Abstract
Using the bacterial K+ channel KcsA as a template, we constructed models of the pore region of the cardiac ryanodine receptor channel (RyR2) monomer and tetramer. Physicochemical characteristics of the RyR2 model monomer were compared with the template, including homology, predicted secondary structure, surface area, hydrophobicity, and electrostatic potential. Values were comparable with those of KcsA. Monomers of the RyR2 model were minimized and assembled into a tetramer that was, in turn, minimized. The assembled tetramer adopts a structure equivalent to that of KcsA with a central pore. Characteristics of the RyR2 model tetramer were compared with the KcsA template, including average empirical energy, strain energy, solvation free energy, solvent accessibility, and hydrophobic, polar, acid, and base moments. Again, values for the model and template were comparable. The pores of KcsA and RyR2 have a common motif with a hydrophobic channel that becomes polar at both entrances. Quantitative comparisons indicate that the assembled structure provides a plausible model for the pore of RyR2. Movement of Ca2+, K+, and tetraethylammonium (TEA+) through the model RyR2 pore were simulated with explicit solvation. These simulations suggest that the model RyR2 pore is permeable to Ca2+ and K+ with rates of translocation greater for K+. In contrast, simulations indicate that tetraethylammonium blocks movement of metal cations.
INTRODUCTION
The release of Ca2+ from intracellular storage organelles, such as the endoplasmic or sarcoplasmic reticulum, is a vital component of cell signaling processes as diverse as muscle contraction and fertilization (Berridge et al., 2003). Pathways for the regulated release of Ca2+ are provided by a family of cation-selective ion channels comprising two related species of channel: the inositol-trisphosphate receptor (InsP3R) and ryanodine receptor (RyR) (Berridge et al., 2003). In this communication, we consider the structures and mechanisms involved in cation selection and translocation in RyR.
Single channel experiments, involving a wide range of permeant and impermeant inorganic and organic cations, have established the following characteristics of ion translocation in RyR. The channel is impermeable to anions but is permeable to a diverse group of cations (Lindsay and Williams, 1991). Discrimination between physiologically relevant cations is limited. The alkaline earth divalents are essentially equally permeant (Tinker and Williams, 1992) as are the group 1a monovalents (Lindsay et al., 1991). However, the relative permeability of divalent cations is ∼6.5-fold greater than monovalents (Tinker and Williams, 1992). Unitary conductance of RyR is very high, reaching ∼1 nS at saturating activities of K+ (Lindsay et al., 1991) and 200 pS at saturating activities of Ba2+ (Tinker and Williams, 1992). Experiments in which the occupancy of the channel has been assessed indicate that RyR appears to be a single-ion channel (Williams et al., 2001).
Information on the dimensions of the pathway through the RyR channel that underlie these properties has been obtained from investigations of the interactions of permeant and impermeant organic cations. The minimum radius of the RyR pore has been estimated as 3.5 Å based on the relative permeability of organic monovalent cations (Tinker and Williams, 1993). Block by bis-quaternary ammonium ions of varying length indicates that the voltage drop across the channel is likely to occur over a distance of ∼10 Å (Tinker and Williams, 1995), and a similar estimate for the pore length has been obtained from the measurement of streaming potentials (Tu et al., 1994a). The overall picture of the pore of the RyR channel that emerges from these studies is that of a short, wide structure that allows phenomenal rates of movement of Ca2+ down a concentration gradient across the reticular membrane to fulfill its role as an effective Ca2+-release channel (Williams et al., 2001).
Our understanding of the mechanisms involved in cation translocation and selection in membrane channels has been advanced dramatically by the determination of the structure of the pore of bacterial K+ channels at atomic resolution (Doyle et al., 1998; Roux and MacKinnon, 1999; Y. Zhou et al., 2001; Morais-Cabral et al., 2001). The pore is formed at the longitudinal axis of the channel homotetramer with each of the monomers contributing two transmembrane helices and components of their connecting extracellular loop. The resulting pore in KcsA, the first of these channels for which structural information was obtained, is 45 Å in length with several clearly defined structural and functional domains. These are, starting from the cytoplasmic entrance, an 18-Å long tunnel termed the internal pore, a cavity ∼10 Å in diameter, a selectivity filter 12 Å in length and 3 Å in diameter, and an extracellular mouth. This structure fulfils the basic requirement for a selective ion channel providing a pathway for the movement of an ion across the energetically hostile environment of the membrane. In addition, the structure achieves near perfect discrimination between ions as closely related as K+ and Na+ by the strict coordination of K+ by backbone carbonyl oxygens of the residues of the selectivity filter. High rates of K+ translocation arise from interactions of ions within this selectivity filter (Y. Zhou et al., 2001; Morais-Cabral et al., 2001; Miller, 2000). From this very brief description of ion translocation in RyR and K+ channels, it is clear that these two processes have very little in common. However, evidence is emerging that might indicate some important similarities in the structural elements that contribute to the pores of these two species of channel.
In the first instance, an amino acid sequence analogous to the signature sequence of K+ channels was identified in the luminal loop connecting the last two transmembrane helices of RyR (Balshaw et al., 1999). Mutations within these sequences, and adjoining residues, produce profound alterations in rates of ion translocation (Zhao et al., 1999; Gao et al., 2000; Du et al., 2001). Together, these observations have led to the proposal that the pore of the RyR Ca2+-release channel could consist of components contributed by the luminal loops of each monomer (Balshaw et al., 1999; Zhao et al., 1999). Further analogies with K+ channels were suggested by comparisons of potential structural elements of the putative pore-forming loop of RyR. In K+ channels, elements of differing secondary structure within the pore-forming loop are arranged in a specific order, the folding of which gives rise to the arrangement seen in the crystal structure of KcsA. Secondary structure predictions for the putative pore-forming loop of RyR indicate that this region contains equivalent structural elements, arranged in the same order, leading to the proposal that the last two transmembrane helices and their linking luminal loop might adopt a similar tertiary structure to that seen in KcsA (Williams et al., 2001; Shah and Sowdhamini, 2001).
In this communication, we have tested this hypothesis by constructing a model incorporating the last two transmembrane helices and the linking luminal loop of each RyR2 monomer using the known tertiary structure of the KcsA K+ channel monomer as a template. In doing so, we have made a quantitative assessment of 1), the probability of the putative pore-forming region of RyR2 adopting a tertiary structure equivalent to that of KcsA, and 2), the probability of the formation of a transmembrane pore by the tetramerization of these monomers. These assessments provide strong support for the validity of the model. We have also investigated the interactions of permeant and impermeant cations with the derived quaternary structure. Finally, we discuss various established characteristics of ion selection and translocation in RyR in the light of structural features emerging from this model.
MATERIALS AND METHODS
The elements of the RyR2 primary structure comprising our model, together with the equivalent elements of KcsA, are shown in Fig. 1. The primary sequence of this region is highly conserved (>90% identity) among RyR isoforms, and as a consequence the model described here can be considered as a model for the pore-forming regions of all RyR isoforms. The residue numbers quoted refer to the rabbit RyR2 sequence. Although the numbering of residues is different, the sequence is identical in all mammalian RyR2 channels.
FIGURE 1.
The primary sequences of (A) the bacterial K+ channel, KcsA, from Streptomyces lividans and (B) the rabbit type-2 ryanodine receptor, RyR2, included in this analysis. The regions identified as the outer helix, pore helix, selectivity filter, and inner helix of both sequences are highlighted by shaded boxes.
The model contains only 2.4% of the total residues in the RyR2 monomer. The transmembrane domains that we have used in the model are TM3 and TM4 of the four transmembrane domain model described by Takeshima et al. (1989); however, these are equivalent to the last two transmembrane domains of other proposed transmembrane domain topologies of RyR (Du et al., 2002).
Building the model of RyR2, step 1
In KcsA, four structurally conserved regions were identified: selectivity filter (Thr-75–Gly-79), pore helix (Tyr-62–Ala-73), inner helix (Leu-86–Thr-112), and outer helix (Ala-28–Glu-51) (Fig. 1). By a combination of sequence analogy and comparison of predicted secondary structure, four corresponding regions in the RyR2 model were identified: selectivity filter (Gly-4826–Asp-4831), pore helix (Met-4806–Arg-4824), inner helix (Ile-4850–Lys-4883), and outer helix (Gln-4768–Arg-4792). Our initial secondary structure predictions indicated that the inner helix contains the residues Ile-4850–Ile-4869; however, to maintain the analogy with KcsA we have extended this region to Lys-4883. The putative selectivity filter of RyR2 contains a motif of residues thought to be analogous to those making up the signature selectivity sequence of K+ channels (Balshaw et al., 1999).
