Abstract
In the preceding, accompanying article, we present models of the structure and voltage-dependent gating mechanism of the KvAP bacterial K+ channel that are based on three types of evidence: crystal structures of portions of the KvAP protein, theoretical modeling criteria for membrane proteins, and biophysical studies of the properties of native and mutated voltage-gated channels. Most of the latter experiments were performed on the Shaker K+ channel. Some of these data are difficult to relate directly to models of the KvAP channel's structure due to differences in the Shaker and KvAP sequences. We have dealt with this problem by developing new models of the structure and gating mechanism of the transmembrane and extracellular portions of the Shaker channel. These models are consistent with almost all of the biophysical data. In contrast, much of the experimental data are incompatible with the “paddle” model of gating that was proposed when the KvAP crystal structures were first published. The general folding pattern and gating mechanisms of our current models are similar to some of our earlier models of the Shaker channel.
INTRODUCTION
Here we extend models of the KvAP channel presented in the accompanying article to the Shaker channel to facilitate comparison of experimental results from Shaker to structural data from KvAP. The general folding pattern of the transmembrane segments of our Shaker models is similar to that of our KvAP models; however, we have introduced some differences to better satisfy the Shaker data and to resolve apparent discrepancies between experimental results obtained from KvAP and Shaker channels. The most important differences involve the magnitude of the movement of S4 and the mechanism by which the voltage sensor is coupled to the activation gate. Also, we have developed tentative models of the S1-S2 and S3-S4 loops that are much longer in Shaker than in KvAP.
METHODS
Most of the methods used to develop and simulate the dynamics of these models are described in the preceding article. Initial homology models of the Shaker structure were developed in two ways: using the Modeler software (Sali and Blundell, 1993), and simply substituting Shaker side chains onto the backbone of the KvAP crystal structures and then minimizing the structures using the molecular mechanics program CHARMM (Brooks et al., 1983) to eliminate atomic overlaps. The results were similar using either method for the regions that could be aligned without insertions or deletions (indels). Other regions; i.e., the loops connecting the helical segments, were modeled manually followed by minimization. The conformations of the extracellular loops were modeled to be consistent with experimental results, with our modeling criteria (see Appendix of accompanying article), and to be relatively stable during a molecular dynamics simulation. For tetrameric structures, some interfacial side chains were manually adjusted before performing the energy minimization step with the constraint of fourfold symmetry. The inner portion of the S6 segments of the open conformation were remodeled to make their structures consistent with experimental results, as explained in the text.
RESULTS
Models of the voltage-sensing domain
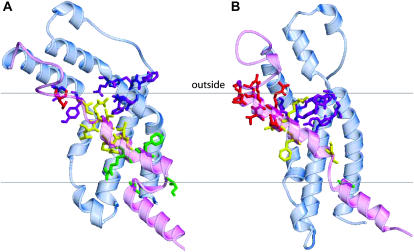
Fig. 1 illustrates a side view of our models of the voltage-sensing domain of Shaker in the open and resting conformations. The domain is positioned and colored in the same way as for KvAP in Fig. 5 of the preceding article. The S1–S4 segments were modeled directly after the crystal structure of the isolated KvAP voltage-sensing domain (Structure 2); however, for reasons explained below, the Shaker S4 was aligned three residues (or one helical screw step) farther toward the C-terminus (i.e., toward the cytoplasmic surface) than in the alignment of Fig. 7 of the accompanying article. The position of S3b was altered slightly from that of KvAP to improve energetically favorable interactions. A break between the S4 and L45 helices was introduced at a hydrophilic region (sequence SRHSKG) that has a low propensity for an α-helix conformation and where indels occur in some sequences (see criterion 11 of the appendix in the preceding article). Transition and resting conformations were generated from the open conformation by moving S4 inwardly along its axis in consecutive helical screw steps (i.e., by aligning the nth residue of the next conformation to the position of the n + 3th residue of the former conformation) (Guy and Seetharamulu, 1986). As in earlier models (Durell et al., 1998; Gandhi and Isacoff, 2002), we propose that the movement between resting and open Shaker conformations involves three helical screw steps; which translate the S4 helix ∼13.5 Å along and rotate it ∼180° about its axis. The positions of S4 in the resting and open conformations of the Shaker channel models correspond to those of putative transition conformations of KvAP; e.g., the position of S4 in the Shaker resting conformation of Fig. 1 B corresponds to that of S4 in the transition KvAP conformation illustrated in Fig. 4 C of the accompanying article.
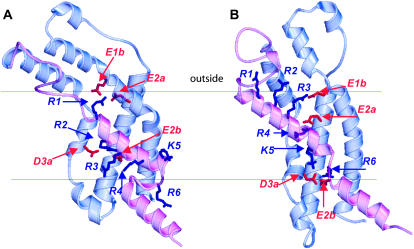
FIGURE 1.
Ribbon representations of our models of the voltage-sensing domain of Shaker in (A) resting and (B) open conformations. The mobile S4 and L45 segments are colored magenta. Positively charged S4 side chains and negatively charged side chains in S1–S3 with which they interact are illustrated in blue and red. The dashed lines represent postulated boundaries of the alkyl phase of the membrane.
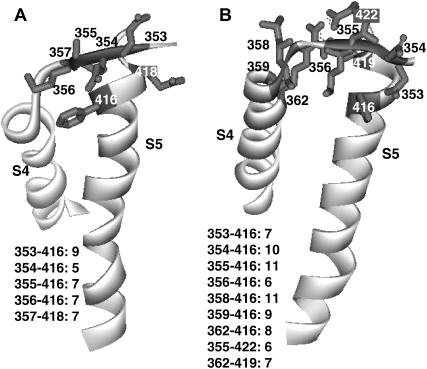
FIGURE 5.
Side view of the S4 and S5 segments showing residue pairs that, when mutated to cysteine, interact in resting (A) and open (B) conformations. Distances to the nearest Å between β-carbons of the residue pairs in these models are indicated in the figure.
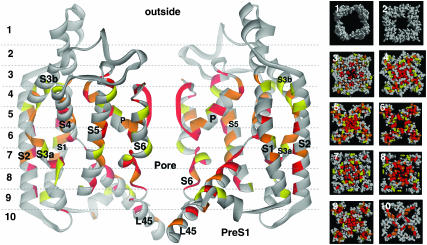
FIGURE 7.
Models of the open conformation colored according to the mutability of residues among eukaryotic Kv channels. Only the most highly conserved residues (red, orange, and yellow) are colored in this figure. The voltage-sensing domain (S1, S2, S3, S4, L45) and pore-forming domain of the side view are from different subunits. Cross sections 5.0 Å thick are illustrated in space-filled representations beside the side view. The numbers indicate the cross-section designated by the dashed lines in the side views. Note that most poorly conserved residues (gray) are on the surface whereas most highly conserved residues are in the core of the protein where they interact with other highly conserved residues. A supplement to this figure illustrates the distribution of the three categories (hydrophobic, hydrophilic, lipid headgroup-favoring) of highly variable residues.
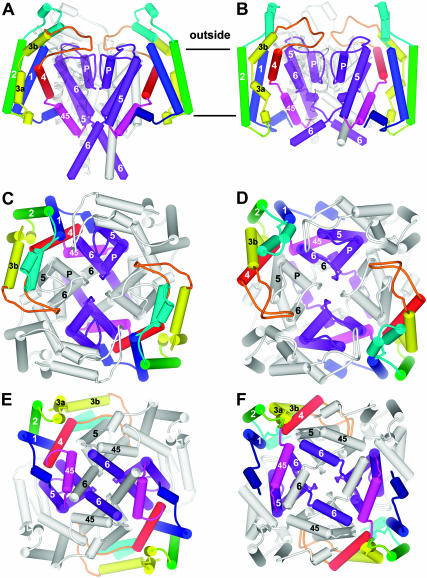
FIGURE 4.
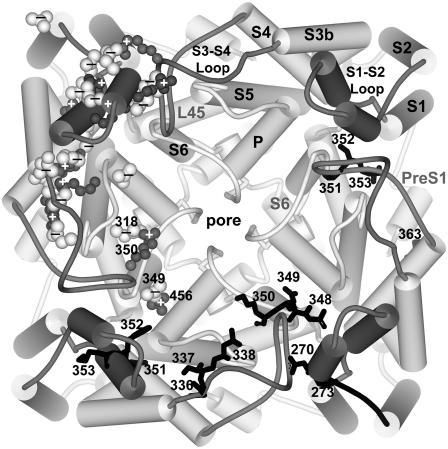
Schematic representation of our Shaker channel models with both transmembrane domains in resting (left) and open (right) conformations. (A and B) Side view. Two subunits are colored, the subunit on the far side is white, and the subunit that would be nearest the viewer has been removed except for a faint ribbon trace of its pore-forming domain. (C and D) All four subunits as viewed from outside the cell. Every other subunit is white. (E and F) View from inside the cell. Color code is PreS1, dark blue; S1, blue; S1-S2 linker, cyan; S2, green; S3, yellow; S3-S4 linker, orange; S4, red; L45, magenta; and S5-P-S6, purple. Cylinders represent α-helices. S1–S6 and P helices are labeled for one subunit in each picture. Stereo views of ribbon representations of these figures, including some “atypically oriented” lipids, are in the supplementary information. Lipids are colored by atom: gray, carbon; red, oxygen; blue, nitrogen; and yellow, phosphorus.
