Water is the major component of all living cells, actually it composes 70% of our bodies. Its properties, very unusual when compared to other solvents, have made possible the life that we know. Water molecules spontaneously dissociate into a hydronium (H3O+) and a hydroxide ion (OH−). This autoprotolysis is a very rare event with the average lifetime of a single H2O molecule being ∼14 h (Eigen, 1964).
However, not all of the H2O molecules in our body are what we call liquid water. H2O molecules can be tightly bound to biological material and are occluded in proteins where they are often involved in catalytic reactions. The membrane protein bacteriorhodopsin (bR) accommodates several of such water-filled cavities (Dencher et al., 2000). Their participation in the light-driven proton translocation, which is the functional task of this molecular machine, is intensively studied. A cavity close to the extracellular membrane surface accommodates a local area network (LAN) of hydrogen-bonded water molecules and amino acid side chains. This LAN houses an excess proton, which is released after photoexcitation of bacteriorhodopsin. Over the recent years, the group of K. Gerwert has specifically addressed the role of this LAN by time-resolved Fourier transform infrared spectroscopy. In this issue of the Biophysical Journal, Garczarek et al. (2004) critically gauged the characteristic infrared (IR) spectroscopic signatures of the excess proton within this LAN. As an excellent scientific practice, they solved the controversy about the nature of the spectral changes in collaboration with the group of M. El-Sayed. Continuum absorbance changes due to the release of the excess proton could be clearly distinguished from photothermal heating artifacts of bR. This is a very important result since the concept of the continuum bands might be applicable to other proton translocating proteins as well.
Considering that almost all known enzymatic mechanisms involve proton transfers, these are issues of major significance for understanding protein function in general. Besides the role per se, the transfer of protons leads to the redistribution of charges in a protein. By these electrostatic means, structural changes of the protein are triggered that may induce changes in affinity to ligands or to interacting proteins.
As a prerequisite for proton translocation, a proton-conducting wire must exist, made of water molecules and/or ionizable amino acid side chains. The remarkably fast proton transfer in water (diffusion constant DH+ = 9.3 × 10−9 m2/s) can be related to the Grotthuss mechanism, where the charge of the proton is displaced along the hydrogen-bonded network of water molecules rather than the mass. Such a mechanism is effective only when the involved hydrogen bonds are easy to break and the cleavage of a single hydrogen bond of liquid water requires only ∼10 kJ/mol.
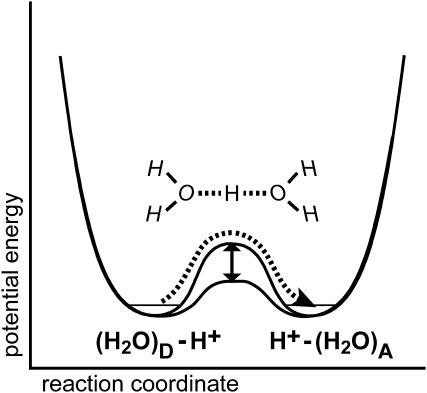
The energetics of proton transfer can be envisaged as a double-well potential where the proton is transferred from a donor (left well in Fig. 1) to the acceptor (right well). The key for fast proton transfer lies in the height of the intermittent barrier. Lowering the barrier by bringing the donor and acceptor molecules in appropriate distance and orientation will accelerate proton transfer. The efficiency of proton transfer is determined by the potential level (free energy) of the donor and the acceptor, respectively. For proton transfer in water, the double-well potential is symmetrical and the barrier is low (Fig. 1). Thus, protons can be rapidly transferred between donor and acceptor. It was Georg Zundel who demonstrated that this “large proton polarizability” gives rise to intense continua in the IR spectra (Zundel, 1992).
FIGURE 1.
Double-well potential showing the energy barrier to overcome for classical proton transfer (dashed line) from the donor molecule (left well) to the acceptor molecule (right well). Lowering the barrier (vertical arrow) accelerates proton transfer.
