Overlap with the target molecule is checked for all atoms on molecule 2 (PC) after each step in the trajectory.
MacroDox determines the closest approach of the two molecules based on a preselected reaction criterion. Closest metal/metal distance was chosen as the reaction criterion to select for electron-transfer-active complexes (Moser et al., 1992, 1995). The point of closest approach (smallest Cu-Fe distance) is recorded for each trajectory allowing us to calculate the number of complexes formed as a function of minimal Cu-Fe distance (Fig. 2). Second order rate constants for complex formation are calculated from the fraction of complexes observed as a function of minimal reaction coordinate (Cu-Fe) distance using equations derived by Northrup et al. (1987a, 1993) from the equations of Smoluchowski. Since we are assuming that the reaction is diffusion limited (Hart et al., 2003), the rate of complex formation (k2) should be equal to the rate constant for electron transfer (ket) (but see the Discussion). Unless otherwise indicated, the number of complexes with minimal center of mass distances of ≤20 Å for the Cu-Fe distance was used to calculate the rate constants; 20 Å was sufficient to include essentially all of the electrostatic complexes formed although excluding those formed solely by random Brownian motion (Fig. 2). MacroDox provides the following information for each trajectory: the structure of the complex at minimal Cu-Fe distance, the 15 closest pairs of charged residues, and the electrostatic interaction energy.
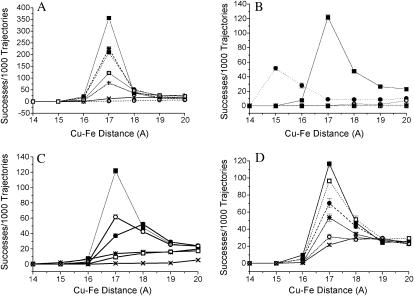
FIGURE 2.
The number of complexes formed in BD simulations of Phormidium cyt f and Phormidium PC under different conditions. (A) The effect of mutation of anionic residues on Phormidium PC on its interaction with Phormidium cyt f. (○) WT PC minus Zn2+; (×) D45A-PC minus Zn2+; (+) D44A + D45A-PC minus Zn2; (□) WT-PC plus Zn2+; (•) D44A + D45A + E54A-PC minus Zn2+.; (*) D44A + D45A + D57A-PC minus Zn2+; and (▪) D44A + D45A + E54A + D57A-PC minus Zn2+. (B) Comparison of cyt f-PC interactions of cyanobacteria with those of green algae. Conditions were as described in the Methods section. (•) Chlamydomonas cyt f interacting with Chlamydomonas PC; (▪) Phormidium cyt f interacting with Phormidium PC in the presence of Zn2+; (*) Phormidium cyt f interacting with Phormidium PC in the presence of Zn2 + in the absence of an electrostatic field; (□) Phormidium cyt f interacting with Chlamydomonas PC; and (○) Chlamydomonas cyt f interacting with Phormidium PC + Zn2+. (C) The effect of mutation of cationic residues on Phormidium PC in the presence of Zn2+ on the number of complexes formed with Phormidium cyt f. (▪) WT-PC; (○) K6A-PC; (•) K35A- PC; (*) K46A-PC; (□) R93A-PC; and (X) R93E-PC. (D) The effect of mutations of Phormidium cyt f on its ability to interact with Phormidium PC in the presence of Zn2+. (▪) WT-cyt f; (□) D42A-cyt f; (•) D63A-cyt f; (*) D187A-cyt f; (○) D122A-cyt f; and (X) E95A-cyt f.
Charge assignments and electrostatic calculations used in MacroDox simulations
Formal charges were assigned to ionizable groups such as amino, carboxyl, guanido, and imidazole groups. This contrasts with the use of partial charges that would assign partial charges to polarizable atoms such as oxygen and nitrogen atoms. Partial charges were not used since they had a negligible effect on the number of complexes formed in the interaction of Chlamydomonas cyt f with Chlamydomonas PC (Gross and Pearson, 2003).
When using formal charges, noninteger values of the charge on PC and cyt f result from the ionization of histidine residues at pH 7. However, H25 on cyt f and H39 and H92 on PC have a zero net charge because they are ligands to the metal center. On the other hand, the sulfur atom of the Cys-89 ligand to the Cu on PC was given a net charge of −1 (Durell et al., 1990), and the Cu atom itself was given a charge of +2. For cyt f, the charges on the heme were as follows: Fe (+2), two ring nitrogen atoms (−1 each), and two propionic acid side chains (−1 each). The pKs were calculated using a modified Tanford-Kirkwood pK algorithm (Matthew, 1985).
Electrostatic calculations were carried out using the Warwicker/Watson finite difference method (Warwicker and Watson, 1982) for solving the linearized Poisson-Boltzmann equation. This algorithm is slightly different from that used in GRASP. Full charges were assigned to the ionized atoms. The center of mass of each protein was placed at the center of a 61 × 61 × 61 grid. The electrostatic field was first iterated over a 3.6-Å grid followed by iteration over a smaller 1.2-Å grid for better accuracy. The choice of grid size has a small (<20%) effect on the rates but no effect on the structure of the complexes formed (Gross and Pearson, 2003). Most importantly, the relative rates of electron transfer are not affected.
Forces, torques, and electrostatic interaction energies
Forces were calculated as described by Northrup et al. (1993). Molecule 1 (cyt f, the target molecule) was given a low internal dielectric constant of 4.0. However, because of computational complexities, molecule 2 (PC) was treated as a set of point charges embedded in a medium of the same dielectric constant and ionic strength as the solvent. Using a high internal dielectric constant for molecule 2 (Gabdoulline and Wade, 1996, 2001; Northrup et al., 1987a; S. H. Northrup, Tennessee Technical University, Cookeville, TN, personal communication, 2002) and neglecting mutual desolvation effects (Elcock et al., 1999; Gabdoulline and Wade, 2001) may cause an overestimate of the reaction rates by as much as 25%. However, desolvation effects are not significant in these simulations (see Discussion). By neglecting these, neither the relative effects of the mutations nor the structures of the complexes formed should be affected (Northrup et al., 1987a; S. H. Northrup, personal communication, 2002). Torques were calculated using dipoles for the moving protein as described by Northrup (1996).
Analysis of the complexes formed
For wild-type (WT) and selected mutant PC-cyt f complexes, 10 complexes were selected for further analysis from the 5000 (5 × 1000) trajectories per experiment. The only criterion used was that the Cu-Fe distance be less than the mean of the peaks shown in Fig. 2.
Close contacts between the two proteins in a given complex were determined using an in-house program that calculates the distance from every atom on molecule 1 (cyt f) to every atom on molecule 2 (PC) and records atom pairs with a distance less than or equal to a preset value. The distance selected for this study was 8 Å to observe the closest contacts although allowing for the fact that the BD complexes were not energy-optimized in any way. To be accepted as a contact, a pair of residues must be ≤8 Å apart in at least eight out of 10 of the complexes analyzed. The structures of these 10 complexes were compared and the 15 closest electrostatic contacts, and the distances between corresponding charged residues were determined as described by Northrup et al. (1993) using the output of the MacroDox program as were electrostatic free energies.
RESULTS
The effect of a Zn2+ ion on the number of complexes formed between Phormidium PC and Phormidium cyt f
In these studies, the number of complexes formed was determined as a function of the minimal Cu-Fe distance observed for Phormidium PC interacting with Phormidium cyt f at 10 mM ionic strength (Fig. 2 A) and pH 7.0. In this plot, the point at 17 Å represents the number of complexes in which the closest approach of the Cu in PC to the Fe in cyt f lies between 16 and 17 Å.
In the crystal structure of Phormidium PC, a Zn2+ ion lies adjacent to D44 and D45. This Zn2+ ion was added to ensure crystallization (Bond et al., 1999). In our initial studies, we removed the Zn2+ ion from the crystal structure. However, only 20 ± 5 complexes were found with Cu-Fe distances ≤ 20 Å. A cutoff Cu-Fe distance of 20 Å was chosen for the calculation of electron transfer rates for the following reasons. First, in the absence of an electrostatic field, the Cu on PC almost never approaches to within 20 Å of the Fe on cyt f. Therefore, we are selecting for electrostatically driven rather than random complexes. Second, 20 Å should be sufficiently close to allow hydrophobic forces to bring PC into a final electron transfer active dock. These assumptions will be discussed below.