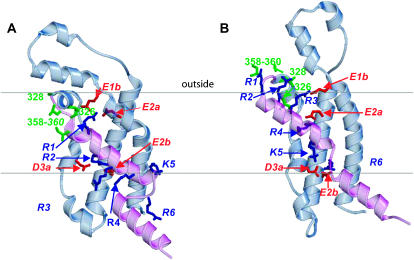
The positions of S4 in our Shaker models were selected to satisfy several results from mutagenesis experiments. Tiwari-Woodruff et al. (2000) found that simultaneous charge reversal mutations of E283K (E2a) and R371E (R4) stabilized the open conformation, whereas a closed conformation was stabilized in the E283K/R368E (E2a/R3) mutant. (The italicized residue numbers in parentheses are the generic numbers for the charged groups of 6TM channels introduced in the accompanying article.) They interpreted these results as indicating that E2a and R4 interact in the open conformation and that E2a and R3 interact in a transition conformation that precedes the opening conformational change. In the models, these interactions occur in the open conformation of Fig. 1 A and in a transition conformation in which S4 is one helical screw step inward from the open conformation (not shown). Papazian et al. (1995) found that the Shaker channel did not express when K374 (K5) or R377 (R6) of S4 were mutated to an uncharged glutamine residue, but that expression was recovered for K374Q/E293Q (K5/E2b) and K374Q/D316Q (K5/D3a) double mutants. They interpreted these results as indicative of electrostatic interactions among these negatively and positively charged residues that likely occur in conformations favored at depolarized potentials. Fig. 1 A shows that these residues form salt bridges in our models of the open conformation.
Several groups have mutated numerous residues in the voltage-sensing domain of voltage-gated channels to cysteine, and have then determined the accessibilities of the introduced cysteines at different voltages to hydrophilic sulfhydryl reagents placed on either side of the membrane (Baker et al., 1998; Larsson et al., 1996; Yang et al., 1996; Gandhi et al., 2003). Results from such studies that use the methanethiosulfonate (MTS) reagents MTSES and/or MTSET on resting and open Shaker channels are illustrated in Fig. 2. Several residues (purple) on the extracellular ends of S1, S2, and S3 are outwardly accessible, and these accessibilities are not affected substantially by membrane voltage. In contrast, the accessibilities of many S4 residues are voltage-dependent. Note that in models of both open and resting channels, all outwardly accessible residues (red) are on the extracellular side of the central barrier, all inwardly accessible residues (green) are on the intracellular side, and that inaccessible residues (yellow) are within the central barrier. (The central barrier is the tightly packed, relatively hydrophobic region where the axes of S4 and S2 cross in Fig. 1.) Thus, our models, which are similar to those proposed by Gandhi et al. (2003), are consistent with these results. Relating these results to the KvAP structure is complicated by the fact that the S1-S2 and S3-S4 loops are much longer in Shaker. However, many of the purple residues in Fig. 2 do align with KvAP residues, and the paddle model predicts that none of these aligned residues should be accessible from the outside in the resting conformation.
FIGURE 2.
Same models as in Fig. 1 illustrating the accessibility of residues to MTSET and/or MTSES reagents (Gandhi and Isacoff, 2002; Larsson et al., 1996). The purple residues on S1, S2, and S3 are accessible from the outside in all conformations. In S4, the red residues are accessible from the outside, the green ones are accessible from the inside, and the yellow ones are inaccessible from either side. The accessibility of most S4 residues depends upon the membrane voltage.
Starace et al. (1997, 2004) mutated the positively charged S4 residues one at a time to histidine in a Shaker mutant that does not conduct ions through the pore-forming domain. They found the following: 1), the R362H (R1) mutant forms a proton pore at hyperpolarized voltages; 2), the R365H (R2) and R368H (R3) mutants can transport H+ ions across the membrane in a voltage-driven manner; 3), the R371H (R4) mutant forms a proton pore at depolarized voltages; and 4), neither the K374H (K5) nor R377H (R6) mutants transport H+ ions or form H+ pores. The formation of proton pores by the R1H and R4H mutants indicates the presence of a pathway across the membrane that is sufficiently polar to conduct H+. This pathway likely has a barrier that prevents proton permeation in the region occupied by R1 in the resting conformation and by R4 in the open conformation. The ability of the R2H and R3H mutants to transport H+ ions across the membrane suggests that these residues cross this barrier during activation and that the pathway through which they move is sufficiently polar to allow the histidine side chain to remain protonated throughout the transition. The lack of effects on proton transport of the K5H and R6H mutants suggests that they do not cross the barrier. Our models possess all of these features, even though these results were not used to develop previous three helical screw step models (Durell et al, 1998). The barrier region corresponds to the central barrier in our models. The hydrophilic crevasses on either side of this zone in the core of the domain should readily allow passage of H+ ions. R1 and R4 are positioned in the central barrier in the resting and open conformations, respectively, where they form a salt bridge to E2a and H-bond to the C-terminus backbone oxygen atoms of the S3a helix. Thus, it is reasonable that replacement of either of these arginine side chains by a smaller histidine side chain that can readily be protonated and deprotonated would allow H+ ions to pass through the membrane. R2 and R3 residues pass through the central barrier region during our models of activation. Histidine side chains at these positions can remain protonated throughout the activation transition because they remain in the polar core of the voltage-sensing domain where they are always near negatively charged residues on S1-S3. K5 and R6 are on the intracellular side of the central barrier in all conformations, explaining why the K5H and R6H mutants do not transport H+ ions.
Ahern and Horn (2004) have analyzed the effect of charged MTS adducts attached to cysteines introduced at positions 362–366 in the initial portion of S4 in Shaker channels. Charged adducts at the positions occupied by arginine (R1 = R362, R2 = R365) contribute to the gating current; however, adducts at adjacent positions (363, 364, and 366) do not. This is true even if the charged adduct is MTSEA, which is deprotonated in a hydrophobic environment. These results suggest that R1 and R2 cross the barrier region and enter an intracellular polar crevasse when the channel deactivates, but that residues 363, 364, and 366, which are on the opposite face of the helix, do not. This concept is consistent with our models, in which all of these residues are exposed on the outside of the open conformation and only the charged S4 residues of this portion of S4 extend into the polar core of the voltage-sensing domain in the resting conformation (residues 363 and 366 are on the face of the tilted helix that is oriented toward the extracellular surface in the resting conformation).
Gonzalez et al. (2001) have examined gating properties of Shaker channels in which the S3-S4 linker is shortened by various amounts. They found that residues 330–360 could be deleted without eliminating voltage-dependent gating, but that longer deletions were not functional. The Δ330–360 mutation reduced the apparent gating charge; however, the Δ330–357 mutation, which we have modeled (see Fig. 3), did not. Recently, Gonzalez et al. (2004) found that in the Δ330–357 mutant, when residues 326, 328, 357, 358, and 359 are mutated to cysteine, they are accessible to extracellular MTSET only in open channels. In contrast, Gandhi et al. (2003) found that residues in S3b were accessible at all voltages in Shaker channels with native S3-S4 linkers. This result is noteworthy because it suggests that the differences in accessibility of S3b residues between KvAP, where they are accessible only at positive voltages (Jiang et al., 2003b), and Shaker channels, where they are accessible at all voltages (Gandhi et al., 2003), may be due to the long S3-S4 loop in Shaker. To demonstrate that our models are consistent with these results, we have modeled open and resting conformations of the Shaker Δ330–357 mutant (see Fig. 3). The open conformation can be accommodated by modifying only a few residues that link S3-S4. In our model of the resting Shaker Δ330–357 mutant, S3b and S4 residues may become inaccessible because the inward movement of S4 causes S3b to swing inward into the alkyl phase, hinging between S3a and S3b. In our models of the native Shaker channel, residues preceding 355 that are accessible in open channels (Larsson et al., 1996) do not need to move very much.
FIGURE 3.
Models of the voltage-sensing domain in resting (A) and open (B) conformations in which residues 330–357 of the S3-S4 linker are deleted. Residues in the Δ330–357 mutant that have been shown to be accessible in only the open conformation (Gonzalez et al., 2004) are colored green. The view and coloring are the same as in Fig. 1.
The extracellular location of S3b in all conformations has been confirmed by mutagenesis experiments with the Hanatoxin spider toxin. Hanatoxin modulates voltage-dependent gating of the drk1 eukaryotic Kv channel by binding to the S3b segment and S3-S4 linker (Li-Smerin and Swartz, 2000, 2001). This binding occurs at all voltages, with slightly higher binding affinity at negative voltages that favor the resting conformation (Lee et al., 2003).