For a heterogeneous proton-conducting chain in a protein, e.g., like that of the extracellular LAN of bR, a series of such potentials may exist for the protonatable groups (peptide backbone, amino acid side chains, and water molecules) where the barrier for proton transfer is low and the lowest potential well determines the localization of the proton. Putting energy into the system, e.g., by light absorption in photosynthetic proteins, will shift the potential wells with respect to each other and proton transfer ensues. If the last member of the chain has the lowest potential, the proton gets trapped in this well. Efficient proton translocation is deducible from the occurrence of broad negative bands in the IR difference spectrum because the proton with its large polarizability is lost.
Protons, although not as small from the standpoint of mass as electrons, are sufficiently light for treating their properties by quantum mechanics. Marcus theory, which has been extremely useful for our current understanding of electron transfer in biological systems, can also be applied to advance our knowledge of the possible pathways for proton transfer (Silverman, 2000). The thermal de Broglie wavelength of the proton is 1.5 Å, which compares well with the distances of proton transfer reactions. An intriguing consequence is that protons may tunnel from a proton donor to the acceptor, i.e., they do not pass the transition state but rather cross the potential energy barrier (Fig. 1). The observation of large kinetic H/D isotope effects and of temperature independent reactions rates at very low temperatures provides experimental evidence for the contribution of tunneling to proton transfer reactions. Advanced Car-Parrinello calculations infer, however, that the proton transfer reaction in water can be described as a classical process—tunneling does not contribute significantly (Marx et al., 1999).
Though water seems to be an ideal mediator for proton transfer in proteins, there is no rule without an exception. In the water transporting channel aquaporin, protons are evidently not transported although a linear chain of hydrogen-bonded water molecules exists. The bipolar organization of the water chain and the electrostatic field at the restriction pore are incompatible with proton translocation across aquaporin (Chakrabarti et al., 2004).
Finally, good scientific work leaves an open end and the work of Gaczareck et al. is inspiring, indeed. Protons released by the extracellular LAN may dwell for a while along the membrane surface before they dissipate into the aqueous bulk medium (Heberle et al., 1994). This reaction, which proceeds on the timescale of several hundred microseconds, is determined by the properties of the membrane surface and the buffer concentration in the medium. Sure enough, the surface of a biological membrane represents a hydrogen-bonded network which is capable of accommodating excess protons. The bioenergetic consequences of this capacity finally culminated in the formulation of the localized variant of chemiosmosis (Williams, 2001). Future studies that aim at revealing the vibrational signature of the polarizable proton along membrane surfaces are highly appreciated and welcome.
References
- Chakrabarti, N., E. Tajkhorshid, B. Roux, and R. Pomès. 2004. Molecular basis of proton blockage in aquaporins. Structure. 12:65–74. [DOI] [PubMed] [Google Scholar]
- Dencher, N. A., H. J. Sass, and G. Büldt. 2000. Water and bacteriorhodopsin: structure, dynamics, and function. Biochim. Biophys. Acta. 1460:192–203. [DOI] [PubMed] [Google Scholar]
- Eigen, M. 1964. Proton transfer, acid-base catalysis and enzymatic hydrolysis. Part I: elementary processes. Angew. Chem. Int. Ed. 3:1–19. [Google Scholar]
- Garczarek, F., J. Wang, M. A. El-Sayed, and K. Gerwert. 2004. The assignment of the different infrared continuum absorbance changes observed in the 3000–1800-cm−1 region during the bacteriorhodopsin photocycle. Biophys. J. 87:2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle, J., J. Riesle, G. Thiedemann, D. Oesterhelt, and N. A. Dencher. 1994. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 370:379–382. [DOI] [PubMed] [Google Scholar]
- Marx, D., M. E. Tuckerman, J. Hutter, and M. Parrinello. 1999. The nature of the hydrated excess proton in water. Nature. 397:601–604. [Google Scholar]
- Silverman, D. N. 2000. Marcus rate theory applied to enzymatic proton transfer. Biochim. Biophys. Acta. 1458:88–103. [DOI] [PubMed] [Google Scholar]
- Williams, R. J. 2001. The structures of organelles and reticula: localised bioenergetics and metabolism. Chem. Biochem. 2:637–641. [DOI] [PubMed] [Google Scholar]
- Zundel, G. 1992. Proton polarizability and proton transfer processes in hydrogen bonds and cation polarizability of other cation bonds. Their importance to understand molecular processes in electrochemistry and biology. Trends Phys. Chem. 3:129–156. [Google Scholar]