When the Zn2+ ion was included, the number of complexes with Cu-Fe distances ≤ 20 Å increased to 225 ± 5 (Fig. 2 A and Table 1). The corresponding electron transfer rates were 5.8 ± 0.6 × 108 M−1 s−1 and 55 ± 3.0 × 108 M−1 s−1, respectively, in the absence and presence of the Zn2+ ion.
TABLE 1.
The effect of the number of charges modified on Phormidium PC on its interaction with Phormidium cyt f
| Mutation* | Net change in charge† | No. of complexes per 1000 trajectories‡ | 10−8 × k2 (M−1 s−1)§ |
|---|---|---|---|
| WT¶ | 0 | 20.4 ± 0.5 | 5.8 ± 0.6 |
| WT (electrostatic field off)¶ | 0 | 2 ± 0.2 | 0.6 ± 0.2 |
| D45(43)A¶ | +1 | 67 ± 1 | 18.9 ± 1.0 |
| D44(42)A + D45(43)A | +2 | 175 ± 6 | 45.2 ± 2.6 |
| WT + Zn2+ | +2 | 225 ± 5 | 55.1 ± 3.0 |
| D44(42)A + D45(43)A + E54(52)A | +3 | 316 ± 6 | 74.9 ± 2.7 |
| D44(42)A + D45(43)A + E57(55)A | +3 | 313 ± 7 | 74.5 ± 3.2 |
| D44(42)A + D45(43)A + E54(52)A + E57(55)A | +4 | 440 ± 2 | 97.2 ± 3.5 |
Mutations were constructed as described in the Methods section. The residue numbers were taken from the PDB file (Bond et al., 1999). The consensus sequence taken from Pearson (2000) is in parentheses. The Zn2+ ion was omitted except where indicated.
Compared to WT minus Zn2+.
Number of complexes formed with Cu-Fe Distances ≤20 Å.
Rates were calculated as described in the Methods section.
Five sets of 5000 trajectories each were carried out for WT (−Zn2+) with the electrostatics on and off and for D45A + Zn2+. Five sets of 1000 trajectories each were completed for the rest.
The number of complexes formed is greater in the presence of the Zn2+ ion, because it increases the magnitude of the positive electrostatic field surrounding R93 (compare Fig. 1, B and C). In the absence of the Zn2+ ion (Fig. 1 C), a negative electrostatic field surrounds D44 and D45 leaving only small positive electrostatic fields surrounding K46, K53, K104, and R93, too small for Phormidium PC to be strongly attracted to the negatively charged cyt f (Fig. 1 A). The Zn2+ ion neutralizes the negative electrostatic field (Fig. 1 B) allowing PC to approach cyt f. This interpretation is supported by the observation that replacing the aspartates at positions 44 and 45 with alanines (i.e., D44A + D45A-PC) in the absence of the Zn+2 ion gave almost the same results as adding the Zn2+ ion to WT PC (Fig. 2 A). Removing three negative charges in the absence of Zn2+ (i.e., D44A + D45A + E54A-PC or D44A + D45A + E57A-PC) increased the number of complexes formed still further. The greatest increase in complex formation was observed for the quadruple mutant (D44A + D45A + E54A + D57A-PC), which shows a very large positive electrostatic field surrounding R93 (Fig. 1 D). Both the addition of the Zn2+ and the progressive removal of negatively charged residues by mutation also increased the computed rates of electron transfer (Table 1). In conclusion, negatively charged residues in the vicinity of R93 inhibited complex formation with cyt f. This inhibition can be relieved either by the addition of the Zn2+ or by mutation of the negatively charged residues.
Comparison of the number of complexes formed for green algal and cyanobacterial cyt f-PC systems
Fig. 2 B compares the number of complexes formed for Chlamydomonas and Phormidium cyt f interacting with PCs from both species. When Phormidium cyt f interacted with Phormidium PC in the presence of the Zn2+ ion, maximal complex formation occurred at 17 Å; in contrast, maximal complex formation occurred at 15 Å for Chlamydomonas cyt f interacting with Chlamydomonas PC. This is because the Cu and Fe are farther apart at the point of contact between the two proteins in the Phormidium system (16 Å) than for the Chlamydomonas system (14 Å). A greater number of complexes with Cu-Fe distances ≤ 20 Å were observed for the Phormidium cyt f interacting with Phormidium PC in the presence of Zn2+ than for Chlamydomonas cyt f interacting with Chlamydomonas PC (Fig. 2 B). The corresponding electron transfer rates were 55.1 ± 0.3 × 108 and 26 ± 3 × 108 M−1 s−1, respectively. In contrast, almost no complexes were formed when cyt f from one species interacted with PC from the other (Fig. 2 B). Thus, the positive electrostatic field of Chlamydomonas cyt f (Pearson et al., 1996) attracts the negative electrostatic field of Chlamydomonas PC (Gross and Pearson, 2003), and the negative electrostatic field of Phormidium cyt f attracts positive charges on Phormidium PC (particularly in the presence of the Zn2+ ion). However, there cannot be cross-reactions between the two species because of electrostatic repulsion. These results, taken together with the observation that almost no complexes are formed in the absence of an electrostatic field (Table 1), show that attractive electrostatic forces are required for complex formation between cyt f and PC in BD simulations. The unit cell of Phormidium PC contains three PC molecules (Bond et al., 1999). All three PC structures gave the same results (not shown).
The effect of mutating positively charged residues on Phormidium PC on complex formation with Phormidium cyt f
The effect of mutation of cationic residues on Phormidium PC in the presence of the Zn2+ cation is shown in Fig. 2 C. K6A and K35A-PC showed moderate inhibition of complex formation, whereas K46A and R93A-PC showed severe inhibition. R93E-PC, having a net change in charge of −2, showed zero activity.
The effect of PC mutants on the number of complexes formed and the corresponding rates of electron transfer in the absence of the Zn2+ ion
The results for mutants of the charged residues on Phormidium PC in the absence of Zn2+ are summarized in Table 2. The mutants can be divided into five classes. For all mutants, the percent change in electron transfer rates paralleled those for complex formation. Class I mutants, including those for which complex formation is ≥200% of the WT PC at 10 mM ionic strength. The greatest stimulation was observed for D44A, D45A, and D10A. E70A, D57A, and E17A also showed over 200% WT complex formation at 10 mM ionic strength. Class II consists of those mutants including E54A, E1A, E73A, and E104A that showed between 130 and 200% WT complex formation at 10 mM ionic strength. D79A and D27A showed less than 30% stimulation of complex formation and were placed in class III. Mutants of cationic residues were divided into two classes (class IV and class V). Class IV included those mutants (K58A, K53A, and K30A) in which complex formation was ≥40% WT PC. The greatest inhibition of complex formation was observed for class V (<40% WT complex formation) that included K100A, K46A, K6A, R93A, and K35A. The locations of those residues whose mutants showed either the greatest stimulation or greatest inhibition are shown in Fig. 3 (left). The pattern of inhibition and stimulation observed at an ionic strength of 100 mM was the same as at 10 mM ionic strength but the magnitude of the effects was smaller. Thus, the effects of the mutations can be observed under conditions (100 mM) that have been used for electron transfer assays (Schlarb-Ridley et al., 2002).
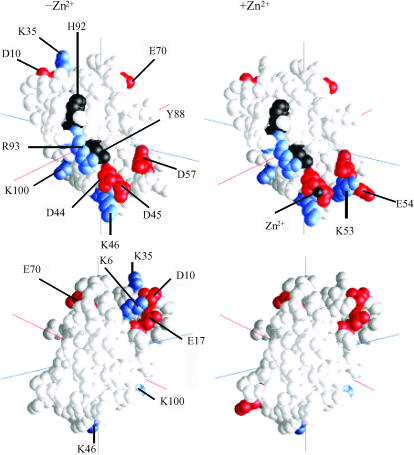
FIGURE 3.