Docking of the voltage-sensing and pore-forming domains
The conformation of the S5, P, and initial part of S6 segments of the pore-forming domain were modeled after the full-length KvAP crystal structure (Structure 1) for the open conformation, and after the KcsA crystal structure (Zhou et al., 2001) for the closed conformations. Two proline residues, P473 and P475, near the middle of S6 likely distort its α-helical structure. Holmgren et al. (1998) have shown that the open conformation of a V476C mutant is stabilized by Cd2+ binding between V476C and H486 of adjacent subunits. Also, Webster et al. (2004) have shown that in a V474C mutant, Cd2+ binds between the introduced cysteines of adjacent subunits with equal affinity in both open and resting conformations, suggesting the V474 remains near the axis of the pore during activation. The latter portion of S6 for the open conformation was modeled by distorting the helix in the vicinity of P473, so that V474 is near the axis of the pore and the side chains of V476 and H486 of the adjacent subunit are near each other (β-carbons of adjacent V476 residues and of V476 and H486 residues of adjacent subunits are 8–9 Å apart in our models). The latter part of S6 was modeled three ways for the resting conformation: 1), after the structure of KcsA; 2), with the inner part of the S6 helices rotated so that the core of the S6 bundle of four helices is comprised of highly conserved hydrophobic residues; and 3), with the S6 helix kinked at residues S479–N480 so that the latter part of S6 extends away from the axis of the pore where its highly conserved residues interact with highly conserved residues of L45 and S5. These alternative models did not alter the rest of the structure significantly, and we do not strongly favor one model over the others. The simplest KcsA-like model is illustrated here (Fig. 4) and was used for the molecular dynamics simulations.
Experimental constraints were used to dock S4, and residues that immediately precede S4, next to S5 (see Fig. 5). Elinder et al. (2001a,b) demonstrated that an electrostatic interaction between the R362 (R1) position on S4 and the E418 residue at the end of S5 occurs at positive voltages. More recently, four laboratories have introduced cysteine pairs into a variety of positions on S4 and S5 (Laine et al., 2003; Gandhi et al., 2003; Neale et al., 2003; Broomand et al., 2003). Laine et al. (2003) showed that disulfide bridges form spontaneously in the R362C/F416C and the R362C/A419C mutants, and that under reducing conditions a Cd2+ binds between these cysteines. These interactions occur between residues of different subunits and only at depolarizing voltages. The interactions are specific since a disulfide bridge is not formed in the double mutants if the location of the cysteine is moved by one residue in either S4 or S5. Gandhi et al. (2003) have shown that a disulfide bridge forms spontaneously in an A355C/E422C mutant, and that under oxidizing conditions disulfide bridges form in F416C mutants in which a second cysteine is introduced at positions 354, 355, 356, 358, 359, or 362 (R1). In contrast, the 360C/F416C and 361C/F416C mutants do not form disulfide bridges. The bridges between residues 354–356C and F416C form at all voltages, but the other bridges occur only when the membrane is depolarized. Broomand et al. (2003) also found that the double mutants R362C/F416C and A359C/F416C form disulfide bridges only at depolarizing voltages and that the double mutant N353C/F416C formed a disulfide bridge at all voltages, but that the double mutants R362C/A417C, R362C/W454C, and R365C/F416C did not form disulfide bridges. Neale et al. (2003) have shown that a disulfide bridge and Cd2+ binding site form in a S357C/E318C mutant only when the membrane voltage is hyperpolarized.
We docked the voltage-sensing domain structures described above onto models of the pore-forming domains and modeled the S3-S4 linker in a manner consistent with these experimental results (see distances between residue pairs in Fig. 5) and with the general position in the membrane predicted for the voltage-sensing domain described in the accompanying article. Residues preceding position 358 (resting) or 359 (open) were given a nonhelical conformation so that cysteines introduced at residues 353–356 could be sufficiently mobile and near F416C for rapid formation of disulfide bridges.
In our initial models of the resting conformation the S1–S3 segments were left in the same location used in modeling the open conformation. In this model, we had to unwind residues 359–361 at the N-terminus of S4 from an α-helical structure to an extended conformation to move S4 inwardly by three helical screw steps while leaving S357 near E318. This makes the location of the N-terminus of the Shaker S4 in the resting conformation correspond to the location of the N-terminus of KvAP's S4 helix in both crystal structures according to the alignment of Jiang et al. (2003a). This type of coiling and uncoiling of the ends of S4 during activation is consistent with the direction of the rotation of the helix in the helical screw mechanism, as has been proposed previously (Guy and Conti, 1990). Although the extended conformation is in the transmembrane region in the resting conformation, it is shielded from the lipid alkyl chains by other segments, and its polar backbone atoms interact with hydroxyl groups of Y323, S411, and Y415. However, this structure was relatively unstable during a molecular dynamics simulation (see below). Therefore we next shifted the voltage-sensing domain extracellularly by 4–7 Å and rotated it counterclockwise about the pore as viewed from the outside. This allowed the resting conformation to be modeled in a manner that satisfied the experimental criteria without unwinding the N-terminus of S4. The simulation of this structure was more stable. The direction of the postulated movement of the S1–S3 segments is consistent with their negatively charged residues moving in response to the change in the electric field; i.e., it is opposite that of S4. Although the helical screw motion of S4 is maintained within the voltage-sensing domain, its motion with respect to the pore-forming domain is more complicated and is reduced in magnitude. With the additional movement of the entire voltage-sensing domain, this model becomes a hybrid of the models in Fig. 1 of the preceding article; i.e., most of the movement involves S4, but the location of the barrier relative to the pore-forming domain also shifts.
Coupling of the voltage-sensor to the activation gate
The activation gate is formed by the inner half of the S6 segment, which swings away from the pore when the channel opens (Jiang et al., 2002). Coupling of the voltage-sensor to the activation gate almost certainly involves the L45 segment that links S4 to S5. L45 is helical in the KvAP crystal structures (Jiang et al., 2003a). The L45 helix is amphipathic, and the hydrophobic face is highly conserved among Kv channels (see Fig. 7 of accompanying article). Conservative mutations of leucines of the hydrophobic face can affect activation gating dramatically (Judge et al., 2002; Lopez et al., 1991; McCormack et al., 1991; Shieh et al., 1997). We modeled the L45 helix as a continuous extension of the S4 helix for KvAP because it has that conformation in the crystal structure of the isolated voltage-sensing domain. However, there is a deletion in the transition region between S4 and L45 in Shaker channels that would likely disrupt the α-helix structure. In our Shaker models of the open conformation, we have modeled R377 (R7) as the C-cap residue of S4 and the N-cap residue of L45; i.e., a break between the two helices occurs at R7 (see Fig. 4). This allows the highly conserved, hydrophobic face of L45 to pack against highly conserved residues of S5 and S6 (see Table 1 for a list of pairwise interactions). The location of the L45 helix is constrained by the necessity to span the relatively long distance between the C-terminus of S4 and the N-terminus of S5. Modeling of L45 in closed conformations is more problematic for the following reasons. Unfortunately, most of the experimental data for the voltage-sensing domain are for the open conformation and for the portions of the protein that are accessible from outside. Also, in the resting conformation, the distance between the C-terminus of S4 and the N-terminus of S5 is relatively short, making it possible to model L45 in numerous ways; e.g., the L45 helix could interact primarily with the S6 and adjacent S5 segments, it could interact primarily with lipids on the cytoplasmic surface of the membrane as illustrated in our KvAP models (Fig. 6 D of the accompanying article), or it could break in the middle, allowing it to extend into the cytoplasm. This situation is complicated further by uncertainties about lipid interactions in the inner half of the membrane. We currently favor models for the resting conformation of Shaker in which the hydrophobic face of L45 still interacts with S5 and S6, albeit it in a different manner from the open conformation (see Fig. 4). These interactions may stabilize closed conformations, preventing opening of the channel until movements of the voltage-sensors disrupt or weaken these interactions. S4 is connected to L45 by a hydrophilic segment (376–381, sequence SRHSKG), which has a low propensity for a helical conformation. Thus, L45 in this location does not have to move during the initial voltage-dependent conformational changes (e.g., first two helical screw steps) that precede opening of the channel; i.e., movements within the flexible 376–381 segment can accommodate movements of the voltage-sensing domain during the initial transitions of activation. Another consideration in favor of this location of L45 involves our hypothesis that the S1–S3 segments of the voltage-sensing domain shift inward toward the cytoplasm relative to the pore-forming domain during activation. If this occurs, then the pore-forming domain may shift toward the cytoplasm during deactivation. This would expose more of the inner portions of S5 and S6 to the cytoplasm in the resting conformation. Interactions with the hydrophobic surface of L45 could shield some of the hydrophobic residues of S5 and S6 from water. The linkage between the L45 and S5 helices is formed by residues 392–395 (sequence SMRE) in all conformations of the illustrated models; however, we have developed alternative models in which the L45 helix is shifted farther inward in the resting conformation and 393–395 become the first three residues of the S5 helix. The alternative model has the advantage of shielding more of the hydrophobic residues of S6 from water if S6 has the KcsA-like conformation illustrated in Fig. 4.