The helical regions were built and placed in the α-helical conformation by setting the Φ- and Ψ- (Ramachandran) torsional angles to values of −57° and −47°, respectively. The inner, outer, and pore helices were aligned in Cartesian space by aligning the backbone atoms of the RyR2 helices to the corresponding KcsA helices. Using the consensus of various secondary structure prediction algorithms (Qian and Sejnowski, 1988; Garnier et al., 1978; Maxfield and Scheraga, 1979) the inner helix was extended. The gaps between the helices were closed by the loop search utility built into the SYBYL suite (Tripos, St Louis, MO). Briefly, the protein database was searched for fragments with sequence homology to the missing amino acid sequences. These fragments were scored for homology by the p-mutation identity matrix (Dayhoff et al., 1978), the lack of steric clashes with structurally conserved regions (helices) of the RyR2 model, and, most importantly, an end-to-end distance that would allow insertion of the loop into the gap without distorting the structurally conserved regions. Interestingly, loops were readily found with excellent matches for end-to-end distance and where the backbone of the loops did not become tangled with each other. The geometry of the loop representing the putative selectivity filter of RyR2 was an excellent match to that of the KcsA selectivity filter, even before a minimization was applied.
Building the model of RyR2, step 2
The completed monomeric unit was then assembled into a tetramer. It was assumed that the relative position of the inner and outer helices would have essentially the same spatial relationship in both KcsA and RyR2. Any differences would be expected to reside primarily in the pore helix and the selectivity filter. Therefore, 19 residues from the inner helical regions of the KcsA monomers were used as templates for RyR2. The peptide backbone of RyR2 (Ile-4850–Ile-4868) was superimposed over the backbone atoms of Gly-88–Val-106 of KcsA. None of the backbone atoms intertwined or had steric clashes. However, some of the side chains did have severe steric clashes with side chains on a neighboring subunit. Most of these were removed by molecular mechanics using the Powell method (Powell, 1977), although a few side-chain interactions remained because of intertwined bonds. Dihedral driver algorithms applied to the trapped side chains removed these clashes.
Testing the ion handling properties of the model RyR2 pore
The simulated system was constructed by centering either KcsA or RyR2 between two water molecules, 100 Å apart, which formed the x axis passing through the center of the pore. The positions of these waters were fixed in all simulations, serving as anchors for movement of the cations. Water molecules were simulated using the TIP model. Various amounts of ions (K+, Ca2+, and tetraethylammonium (TEA+)) were added, and the system was solvated using TIP waters and Silverware software (Tripos). A distance constraint was applied on one ion (designated the probe ion) to bias the diffusion toward the anchor on one or the other side of the pore. The force used follows a simple reciprocal distance squared relationship (1/Å2) such as seen in Coulomb's law. Therefore, the force on the ion decreases as the ion approaches the target. The constraint simulates experimental measurements on channels in symmetrical ionic conditions where the driving force is provided by an exogenous transmembrane voltage. In addition, the constraint permitted the simulated transit to occur in reasonable periods of computer time. In all cases, ion movement was measured through the pore in both directions: thus, the probe ion alternately first encountered the cytosolic and luminal ends of the selectivity filter. All other ions in the simulation were unconstrained (i.e., they are free to move in any direction, influenced by only the protein).
Limitations
It should be noted that as only a small portion of the total amino acid sequence of RyR2 forms the model, factors such as electrostatic forces from the omitted regions of the channel that may influence absolute rates of ion translocation are not accounted for in our simulations. Similarly, the omission of an as yet undefined number of transmembrane helices may limit the ability to model conformers of the two transmembrane helices incorporated in the model.
RESULTS
Comparison of the helical components of the KcsA and RyR2 monomers
In the following sections, we compare features of the helical components of the K+ channel template and the RyR2 model. Various parameters are summarized in Table 1.
TABLE 1.
Comparison of physicochemical parameters of the helical elements of KcsA and the RyR2 model
| KcsA | RyR2 | |
|---|---|---|
| (a) Inner helix | ||
| Sequence identity to KcsA (%) | Not applicable | 18.5 |
| Predicted α-helical content (%) | 12 | 48 |
| Hydrophobicity | ||
| Kyte-Doolittle (average (range)) | 1.77 (from −4.5 to +4.5) | 1.01 (from −4.5 to +4.5) |
| Octanol–water (average (range)) | 0.86 (from −1.32 to +2.51) | 0.99 (from −1.32 to +2.09) |
| Surface area (Å2) | 1818 | 2592 |
| Hydrophobic surface area (% total) | 33 | 40 |
| Electrostatic charge (kcal/mol) | From −55 to +273 | From −298 to +93 |
| Positive surface area (% total) | 94 | 14 |
| (b) Outer helix | ||
| Sequence identity to KcsA (%) | Not applicable | 16.7 |
| Predicted α-helical content (%) | 55 | 20 |
| Hydrophobicity | ||
| Kyte-Doolittle (average (range)) | 2.07 (from −3.5 to +4.5) | 1.78 (from −4.5 to +4.5) |
| Octanol–water (average (range)) | 0.92 (from −0.79 to +2.04) | 0.92 (from −0.79 to +2.04) |
| Surface area (Å2) | 1528 | 1959 |
| Hydrophobic surface area (% total) | 38 | 46 |
| Electrostatic charge (kcal/mol) | From −203 to +103 | From −70 to +139 |
| Positive surface area (% total) | 26 | 90 |
| (c) Pore helix | ||
| Sequence identity to KcsA (%) | Not applicable | 8.3 |
| Predicted α-helical content (%) | 2.8 | 39 |
| Hydrophobicity | ||
| Octanol–water (average(range)) | 0.80 (from −1.32 to +2.51) | 0.98 (from −1.32 to +2.09) |
| Surface area (Å2) | 936 | 1475 |
| Hydrophobic surface area (% total) | 41 | 32 |
| Electrostatic charge (kcal/mol) | From −175 to +162 | From −175 to +162 |
| Positive surface area (% total) | 60 | 49 |
Inner helix
The putative inner helix of RyR2 shares 18.5% sequence identity with the inner helix of KcsA. Secondary structure predictions (Garnier et al., 1978; Maxfield and Scheraga, 1979; Qian and Sejnowski, 1988) indicate an average α-helix component of 48% for this region of RyR2 compared to 12% for the inner helix of KcsA. The putative inner helix of RyR2 has a much greater total surface area than the equivalent region of KcsA. Forty percent of the RyR2 helix is hydrophobic compared to 33% with KcsA (MOLCAD, Tripos; see Ghose and Crippen, 1986; Ghose et al., 1998; Heiden et al., 2004, for principles involved), and in both helices the C-terminus is more negative than the N-terminus. However, the RyR2 inner helix has a much greater overall negative electrostatic charge and a lower positive surface area than the inner helix of KcsA.
Outer helix
The putative outer helix of RyR2 shares 16.7% sequence identity with the outer helix of KcsA. The RyR2 helix has a lower predicted average α-helix component (20%) than the outer helix of KcsA (55%). Using the Kyte-Doolittle scale, the RyR2 helix is less hydrophobic than the KcsA helix with the N-terminus of both helices being more hydrophilic than the C-terminus. However, no differences were found when the two helices were compared using the octanol-water transfer free energy scale. In RyR2, the surface of the outer helix is 46% hydrophobic compared to 38% for the equivalent structure in KcsA. The RyR2 outer helix also has a large positive surface area (90% total area) with charge distributed over its entire length (with the largest area at the C-terminus due to the presence of an arginine), whereas the KcsA helix has a much lower positive surface area (26% total area) with the positive surface restricted to the N-terminus.
Pore helix
The pore helices of RyR2 and KcsA share only 8.3% sequence identity and have an average α-helix content of 39% and 2.8%, respectively. Kyte-Doolittle analysis of hydrophobicity was not performed as the pore helix of KcsA is too short to yield meaningful information. However, the hydrophobicities of both helices are similar using the octanol-water transfer free energy scale. In RyR2, the surface of the pore helix is 32% hydrophobic compared to 41% for the pore helix of KcsA. In both cases, hydrophobic regions are distributed evenly over the surface of the structure. We observed no striking bias in electrostatic potential in either pore helix, although the KcsA pore helix has a slightly larger positive surface area (60% total surface area) than the pore helix of RyR2 (49% total surface area).
A comparison of the KcsA and RyR2 tetramers
General features of the model
We have used the crystal structure of KcsA as a template in the construction of our model of the RyR2 pore and for quantitative comparisons of the physicochemical features of the model. It should be noted that the KcsA structure was determined in the presence of K+. The RyR2 target is modeled and refined without ions and with implicit solvent. Subsequently, explicit solvation was included with (1–13 K+) and without ions. The addition of water and ions caused small changes in the dimensions of the selectivity filter and the positions of the luminal loops. The peptide backbone of the helices moved only slightly in molecular dynamics simulations (<0.1 Å root mean-square (rms)). Changes are due to charge neutralization (e.g., luminal loops), ionic cross-linking (e.g., Glu-4832), van der Waals interactions with solvent, and hydrogen bonding to solvent.