Location of the mutations on Phormidium PC. Only those mutants having the greatest effects are shown. In the absence of the Zn2+ ion (Table 2), results are shown for class I (greatest stimulation) and class V (greatest inhibition; in the absence of Zn2+ (Table 3), results are shown for class II (greatest stimulation) and classes IV and V (greatest inhibition). (Black) Y88, H92, and the Zn2+ (when present); (red) anionic residues; and (blue) cationic residues. (Left) −Zn2+ and (right) +Zn2+. (Top) Front and (bottom) back of the PC molecules.
The effects of the mutations were also determined in the presence of the Zn2+ ion (Table 3). The total number of complexes formed and the reaction rates were greater in the presence of the Zn2+ ion at both 10 and 100 mM ionic strength. As in the absence of Zn2+, the pattern of inhibition was the same at both 10 and 100 mM. Thus, as in the absence of the Zn2+ ion, the mode of interaction of PC with cyt f is the same at 10 and 100 mM ionic strength.
The location on the PC molecule of those mutants that showed greatest stimulation and inhibition in the presence of the Zn2+ ion is shown in Fig. 3 (right). PC mutants that showed the greatest effects on complex formation both in the presence and absence of the Zn2+ ion included D10, E17, D44, D45, K46, D57, E70, R93, and K100. Residues on PC that show the greatest effects on complex formation both in the presence and absence of the Zn2+ are located in two clusters: D44, D45, K46, D57, R93, and K100 are located on the lower part of the front face of PC as presented in Fig. 3, and D10, E17, and E70 are located on the upper portion of the back of the PC molecule. In the absence of the Zn2+ ion, two additional residues on the upper portion of the back of the PC molecule also show large effects. These are K6 and K35. In contrast, there are two residues that show greater effects in the presence of the Zn2+ ion, namely, K53 and E54 located on lower front side of the PC molecule. These results suggest that the binding site on PC for cyt f is somewhat different in the presence and absence of the Zn2+ ion.
The effect of mutation of residues on Phormidium cyt f on its interaction with Phormidium PC
D→A and E→A mutants of cyt f were used to map the binding site on cyt f for PC both in the presence and the absence of the Zn2+ ion. Of the five cationic residues (K58, K65, K66, K187, and R209) that contribute to the positive electrostatic field on turnip cyt f, only K66 is conserved in cyanobacterial cyt fs (Pearson, 2000). Instead, the surface of Phormidium cyt f is covered by a series of anionic residues that contribute to a diffuse negative electrostatic field as shown in Fig. 1 A.
The effect of Phormidium cyt f mutations on its interaction with Phormidium PC in the presence of the Zn2+ ion is shown in Figs. 2 D and 4 A as well as Table 4. Fig. 2 D shows that mutations E95A and D122A caused severe inhibition of complex formation, whereas mutations D187A and D63A caused only moderate inhibition, and D42A had very little effect.
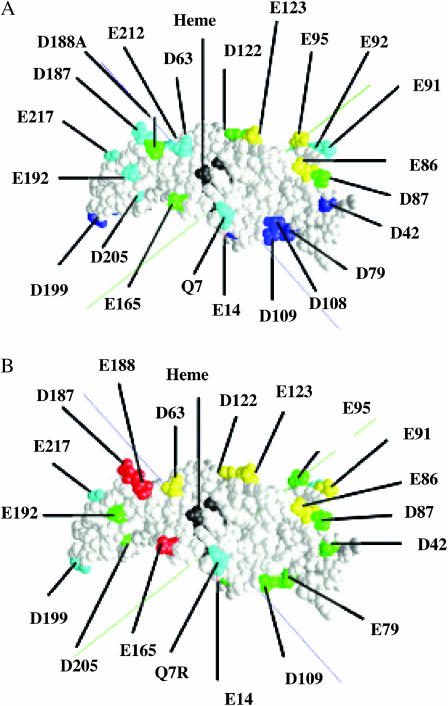
FIGURE 4.
Phormidium cyt f mutations. Data were taken from Tables 4 and 5. Class I (>80% WT; dark blue); class II (59–77% WT; cyan); class III (50–59% WT; green); class IV (44–48% WT; yellow); class V (<40% WT; red); and heme (black). (A) Plus Zn2+ and (B) minus Zn2+.
Table 4 shows the effect of mutation of all anionic residues on the front or heme side of cyt f plus a selection of residues on the backside. The mutations listed in Table 4 fall into four classes. Those mutants for which complex formation is ≥80% control fall into class I; those for which complex formation lies between 59 and 77%, between 50 and 59%, and between 44 and 48% WT control fall into classes II, III, and IV, respectively. The greatest inhibition (class IV) occurs for cyt fs containing mutants E86A, E95A, and E123A. Class III mutants include E165A, D188A, D87A, and D122A. Only one of these, D188A, is located in the region of the positive patch on Chlamydomonas cyt f. Instead, they are clustered at the opposite end of the cyt f molecule (Fig. 4 A). Cyt fs with mutations D63A and D187A showed moderate inhibition of complex formation and fall into class II. Cyt f mutants of residues on the backside of cyt f all fall into classes I and II. These include E92A, E34A, D205A, and E96A.
The effect of the various cyt f mutations was also examined for Phormidium PC in the absence of the Zn2+ ion (Table 5). A different pattern of inhibition was observed. First, the percent inhibition of complex formation was greater in the absence of the Zn2+ ion than in its presence. Second, the pattern of inhibition was different. The greatest inhibition was observed for cyt f mutants E165A, D187A, and D188A, although residues E86, E95, and E123 also showed significant inhibition. This pattern of inhibition is very different than that observed in the presence of the Zn2+ ion in which residues E165A, D187A, and D188A fall into classes II and III. It is also reminiscent of the inhibition pattern observed for mutants of Chlamydomonas cyt f, in which case mutations of residue 187 and 188 caused severe inhibition although mutation of residue 165 had little effect (Gross and Pearson, 2003). Thus, the presence of Zn2+ shifts the location of the binding site for PC on the cyt f molecule from D187, D188, and D165 on the small domain to E86, E95, and E123 on the large domain. A comparison of Fig. 4 A (PC + Zn2+) with Fig. 4 B (PC − Zn2+) shows this shift in binding site on cyt f.
To study this effect further, we examined the structure of the complexes formed between Phormidium cyt f and Phormidium PC. Note that all of the residues on Phormidium cyt f involved in PC binding both in the presence and absence of the Zn2+ ion are on the outside of the cytochrome b6f complex (Kurisu et al., 2003).
The structure of complexes formed between Phormidium cyt f and Phormidium PC
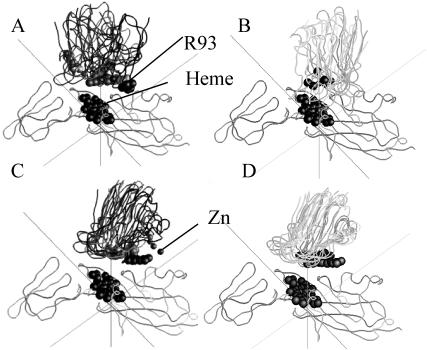
Ten complexes for both the WT (both plus and minus Zn2+) and the quadruple mutant (D44A + D45A + E54A + D57A) minus Zn2+ were chosen for further analysis as described in the Methods section. The homogeneity (or lack thereof) of the complexes formed is depicted in Fig. 5 that shows the peptide backbone, R93, the Cu, and the Zn2+ ion (if present) for 5 of the 10 complexes chosen for each case.
FIGURE 5.
Heterogeneity of Phormidium cyt f-Phormidium PC BD complexes formed under different conditions. Five of the 10 complexes that were selected for detailed analysis as described in the Methods section are depicted unless otherwise indicated. The backbone ribbons were superimposed using GRASP. The heme groups on cyt f and the Cu and Zn ions as well as R88 are shown for PC. The ionic strength was 10 mM. (A) WT-PC minus Zn2+; (B) Phormidium cyt f-Phormidium PC complexes from Crowley et al. (2001). Twenty-five complexes were supplied by M. Ubbink. Five of these were chosen at random for display. Conditions were 10 mM sodium phosphate buffer (pH 6.0) + 1 mM Na-absorbate; (C) WT-PC plus Zn2+; and (D) (D42A + D43A + E52A + D57A-PC)-minus Zn2+.