TABLE 2.
Distances from LRET, tethered TEA, and models
| Residue No. | Method | Conformation | Rexp | Rmod |
|---|---|---|---|---|
| 270 | LRET | R | 22.5 | 22 |
| LRET | O | 22.5 | 26 | |
| 273 | LRET | R | 23.5 | 23 |
| LRET | O | 23.5 | 29 | |
| 346 | LRET | R | 18.5 | 20 |
| LRET | O | 21 | 22 | |
| 351 | LRET | R | 20.5 | 20 |
| LRET | O | 21 | 17 | |
| 352 | LRET | R | 20 | 25 |
| LRET | O | 20 | 21 | |
| 353 | LRET | R | 22 | 26 |
| LRET | O | 21 | 23 | |
| 363 | LRET | R | 22.5 | 23 |
| LRET | O | 22.5 | 29 | |
| 425 | LRET | R | 15 | 13 |
| LRET | O | 15 | 13 | |
| 251 | TEA | O | 30 | 25 |
| 252 | TEA | O | 30 | 23 |
| 253 | TEA | O | 30 | 27 |
| 334 | TEA | O | 30 | 25 |
| 335 | TEA | O | 30 | 27 |
| 336 | TEA | O | 30 | 26 |
| 348 | TEA | O | 17-18 | 18 |
| 349 | TEA | O | 17-18 | 16 |
| 350 | TEA | O | 17-18 | 14 |
The distances, in angstroms, for the LRET (Cha et al., 1999) data are from the axis of the channel. The model distances for the tethered TEA (Blaustein et al., 2000) data are from the center of a TEA molecule that was positioned on the axis and as far into the pore as possible without substantial molecular overlap. The conformation is indicated as open (O) or resting (R).
FIGURE 6.
View from the outside of the open conformation illustrating models of the S1-S2 and S3-S4 loops. Segments are labeled at the top; the cylinders represent helices. Stick representations of residues on the subunit on the right to which LRET probes have been attached and on the bottom subunit to which TEA analogs have been tethered are black. Ball representations of positively (dark gray) and negatively (white) charged side chains of the loops are shown in the subunit on the left. All positively charged residues of the loops form salt bridges with negatively charged residues.
Atypical lipid conformations in the cytoplasmic leaflet
L45 packs more tightly against the pore-forming domain in our Shaker models than in our KvAP models, which leaves less room for lipids to be bound between the two domains for the open conformation. However, for the resting conformation it may still be energetically favorable for some lipids to bind between the two domains. In our models, some alkyl chains of these lipids have an atypical orientation parallel to the S4 helix. The atypical orientation of these cytoplasmic lipids may be facilitated by the tilted nature of the S4 helix and the fact that the cross-sectional area of the channel is substantially larger at the extracellular surface than at the intracellular surface; i.e., the alkyl chains of several of these lipids may not interact with alkyl chains of lipids from the extracellular leaflet. The alkyl chains of lipids positioned between S3 and S5 extend into the hydrophobic phase of the membrane, whereas their headgroups comprise part of the lining of the inner crevasse (see supplementary material, Figs. 1 and 4). In addition, in some of our models, alkyl chains of lipids pack between the hydrophobic surfaces of the voltage-sensing and the pore-forming domains, whereas their headgroups bind to positively charged S4 residues. The potential importance of negatively charged lipids is suggested by the abundance of positively charged residues at the cytoplasmic interface; e.g., negatively charged headgroups of lipids bind to K212, R227, R297, R309, R365 (R2), R368 (R3), R371 (R4), K374 (K5), K380 (K7), R387, R395, and the N-termini of the Pre-S1 and L45 helices in some of our models of the resting conformation. One working hypothesis is that during activation, lipids that are sandwiched between the domains remain attached to S4 as it rotates and translates until they reach the lipid-lined portion of the crevasse that is located between S3 and S5. When that occurs, the lipid detaches from S4 and becomes part of the crevasse lining and another lipid that formed part of the crevasse lining diffuses into the inner leaflet. Thus, each helical screw step removes one lipid and one positively charged S4 residue from the interface between the voltage-sensing and pore-forming domains. After three helical screw steps, all lipids that were located between the domains have been removed and the channel can open. The advantages of this model are that the positively charged residues of S4 are always in the proximity of negatively charged groups (lipid headgroup or negatively charged residue of S1–S3), there is less breakage and reformation of electrostatic interactions between lipid headgroups and charged residues of S4, and the dynamic lipid alkyl chains act as a lubricant that reduces barriers to S4 movement. The manner in which the two domains interact will be influenced by the number of lipids that are sandwiched between the domains and/or that line the crevasses; i.e., the number of lipids can be increased by swinging the cytoplasmic portion of the voltage-sensing domain away from the pore-forming domain. A model with eight negatively charged lipids per subunit is illustrated in Fig. 1 of the supplement. However, we consider this hypothesis of lipid interaction as highly tentative because we know of no studies that indicate strong dependency of voltage-gating of Shaker channels on composition of the lipid bilayer. Thus, if such lipid interactions with the voltage sensor occur, they are likely nonspecific.
The pre-S1 segment
In our models, S4 does not span the entire transmembrane region in either open or resting conformations. We believe that it would be energetically unfavorable for the C-terminus of S4 to be exposed solely to lipid alkyl chains. To avoid this, we have modeled the segment immediately preceding S1 as an α-helix that lies on the cytoplasmic surface of the membrane (see Fig. 4). This putative pre-S1 helix is amphipathic and its hydrophobic face is modeled to interact with lipid alkyl chains. The helix forms part of the barrier between the lipids and the polar interior of the voltage-sensing domain and/or the C-terminus of S4. Some of its hydrophobic residues (V213, W214, and F217) are conserved, suggesting that they interact with other conserved residues (criterion 12 in Appendix of accompanying article). In the model of the resting conformation, these residues interact with the C-terminus of S4, which is composed of conserved residues. However, when S4 moves outward during activation, a gap forms in the wall of the inner crevasse of the open conformation. We propose that lipids fill this gap with their headgroups forming part of the polar wall of the crevasse. This lipid lining is on the opposite side of the voltage-sensing domain from the lipid lining postulated for closed channels and involves lipid interactions with pre-S1 and S1 (see Fig. 4, E and F). As in the model of the resting conformation, the headgroups of these “atypical” lipids would be farther into the transmembrane region than those of normally oriented lipids of the cytoplasmic leaflet. Note that the pre-S1 portion of our Shaker models is highly tentative because we know of no experimental studies of this segment, and no analogous segment exists in some other families of 6TM type channels.
Models of the extracellular loops
The loop segments that connect S1 to S2 and S3 to S4 are >20 residues long in the Shaker channel, and are poorly conserved among eukaryotic Kv channels. Long, poorly conserved loops are notoriously difficult to model. However, numerous mutagenesis experiments have been performed on residues in these segments. To demonstrate that our models of the transmembrane domains can be consistent with almost all of these experimental results, we have developed highly tentative models of these loop segments (see Fig. 6).
The segment following S1 was modeled as an α-helix because scanning mutagenesis experiments identified an α-helical periodicity in the analogous segment in drk1 (Li-Smerin et al., 2000b), and because this segment has a reasonably high propensity for a helical conformation based on statistical studies of known protein structures. Most of the remaining loop residues were modeled as random coils because they have many residues with high propensities for coiled structures (e.g., proline, glycine, serine, threonine, asparagine, and aspartic acid) and because multisequence alignments of eukaryotic Kv channels have many indels for these segments (see criteria 6 and 11 in Appendix of accompanying article). Initial models of these putative coiled regions were modeled manually to satisfy our modeling criteria for energetically favorable residue-residue interactions (see criterion 4 in Appendix of accompanying article). Some adjustments were made after initial molecular dynamic simulations to increase the stability of the loops.