The model of the RyR2 pore (Fig. 2) contains the same structural elements as KcsA, and the overall topography of these elements is similar. The KcsA structure is highly symmetrical because it was solved from a crystal. However, the RyR2 tetramer is asymmetrical because the structure is based on energy calculations in which all regions are allowed independent motion, as they would be in solution. In addition, the KcsA structure (1BL8) is in a closed conformation with the overlap of the four inner helices forming a barrier to the movement of ions or gate. Likewise the RyR2 model has an equivalent overlap at the cytosolic entrance to the structure that may represent a gate. As a consequence, we assume that the RyR2 model represents the closed conformation of this channel.
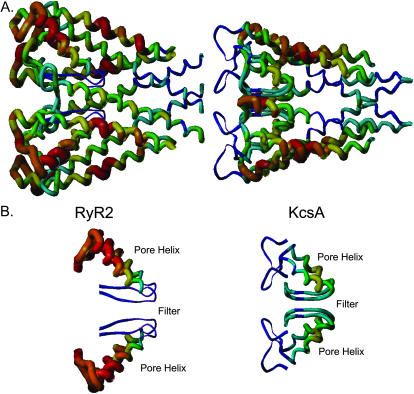
FIGURE 2.
Schematic tube diagram of (A) the RyR2 model and (B) KcsA. The individual structural elements that comprise the pore-forming regions of these two structures have been colored as follows: outer helix (blue); pore helix (red); selectivity filter (green); and inner helix (cyan). For purposes of clarity, just two of the four monomers are shown.
Both the KcsA template and RyR2 model have a cytosolic cavity lined by residues of the inner helices. In both channels, the pore helices are orientated so that the helix dipole is directed into the cytosolic cavity. The C-terminal ends of the pore helices are located at equivalent positions at the extracellular (KcsA) or luminal (RyR2) end of this cavity. The longer RyR2 pore helix extends further into space at the luminal face of the pore.
The loops linking the pore helices and inner helices of KcsA contain residues essential for K+ selectivity (TVGYGD) and form the selectivity filter of this channel. The equivalent loops in the RyR2 model contain analogous residues (GGGIGD) and form an analogous structural element.
The principal differences between the KcsA template and the RyR2 pore model are a), the arrangement of the loops at the luminal face of the structure that link the outer helix to the pore helix and the pore helix to the putative selectivity filter and b), the shape of the selectivity filter.
Quantitative comparisons
We have examined the plausibility of our model by carrying out a quantitative comparison of a variety of its features with equivalent features of KcsA.
Probability of fold. The probability of a fold can be estimated by the use of a pseudoenergy function (i.e., the probability that an amino acid residue will be located in a particular environment based on observations of solved protein structures; Leach, 2001). Fig. 3 A compares the pseudo- (statistical) energy functions (MatchMaker, Tripos; Godzik et al., 1992; Godzik and Skolnick, 1992) at various positions along the peptide backbone of the two structures.
FIGURE 3.
(A) Statistical energy functions of the RyR2 model compared to KcsA. Colors represent the statistical energies at various positions along the peptide backbone: purple is the energetically most favorable, whereas red is the energetically least favorable. The width of the tubes conveys similar information: narrow regions are energetically favorable whereas wide regions are energetically less favorable. Both structures are orientated such that the cytosolic side is on the right. (B) The trajectories of the selectivity filters and pore helices of the two structures are shown in more detail. Color coding is as described for A.
Interestingly, the putative selectivity filter region of the RyR2 model is considerably more favorable than that of the KcsA template. In contrast, the areas near the luminal face of RyR2 are considerably less favorable than the corresponding extracellular regions of KcsA. The overall (average) empirical energy of the two molecules is quite close with the RyR2 tetramer (+0.01 kT) being slightly more favorable than KcsA (+0.02 kT). It is important to note that these are both membrane proteins and not all the probable transmembrane segments of RyR2 are included in the model. Therefore, the many positive values (Fig. 3 A, red areas) present in the RyR2 model are likely to reflect interactions omitted from the model. Fig. 3 B compares the trajectories of the pore helices and selectivity filters of the two structures. The energy range for KcsA is from −0.37 to +0.46 kT. In comparison, the range for RyR2 is from −0.35 to +0.5 kT. Clearly, the pore helix is much longer in our model of RyR2 than in the crystal structure of KcsA, and the luminal ends of the RyR2 pore helices are somewhat less favorable than the equivalent structures in KcsA.
Strain energy. Molecular mechanics and dynamics use an empirical energy function known as a force field to model the conformation of a molecule. The average per residue strain energy (Amber7 force field) of the 1BL8 structure of KcsA (template) is −10.4 kcal/mol compared to −10.3 kcal/mol for the RyR2 model. The distribution of strain energies is essentially identical in both structures (data not shown).
Solvation free energy. Solvation free energies of the two structures are shown in Fig. 4 A. The average solvation free energy of KcsA is 0.315 kcal/mol (range from −2.980 to +1.854; positive is unfavorable hydration) and RyR2 is 0.309 kcal/mol (range from −2.812 to +1.784). Purple is the most negative (strongest solvation), whereas the red is the most positive. A more detailed comparison of the selectivity filters and pore helices of the two structures is given in Fig. 4 B. These comparisons provide further evidence that the fold of the putative RyR2 pore is equivalent to the known structure of KcsA.
FIGURE 4.
(A) The solvation free energy of the RyR2 model compared to KcsA. The peptide backbone of the two structures has been colored so that purple represents the most negative solvation free energy (i.e., strongest solvation) and red the most positive (i.e., unfavorable hydration). Both structures are orientated such that the cytosolic side is on the right. (B) The trajectories of the selectivity filters and pore helices of the two structures are shown in more detail. Color coding is as described for A.
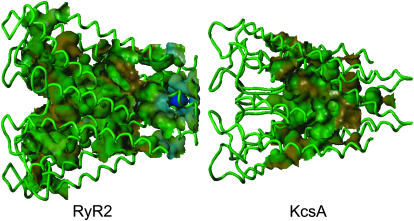
Solvent accessible surfaces of the RyR2 pore model. Fig. 5 compares the internal volumes that can be occupied by water. It is noticeable that although water is excluded from the narrow selectivity filter of KcsA, the solvent has access to the entire length of the much larger selectivity filter of the RyR2 model. The hydrophobicities of the surfaces lining the pathways (Fig. 6) are, in general, similar with one major exception. It is noticeable that residues at the cytosolic entrance of RyR2 (at the gate in this putative closed structure) are significantly less hydrophobic (from −0.21 to +0.12) than the equivalent residues in KcsA (from −0.087 to +0.13). In addition, there is a notable difference between RyR2 and KcsA in the electrostatic potential projected on the channel (Fig. 7). The ratio of acid to basic residues is 1.9 for RyR2 in comparison to a ratio of 0.8 for KcsA. The large number of acid residues and the long-range effect of electrostatic charge make both the cytosolic and luminal faces of the RyR2 pore model more negative than the equivalent regions of the KcsA template. Since only a small fraction of the RyR2 is modeled here, a quantitative assignment of the electrostatic nature of the lining of the pore cannot be made. However, the close proximity of the large number of acidic amino acids near the RyR2 pore will most likely make the surfaces of the RyR2 pore more negative than the corresponding regions of KcsA.
FIGURE 5.
The water accessible areas (Connolly channel as implemented in SYBYL) of the RyR2 model compared to KcsA. The internal volume that can be occupied by water is colored green. Note that water can transverse the entire length of the predicted RyR2 pore, whereas it cannot get into the selectivity filter of KcsA (1BL8). Both structures are orientated such that the cytosolic side is on the right.
FIGURE 6.
The hydrophobicity projected onto the water accessible surface of the RyR2 model compared to KcsA. Hydrophobicity of the pore-lining residues is symbolized as colors: (brown) the most hydrophobic, (green) borderline hydrophobic, and (blue) the most polar. Both structures are orientated such that the cytosolic side is on the right.
FIGURE 7.
The electrostatic potential projected onto the water accessible surface of the RyR2 model compared to KcsA. Electrostatic potential is symbolized as colors: (blue) the most negative potential and (red) the most positive. Both structures are orientated such that the cytosolic side is on the right.
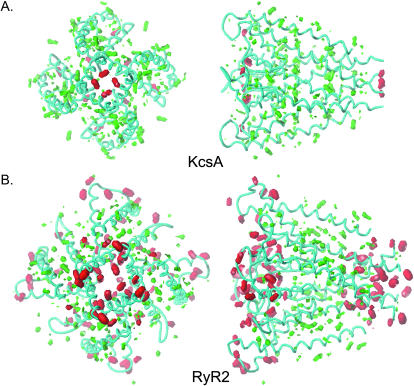
Hydrophobic, polar, acid, and base moments of KcsA and the RyR2 model. Although the RyR2 model has far more polar potential than KcsA, the distribution of the polar and hydrophobic areas is essentially identical in the two channels (Fig. 8). In addition, in both channels, the hydrophobic potential is similar, and the lining of the pore is more polar than hydrophobic (Fig. 8). Although the residues lining the pore may be hydrophobic, the polar peptide backbone makes an important contribution to the character of the pore.