In the absence of the Zn2+ ion, the complexes were random in orientation (Fig. 5 A) in agreement with the results of Crowley et al. (2001) in their NMR study of the interaction of Phormidium cyt f with Phormidium PC in the absence of Zn2+. The structures of 5 of the 25 complexes supplied by M. Ubbink are shown in Fig. 5 B for comparison (note that the NMR studies were carried out in 10 mM phosphate buffer, pH 6.0). Some of complexes observed in both the BD and NMR studies had orientations similar to that observed for NMR experiments of the interaction of spinach PC with turnip cyt f (Ubbink et al., 1998) and in BD simulations of the interactions between poplar PC with turnip cyt f (Pearson and Gross, 1998) and for Chlamydomonas PC interacting with Chlamydomonas cyt f (Gross and Pearson, 2003). These results are consistent with the observation that cyt f mutants D187A and D188A showed severe inhibition in the absence of the Zn2+ ion. Mutants E86A, E95A, and E123A show a lesser but still significant degree of inhibition that is consistent with some of the complexes facing the large domain (see below).
In the presence of the Zn2+ ion, the complexes were much more homogeneous in orientation (Fig. 5 C) and were turned so that the portion of PC containing R93, D44, and D45 faced the large domain on cyt f. This is consistent with the mutation data that showed the greatest inhibition for the cyt f mutants E86A, E95, and E123A . The quadruple mutant showed even greater homogeneity of complex formation (Fig. 5 D) consistent with the larger positive electrostatic field surrounding R93 on PC (Fig. 1 D).
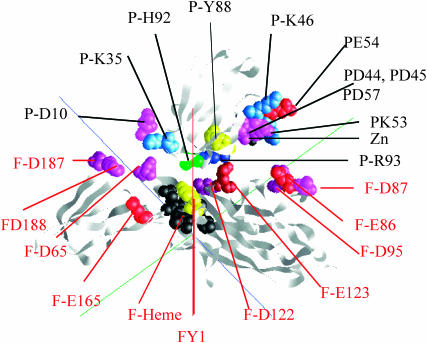
A typical complex observed for the interaction between Phormidium cyt f and Phormidium PC in the presence of Zn2+ is shown in Fig. 6. As suggested from the mutation studies discussed above, R93, D44, and D45 on PC face the large domain on cyt f where they interact with anionic residues on cyt f including E86, E95, and E123.
FIGURE 6.
The complex formed between Phormidium cyt f and Phormidium PC in the presence of Zn2+. A representative complex was selected from the 10 chosen for detailed analysis as described in the Methods section. The peptide backbone is shown as well as selected residues. The cyt f heme (black); cyt f-Y1 and PC-Y83 (yellow); Arg (dark blue); Lys (light blue); Glu (red); and Asp (magenta). Cyt f residues are labeled in red; PC residues are labeled in black.
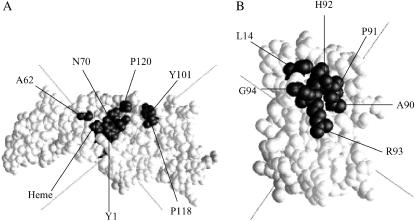
Close contacts for the WT with Zn2+ are presented in Table 6. It was impossible to analyze the WT complexes formed in the absence of the Zn2+ ion for close contacts because of the heterogeneity of the complexes formed. Close contacts are defined as those pairs of residues that are ≤8 Å apart in at least 8 out of 10 of the complexes chosen. This method was also used to examine complex formation for the interaction of Chlamydomonas cyt f with Chlamydomonas PC (Gross and Pearson, 2003). Residues on cyt f that form close contacts with WT PC are Y1, A62, N70, Y101, P118, and P120. Note that all of the residues except Y101 and N63 are nonpolar, and Y101 is partially nonpolar. The location of these residues on cyt f is shown in Fig. 7 A. Note that the footprint of Phormidium PC on Phormidium cyt f is situated further toward the large domain than is the case for the footprint of Chlamydomonas PC on Chlamydomonas cyt f (Gross and Pearson, 2003).
TABLE 6.
Close contacts for Phormidium cyt f -PC Brownian dynamics complexes
| PC | |
|---|---|
| Cyt f* | WT plus Zn2+ |
| Y1 | P91(86) and H92(87) |
| A62 | L14(12) |
| N70 | G94(89) |
| Y101 | R93(88) |
| P118 | A90(85), P91(86), H92(87), R93(88), and G94(89) |
| P120 | G94(89) |
Ten complexes were chosen at random as described in the Methods section. Residues were designated to be part of a close contact pair if they were found to be <8 Å apart in at least 8 of the 10 complexes examined.
FIGURE 7.
Location of the binding sites on Phormidium cyt f and PC. The close contacts listed in Table 6 were mapped onto cyt f (A) and PC (B). Contacts found in >8 of the 10 selected complexes are shown. The cyt f heme is also shown.
The close contacts on PC with the Zn2+ ion include L14, A90, P91, H92, R93, and G94. These residues have a strong hydrophobic character in agreement with the NMR results of Crowley et al. (2001, 2002) in the absence of the Zn2+ ion. Also, all of these residues with the exception of L14 are located on one segment of the polypeptide chain. The location of these residues on PC is shown in Fig. 7 B.
No charge pairs were observed with distances ≤8 Å. For this reason, electrostatic interactions were investigated by examining the residues on both cyt f and PC that appear in the list of 15 closest electrostatic contacts (one of the outputs of the program MacroDox) for the 10 chosen complexes. The results are shown in Table 7. A contact was recorded if it was observed in at least 5 out of 10 cases for either WT PC minus Zn2+ or WT PC plus Zn2+ complexes.
TABLE 7.
Electrostatic contacts in Brownian dynamics complexes formed between Phormidium cyt f and Phormidium PC
| Specific contacts*
|
No. of complexes | Distance between charged pairs (Å)† | ||
|---|---|---|---|---|
| Condition | Cyt f | PC | ||
| −Zn2+ | ||||
| D63 | K35(33) | 6 | 11.1 | |
| D188 | K35(33) | 7 | 10.8 | |
| Heme | K35(33) | 7 | 13.4 | |
| +Zn2+ | ||||
| E86 | D45(43) | 5 | 12.7 | |
| K46(44) | 6 | 11.7 | ||
| K53(51) | 6 | 9.5 | ||
| D57(55) | 5 | 12.4 | ||
| Zn2+ | 9 | 12.1 | ||
| E95 | D45(43) | 5 | 12.2 | |
| K46(44) | 5 | 10.8 | ||
| Zn2+ | 5 | 11.4 | ||
| E123 | R93(88) | 6 | 7.2 | |
| D188 | K35(33) | 5 | 10.4 | |
Taken from the list of the 15 closest electrostatic contacts for 10 complexes chosen at random as described in the Methods section. Only those contacts that were found in at least 5 out of 10 complexes for at least one condition (minus Zn2+ or plus Zn2+) were chosen.
Calculated for the number of complexes listed as described in the Methods section.
In the absence of the Zn2+ ion, only D63, D188, and the heme on cyt f were involved in electrostatic contacts with PC. These results are consistent with the mutant data presented in Table 4. K35 was the only residue on PC in close electrostatic contact with residues on cyt f. The D188-K35 contact is also observed in the presence of the Zn2+ ion, but the D63-K35 and heme-K35 contacts are not.
In the presence of the Zn2+ ion, E86 interacted with D45, K46, K53, D57, and the Zn2+ ion; E95 interacts with K46, K53, and the Zn2+; ion E123 interacts with R93; and D188 interacts with K35. The residues on both cyt f and PC that are found in the electrostatic contacts (Table 7) are the same as those that showed the greatest effect upon mutation. These results, taken together, provide evidence that the structure shown in Fig. 6 is correct.