The loop conformations were further constrained by requiring that residues be located in a manner consistent with two types of experiments. In the first type, distances from the axis of the channel were calculated by fluorescence resonance energy transfer (FRET) (Glauner et al., 1999) and lanthanide-based resonance energy transfer (LRET) (Cha et al., 1999) methods, in which fluorescent probes are attached to cysteine residues introduced at specific positions. Unfortunately, the large size of the probes (e.g., the dimensions of the donor probe in LRET are ∼8 × 8 × 13 Å) may lead to substantial differences between the experimentally determined distances between the chromophores and the distances between β-carbons of the residues to which the probes are anchored. Also, their large size may lead to distortions of the protein backbone. Furthermore, distances calculated by the two laboratories for residues 352–354 do not agree well; those calculated by Glauner et al. (1999) are all substantially greater (∼1.9× larger) than those calculated by Cha et al. (1999), and the direction of motion during activation is opposite for residues 352 and 354. The LRET values are more consistent with our models. Our criterion for consistency is that the distance of the β-carbon from the center of the pore is within 5 Å of that calculated by Cha et al. (1999). Both laboratories detect only small movements of residues 352–354 relative to the center of the pore during activation. The relatively small magnitude of the motion of these residues predicted by the LRET and FRET experiments is consistent with the observation that cysteine residues introduced into positions 353–356 can form disulfide bridges to a cysteine introduced at position 416 in both the open and resting conformations (Gandhi et al., 2003; Broomand et al., 2003). Cha et al. (1999) and Glauner et al. (1999) attempted to relate these small motions to the movement of the S4 helix by assuming that these residues are part of the S4 helix. However, residues 352–354 have a coiled backbone conformation in our models and they do not move much during activation. Also, very little motion was detected for a probe attached to residue 363 (Cha et al., 1999), which is in the S4 helix in our models and does experience substantial radial movement in our models. Cha et al., 1999 assert that their method does not detect any substantial movements of any of the analyzed positions orthogonal to the plane of the membrane. However, this movement is only 0.4 Å outward relative to the pore-forming domain for residue 363 β-carbon during activation in models of Fig. 4. This small orthogonal motion is due to the 180° rotation about the axis of the very tilted S4 helix (residue 363 is on the face oriented toward the extracellular phase in the resting conformation, and oriented toward the cytoplasm in the open conformation), and the inward movement of the S1–S3 segments of the voltage-sensing domain. The distance for residue 363 in the open conformation was the only substantial discrepancy with the LRET data that we were unable to eliminate without introducing inconsistencies with other experimental results or distorting the α-helical conformation of this portion of S4; however, all LRET distances were satisfied by our models of the resting conformation (see Table 2). The apparent lack of movement detected by LRET for residue 363 may be an experimental artifact because numerous studies indicate a substantial movement of R362 (R1) during activation (Elinder et al., 2001a,b; Laine et al., 2003; Broomand et al., 2003; Larsson et al., 1996).
TABLE 1.
Postulated interactions of L45 residues with S5 or S6 residues (β-carbons <7 Å apart)
| L45 residues | S5 or S6 residues |
|---|---|
| Model of open conformation | |
| H378 | L403, F404, I464 |
| S379 | L403, L399, I400 |
| G481 | A465* |
| L382 | A465*, L468*, P473, V474 |
| Q383 | L396 |
| L385 | L468* |
| G386 | L472*, I477 |
| L389 | L472*, F481 |
| S389 | L398* |
| M393 | F484, Y485 |
| Resting conformation (model 1 of Fig. 4) | |
| G381 | I464* |
| L382 | L399, I400, L403, T469, L472, P473 |
| L385 | I464*, V467*, L468*, P473 |
| G386 | L496, P473 |
| T388 | L468* |
| L389 | L468*, A472*, L472*, P473, I477 |
| S392 | L396*, L399*L472*, |
| Resting conformation (model 2, L45 more inward) | |
| G381 | L396, P473 |
| L382 | M393, L396, V476 |
| L385 | V476, I477, N480 |
| G386 | N480 |
| T388 | L472*, P475*, I477, N480 |
| L389 | N480, F481, F484 |
Bold residues are conserved among Kv channels (red, orange, and yellow in Fig. 7 of accompanying article).
Interactions are within the same subunit. Residues were considered proximal if the distance between β-carbons was <7 Å.
Extracellular tetraethylammonium (TEA) blocks Shaker channels by binding in the outer entrance of the pore (MacKinnon and Yellen, 1990). Blaustein et al. (2000) synthesized TEA derivatives to which a sulfhydryl reagent is tethered by an alkyl chain of variable lengths. They then attached these probes to specific positions in the loop segments by introducing cysteine residues. The distance of the anchor residue from the entrance of the pore was then approximated from the length of the shortest tether for which substantial blockade was observed. Residues Q348, D349, and K350 in the S3-S4 linker were found to be quite close (∼17–18 Å) from the pore's outer entrance. To satisfy these data, we positioned these residues between the “turrets” formed by the S5-P loops. This allows salt bridges to form between D349 and K456 of S6 and between K350 and E418 of S5 (see Fig. 6). Residues in the S1-S2 loop and near the end of S3 were found to be further from the pore (∼30 Å). These distances are somewhat larger than in our models (see Table 2), but we consider the differences to be within the accuracy of the technique.
The loops were also modeled to optimize energetically favorable salt bridges (see Fig. 6) and hydrophobic-hydrophobic interactions. In our models of the open conformation, all positively charged residues of the loops form at least one salt bridge to a negatively charged group (see Fig. 6); however, in models of the resting conformation there are more negatively charged than positively charged residues, and thus a few of these do not form salt bridges. Also, glycosylation sites at N259 and N263 are on the outermost portion of the S1-S2 loop in the region linking the two helices, consistent with the finding that channel functional properties remain normal in a nonglycosylated N259Q/N263Q double mutant (Khanna et al., 2001). The loop structures illustrated in Figs. 5 and 6 were substantially more stable during a molecular dynamics simulation than some alternative conformations that we simulated; however, residues 340–350 were still quite dynamic.
In spite of the ability of our loop models to satisfy almost all experimental results and modeling criteria, we expect them to still have substantial errors due to the large number of conformational degrees of freedom. Nonetheless, they demonstrate the consistency of our models of the basic folding pattern of the protein with most experimental results, and serve as a starting point for additional refinement as more data become available.
Consistency of models with residue tolerances
Li-Smerin et al. (2000a) measured the effects on activation gating of mutating residues on the outer surface of the pore-forming domain of the Shaker channel modeled after the KcsA crystal structure. They found that most residues where mutations affect the gating or expression of the protein reside in strips that are tilted ∼43° relative to the axis of the pore. S4 and L45 in our models abut closely to these “intolerant” strips. Additional experimental studies of the effects of mutations in the S1–S4 segments on activation gating have been performed (Li-Smerin et al., 2000b; Hong and Miller, 2000; Monks et al., 1999). Almost all tolerant residues (those for which mutations have little effect) of the voltage-sensing domain are on the outer surface in our models, where they should interact with lipids (see Fig. 8 A of the accompanying article). These results are consistent with residue mutabilities calculated from a multisequence alignment (unpublished) that we have made of many eukaryotic Kv channels; i.e., Fig. 7 illustrates that almost all highly conserved residues (red, orange, and yellow) are in the core of the protein and where they interact with other highly conserved residues. Likewise, almost all highly mutable residues are on the surface of the protein; mutable positions composed of hydrophobic residues are positioned where they should interact with lipid alkyl chains, those that are predominantly hydrophilic are located in the aqueous phase, and those that have characteristics typical of interactions with lipid headgroups tend to cluster at the lipid interfaces (see supplement to Fig. 7). Most of the transmembrane helices (S1, S2, S4, S5, and the initial part of S6) have poorly-conserved hydrophobic faces that are exposed to lipids in our models. The pattern of conservation on S4 is especially interesting and differs from the pattern for prokaryotic Kv sequences. Residues A359, I360, V363, L366, V367, and F370 comprise a poorly conserved, hydrophobic face on S4 that we predict should be exposed to lipid alkyl chains. In our model of the open conformation, these residues are exposed to lipid alkyl chains in the outer half of the transmembrane region at the interface between the two domains (see supplement to Fig. 7). When S4 moves inward to its resting conformation, it rotates about its axis by 180°, placing these residues on the opposite side of the interface between the two domains where they are still exposed to lipid alkyl chains. Awareness that S4 is exposed to lipids in our models, and that lipids of the cytoplasmic leaflet may pack between the two domains, is important because it has been incorrectly asserted that conventional models require S4 to be completely surrounded by protein (Jiang et al., 2003b). This stipulation has been interpreted to be inconsistent with the finding that the channel can still gate when biotin is attached to sites on S4 that apparently traverse most of the transmembrane region.
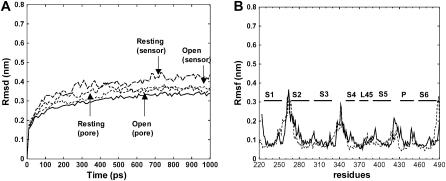
FIGURE 8.
Results of molecular dynamics simulations of our models of the resting and open conformations of the Shaker channel embedded in a phosphatidylethanlomine lipid bilayer with water on each side and K+ ions in the selectivity filter. (A) The RMSD of the α-carbons from the starting models of the pore-forming domain of the open (solid line) and resting (thin-dashed line), and the voltage-sensing domain of open (dashed line) and resting (long-dashed line) models. (B) The RMSF for the models of the open (solid line) and resting (dashed line) models during the last 500 ps of the simulations.