FIGURE 8.
The hydrophobic and polar moments of the RyR2 model compared to KcsA (HINT as implemented in SYBYL). In both cases the peptide backbone is shown in cyan. Both KcsA and RyR2 are contoured at the same potentials (red: polar, contoured at −56; and green: hydrophobic, contoured at +28). The volumes enclosed are proportional to the value of property. The left-hand panels of A and B are orientated such that the structures are viewed from the cytosol. In the right-hand panels, the cytosolic ends of the structures are on the right. Both RyR2 and KcsA are on the same scale so that volumes can be compared directly.
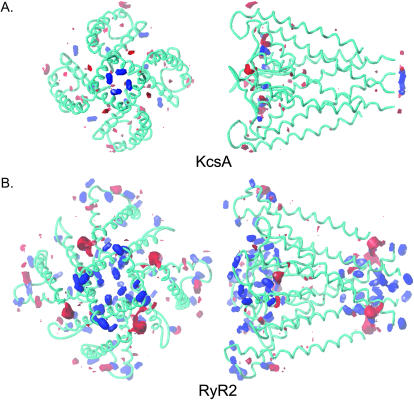
In both structures, the lining of the pore is overwhelmingly basic at pH 7.0 (Fig. 9). Both the RyR2 model and KcsA contain rings of negative charge at the cytosolic and luminal/extracellular ends of the pore, but the amount of charge is much higher overall in RyR2. Although the anionic character of the two channels is quantitatively very different, the distribution of acid and base moments are almost identical. KcsA and RyR2 both have high charge density at the faces of the structures that would be in contact with aqueous environments and little charge in the transmembrane portion of the structure.
FIGURE 9.
The acid/base potential of the RyR2 model compared to KcsA (HINT as implemented in SYBYL). In both structures, the contours demonstrate the condition of amino acid residues at pH 7.0. Conjugate acids under these conditions, such as Lys and Arg, are contoured red (at −18), and conjugate bases, such as Glu and Asp are contoured blue (at +56). Acids and bases were calculated using the Lewis acid definition to give the most general description of the properties of KcsA and RyR2. At pH 7.0 the carboxylates will be Lewis bases because they are capable of donating electrons to a proton (a Lewis acid). The Lewis acid/base contours also describe the tendency of the groups to form interactions with metal ions. The volumes enclosed are proportional to the value of the property. The left-hand panels of A and B are orientated such that the structures are viewed from the cytosol. In the right-hand panels, the cytosolic ends of the structures are on the right. Both RyR2 and KcsA are contoured on the same scale so that volumes can be compared directly.
The preceding comparison of the physicochemical properties of the RyR2 model and KcsA indicate that the RyR2 fold is consistent with the established structure of the K+ channel template.
Plausibility of the RyR2 pore model
Testing the ion handling properties of the model RyR2
The data described above supports our use of KcsA as a valid template for the folding of the putative pore-forming region of the RyR2 channel. However, is the model consistent with experimental observations of ion translocation in the channel? To test this, we performed a series of simulations of ion flow through the RyR2 pore model using KcsA to calibrate the system. These simulations are not intended to replicate electrochemical potential driven ion flow under physiological conditions but to show that the RyR2 model qualitatively simulates experimental findings and to identify landmark interactions between permeant and impermeant ions and amino acids in the pore region of RyR2. We kept the computational algorithm as simple and straightforward, and the computational times as short, as practical.
Simulations of ion flow in water. To initially calibrate the molecular dynamics simulations, we examined the effect of exogenous force on the velocity of K+ and Ca2+ through water without the complications of the channel. As the force on K+ increases from 0.1 to 1.0 kcal/Å2, the velocity rises to a maximum of 38–39 Å/ps after which velocity becomes independent of the applied force. Most likely this indicates that the rate-determining step is the diffusion of the water molecules from the path of the moving ion. The average velocity of K+, measured between the 10- and 90-Å marks, is roughly proportional to the applied force. When compared at the same applied force, the velocity of K+ is consistently faster than Ca2+. The ratio of the velocities varies between 1.6 and 1.8 in qualitative agreement with the larger (2.5-fold) diffusion coefficient of K+ (Lide, 2002).
Simulations of K+ movement in KcsA. At low applied forces (0.2–0.3 kcal/Å2) and in the absence of any other ions, a single K+ moving from the cytosolic side of the channel interacts with residues of the inner helix crossover region (Thr-112, Val-115, Gly-116, and Glu-118) and does not traverse the pore within the 10-ps simulation period. At forces of 0.5 and 1.0 kcal/Å2, K+ traverses the pore at velocities similar to those seen in bulk water and interacts with residues at the inner helix crossover (Val-115) and at the cytosolic entrance of the selectivity filter (Thr-75). Simulations were also performed in the presence of excess K+ ions. Twelve additional K+ ions were added. These ions were unconstrained at all times and allowed to diffuse freely. The system was first equilibrated by an extended molecular dynamics run until the potential and total energies were constant and the radius of gyration of the K+ ions oscillated about an average. Interestingly, K+ ions entered the selectivity filter and spontaneously inserted themselves into the same positions occupied in the crystal structure of KcsA (Doyle et al., 1998). One K+ is coordinated by four Tyr-78 through the peptide oxygens. Another K+ is coordinated by four Val-76 in the middle of the selectivity filter, again through the peptide oxygens. The interactions of a third K+ are a bit more complex. A mix of peptide and side-chain oxygens of Thr-75 and peptide oxygens of Thr-74 forms a binding site at the cytosolic end of the selectivity filter. Within the selectivity filter, K+ ions are interspersed with water molecules. In the initial and final states of the simulation, the atoms are lined up as water–K+–water–K+–water–K+–water. These K+ ions are tightly bound and do not move unless displaced by one of the moving cations.
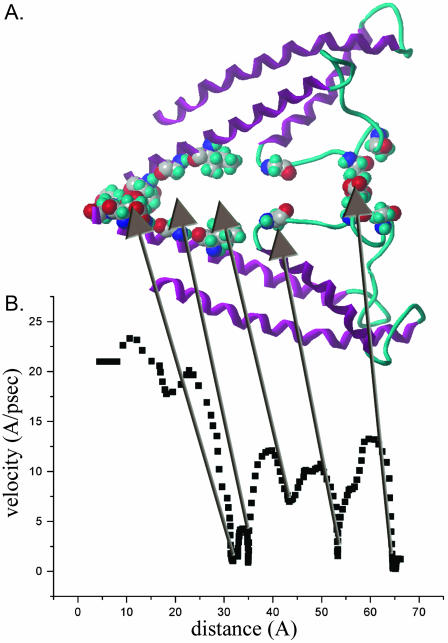
Fig. 10 is an example of the relationship between the velocity of a probe K+ and distance as it is pulled through KcsA pore (1BL8) in the presence of explicit solvation and 12 bystander K+ ions (dispersed throughout the volume used for simulation). Kinetically important residues are illustrated as space fill amino acid residues (hydrogens omitted) in Fig. 10 A. These residues are located within 3 Å of the K+ during the dips in velocity shown in B. Combining a number of velocity profiles such as that in B, we have identified the most significant kinetic barriers at (from left to right, cytosol to extracellular) Glu-118, Gly-116, Val-115, Thr-112, Ala-111, Thr-74, Thr-75, Gly-77, Tyr-78, and Gly-79. The major dip in the 20–30-Å region is due to interactions with residues at the inner helix crossover (Val-115 and environs). The major dip in velocity at 52 Å is the result of interactions with Thr-75 at the cytosolic entrance of the selectivity filter. The dip in velocity at the 66-Å mark is due to interactions at the extracellular end of the selectivity filter (Gly-79). Both of these dips appear to represent dehydration and/or rehydration of the K+ as it enters or leaves the selectivity filter. The dip in velocity at the 85-Å mark occurs outside of the KcsA pore. At this point, the K+ is fully solvated. This dip is consistently observed under a wide variety of conditions. Presumably the transient decrease in velocity is due to electrostatic interactions with the KcsA protein. The extreme left of the trace contains the initial acceleration of K+; at the extreme right, the force on the K+ falls below that of thermal motion.
FIGURE 10.
Molecular dynamics simulation of a single K+ ion as it is pulled though the KcsA pore in the presence of explicit solvation and 12 bystander K+ ions. Kinetically important residues (located within 3 Å of the K+ ion during the dips in velocity shown in B) are illustrated as space fill amino acid residues. Velocity profiles, such as that in B, identify the most significant kinetic barriers at (from left to right, cytosol to extracellular) Glu-118, Gly-116, Thr-112, Ala-111, Thr-74, Thr-75, Gly-77, Try-78, and Gly-79. See text for further details.