An inspection of Fig. 6 shows than one possible electron transfer pathway would be from the Fe to the Y1 ligand to the heme on cyt f followed by electron transfer to the H92 ligand to the Cu on PC and finally to the Cu atom itself in an outer sphere electron transfer mechanism. The closest Y1-H92 distance in the 10 complexes studied was 10.2 ± 4.5 Å in the absence of the Zn2+ ion and 6.4 ± 0.8 Å in its presence, well within the limits of electron transfer postulated by Moser et al. (1992, 1995).
DISCUSSION
Evaluation of the MacroDox program
MacroDox is a very useful program for examining electrostatic interactions between proteins. In particular, it allows one both to predict the results of various mutations and to determine the structure and other properties of the complexes formed. These, in turn, provide the basis for further experiments. One advantage is that MacroDox includes random, diffusional factors as well as electrostatic components. It also requires relatively little computational time per simulation so that a large number of conditions (mutations, different ionic strengths, etc.) can be studied in a brief time. Steric effects can also be examined since both molecules have irregular geometrical shapes taken from the PDB files. For example, MacroDox was able to distinguish between wild-type and the Y10G mutant of Prochlorothrix PC (Gross, 2001). However, there are some serious limitations. First, both molecules are rigid, allowing no conformational changes in either molecule as they approach to form a complex. However, it does allow one to examine the effects of a particular predetermined conformation. Second, the Tanford-Kirkwood algorithm for calculating charges and pKs does not work well when the charges on a particular molecule are close enough to affect each other's protonation state. Third, hydrophobic interactions are not explicitly included. Fourth, reaction rates may be overestimated because molecule 2 (PC) lacks a low dielectric center (Northrup et al., 1993; Gabdoulline and Wade, 2001) and desolvation effects are neglected (Elcock et al., 1999). Desolvation effects will be negligible in our simulations because the charged residues never get close enough in the complexes to be desolvated since the average distance between charged pairs in a complex is 11 Å (Table 7). Moreover, desolvation effects should not affect the relative effects of mutations to within the ±10% standard deviation observed in the simulations (S. H. Northrup, personal communication, 2002).
Agreement of BD simulations and experimental results
The question arises as to how well the BD results agree with experimentally determined rate constants. Table 8 compares the BD results with the experimental results of Schlarb-Ridley et al. (2002) for the interaction of Phormidium cyt f with WT and mutant Phormidium PCs and with those of Hart et al. (2003) for the interaction of Phormidium cyt f mutants with wild-type Phormidium PC in the absence of the Zn2+ ion and at ∼100 mM ionic strength.
TABLE 8.
Comparison of rates of electron transport calculated from BD simulations with experimental values
| BD simulation†
|
||||||
|---|---|---|---|---|---|---|
| Experimental*
|
18-Å cutoff
|
20-Å cutoff
|
||||
| Mutant | 10−6 × k2† (M−1 s−1) | % WT | 10−6 × k2 (M−1 s−1) | % WT | 10−6 × k2 (M−1 s−1) | % WT |
| PC mutants | ||||||
| WT | 47 | 100 | 80 | 100 | 336 | 100 |
| D45(43)A‡ | 59 | 126 | 147 | 184 | 501 | 149 |
| K46(44)A | 31 | 66 | 41 | 52 | 227 | 68 |
| K53(51)A | 33 | 70 | 48 | 60 | 232 | 69 |
| R93(88)A | 19 | 40 | 29 | 37 | 152 | 45 |
| R93(88)Q | 21 | 45 | 19 | 24 | 107 | 32 |
| R93(88)E | 10 | 21 | 18 | 22 | 113 | 33 |
| Cyt f mutants | ||||||
| WT | 40 | 100 | – | – | 324 | 100 |
| Q7R | 29 | 73 | – | – | 236 | 73 |
| Q7E | 40 | 100 | – | – | 372 | 115 |
| D63A | 31 | 78 | – | – | 248 | 77 |
| E165A | 39 | 98 | – | – | 217 | 67 |
| D187A | 38 | 95 | – | – | 215 | 66 |
| D188A | 42 | 105 | – | – | 207 | 64 |
| D192A | 41 | 103 | – | – | 223 | 68 |
PC results taken from Schlarb-Ridley et al. (2002); cyt f results taken from Hart et al. (2003).
A total of 10,000 trajectories were completed for WT and mutant PCs at 100 mM ionic strength in the absence of Zn2+. Rates of electron transfer (k2) were calculated as described in the Methods sections for reaction coordinate cutoff distances of 18 and 20 Å. Rates obtained in the absence of the electrostatic field were not subtracted from the values shown to compare with the experimental values that would include nonelectrostatic contributions.
Mutants were constructed as described in the Methods section. Residue numbers are taken from the consensus sequence (Pearson, 2000) with residue numbers from the PDB file of Phormidium PC sequence in parentheses.
The reaction rates for WT PC are greater for the BD simulations than for experiments. There are at least three reasons for this. First, the rate constants for the simulations are overestimated due to the absence of a low internal dielectric constant for PC and the desolvation effects discussed above. Second, no attempt was made to correct the BD reaction rates for attenuation due to the distance between the metal centers that would decrease the measured reaction rates (Moser et al., 1992, 1995). Note, however, as discussed above, the effective electron transfer distance could be as low as 6–11 Å if the rate limiting step is the transfer of the electron between Y1 on cyt f and H87 on PC. Third, the calculated rates of electron transfer are a function of the reaction coordinate cutoff distance. For example, for the 20-Å cutoff distance, all complexes showing a Cu-Fe distance of less than 20 Å at closest approach were used to calculate the rates. The assumption is that the Cu in PC in all of these complexes will eventually come close enough to the Fe atom on cyt f to receive an electron. This is based on the assumption of a two-step reaction model: First, electrostatic forces would bring PC to within 20 Å of cyt f, at which point short range forces such as hydrophobic interactions bring the two molecules to a final electron-transfer-active dock. Electrostatic forces also help to orient PC within the complexes (compare Fig. 4, A with B). The use of smallest Cu-Fe distance as the reaction criterion also brings hydrophobic residues on PC close to those on cyt f. The use of the 20-Å reaction criterion is almost certainly an overestimate. If 18 Å is used instead, the calculated rate is decreased to 80 × 10 6 M−1 s−1, which is comparable to the experimental value of 47 × 10 6 M−1 s−1.
Please note, however, that the percent of WT electron transport observed for the various mutants is essentially independent of the reaction criterion cutoff used. There is good agreement between the percent of WT determined experimentally and from simulations for the PC mutants except for D45A in which the percent stimulation is significantly greater for the simulated than for the experimental rates. The difference between calculated and experimental results for D45A-PC is greater than the error in either the BD simulations or the experiments. There may be something else besides electrostatic effects that limit rapid experimental electron transfer rates.
The results for the cyt f mutants are more complicated. Computational and experimental results agree fairly well for the Q7R and D63A mutants. These residues are located on the large domain of cyt f. In contrast, there is much less inhibition of electron transfer rates in experiments than in simulations for the residues located on the small domain of cyt f: namely, D187A, D188A, and D192A. One explanation is that the conformation of Phormidium cyt f may be different in solution than in the crystal structure. In particular, there may be a slight difference in the hinge angle between the two domains. Nonetheless, the close agreement observed for the PC mutants shows that BD simulations can be used to predict the effects of mutations.
Several factors may be important in complex formation including simple diffusion, electrostatic forces, and hydrophobic interactions. In the MacroDox simulations, simple diffusion is not important at low ionic strength, particularly in the presence of the Zn2+ ion as indicated by the small number of complexes formed in the absence of the electrostatic field. However, it is responsible for ∼12% of the complexes formed in the absence of the Zn2+ ion at 100 mM ionic strength.
The role of electrostatic interactions in complex formation
Several lines of evidence indicate that the interaction of Phormidium cyt f with Phormidium PCs is a function of the charge configurations on both molecules both in the presence and absence of the Zn2+ ion. First, Phormidium cyt f with a net negative charge of −13 and a large negatively charged electrostatic field (Fig. 1 A) does not interact with negatively charged Chlamydomonas PC. Furthermore, Phormidium PC, which has a large positive electrostatic field (Fig. 1 B) replacing the negatively charged electrostatic field on green algal and higher plant PCs, does not interact with Chlamydomonas cyt f that, like which turnip cyt f (Pearson et al., 1996), has a large positive electrostatic field.