Molecular dynamics simulations
After developing the models as described above, we performed molecular dynamics simulations of the structures embedded in a phosphatidylethanlomine lipid bilayer, as described in the accompanying article. The root mean square deviations (RMSDs) of these models were somewhat greater than for our models of KvAP (see Fig. 8 A). However, most of this increase was due to the long extracellular loops in the Shaker models, which were substantially less stable than the transmembrane segments (see Rmsf (root mean square fluctuation) in Fig. 8 B). Such instability is typical of long flexible loops, but is likely increased by modeling errors. Also, small errors in modeling the transmembrane segments are likely since no portion of the Shaker models is based directly on a Shaker crystal structure. The first model of the resting conformation (with S1–S3 in the same location as in the model of the open conformation) that we simulated was relatively unstable; however, the RMSD and RMSF values were much lower for the second to fourth simulations for models in which S1–S3 are shifted inward relative to the pore forming domain (see Fig. 8 for results of our last simulation). The starting models for these simulations differed primarily in the specific location of the voltage-sensing domain, position of the L45 helix and the conformation of its linker to S4 and S5, the number and locations of atypical lipids that were positioned so that their negatively charged headgroups bind to positively charged R3, R4, and K5 side chains, and the conformations of the S3-S4 linker. We do not believe that these molecular dynamic simulation results identify which of the last three relatively similar models is best, because the results are comparable. However, we do interpret the relative stability of the transmembrane segments during the simulations to be supportive of the general folding pattern of our models.
Comparisons to other models
Current models of the voltage-sensing mechanism can be classified into three categories.
Models such as those presented here, in which the S4 helix moves through the transmembrane region. Positively charged groups on S4 remain in a polar environment in which they can interact with negatively charged residues of S1–S3 and other polar atoms of S1–S3, negatively charged lipid headgroups, and water in transmembrane crevasses. These models have a single barrier that separates inwardly and outwardly accessible residues on S4.
Models such as the “transporter” model of Starace and Bezanilla (2004), in which S4 does not move much during activation (see Fig. 1 of the accompanying article). These models are similar to the first category in that positive charges of S4 can always be in a polar environment. The primary difference is that these models have two barriers that control internal and external accessibility of the S4 residues. In the resting conformation the more interior barrier is open and the more exterior barrier is closed, so that most of the charged S4 residues are accessible from the inside, whereas in the open conformation the interior barrier is closed and the external barrier is open.
In the “paddle” model of Jiang et al. (2003a,b), the S3b–S4 paddle remains intact and moves through the lipid phase of the membrane during activation. There are no apparent water-filled crevasses or short protein barriers in these models, and the positively charged residues of S4 would be exposed to lipid alkyl chains during the transition of activation.
Table 3 lists whether or not we consider the results of various experiments to be consistent with each of these models. Comparisons of the consistency of these models with experimental results and with the KvAP crystal structures are subjective because ours are the only models for which we have coordinates. Although Fig. 5 of Jiang et al. (2003b) has a ribbon representation of their model; they describe their model as being so dynamic that it can be consistent with almost any result. For example, the concept of a dynamic structure is evoked to explain why the S1-S2 loop can be near the intracellular surface in their illustration even though it is glycosylated and thus in the extracellular aqueous phase in Shaker channels (Khanna et al., 2001), why the S4 segment can be on the periphery of the protein far from the S5 segment in their illustration even though experiments in Shaker indicate that these segments interact (Elinder et al., 2001a,b), and why a cysteine introduced into S4 can cross-link S4 segments of different subunits (Aziz et al., 2002) even though these segments are far apart in all illustrations of their models. Thus, we have classified the inconsistencies with the paddle model into two categories, those that are inconsistent only with the explicit ribbon model (indicated by an asterisk), and those inconsistent with the more general concept that the paddle is on the periphery of the protein where it diffuses through the lipid phase of the membrane during activation. We agree that the voltage-sensing domain is probably highly dynamic and that many transition configurations may occur during activation. Thus, caution should be used in interpreting data involving irreversible interactions (e.g., disulfide bridge formation, MTS binding to cysteines, and biotin/avidin binding). This is especially true when rates of these interactions are not determined, since the data could reflect results from several different conformations, some of which may be infrequent perturbations that differ substantially from the principal conformational states. Thus, some experimental results may be misleading or misinterpreted, and some data may come from multiple conformational states. Nonetheless, we think that most experimental data are consistent with the concept that at very negative or very positive voltages, the Shaker channel has a relatively small number of principal conformation states in which it exists most of the time. Some data cannot be satisfied because they are contradictory. For example, FRET (Cha et al., 1999) and LRET (Glauner et al., 1999) distances calculated for the same residues differ markedly. Furthermore, potential differences in the structures and gating mechanisms of KvAP and Shaker channels complicate evaluations of the consistency of models with the data. Thus, models should be evaluated by whether they are consistent with the preponderance of the data, since no single model can satisfy all of the data.
TABLE 3.
Consistency of models with experimental results
| Experiment (in Shaker unless indicated otherwise) | Our current | Transporter | Paddle |
|---|---|---|---|
| Biotin/avidin accessibility in KvAP (Jiang et al., 2003b) | yes | no | yes |
| S1 C-terminus, S2 N-terminus, and S1-S2 loop accessible from the outside in all conformations. (Gandhi et al., 2003) | yes | yes | no* |
| S3b and S3-S4 linker is accessible from outside in all conformations (Gandhi et al., 2003) | yes | yes | no |
| Hanatoxin binds to S3b of both open and resting conformations (Lee et al., 2003; Li-Smerin and Swartz, 2000, 2001) | yes | yes | no* |
| The Δ330–357 mutant gates normally, but residues on S3b and S4 are accessible to MTSET only at positive voltages (Gonzalez et al., 2004) | yes | ? | yes |
| N-terminus of S4 interacts with C-terminus of S5 at positive voltages (Elinder et al., 2001a,b; Laine et al., 2003; Gandhi et al., 2003; Broomand et al., 2003) | yes | yes | no* |
| S357C/E318C residues are proximal at negative voltages (Neale et al., 2003) | yes | yes | no |
| Cys mutants of S3-S4 loop residues 353–356 form disulfide bridges with S5 residue F416C at all voltages (Gandhi et al., 2003; Broomand et al., 2003) | yes | yes | no |
| Disulfide bridge cross-links S4 segments of different subunits (Aziz et al., 2002) | no | no | ? |
| R362H (R1) and R371H (R4) mutants form proton pores at negative and positive voltages (Starace et al., 1997; Starace and Bezanilla, 2004) | yes | yes | no |
| R365H (R2) & R368H (R3) mutants transport protons during activation/deactivation (Starace et al., 1997; 2004) | yes | yes | no |
| Voltage-dependent accessibility of S4 cysteine mutants to MTS reagents (Baker et al., 1998; Larsson et al., 1996; Yang et al., 1996; Gandhi et al., 2003) | yes | ? | no |
| Charges of adducts contribute to the gating current for R362C (R1) and R365C (R2) mutants but do not when adducts are at positions 363, 364, or 366 (Ahern and Horn, 2004) | yes | ? | no |
| R362 (R1) and R365 (R2) interact with E283 (E2a) in activated and transition conformations (Tiwari-Woodruff et al., 2000) | yes | ? | ? |
| K374 (K5) and R377 (R6) interact with E293 (E2b) and D316 (D3a) (Papazian et al., 1995) | yes | ? | ? |
| Distances of residues from the axis of the pore determined by LRET (Cha et al., 1999) | Almost all | yes | no |
| Tethered TEA analogs (Blaustein et al., 2000) | yes | ? | ? |
| Tolerance of S1–S3 residues to mutations (Li-Smerin et al., 2000b; Hong and Miller, 2000; Monks et al., 1999) | yes | ? | no* |
| Tolerance of residues on the outer surface of the pore-forming domain to mutations (Li-Smerin et al., 2000a) | yes | ? | ? |
Our current is the model presented in this article; Transporter is the model of Starace and Bezanilla (2004), in which the position of barriers that control access to crevasses changes during activation but S4 does not move much; and the Paddle model is that of Jiang et al. (2003a,b). An asterisk indicates that the assessment is only for the more explicit depiction of the paddle model in Fig. 5 of Jiang et al. (2003b). The question marks indicate uncertainty of the assessment.
Clearly, the more traditional models are more consistent with the data from Shaker than is the paddle model. Even the most liberal interpretation of the paddle model cannot explain why histidine substitutions (Starace et al., 1997) or MTSEA adducts (Ahern and Horn, 2004) on S4 remain protonated during gating (the relatively neutral intrinsic pKA values of these groups would lead to deprotonation when in a hydrophobic environment as proposed for transition conformations of the paddle model), why proton pores are formed by R362H (R1) and R371H (R4) mutants at negative and positive voltages, respectively (Starace et al., 1997; Starace and Bezanilla, 2004) (the paddle model does not specify a polar transmembrane pathway for proton permeation that would be blocked by R1 in the resting conformation), why charged adducts at positions adjacent to R1 and R2 do not contribute to gating (Ahern and Horn, 2004), or why residues near the beginning of S4 interact with the C-terminus end of S5 (Laine et al., 2003; Gandhi et al., 2003; Neale et al., 2003; Broomand et al., 2003) and remain accessible from the outside at all voltages (Gandhi et al., 2003). Models in which S4 does not move much may be consistent with much of the Shaker data, but are inconsistent with the biotin/avidin data from KvAP (Jiang et al., 2003b), provided that these data are in fact due to interactions with principal conformations instead of rare perturbations (reaction rates and voltage dependencies were not reported for inward accessibility of avidin to biotin adducts). Also, we have been unable to develop a viable transporter type model of activation gating starting from the S1–S3 structure of the isolated voltage-sensing domain of KvAP. The difficulty in doing so involves the location of the central barrier and polar transmembrane pathway through the voltage-sensing domain. In our model of the open conformation, which is supported by a substantial amount of experimental data, the barrier occupied by R4 is already near the extracellular surface (in the schematic of the transporter model (Starace and Bezanilla, 2004) the barrier is near the intracellular surface for the open conformation) and R1 is far from the only peripheral polar pathway (core of the S1–S4 domain) through which protons could flow through the membrane in histidine mutants. Thus, substantial movement of S4 within the voltage-sensing domain is required to place R1 in a polar pathway when the channel is at rest. However, in our models of Shaker, the barrier may move some and the size of the inner crevasse may be substantially larger in the resting conformation; i.e., during activation negatively charged S1–S3 segments may move toward the cytoplasm and the cytoplasmic side of the voltage-sensing domain may swing toward the pore as the positively charged S4 segment moves outward. This movement, coupled with the 180° rotation of the tilted S4 helix, results in little transmembrane movement of some S4 residues relative to the pore-forming domain. For example, the outward movement of the β-carbons of V363 and F370 during activation is negligible (∼0.4 Å) in our models in which S1–S3 move ∼7 Å inward relative to the pore-forming domain during activation. Thus, experiments indicating little transmembrane movement of probes attached to some S4 residues are not necessarily inconsistent with our models.