Simulations of K+ flow in RyR2. In the absence of additional nonprotein ions, a single K+ ion moved through the channel with a velocity comparable to that through water and KcsA at applied forces between 0.5 and 1.0 kcal/Å2. A major dip in the velocity occurred at Glu-4832, located at the luminal entrance of the selectivity filter, as the result of an interaction with the side-chain carboxyl groups of these residues. As the applied force is decreased, major interactions with Gly-4827, Ala-4837, Ile-4869, Asp-4877, Glu-4880, and Gln-4881 are identified. The latter four amino acids are located in the C-terminal half of the putative inner helix and are assumed to represent a physical barrier equivalent to the gate formed by the crossover of inner helices in KcsA.
As with KcsA, additional simulations were performed in which the RyR2 model was soaked in water containing 12 additional K+ ions. The system was equilibrated by using molecular dynamics with no constraints to movement of the additional K+ ions. Two K+ ions migrated into the selectivity filter where they came in van der Waals contact with the ring of Glu-4832. One was located on the luminal side and the other on the selectivity filter side of the glutamyl residues. Similarly to KcsA, the K+ in the selectivity filter remained in place until displaced by another cation.
Using these conditions, residues forming kinetic barriers for K+ in the RyR2 model were identified as shown in Fig. 11. Kinetically important residues are illustrated as space fill amino acid residues in A. These residues are located within 3 Å of the K+ during the dips in velocity shown in B. Combining a number of velocity profiles such as that in B, we have identified the most significant kinetic barriers (from left to right, cytosol to lumen) at Gln-4881, Glu-4880, Asp-4877, Gly-4873, Ile-4869, Gly-4827, Glu-4832, and Ala-4837. Referring specifically to B, the small dip at 18 Å is just outside of the pore (at the cytosolic side of the inner helix crossover). The K+ is completely hydrated, and this slowing probably arises from electrostatic interactions with RyR2. The major dip at 32 Å is due to interactions with Asp-4877, and during the dip at 35 Å the hydrated K+ makes a water bridge to Gly-4873. The velocity minimum at 43 Å is also in the vestibule, approximately at the apex of the angle formed by the pore helices with a water bridge of the hydrated K+ to Ile-4869. The dip in velocity at 53 Å is the result of a dalliance in the selectivity filter with Gly-4827. As the K+ ion moves through the selectivity filter, it appears to undergo partial dehydration having only one water of hydration at some points in its translocation. The major dip in velocity at 65 Å is the result of interactions with carboxyl groups of Glu-4832 at the luminal end of the selectivity filter. The initial acceleration of K+ is shown on the extreme left of B, and the extreme right of the plot shows the slow movement of K+ away from the ring of Glu-4832.
FIGURE 11.
Molecular dynamics simulation of a single K+ ion as it is pulled though the RyR2 pore in the presence of explicit solvation and 12 bystander K+ ions. Kinetically important residues (located within 3 Å of the K+ ion during the dips in velocity shown in B) are illustrated as space fill amino acid residues. Velocity profiles, such as that in B, identify the most significant kinetic barriers (from left to right, cytosol to lumen) at Gln-4881, Glu-4880, Asp-4877, Gly-4873, Ile-4869, Gly-4827, Glu-4832, and Ala-4837. See text for further details.
Simulations of Ca2+ flow in RyR2. Considerably fewer simulations have been performed with Ca2+ with explicit solvent in RyR2. However, the picture that emerges is essentially that seen in Fig. 11 for K+. As judged by the velocity profiles, Ca2+ interacts with the same amino acid residues as K+, with Glu-4832, Asp-4877, and Gln-4881 having the greatest effect on ion velocity. K+ moves through the model RyR2 pore faster than Ca2+ under otherwise identical conditions. For example, at an applied force of 1 kcal/Å2, K+ moves through the model pore with an average velocity of 30 Å/ps, whereas Ca2+ does not pass through the pore within the 10-ps simulation period. Doubling the applied force to 2 kcal/Å2 causes K+ to move through the RyR2 model pore with an average velocity of 60 Å/ps compared to an average velocity of Ca2+ of 26 Å/ps. These values are consistent with the experimentally observed higher conductance of K+ compared to Ca2+ in single, voltage-clamped RyR2 channels (Williams et al., 2001).
During excitation-contraction coupling, Ca2+ is translocated through RyR from the sarcoplasmic reticulum lumen to the cytosol. Simulations of Ca2+ flux from the luminal side of the model indicate that the Ca2+ remains hydrated as it passes through the selectivity filter. However, the number of waters in the inner hydration shell varies during its passage depending on the strength of the interaction with the amino acids of the selectivity filter. Hydrated Ca2+ has a strong interaction with the luminal mouth of the selectivity filter. At this point two or more of the four glutamic acid residues in the tetramer (Glu-4832) interact strongly with the cation displacing from three to four of the six waters hydrating the Ca2+. The interaction with Glu-4832 is maintained as the cation passes Ile-4829 and is ultimately broken as the cation leaves the filter at Gly-4826. The selectivity filter changes shape as the ion passes through, but these changes are small and the basic shape remains constant throughout the simulation. On leaving the filter region of the model, hydrated Ca2+ travels freely through the cytosolic cavity before being effectively stopped by interactions with Gln-4881 at the inner helix crossover.
Simulation of TEA+ block in RyR2. In 10-ps simulations, in the absence of any bystander ions, the organic cation, TEA+, does not move readily through the model RyR2 pore at an applied force of 1 kcal/Å2, due to strong interactions with Glu-4832, Asp-4870, Gly-4873, Glu-4874, Asp-4877, and Gln-4881 (the location in the model of residues highlighted in this and subsequent sections can be seen in Fig. 11).
Previous investigations of the influence of TEA+ on K+ translocation in individual RyR2 channels have demonstrated a reduction in unitary current amplitude at positive holding potentials but not at negative holding potentials when TEA+ is present on both sides of the channel (Lindsay et al., 1991). The attenuation of current amplitude becomes more marked as holding potential becomes more positive. These observations indicate that the reduction in current amplitude at positive holding potential results from a voltage-dependent block of K+ translocation by TEA+ acting from the cytosolic side of the channel. The lack of influence of TEA+ at negative holding potentials indicates that this cation is not an effective blocker of K+ translocation from the luminal side of the channel. We wished to see if the model of the RyR2 pore could qualitatively simulate these effects.
Typical experimental conditions used to detect TEA+ block involve symmetrical K+ and TEA+ in a ratio of 10:1 (Lindsay et al., 1991). We carried out simulations which contained 1 TEA+ and 10 K+ in addition to the RyR2 model and explicit solvent. A distance constraint was applied to one of the K+. In some cases, a distance constraint (equal in magnitude and direction to that applied to the K+) was applied to the TEA+. The effect was the same in both cases. To simulate the effect of TEA+ on the cytosolic to luminal translocation of K+, the TEA+ was initially positioned at Asp-4870 and constraints removed. The probe K+ was prepositioned at the cytosolic anchor, and a distance constraint of 0.3 kcal/Å2 was applied to pull the K+ toward the luminal anchor. The total simulation time was increased to 40 ps. Under these conditions, the TEA+ quickly (<10 ps) moved between two of the helices in the vestibule and no longer coordinated with all four Asp-4870 residues. Although the TEA+ remained near two of the Asp-4870, this movement created a path through which the K+ could readily move.
To simulate the effect of TEA+ on K+ flux from lumen to cytosol, the TEA+ was prepositioned at the far luminal edge of the selectivity filter (Asp-4835). A distance constraint of 0.3 kcal/Å2 was applied to pull the probe K+ from the luminal to the cytosolic anchor. Under these conditions, the TEA+ did not block the movement of K+. During the simulation the TEA+ diffuses away from the center of the channel and becomes buried between two of the chains. Again, this movement is sufficient to create a clear path for the K+.
A second pair of simulations was performed with the TEA+ prepositioned in the cytosolic entrance of the selectivity filter (within 4 Å of Gly-4826) and constraints removed. The forced march of the probe K+ was initiated as before with a distance constraint of 0.3 kcal/Å2. Potassium movement from cytosol to lumen was blocked for the entire 40-ps simulation. During the simulation the TEA+ became centered in the selectivity filter and moved closer to the ring of Glu-4832. In contrast, the velocity of K+ from lumen to cytosol was slowed by less than 10% by the presence of the TEA+ at the cytosolic entrance of the selectivity filter.
These data are consistent with the experimental observation that TEA+ blocks K+ translocation much more effectively from the cytosolic than the luminal side of the RyR2 pore. In addition, these simulations indicate that cytosolic TEA+ must enter the selectivity filter to produce an effective block of cytosolic to luminal flux of K+.