Second, mutants of Phormidium PC that decrease the number of negative charges, such as D45A, D44A + D43A, the triple mutants such as D44A + D45A + E54A, and the quadruple mutant D44A + D45A + E54A + D57A, all increase the number of complexes formed (Fig. 2 A). In contrast, those that decrease the number of positive charges, such as R93A and K46A, inhibit complex formation. These results agree with the experimental results of Schlarb-Ridley et al. (2002; see Table 8 for a comparison). Conversely, mutants of negatively charged residues on Phormidium cyt f also inhibit complex formation.
Third, the number of complexes formed and reaction rates are dependent on the ionic strength of the medium (Tables 2 and 3) in agreement with the kinetic studies of Schlarb-Ridley et al. (2002) but not with the results of NMR complex formation (Crowley et al., 2001). The ionic strength dependence was observed both the in presence and absence of the Zn2+ ion.
However, the strength of the electrostatic free energy is larger in magnitude in the presence of the Zn2+ ion than in its absence. The average electrostatic free energies for the 10 complexes studied were −8.2 ± 0.8 kcal/mol−1 in the presence of the Zn2+ ion and 2.5 ± 0.7 kcal/mol−1 in its absence. The small electrostatic free energy observed in the absence of the Zn2+ ion is consistent with the experimental results of Schlarb-Ridley et al. (2002) who observed small electrostatic effects. Even in the presence of the Zn2+ ion, the electrostatic free energy of Phormidium cyt f-Phormidium PC interactions was less than the 10.5 ± 0.5 kcal/mol−1 observed by Gross and Pearson (2003) for Chlamydomonas cyt f-Chlamydomonas PC complexes (see Gross and Pearson, 2003, for a discussion of the calculation of electrostatic free energies).
The role of the Zn2+ ion in complex formation
The question arises as to the role of the Zn2+ ion in complex formation. Although the Zn2+ ion was used to crystalize Phormidium PC, there is as yet no evidence that it is found in vivo. However, this does not say that divalent cations such as Ca2+ may not be involved in regulating electron transfer. It would be interesting to see how bound divalent cations (i.e., those that stimulate electron transfer when present at very low concentrations) affect experimental rates of electron transfer and complex formation between Phormidium PC and cyt f. The Zn2+ ion studies also provide an interesting model system for studying the effects of electrostatic fields and complex orientation on rates of electron transfer.
Mapping of binding regions on Phormidium cyt f and PC
The mutant studies also allowed us to map the binding regions on both PC and cyt f (Figs. 3. and 4). In the case of PC, the mutant studies show two regions that are involved in its interaction with cyt f. One is on the front side of the PC molecule as presented in Fig. 3 centered on D44 and D45 (and the Zn2+ ion when present). It also includes D79, R93, K46, and K100. In the presence of the Zn2+ ion, residues K53 and E54 are also involved. An examination of the structure of the complex formed between PC and cyt f in the presence of the Zn2+ ion also implicates these residues (Fig. 6 and Table 7). Thus, both methods (mutant studies and an examination of the complexes formed) lead to the conclusion that these residues are involved in complex formation.
The second region is located on the backside of the PC molecule near the top and includes D10, E17, and E70 on PC. K35 and K6 on PC are also involved for complexes formed in the absence of the Zn2+ ion. This region appears to be the more important of the two in the absence of the Zn2+ ion, which is consistent with the superimposed structures of the complexes shown in Fig. 5 A in which the top of the PC molecule interacts with cyt f.
The role of hydrophobic interactions
Although hydrophobic interactions are not explicitly present in MacroDox simulations, the use of the smallest Cu-Fe distance as the reaction criterion (to select for electron-transfer-active complexes) also selects for complexes that show significant hydrophobic interactions as shown in Table 6 for complexes formed in the presence of the Zn2+ ion. Although not shown, this is also true for complexes formed in the absence of the Zn2+ ion and agrees with the experimental results of Crowley et al. (2001). Hydrophobic interactions have also been shown to be important in NMR studies of the interaction of turnip cyt f with spinach PC (Ubbink et al., 1998; Ejdeback et al., 2000) and for Phormidium cyt f interacting with Prochlorothrix PC (Crowley et al., 2002). BD studies of the interactions of turnip cyt f with either poplar (Pearson and Gross, 1998) or spinach PC (Nelson et al., 1999) and Chlamydomonas cyt f with Chlamydomonas PC also implicated hydrophobic interactions, although hydrophobic interactions are not explicitly treated in the present BD simulations.
The binding site on cyt f for PC
The binding site on cyt f for Phormidium PC is different in the presence and absence of the Zn2+ ion. In its presence, the east face of Phormidium PC, containing R93, D44, D45, and the Zn2+ ion, faces toward the large domain on cyt f containing E86, D95, and E123 as documented by both the mutant studies (Table 4) and close electrostatic contacts found in the complexes (Table 7). This orientation is different from that observed in algae (Gross and Pearson, 2003) in which the negatively charged residues on the east face of PC interact with residues K65, K188, and K189 on the small domain of cyt f. Interactions between anionic residues on PC and those of the small domain of cyt f were also observed in the NMR studies of complexes formed between turnip cyt f and spinach PC (Ubbink et al., 1998; Ejdeback et al., 2000). In contrast, in the absence of the Zn2+ ion, Phormidium PC interacts with anionic residues on the small domain of cyt f including E165, D187, and D188 as shown by the mutant studies (Table 5).
In the presence of the Zn2+ ion, the PC molecules in the complexes formed have uniform orientations (Fig. 5 C). PC also showed a uniform orientation in BD studies of complexes formed between Chlamydomonas cyt f and Chlamydomonas PC (Gross and Pearson, 2003), turnip cyt f and poplar (Pearson and Gross, 1998) and spinach (Nelson et al., 1999) PC, as well as in experimental studies of complex formation between turnip cyt f and spinach PC (Ubbink et al., 1998). In algae and higher plant PCs, negatively charged residues on the east face of PC in the vicinity of E43 and D44 interact with a line of highly conserved positively charged residues on cyt f. These PCs show two important characteristics: large electrostatic fields on the east face and an asymmetric distribution of charged residues on both PC and cyt f.
Moreover, in the absence of the Zn2+ ion, the orientation of Phormidium PC on Phormidium cyt f is heterogeneous for both the NMR complexes (Crowley et al., 2001; Fig. 5 B) and the BD simulations (Fig. 5 A). The electrostatic field is weaker and the distribution of charges on PC is less asymmetric. Decreasing the number of negative charges on the east face of Phormidium PC (by mutating negatively charged residues to alanine) increased the magnitude of the electrostatic field (Fig. 1 D), the degree of asymmetry of the charge distribution in the vicinity of residue 44, and the homogeneity of the complexes formed (Fig. 5 D). Thus, electrostatic forces may have two roles in complex formation: 1), To bring the two molecules together and 2), to ensure a correct orientation provided there is an asymmetric distribution of the charges on both molecules. However, it is not known whether homogeneity of orientation of the two molecules within the complex enhances the rate of electron transfer.
Note that the binding sites for PC on Phormidium cyt f, both in the presence and absence of the Zn2+ ion, are located on the top portion of the cyt f molecule as presented in Fig. 6. This portion of the cyt f molecule faces the solvent in the structure of the cyanobacterial cyt b6f complex determined by Kurisu et al. (2003). Thus, binding of PC at either site is permitted.
Crowley and Ubbink (2003) have proposed that electrostatic interactions bring PC into an encounter complex with cyt f, whereas hydrophobic interactions are involved in forming the final electron-transfer-active complexes. The question arises as to whether the complexes that we observed are encounter complexes, final electron transfer complexes, or something in between. We do not believe that these are initial encounter complexes that are probably similar to complex I observed by Pearson et al. (1996) in which the electrostatic interactions are greater than observed here but the Cu-Fe distance is much larger making electron transfer impossible. Our complexes are not final electron transfer complexes either because complex formation has not been optimized in any way. In an experimental situation, hydrophobic interactions would promote the formation of the final electron-transfer-active dock. However, the BD complexes obtained are probably very close to the final electron transfer complexes since the Y1-H92 distance is only 10.2 ± 4.5 Å in the absence of the Zn2+ ion and 6.4 ± 0.8 Å in its presence. Thus, we can conclude that electrostatic effects are important both for forming the initial encounter complex and the final electron-transfer-active complex, whereas hydrophobic effects are only involved in the producing the final electron-transfer-active complexes.