Many atomically explicit models can be developed for each of the three general categories of models described above. Numerous specific models of the first categories have been published by our group as well as others. Below we compare some of these models to our current models. Gandhi et al. (2003) and Broomand et al. (2003) have recently proposed models of the Shaker channel that are similar to ours. They also postulate that the crystal structure of the isolated KvAP voltage-sensing domain has a native fold that corresponds to the open conformation, and Gandhi et al. (2003) have docked it onto the pore-forming domain in a manner similar to our models. Both models of the voltage-dependent transition are also similar to ours in that S4 moves along its axis by a substantial amount (via the helical-screw mechanism in the Broomand et al. (2003) model), and S3b remains exposed on the extracellular surface when the channel deactivates. A major difference in our models of the Shaker open conformation is that we place S4 one helical screw step (or three residues) more inward to be more consistent with experimental results. We also include models of the segments that link S1 to S2, S3 to S4 and S4 to S5, most of which were omitted in the other models (Broomand et al. (2003) included L45). Gandhi et al. (2003) and Broomand et al. (2003) did not attempt to model the KvAP gating mechanism or explain why its S3b segment becomes relatively inaccessible in the resting conformation, and did not attempt to explain how movement of S4 is coupled to the opening of the gate. The orientation of the voltage-sensing domain relative to the pore-forming domain is markedly different in the Broomand et al. (2003) model; i.e., it is rotated with respect to the membrane's normal by ∼90° relative to our model so that S3b fits between the P and S6 helices of adjacent subunits, S2 interacts with S5 of the same subunit, and S4 is substantially more exposed to the lipid. Although we cannot exclude this orientation, there are several reasons why we do not favor it: 1), as best we can tell without coordinates, it places residues 360 and 361 nearer residue 416 (the 360C/416C and 361C/416C double mutants do not form disulfide bridges (Gandhi et al., 2003)); 2), it is difficult to envision how S357 can be near E418 (Neale et al., 2003) in the resting conformation of this model; 3), it exposes more highly conserved residues to lipids; 4), it buries more poorly conserved residues between the subunits; and 5), it orients the polar crevasses of the voltage-sensing domain toward the bulk lipid phase instead of toward the pore-forming domain.
Laine et al. (2003) have proposed an alternative model for the open conformation of Shaker, in which S4 docks on the exterior of the pore-forming domain in a manner quite different from our model. In both models of the open conformation, R362 is near F416 and A419 of an adjacent subunit, as suggested by their experimental results. However, they tilt S4 by about −15° relative to the axis of the pore in the direction opposite to the tilt in our models. If S4 of subunit I interacts with S5 of subunit II, then when the channel is viewed from the extracellular side, subunit II is on the clockwise side of subunit I in their model but is on the counterclockwise side in our model. In contrast to our models, the tilt and location of S4 in their models does not correspond to the intolerant stripes on the pore-forming domain reported by Li-Smerin et al. (2000a). Also, their model did not consider the crystal structures of KvAP. We were unable to orient S4 as in their models and also model S1–S3 after the KvAP crystal Structure 2 in a reasonable manner; e.g., we would have to rotate S1–S3 by ∼85° relative to the plane of the membrane. This would place the partially charged termini of the S1 and S2 helices in the hydrophobic transmembrane region and expose numerous charged residues to the lipids, which would be energetically unfavorable. The principal advantage of their model over ours is that it is consistent with the distance of residue 363 from the axis of the open channel as calculated from the LRET measurements (Cha et al., 1999). They did not attempt to model other conformations.
We have been using a long-term iterative approach in modeling these channels ever since the first Na+ channel sequence was published. Each new generation of models is intended to be more accurate and more soundly based than earlier versions. Our strategy is to work with the larger scientific community to achieve molecularly precise models that are soundly based on a wealth of data. The last Shaker model of our group (Durell et al., 1998) preceded determination of any K+ channel crystal structures. Our current models have many features similar to our earlier models, but also have important differences. The comparison below is intended to convey our preconceived biases before developing our current generation of models and to illustrate what can, and cannot, be predicted in the absence of crystal structures. The following features were similar to those of our current model: 1), the secondary structures; 2), the location of the S5, P, and S6 helices at the extracellular surface; 3), the chirality of the bundle of S1–S4 helices; 4), the three-helical screw step motion of S4 during activation; 5), the tilt of S4 with respect to the membrane; 6), formations of numerous salt bridges of positively charged S4 residues to negatively charged residues on S1–S3; 7), the general location of the S1–S4 voltage-sensing domain on the exterior of the pore-forming domain; 8), involvement of S4 in most interactions between the two domains; 9), exposure of poorly-conserved (or tolerant) faces of S1, S2, and S3 to lipids; and 10), the translation of the inner half of the transmembrane region by the L45 helix when the channel is open. Although these gross features are similar, many details differ. For example, although the selectivity filter (the TVGYG segment) of our 1995 models (Guy and Durell, 1995) was very similar to that of crystal structures (Doyle et al., 1998; Jiang et al., 2002; Kuo et al., 2003), it was altered to an incorrect conformation in subsequent models to better satisfy some mutagenesis data (Durell et al., 1998). The activation gate formed by S6 was also modeled incorrectly and numerous fine details of the S1–S4 bundle differed form the crystal structure of the isolated voltage-sensing domain of KvAP. Our current models are much less speculative than the last generation because they are derived by homology modeling from crystal structures, and are constrained by substantially more mutagenesis data. However, all current models are still tentative, and more data are required to analyze their validity.
CONCLUSIONS
Here and in the preceding article, we present models that, as best we can tell, are derived from undistorted portions of the KvAP crystal structures, and that are energetically, evolutionarily, and experimentally sound. Our models have many features that have been proposed previously. In contrast, the paddle model of Jiang et al. (2003a,b) is based on distorted portions of the full-length KvAP crystal structure and experiments that could trap the protein in perturbed conformations by exposing it for long durations to molecules that bind almost irreversibly. The paddle model is inconsistent with many experimental results and with energetic and evolutionary modeling criteria that have served us well in the past. Thus, proclamations that there are “very few existing studies on eukaryotic Kv channels that cannot be understood in terms of the gating mechanism of Jiang and colleagues” (Miller, 2003), that previously proposed models for the movement of S4 are “almost certainly wrong” (Sigworth, 2003), and that “the structure of KvAP's voltage sensor … is a wonderful end to a 50-year-old mystery” (Sigworth, 2003) are unwarranted and premature.