DISCUSSION
Although it would appear that the pore of the RyR channel shares a range of structural features with the pore of K+ channels, the mechanisms underlying ion selection and ion translocation in these two species of channel are very different (Williams et al., 2001). In the absence of a crystal structure for RyR, we have reasoned that the construction and characterization of a model of the putative pore-forming region of RyR2 would provide us with information on both the probable structure of the pore and insights into the mechanisms involved in ion translocation and selection in RyR. Comparison of the model structure with that of KcsA will highlight how nominally similar structures produce pores with enormously different properties of ion discrimination and rates of ion translocation.
Quantitative assessment of the plausibility of the RyR2 pore model
Several factors support the plausibility of the RyR2 pore model proposed here. First, the statistical energy function of the RyR2 model closely parallels that of the KcsA template. This indicates that the assignment of the RyR2 amino acid sequence to the helical portions of the KcsA template (the thread, if you will) is essentially correct. Second, the RyR2 model is a stable structure: unrestrained molecular dynamics simulations of the model gave peptide backbone root mean-square deviations no greater than that of the KcsA template (data not shown). Third, the striking similarity in distributions of hydrophobic, polar, acid and base moments between the two structures argues in favor of the validity of the model.
The ability of the RyR2 model to simulate experimental observations with both individual ionic species and combinations of permeant and impermeant ions also supports the plausibility of the model. The system was calibrated by carrying out simulations of K+ interactions within the KcsA pore. The demonstration of K+ translocation and the identification of interactions between K+ and specific residues within the KcsA pore demonstrate the usefulness of the simulation protocols used in this communication. Simulations in the RyR2 model suggest that Ca2+ and K+ can pass through the putative pore and that, under appropriate conditions, TEA+ is a blocker of K+ flux. These findings are in qualitative agreement with observations of ion movement in single, voltage-clamped RyR2 channels (Williams et al., 2001).
General features of the model of the RyR2 pore
In keeping with our conclusion that KcsA provides an excellent template for the pore-forming region of RyR2, the overall structure and the arrangement of contributing elements of the model closely resembles the known structure of KcsA. The model indicates that, in agreement with conclusions drawn from recent experimental evidence (Zhao et al., 1999; Anyatonwu et al., 2003), each functional RyR2 Ca2+-release channel contains a single pore formed at the longitudinal axis of a tetramer. In the model each monomer contributes two transmembrane helices and a linking luminal loop that folds into the membrane to form a pore helix and a region equivalent to the selectivity filter of KcsA. In the full-length RyR, this structure will be surrounded by other transmembrane helices just as regions equivalent to the pore-forming structure of KcsA are surrounded by other transmembrane helices in more complex voltage-dependent K+ channels (Shealy et al., 2003).
Various structural domains of the KcsA channel have been shown to contribute to the overall function of the molecule as an ion channel. In the following sections, we will compare the structure and function of these regions with equivalent regions in the RyR2 pore model and, where appropriate, discuss correlations between structural features of the model and established ion handling properties of RyR2.
The gate
As in KcsA, an ion entering the RyR2 model from the cytosolic side of the membrane would initially encounter the junction of the four inner helices. In agreement with simulations of cation flux in KcsA, simulations in the RyR2 model demonstrate that residues in this C-terminal region of the inner helix provide sites of interaction for cations and that the region of inner helix crossover acts as a barrier to cation translocation. By analogy with the established structure of K+ channels (Doyle et al., 1998; Jiang et al., 2002; Kuo et al., 2003), it seems reasonable to conclude that this crossover region of the inner helices could be considered as a gate and that the model of the RyR2 pore is in a closed conformation.
The cytosolic cavity or vestibule
A fundamental role of a channel protein is to provide a mechanism that overcomes the inherent destabilization of an ion in the low dielectric environment of the membrane. In KcsA such a mechanism is achieved by effectively bringing the bulk solution into the interior of the membrane. The cavity or vestibule, lined with residues of the four inner helices, provides a water-filled space, contiguous with the cytoplasm, at the center of the membrane (Doyle et al., 1998). The structure of KcsA determined in the presence of permeant cations has a hydrated cation in this cavity. The cation is further stabilized by helix dipoles arising from the pore helices of each monomer focused at the cavity (Doyle et al., 1998; Roux and MacKinnon, 1999). It seems probable that the cavity and pore helices of the RyR2 model fulfill similar roles to their counterparts in KcsA. As a consequence, we would expect the cavity of RyR2 to be contiguous with the cytosolic solution in an open conformation of the channel and to contain at least one hydrated cation stabilized by the surrounding water and helix dipoles.
Experimental support for the existence of a cytosolic cavity in RyR2 comes from the demonstration of block by K+ channel N-type inactivation peptides (Mead et al., 1998) and large tetraalkylammonium cations, such as tetrabutylammonium (Tinker et al., 1992b,c). Structural studies in K+ channels have demonstrated that block by these ligands involves interactions with hydrophobic residues of the inner helix lining the cytosolic cavity (M. Zhou et al., 2001).
It is also worth noting the existence of a motif (G(4866)LIIDA(4871)) in the inner helix of the RyR2 model that is analogous to the gating hinge motif (GXXXXA) recently identified in the crystal structure of the open conformation of the Methanobacterium thermoautrophicum K+ (MthK) channel and present in the inner helices of a range of K+ channels (Jiang et al., 2002; Shealy et al., 2003). Transition from the closed to open conformation in K+ channels involves a bending at the glycine hinge by ∼30°. In the open configuration, the apposition of an alanine residue from each of the four inner helices forms the narrowest portion of the open cavity. The existence of this motif in the inner helix of the RyR2 pore model highlights another feature common to K+ channels and RyR2 and provides independent circumstantial support for the validity of the model. It appears probable that gating of the RyR2 channel involves structures and mechanisms similar to those identified in K+ channels. As a consequence, in the open conformation of RyR, the cytosolic mouth of the channel would be lined with residues of the putative inner helices.
Although our comparison of the inner helices of KcsA and the RyR2 model has revealed important correlations in the distribution of acidic residues in their C-terminal regions, there are very significant quantitative differences. In the RyR2 model, this region of the inner helix is considerably more acidic than its counterpart in KcsA. Based on the arguments set out above, it is probable that the cytosolic mouth of the open RyR channel would have a significant net negative charge. Experimental evidence for fixed negative charge in the cytosolic mouth of the RyR2 pore has been provided by experiments using Shaker N-type inactivation peptides as blockers. An increase in the net charge of the inactivation peptide from +3 to +7 results in a 500-fold increase in the rate of association of the peptide with its site of interaction at the cytosolic side of the RyR2 channel indicating the involvement of an electrostatic component in the association of the polycation blocker with this site (Mead et al., 1998). The potential contribution that this concentrated area of negative charge may make to RyR channel function will be considered in a later section of this discussion.
The selectivity filter
In K+ channels, discrimination between cations takes place in a selectivity filter located at the extracellular end of the pore and formed by the apposition of signature sequence residues from each of the monomers (Doyle et al., 1998; Morais-Cabral et al., 2001; Y. Zhou et al., 2001). The residues are arranged so that the filter is lined with backbone carbonyl oxygens and the dimensions of the structure are maintained by interactions between side-chain groups of the signature sequence residues and residues of the pore helix. The K+ channel selectivity filter provides a perfect environment for the dehydration, coordination, and rehydration of K+ by rings of carbonyl oxygens. The coordination of K+ within the selectivity filter replicates exactly the coordination of the cation in the cavity by water molecules in the inner hydration shell; as a consequence, cations as similar to K+ as Na+ are effectively excluded from the filter. In addition to providing a mechanism for ion discrimination, the selectivity filter maximizes rates of ion translocation in K+ channels. In the open channel, in the presence of physiological K+ activities, the selectivity filters of K+ channels are occupied by two cations separated by a molecule of water (Morais-Cabral et al., 2001). Net movement of K+ out of the channel occurs when a third ion enters and ejects an ion at the opposite end of the filter.
A comparison of the ion handling properties of K+ channels and RyR suggests that the mechanisms governing ion discrimination and translocation in the two species of channel must be very different. Whereas K+ channels show exquisite powers of discrimination, RyR is permeable to a wide range of inorganic and organic monovalent and divalent cations, with a relative permeability of Ca2+ to K+ of only 6.5 (Tinker and Williams, 1992). Can we gain insights into the mechanisms underlying ion discrimination and translocation from the model of the RyR2 pore?