CONCLUSIONS
BD simulations show that electrostatic interactions are important for complex formation between Phormidium cyt f and Phormidium PC. However, the exact nature of the complexes formed depends on the presence of the Zn2+ observed in the crystal structure. In the absence of the Zn2+ ion, the orientation of the complexes formed is random with R93, D44, and D45 on PC in many of them oriented so that they face the small domain on cyt f containing D187 and D188. The addition of the Zn2+ ion greatly increases the number of complexes formed and also shifts the orientation of PC in the complexes so that R93, D44, and D45 on PC face the large domain on cyt f containing E86, E95, and E123. The complexes are also more homogeneous in orientation. Hydrophobic interactions stabilize the complexes both in the presence and absence of the Zn2+ ion.
Acknowledgments
The author thanks Mr. Irving Rosenberg for his assistance in carrying out the MacroDox simulations and Dr. M. Ubbink for the NMR structures of the Phormidium cyt f-Phormidium PC complexes. The author also thanks Esmael Haddanian, Kathryn Thomasson, Dan Davis, and George Bullerjahn for their helpful discussions and careful reading of the manuscript.
References
- Anderson, G. P., D. G. Sanderson, C. H. Lee, S. Durell, L. B. Anderson, and E. L. Gross. 1987. The effect of ethylene diamine chemical modification of plastocyanin on the rate of cytochrome f oxidation and P-700+ reduction. Biochim. Biophys. Acta. 894:386–398. [DOI] [PubMed] [Google Scholar]
- Berman, H. M., J. Westboro, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, C. S., D. S. Bendall, H. C. Freeman, J. M. Guss, C. J. Howe, M. J. Wagner, and M. C. Wilce. 1999. The structure of plastocyanin from the cyanobacterium Phormidium laminosum. Acta Crystal. D55:414–421. [DOI] [PubMed] [Google Scholar]
- Carrell, C. J., B. G. Schlarb, D. S. Bendall, C. J. Howe, W. A. Cramer, and J. L. Smith. 1999. Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum. Biochemistry. 38:9590–9599. [DOI] [PubMed] [Google Scholar]
- Chi, Y. I., L. S. Huang, Z. Zhang, J. G. Fernandez-Velasco, and E. A. Berry. 2000. X-ray structure of a truncated form of cytochrome f from Chlamydomonas reinhardtii. Biochemistry. 39:7689–7701. [DOI] [PubMed] [Google Scholar]
- Cramer, W. A., G. M. Soriano, M. Ponomarev, D. Huang, H. Zhang, S. E. Martinez, and J. L. Smith. 1996. Some new structural aspects and old controversies concerning the cytochrome b6f complex of oxygenic photosynthesis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 47:477–508. [DOI] [PubMed] [Google Scholar]
- Crowley, P. B., G. Otting, B. G. Schlarb-Ridley, G. W. Canters, and M. Ubbink. 2001. Hydrophobic interactions in a cyanobacterial plastocyanin-cytochrome f complex. J. Am. Chem. Soc. 123:10444–10453. [DOI] [PubMed] [Google Scholar]
- Crowley, P. B., and M. Ubbink. 2003. Close encounters of the transient kind: protein interactions in the photosynthetic redox chain investigated by NMR spectroscopy. Acc. Chem. Res. 36:723–730. [DOI] [PubMed] [Google Scholar]
- Crowley, P. B., N. Vintonenko, G. S. Bullerjahn, and M. Ubbink. 2002. Plastocyanin-cytochrome f interactions: the influence of hydrophobic patch mutations studied by NMR spectroscopy. Biochemistry. 41:15698–15705. [DOI] [PubMed] [Google Scholar]
- De Rienzo, F., R. R. Gabdoulline, M. C. Menziani, P. G. De Benedetti, and R. C. Wade. 2001. Electrostatic analysis and Brownian dynamics simulation of the association of plastocyanin and cytochrome f. Biophys. J. 81:3090–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell, S. R., J. K. Labanowski, and E. L. Gross. 1990. Modeling of the electrostatic potential field of plastocyanin. Arch. Biochem. Biophys. 277:241–254. [DOI] [PubMed] [Google Scholar]
- Ejdeback, M., A. Bergkvist, B. G. Karlsson, and M. Ubbink. 2000. Side-chain interactions in the plastocyanin-cytochrome f complex. Biochemistry. 39:5022–5027. [DOI] [PubMed] [Google Scholar]
- Elcock, A. H., R. R. Gabdoulline, R. C. Wade, and J. A. McCammon. 1999. Computer simulation of protein-protein kinetics: acetylcholinesterase-fasciculin. J. Mol. Biol. 291:149–162. [DOI] [PubMed] [Google Scholar]
- Ermak, D. L., and J. C. McCammon. 1978. Brownian dynamics with hydrodynamic interactions. J. Phys. Chem. 69:1352–1360. [Google Scholar]
- Freeman, H. C. 1981. Electron transfer in “blue” copper proteins. Coord. Chem. 21:29–52. [Google Scholar]
- Gabdoulline, R. R., and J. C. Wade. 1996. Effective charges for macromolecules in solvent. J. Phys. Chem. 100:3868–3878. [Google Scholar]
- Gabdoulline, R. R., and J. C. Wade. 1998. Brownian dynamics simulation of protein-protein diffusional encounter. Methods. 14:329–341. [DOI] [PubMed] [Google Scholar]
- Gabdoulline, R. R., and J. C. Wade. 2001. Protein-protein association: investigation of factors influencing association rates by Brownian dynamics simulations. J. Mol. Biol. 9:1139–1155. [DOI] [PubMed] [Google Scholar]
- Gong, E. S., J. Q. Wen, N. E. Fisher, S. Young, C. J. Howe, D. S. Bendall, and J. C. Gray. 2000. The role of individual lysine residues in the basic patch on turnip cytochrome f for the electrostatic interactions with plastocyanin in vitro. Eur. J. Biochem. 267:3461–8. [DOI] [PubMed] [Google Scholar]
- Gray, J. C. 1992. Cytochrome f: structure, function and biosynthesis. Photosynth. Res. 34:359–374. [DOI] [PubMed] [Google Scholar]
- Gross, E. L. 1993. Plastocyanin: structure and function. Photosynth. Res. 37:103–116. [DOI] [PubMed] [Google Scholar]
- Gross, E. L. 1996. Plastocyanin: structure, location, diffusion, and electron transfer mechanisms. In Oxygenic Photosynthesis: The Light Reactions. D. Ort and C. Yocum, editors. Kluwer Academic Publishers, Dordrecht, The Netherlands. 413–429.
- Gross, E. L. 2001. A Brownian dynamics study of evolutionary changes in the electrostatic interactions between plastocyanin and cytochrome f in cyanobacteria and green plants. In Proceedings of the 12th International Congress on Photosynthesis. CSIRO Publishing, Brisbane, Australia. S11–014.