The KvAP crystal structures of Jiang et al. (2003a) will likely greatly advance our understanding of the native structures of voltage-gated channels if they can be interpreted correctly. Although these structures make current models much less ambiguous and more soundly based than earlier models, numerous features are still ambiguous and have not been experimentally verified. A supplement to this article lists major unresolved issues about the structure and gating mechanism of these channels, and suggests how some of our predictions can be tested experimentally. These suggestions all involve techniques that have already been used in studying these channels. Additional structural studies of other channels and/or using other techniques, such as electron microscopy (Sokolova et al., 2001, 2003), that do not remove the protein from the lipid bilayer are underway. We are optimistic that such structural studies, combined with the types of hypothesis-based experiments suggested in the supplement and additional computational analyses, will soon succeed in determining the correct atomically precise models of the structures and functional mechanisms of voltage-gated channels.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
References
- Ahern, C. A., and R. Horn. 2004. Specificity of charge-carrying residues in the voltage sensor of potassium channels. J. Gen. Physiol. 123:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, Q. H., C. J. Partridge, T. S. Munsey, and A. Sivaprasadarao. 2002. Depolarization induces intersubunit cross-linking in a S4 cysteine mutant of the Shaker potassium channel. J. Biol. Chem. 277:42719–42725. [DOI] [PubMed] [Google Scholar]
- Baker, O. S., H. P. Larsson, L. M. Mannuzzu, and E. Y. Isacoff. 1998. Three transmembrane conformations and sequence-dependent displacement of the S4 domain in shaker K+ channel gating. Neuron. 20:1283–1294. [DOI] [PubMed] [Google Scholar]
- Blaustein, R. O., P. A. Cole, C. Williams, and C. Miller. 2000. Tethered blockers as molecular ‘tape measures’ for a voltage-gated K+ channel. Nat. Struct. Biol. 7:309–311. [DOI] [PubMed] [Google Scholar]
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy minimization and dynamic calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- Broomand, A., R. Mannikko, H. P. Larsson, and F. Elinder. 2003. Molecular movement of the voltage sensor in a K channel. J. Gen. Physiol. 122:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, A., G. E. Snyder, P. R. Selvin, and F. Bezanilla. 1999. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 402:809–813. [DOI] [PubMed] [Google Scholar]
- Doyle, D. A., C. J. Morais, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Durell, S. R., Y. Hao, and H. R. Guy. 1998. Structural models of the transmembrane region of voltage-gated and other K+ channels in open, closed, and inactivated conformations. J. Struct. Biol. 121:263–284. [DOI] [PubMed] [Google Scholar]
- Elinder, F., P. Arhem, and H. P. Larsson. 2001a. Localization of the extracellular end of the voltage sensor S4 in a potassium channel. Biophys. J. 80:1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinder, F., R. Mannikko, and H. P. Larsson. 2001b. S4 charges move close to residues in the pore domain during activation in a K channel. J. Gen. Physiol. 118:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, C. S., E. Clark, E. Loots, A. Pralle, and E. Y. Isacoff. 2003. The orientation and molecular movement of a k(+) channel voltage-sensing domain. Neuron. 40:515–525. [DOI] [PubMed] [Google Scholar]
- Gandhi, C. S., and E. Y. Isacoff. 2002. Molecular models of voltage sensing. J. Gen. Physiol. 120:455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner, K. S., L. M. Mannuzzu, C. S. Gandhi, and E. Y. Isacoff. 1999. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 402:813–817. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C., R. Morera, F. Munoz, E. Rosenmann, O. Alvarez, and R. Latorre. 2004. Displacement of S3 segment during activation of Shaker K+ channel with a short S3–S4 linker. Biophys. J. 86:123a. [Google Scholar]
- Gonzalez, C., E. Rosenman, F. Bezanilla, O. Alvarez, and R. Latorre. 2001. Periodic perturbations in Shaker K+ channel gating kinetics by deletions in the S3–S4 linker. Proc. Natl. Acad. Sci. USA. 98:9617–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, H. R., and F. Conti. 1990. Pursuing the structure and function of voltage-gated channels. Trends Neurosci. 13:201–206. [DOI] [PubMed] [Google Scholar]
- Guy, H. R., and S. R. Durell. 1995. Structural models of Na+, Ca2+, and K+ channels. Soc. Gen. Physiol. Ser. 50:1–16. [PubMed] [Google Scholar]
- Guy, H. R., and P. Seetharamulu. 1986. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. USA. 83:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, M., K. S. Shin, and G. Yellen. 1998. The activation gate of a voltage-gated K+ channel can be trapped in the open state by an intersubunit metal bridge. Neuron. 21:617–621. [DOI] [PubMed] [Google Scholar]
- Hong, K. H., and C. Miller. 2000. The lipid-protein interface of a Shaker K(+) channel. J. Gen. Physiol. 115:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003a. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., V. Ruta, J. Chen, A. Lee, and R. MacKinnon. 2003b. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 423:42–48. [DOI] [PubMed] [Google Scholar]
- Judge, S. I., J. Z. Yeh, J. E. Goolsby, M. J. Monteiro, and C. T. Bever, Jr. 2002. Determinants of 4-aminopyridine sensitivity in a human brain kv1.4 k(+) channel: phenylalanine substitutions in leucine heptad repeat region stabilize channel closed state. Mol. Pharmacol. 6:913–920. [DOI] [PubMed] [Google Scholar]
- Khanna, R., M. P. Myers, M. Laine, and D. M. Papazian. 2001. Glycosylation increases potassium channel stability and surface expression in mammalian cells. J. Biol. Chem. 276:34028–34034. [DOI] [PubMed] [Google Scholar]
- Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- Laine, M., M. C. Lin, J. P. Bannister, W. R. Silverman, A. F. Mock, B. Roux, and D. M. Papazian. 2003. Atomic proximity between S4 segment and pore domain in Shaker potassium channels. Neuron. 39:467–481. [DOI] [PubMed] [Google Scholar]
- Larsson, H. P., O. S. Baker, D. S. Dhillon, and E. Y. Isacoff. 1996. Transmembrane movement of the shaker K+ channel S4. Neuron. 16:387–397. [DOI] [PubMed] [Google Scholar]
- Lee, H. C., J. M. Wang, and K. J. Swartz. 2003. Interaction between extracellular Hanatoxin and the resting conformation of the voltage-sensor paddle in Kv channels. Neuron. 40:527–536. [DOI] [PubMed] [Google Scholar]
- Li-Smerin, Y., D. H. Hackos, and K. J. Swartz. 2000a. A localized interaction surface for voltage-sensing domains on the pore domain of a K+ channel. Neuron. 25:411–423. [DOI] [PubMed] [Google Scholar]
- Li-Smerin, Y., D. H. Hackos, and K. J. Swartz. 2000b. Alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J. Gen. Physiol. 115:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin, Y., and K. J. Swartz. 2000. Localization and molecular determinants of the Hanatoxin receptors on the voltage-sensing domains of a K(+) channel. J. Gen. Physiol. 115:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin, Y., and K. J. Swartz. 2001. Helical structure of the COOH terminus of S3 and its contribution to the gating modifier toxin receptor in voltage-gated ion channels. J. Gen. Physiol. 117:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, G. A., Y. N. Jan, and L. Y. Jan. 1991. Hydrophobic substitution mutations in the S4 sequence alter voltage-dependent gating in Shaker K+ channels. Neuron. 7:327–336. [DOI] [PubMed] [Google Scholar]
- MacKinnon, R., and G. Yellen. 1990. Mutations affecting TEA blockade and ion permeation in voltage-activated K+ channels. Science. 250:276–279. [DOI] [PubMed] [Google Scholar]
- McCormack, K., M. A. Tanouye, L. E. Iverson, J.-W. Lin, M. Ramaswami, T. McCormack, J. T. Campanelli, M. K. Mathew, and B. Rudy. 1991. A role for hydrophobic residues in the voltage-dependent gating of Shaker K+ channels. Proc. Natl. Acad. Sci. USA. 88:2931–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. 2003. A charged view of voltage-gated ion channels. Nat. Struct. Biol. 10:422–424. [DOI] [PubMed] [Google Scholar]
- Monks, S. A., D. J. Needleman, and C. Miller. 1999. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J. Gen. Physiol. 113:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale, E. J., D. J. Elliott, M. Hunter, and A. Sivaprasadarao. 2003. Evidence for intersubunit interactions between S4 and S5 transmembrane segments of the Shaker potassium channel. J. Biol. Chem. 278:29079–29085. [DOI] [PubMed] [Google Scholar]
- Papazian, D. M., X. M. Shao, S. A. Seoh, A. F. Mock, Y. Huang, and D. H. Wainstock. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301. [DOI] [PubMed] [Google Scholar]
- Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- Shieh, C. C., K. G. Klemic, and G. E. Kersch. 1997. Role of transmembrane segment S5 on gating of voltage-dependent K+ channels. J. Gen. Physiol. 109:767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth, F. J. 2003. Structural biology: Life's transistors. Nature. 423:21–22. [DOI] [PubMed] [Google Scholar]
- Sokolova, O., A. Accardi, D. Gutierrez, A. Lau, M. Rigney, and N. Grigorieff. 2003. Conformational changes in the C terminus of Shaker K+ channel bound to the rat Kvbeta2-subunit. Proc. Natl. Acad. Sci. USA. 100:12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova, O., L. Kolmakova-Partensky, and N. Grigorieff. 2001. Three-dimensional structure of a voltage-gated potassium channel at 2.5 nm resolution. Structure (Camb). 9:215–220. [DOI] [PubMed] [Google Scholar]
- Starace, D. M., and F. Bezanilla. 2004. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 427:548–553. [DOI] [PubMed] [Google Scholar]
- Starace, D. M., E. Stefani, and F. Bezanilla. 1997. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 19:1319–1327. [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff, S. K., M. A. Lin, C. T. Schulteis, and D. M. Papazian. 2000. Voltage-dependent structural interactions in the Shaker K(+) channel. J. Gen. Physiol. 115:123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, S. M., D. Del Camino, J. P. Dekker, and G. Yellen. 2004. Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges. Nature. 428:864–868. [DOI] [PubMed] [Google Scholar]
- Yang, N., A. L. George, Jr., and R. Horn. 1996. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 16:113–122. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.