- Gross, E. L., and D. C. Pearson, Jr. 2003. Brownian dynamics simulations of the interaction of Chlamydomonas cytochrome f with plastocyanin and cytochrome c6. Biophys. J. 85:2055–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss, J. M., H. D. Bartunik, and H. C. Freeman. 1992. Accuracy and precision in protein structure analysis: restrained least-squares refinement of the structure of poplar plastocyanin at 1.33 Å resolution. Acta Crystalogr. B. 48:790–811. [DOI] [PubMed] [Google Scholar]
- Hart, S. E., B. G. Schlarb-Ridley, C. Delon, D. S. Bendall, and C. J. Howe. 2003. Role of the charges on cytochrome f from the cyanobacterium Phormidium laminosum in its interactions Plastocyanin. Biochemistry. 42:4829–4836. [DOI] [PubMed] [Google Scholar]
- Harvey, S. C. 1989. Treatment of electrostatic effects in macromolecular modeling. Proteins. 5:78–92. [DOI] [PubMed] [Google Scholar]
- Hauska, G., E. Hurt, N. Gabellini, and W. Lockau. 1983. Comparative aspects of the quinol-cytochrome c/plastocyanin oxidoreductases. Biochim. Biophys. Acta. 726:97–133. [DOI] [PubMed] [Google Scholar]
- Hope, A. B. 2000. Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms. Biochim. Biophys. Acta. 1456:5–26. [DOI] [PubMed] [Google Scholar]
- Kannt, A., S. Young, and D. S. Bendall. 1996. The role of acidic residues of plastocyanin in its interaction with cytochrome f. Biochim. Biophys. Acta. 1277:115–126. [PubMed] [Google Scholar]
- Kurisu, G., H. Zhang, J. L. Smith, and W. A. Cramer. 2003. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 302:1009–1014. [DOI] [PubMed] [Google Scholar]
- Lee, B. H., T. Hibino, T. Takabe, P. J. Weisbeek, and T. Takabe. 1995. Site-directed mutagenetic study on the role of negative patches on silene plastocyanin in the interactions with cytochrome f and photosystem I. J. Biochem. (Tokyo). 117:1209–1217. [DOI] [PubMed] [Google Scholar]
- Martinez, S. E., D. Huang, M. Ponomarev, W. A. Cramer, and J. L. Smith. 1996. The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain. Protein Sci. 5:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, S. E., D. Huang, A. Szczepaniak, W. A. Cramer, and J. L. Smith. 1994. Crystal structure of the chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure. 2:95–105. [DOI] [PubMed] [Google Scholar]
- Matthew, J. B. 1985. Electrostatic effects in proteins. Annu. Rev. Biophys. Biophys. Chem. 14:387–417. [DOI] [PubMed] [Google Scholar]
- McCammon, J. C., and S. C. Harvey. 1987. Dynamics of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, UK.
- Morand, L. Z., M. K. Frame, K. K. Colvert, D. A. Johnson, D. W. Krogmann, and D. J. Davis. 1989. Plastocyanin cytochrome f interaction. Biochemistry. 28:8039–8047. [DOI] [PubMed] [Google Scholar]
- Moser, C. C., J. M. Keske, K. Warncke, R. S. Farid, and P. L. Dutton. 1992. Nature of biological electron transfer. Nature. 355:796–802. [DOI] [PubMed] [Google Scholar]
- Moser, C. C., C. C. Page, R. Farid, and P. L. Dutton. 1995. Biological electron transfer. J. Bioenerg. Biomembr. 27:263–274. [DOI] [PubMed] [Google Scholar]
- Nelson, N., D. C. Pearson, Jr., and E. L. Gross. 1999. The interaction of plastocyanin with cytochrome f: a Brownian dynamics study. In Photosynthesis: Mechanisms and Effects, Vol. 3. G. Garab, editor. Kluwer Academic Publishers, Dordrecht, The Netherlands. 1493–1498.
- Nicholls, A., and B. Honig. 1991. A rapid finite-difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 12:435–445. [Google Scholar]
- Northrup, S. H. 1996. Theoretical simulation of protein-protein interactions. In Cytochrome c: A Multidiciplinary Approach. R. A. Scott and A. G. Mauk, editors. University Science Publishers, Sausalito, CA. 543–570.
- Northrup, S. H. 1999. MacroDox v.2.3.1: Software for the Prediction of Macromolecular Interaction. Tennessee Technological University, Cookeville, TN.
- Northrup, S. H., J. O. Boles, and J. C. L. Reynolds. 1987a. Electrostatic effects in the Brownian dynamics of association and orientation of heme proteins. J. Phys. Chem. 91:5991–5998. [Google Scholar]
- Northrup, S. H., J. O. Boles, and J. C. Reynolds. 1988. Brownian dynamics of cytochrome c and cytochrome c peroxidase association. Science. 241:67–70. [DOI] [PubMed] [Google Scholar]
- Northrup, S. H., J. C. Luton, J. O. Boles, and J. C. L. Reynolds. 1987b. Brownian dynamics simulation of protein association. J. Comput. Aided Mol. Des. 1:291–311. [DOI] [PubMed] [Google Scholar]
- Northrup, S. H., K. A. Thomasson, C. M. Miller, P. D. Barker, L. D. Eltis, J. G. Guillemette, S. C. Inglis, and A. G. Mauk. 1993. Effect of charged amino acid mutations on the bimolecular kinetics of reduction of yeast iso-1-ferricytochrome c by bovine ferrocytochrome b5. Biochemistry. 32:6613–6623. [DOI] [PubMed] [Google Scholar]
- Pearson, D. C., Jr. 2000. Brownian dynamics study of the interaction between cytochrome f and mobile electron transfer proteins. PhD dissertation. The Ohio State University, Columbus, OH.
- Pearson, D. C., Jr., and E. L. Gross. 1998. Brownian dynamics study of the interaction between plastocyanin and cytochrome f. Biophys. J. 75:2698–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, D. C., Jr., E. L. Gross, and E. S. David. 1996. The electrostatic properties of cytochrome f: implications for docking with plastocyanin. Biophys. J. 71:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, L., and N. M. Kostic. 1993. Importance of protein rearrangement in the electron-transfer reaction between the physiological partners cytochrome f and plastocyanin. Biochemistry. 32:6073–6080. [DOI] [PubMed] [Google Scholar]
- Redinbo, M. R., D. Cascio, M. K. Choukair, D. Rice, S. Merchant, and T. O. Yeates. 1993. The 1.5-Å crystal structure of plastocyanin from the green alga Chlamydomonas reinhardtii. Biochemistry. 32:10560–10567. [DOI] [PubMed] [Google Scholar]
- Redinbo, M. R., T. O. Yeates, and S. Merchant. 1994. Plastocyanin: structural and functional analysis. J. Bioenerg. Biomembr. 26:49–66. [DOI] [PubMed] [Google Scholar]
- Schlarb-Ridley, B. G., D. S. Bendall, and C. J. Howe. 2002. Role of electrostatics in the interaction between cytochrome f and plastocyanin of the cyanobacterium Phormidium laminosum. Biochemistry. 41:3279–3285. [DOI] [PubMed] [Google Scholar]
- Sigfridsson, K. 1998. Plastocyanin, an electron-transfer protein. Photosynth. Res. 57:1–28. [Google Scholar]
- Soriano, G. M., M. V. Pomamarev, R. A. Piskorowski, and W. A. Cramer. 1998. Identification of the basic residues of cytochrome f responsible for electrostatic docking interactions with plastocyanin in vitro: relevance to the electron transfer reaction in vivo. Biochemistry. 37:15120–15128. [DOI] [PubMed] [Google Scholar]
- Sykes, A. G. 1991. Plastocyanin and the blue copper proteins. Struct. Bond. 75:175–224. [Google Scholar]
- Takabe, T., and H. Ishikawa. 1989. Kinetic studies on a cross-linked complex between plastocyanin and cytochrome f. J. Biochem. (Tokyo). 105:98–102. [DOI] [PubMed] [Google Scholar]
- Takabe, T., K. Takenaka, H. Kawamura, and Y. Beppu. 1986. Charges on proteins and distances of electron transfer in metalloprotein redox reactions. J. Biochem. (Tokyo). 99:833–840. [DOI] [PubMed] [Google Scholar]
- Takenaka, K., and T. Takabe. 1984. Importance of local positive charges on cytochrome f for electron transfer to plastocyanin and potassium ferricyanide. J. Biochem. (Tokyo). 96:1813–1821. [DOI] [PubMed] [Google Scholar]
- Ubbink, M., M. Ejdebäck, B. G. Karlsson, and D. S. Bendall. 1998. The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure. 6:323–335. [DOI] [PubMed] [Google Scholar]
- Warwicker, J., and H. C. Watson. 1982. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J. Mol. Biol. 157:671–679. [DOI] [PubMed] [Google Scholar]