An initial comparison of the structural features of the selectivity filters of KcsA and the RyR2 model reveal some very important differences. Despite the apparent homology in this region, the dimensions of the pathway and arrangement of the component residues are very different. Whereas the selectivity filter in KcsA is essentially a symmetrical tube of radius 1.5 Å lined with backbone carbonyl oxygens, the equivalent region of the RyR2 model is a considerably more open structure. Consistent with this, we have demonstrated that water has access to this region of the RyR2 model although it is excluded from the selectivity filter of KcsA. The arrangement of residues making up this region of RyR2 is less ordered than that of the equivalent region of KcsA. In KcsA, interactions between ion and selectivity filter are largely through the oxygen atoms of the peptide backbone (Doyle et al., 1998; Morais-Cabral et al., 2001; Y. Zhou et al., 2001). In simulations in the RyR2 model, interactions between peptide oxygens and transient ions are also observed. However, the major interactions are between the side-chain carboxylates of the four Glu-4832. The side chains of Ile-4829 and Asp-4831 help form the walls of the RyR2 selectivity filter. The Ile-4829 side chain is partially exposed to solvent; the other part is stabilized by interactions with the side chain of Val-4823 (on the pore helix). The side chain of Asp-4931 alternates between two positions. In one conformer, the carboxyl is exposed to water. In the other conformer, the carboxyl forms a hydrogen bond with the phenol group of Tyr-4841. Simulations also demonstrate that, during translocation through the RyR2 pore, both Ca2+ and K+ are slowed as the result of a series of interactions with residues making up the region equivalent to the selectivity filter in KcsA. In both cases, the strongest interaction is with the ring of Glu-4832 residues at the luminal end of the filter.
In contrast to KcsA, our simulations demonstrate that permeant cations remain partially hydrated as they move through the selectivity filter of RyR2 with the filter catalyzing ion movement by replacing some of the inner sphere waters of hydration. The retention of components of the inner hydration shell during translocation is entirely consistent with the high permeability of the alkaline earth divalent cations, including Mg2+, in this channel (Tinker and Williams, 1992). The low permeability of Mg2+ in other cation-selective channels is thought to reflect the large amounts of energy required to dehydrate this cation. In the past we have suggested that Mg2+ may be translocated in RyR without complete dehydration (Tinker and Williams, 1992); simulations in the model of the selectivity filter of RyR2 would appear to support this proposal.
It is also important to note that the relative rates of translocation of Ca2+ and K+ in simulations in the RyR2 pore model are consistent with rates monitored experimentally and reflect differences in the strength of interactions of the two cations with residues in the model filter.
Experiments with individual RyR2 channels have demonstrated that the translocation of K+ can be blocked by TEA+ (Lindsay et al., 1991). Block is both concentration and voltage dependent, the latter observation indicating that the site of interaction of the blocking cation is within the voltage drop across the channel. These experiments have also established that TEA+ can only block from the cytosolic side of the channel. Simulations, in which we have gone some way toward reproducing the relative proportions of K+ and TEA+ present in the experimental situation, qualitatively reproduce our earlier experimental observations. Luminal TEA+ does not block K+ translocation, whereas cytosolic TEA+ does block K+ translocation, but to do this TEA+ must first enter the selectivity filter region of the model. The simulations also identify interactions of TEA+ with Glu-4832 residues as potentially important in the blocking reaction.
These findings correlate extremely well with data arising from investigations of the influence of transmembrane voltage on cytosolic TEA+ block in RyR2. These experiments indicate that the site of interaction is located 90% into the voltage drop across the channel from its cytosolic origin (Lindsay et al., 1991). Such a location would be entirely consistent with the position of Glu-4832 at the luminal end of the selectivity filter in the model.
The luminal mouth of the RyR2 pore model
Our comparison of the luminal mouth of the RyR2 model with the equivalent extracellular mouth of KcsA indicates a remarkable similarity in the nature of the components contributing to these regions. Both are predominantly polar and acidic; however, quantitative comparisons indicate that the relative charge is much greater in RyR2.
Considerable experimental evidence supports the existence of a net negative potential at the luminal mouth of the RyR2 channel. Tu et al. (1994b) demonstrated that the addition of carboxyl-neutralizing chemical modifiers to the luminal face of RyR reduced rates of luminal to cytosolic ion translocation. The polycation neomycin has also been shown to block K+ translocation in RyR when present, in nanomolar concentrations, in the solution at the luminal side of the channel, and it has been proposed that block involves interactions of the polycation with fixed negative charge at the luminal mouth of the channel (Mead and Williams, 2002). More recently, the regulatory protein triadin has been shown to interact with specific negatively charged residues that would be located in the luminal end of the pore helix and the loop connecting the pore helix to the inner helix of the RyR1 isoform (equivalent to D4808, D4837, and E4838 in this model) (Lee et al., 2004).
Potential contributions of acidic residues in the cytosolic and luminal mouths of the pore to RyR channel function
Although maximal unitary conductance in K+ channels is high (ranging from 50 to 250 pS) (Latorre and Miller, 1983), unitary conductance of K+ in RyR is much higher, saturating at ∼1 nS (Lindsay et al., 1991). Recent work by Nimigean et al. (2003) has highlighted a role for acidic residues at the C-terminus of K+ channel inner helices in the electrostatic tuning of conductance. Assuming that channel opening involves movement of the four inner helices at the gating hinge described in an earlier section of this discussion, the residues of the C-terminal regions of these inner helices will form the intracellular mouth of the open K+ channel. Nimigean et al. (2003) demonstrated that the introduction of a ring of negative charge in this region of KcsA increases unitary conductance. As a corollary, removal of conserved negative charge at equivalent locations in a high-conductance K+ channel produced a decrease in conductance. It was proposed that the density of negative charge in the mouth of the open channel altered the electrical potential and hence the local K+ concentration.
The high densities of negative charge identified at both the cytosolic and luminal mouths of the RyR2 pore model will more than likely contribute to the phenomenal rates of translocation of cations achieved by these channels. Measurements of unitary current amplitude at high holding potentials indicate that the entry of cations to the RyR pore is not limited by diffusion; at a holding potential of 100 mV, single channel current is 20 pA with Ba2+ as the charge carrier (Tinker and Williams, 1992) and 70 pA with K+ (Lindsay et al., 1994). Consistent with this proposal, the size of the first barrier for ion entry in a rate theory model that provides a good description of ion handling in RyR is significantly smaller than would be predicted from calculated, diffusion-limited rates (Tinker et al., 1992a). A high density of acidic residues, giving rise to significant negative electrostatic potentials, at both the luminal and cytosolic mouths of the model of the RyR2 pore could provide a mechanism for overcoming the limitations of diffusion and ensuring a high rate of delivery of cations to the RyR pore.
Simulations of cation movement in the RyR2 model indicate that the putative selectivity filter is likely to contribute very little to the process of discrimination between physiologically relevant cations, such as Ca2+ and K+. If RyR2 does not have a classical selectivity filter, we must find an alternative mechanism to account for the ability of the RyR pore to exclude anions and to discriminate, to some extent, between divalent and monovalent inorganic cations. It is logical to propose that the presence of a high density of fixed negative charge at both mouths of the RyR2 pore would be sufficient to deny access to anions. A high density of negative charge could also contribute to, or possibly even account for, the relative permeabilities of divalent and monovalent cations in RyR. Investigations of cation discrimination in physical systems such as zeolites (Sherry, 1969) and glass electrodes (Truesdell and Christ, 1967) indicate that selection between divalent and monovalent cations is governed by charge density. Systems with closely packed negative charge select for divalents over monovalents.
Therefore, it is reasonable to hypothesize that the high densities of acidic residues identified at both the cytosolic and luminal mouths of the RyR2 model provide not only a means for maximizing cation conductance in RyR2 but also a mechanism for selecting cations over anions and the necessary discrimination between divalent and monovalent cations.
In conclusion, we have demonstrated that the known structure of the bacterial K+ channel KcsA provides an extremely plausible template for the pore-forming region of the RyR2 channel. The correlation between experimental data and simulations in the RyR2 pore model demonstrate the enormous potential of the model in providing information on both the individual residues involved in interactions with permeant and impermeant cations and the mechanisms underlying these interactions. The model provides us with a theoretical framework within which we can interpret the consequences of mutations of specific residues within the various structural elements of the putative RyR pore; at the same time, the information provided by these functional studies will be used to refine the model.
Acknowledgments
We are grateful to Drs. Wayne Chen, Kishani Ranatunga, Fiona Mead, Bhavna Tanna, and Mark Bannister for very useful discussions on the structure of the RyR pore and to Barrett Abel and Robert Weiss for technical assistance.
Work in Professor Welch's laboratory was supported by funds from the National Science Foundation (MCB 9817605) and that in Professor Williams' laboratory by funds from the British Heart Foundation (RG/03/003).
References
- Anyatonwu, G. I., E. D. Buck, and B. E. Ehrlich. 2003. Methanethiosulfonate ethylammonium block of amine currents through the ryanodine receptor reveals single pore architecture. J. Biol. Chem. 278:45528–45538. [DOI] [PubMed] [Google Scholar]
- Balshaw, D., L. Gao, and G. Meissner. 1999. Luminal loop of the ryanodine receptor: a pore-forming segment? Proc. Natl. Acad. Sci. USA. 96:3345–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4:517–529. [DOI] [PubMed] [Google Scholar]
- Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure. M. O. Dayhoff, editor. National Biomedical Research Foundation, Silver Spring, MD. 345–52.
- Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. L. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Du, G. G., X. H. Guo, V. K. Khanna, and D. H. MacLennan. 2001. Functional characterization of mutants in the predicted pore region of the rabbit cardiac muscle Ca2+ release channel (ryanodine receptor isoform 2). J. Biol. Chem. 276:31760–31771. [DOI] [PubMed] [Google Scholar]
- Du, G. G., B. Sandhu, V. K. Khanna, X. H. Guo, and D. H. MacLennan. 2002. Topology of the Ca2+ release channel of skeletal muscle sarcoplasmic reticulum (RyR1). Proc. Natl. Acad. Sci. USA. 99:16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L., D. Balshaw, L. Xu, A. Tripathy, C. L. Xin, and G. Meissner. 2000. Evidence for a role of the lumenal M3–M4 loop in skeletal muscle Ca2+ release channel (ryanodine receptor) activity and conductance. Biophys. J. 79:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97–120. [DOI] [PubMed] [Google Scholar]
- Ghose, A. K., and G. M. Crippen. 1986. Atomic physicochemical parameters for 3-dimensional structure-directed quantitative structure-activity-relationships.1. Partition-coefficients as a measure of hydrophobicity. J. Comput. Chem. 7:565–577. [DOI] [PubMed] [Google Scholar]
- Ghose, A. K., V. N. Viswanadhan, and J. J. Wendoloski. 1998. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: an analysis of ALOGP and CLOGP methods. J. Phys. Chem. 102:3762–3772. [Google Scholar]
- Godzik, A., A. Kolinski, and J. Skolnick. 1992. Topology fingerprint approach to the inverse protein folding problem. J. Mol. Biol. 227:227–238. [DOI] [PubMed] [Google Scholar]
- Godzik, A., and J. Skolnick. 1992. Sequence-structure matching in globular proteins: application to supersecondary and tertiary structure determination. Proc. Natl. Acad. Sci. USA. 89:12098–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden, W., G. Moeckel, and J. Brickmann. 2004. A new approach to the display of local lipophilicity/hydrophilicity mapped on molecular surfaces. J. Comput. Aided Mol. Des. 7:503–514. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. X., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- Latorre, R., and C. Miller. 1983. Conduction and selectivity in potassium channels. J. Membr. Biol. 71:11–30. [DOI] [PubMed] [Google Scholar]
- Leach, A. R. 2001. Molecular Modelling: Principles and Applications. Pearson Education EMA, Harlow, Essex, UK.
- Lee, J. M., S. H. Rho, D. W. Shin, C. Cho, W. J. Park, S. H. Eom, J. Ma, and D. H. Kim. 2004. Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J. Biol. Chem. 279:6994–7000. [DOI] [PubMed] [Google Scholar]
- Lide, D. R. 2002. CRC Handbook of Chemistry and Physics, 83rd ed. CRC Press, Boca Roton, FL.
- Lindsay, A. R. G., S. D. Manning, and A. J. Williams. 1991. Monovalent cation conductance in the ryanodine receptor-channel of sheep cardiac muscle sarcoplasmic reticulum. J. Physiol. 439:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A. R. G., A. Tinker, and A. J. Williams. 1994. How does ryanodine modify ion-handling in the sheep cardiac sarcoplasmic reticulum Ca2+-release channel? J. Gen. Physiol. 104:425–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A. R. G., and A. J. Williams. 1991. Functional characterisation of the ryanodine receptor purified from sheep cardiac muscle sarcoplasmic reticulum. Biochim. Biophys. Acta. 1064:89–102. [DOI] [PubMed] [Google Scholar]
- Maxfield, F. R., and H. A. Scheraga. 1979. Improvements in the prediction of protein backbone topography by reduction of statistical errors. Biochemistry. 18:697–704. [DOI] [PubMed] [Google Scholar]
- Mead, F. C., D. Sullivan, and A. J. Williams. 1998. Evidence for negative charge in the conduction pathway of the cardiac ryanodine receptor channel provided by the interaction of K+ channel N-type inactivation peptides. J. Membr. Biol. 163:225–234. [DOI] [PubMed] [Google Scholar]
- Mead, F. C., and A. J. Williams. 2002. Block of the ryanodine receptor channel by neomycin is relieved at high holding potentials. Biophys. J. 82:1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. 2000. Ion channels: doing hard chemistry with hard ions. Curr. Opin. Chem. Biol. 4:148–151. [DOI] [PubMed] [Google Scholar]
- Morais-Cabral, J. H., Y. F. Zhou, and R. MacKinnon. 2001. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 414:37–42. [DOI] [PubMed] [Google Scholar]
- Nimigean, C. M., J. S. Chappie, and C. Miller. 2003. Electrostatic tuning of ion conductance in potassium channels. Biochemistry. 42:9263–9268. [DOI] [PubMed] [Google Scholar]
- Powell, M. J. D. 1977. Restart procedures for the conjugate gradient method. Math. Prog. 12:241–254. [Google Scholar]
- Qian, N., and T. J. Sejnowski. 1988. Predicting the secondary structure of globular-proteins using neural network models. J. Mol. Biol. 202:865–884. [DOI] [PubMed] [Google Scholar]
- Roux, B., and R. MacKinnon. 1999. The cavity and pore helices the KcsA K+ channel: electrostatic stabilization of monovalent cations. Science. 285:100–102. [DOI] [PubMed] [Google Scholar]
- Shah, P. K., and R. Sowdhamini. 2001. Structural understanding of the transmembrane domains of inositol triphosphate receptors and ryanodine receptors towards calcium channeling. Protein Eng. 14:867–874. [DOI] [PubMed] [Google Scholar]
- Shealy, R. T., A. D. Murphy, R. Ramarathnam, E. Jakobsson, and S. Subramaniam. 2003. Sequence-function analysis of the K+-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys. J. 84:2929–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry, H. S. 1969. The ion-exchange properties of zeolites. In Ion Exchange. J. Marinsky, editor. Marcel Dekker, New York. 89–133.
- Takeshima, H., S. Nishimura, T. Matsumoto, H. Ishida, K. Kangawa, N. Minamino, H. Matsuo, M. Ueda, M. Hanaoka, T. Hirose, and S. Numa. 1989. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 339:439–445. [DOI] [PubMed] [Google Scholar]
- Tinker, A., A. R. G. Lindsay, and A. J. Williams. 1992a. A model for ionic conduction in the ryanodine receptor-channel of sheep cardiac muscle sarcoplasmic reticulum. J. Gen. Physiol. 100:495–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker, A., A. R. G. Lindsay, and A. J. Williams. 1992b. Block of the sheep cardiac sarcoplasmic reticulum Ca2+-release channel by tetraalkyl ammonium cations. J. Membr. Biol. 127:149–159. [DOI] [PubMed] [Google Scholar]
- Tinker, A., A. R. G. Lindsay, and A. J. Williams. 1992c. Large tetraalkyl ammonium cations produce a reduced conductance state in the sheep cardiac sarcoplasmic reticulum Ca2+-release channel. Biophys. J. 61:1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker, A., and A. J. Williams. 1992. Divalent cation conduction in the ryanodine receptor-channel of sheep cardiac muscle sarcoplasmic reticulum. J. Gen. Physiol. 100:479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker, A., and A. J. Williams. 1993. Probing the structure of the conduction pathway of the sheep cardiac sarcoplasmic reticulum calcium-release channel with permeant and impermeant organic cations. J. Gen. Physiol. 102:1107–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker, A., and A. J. Williams. 1995. Measuring the length of the pore of the sheep cardiac sarcoplasmic reticulum calcium-release channel using trimethylammonium ions as molecular calipers. Biophys. J. 68:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell, A. H., and C. L. Christ. 1967. Glass electrodes for calcium and other divalent cations. In Glass Electrodes for Hydrogen and Other Cations. G. Eisenman, editor. Marcel Dekker, New York. 293–321.
- Tu, Q., P. Vélez, M. S. Brodwick, and M. Fill. 1994a. Streaming potentials reveal a short ryanodine-sensitive selectivity filter in cardiac Ca2+ release channel. Biophys. J. 67:2280–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Q., P. Velez, M. Cortes-Gutierrez, and M. Fill. 1994b. Surface charge potentiates conduction through the cardiac ryanodine receptor channel. J. Gen. Physiol. 103:853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A. J., D. J. West, and R. Sitsapesan. 2001. Light at the end of the Ca2+-release channel tunnel: structures and mechanisms involved in ion translocation in ryanodine receptor channels. Q. Rev. Biophys. 34:61–104. [DOI] [PubMed] [Google Scholar]
- Zhao, M. C., P. Li, X. L. Li, L. Zhang, R. J. Winkfein, and S. R. W. Chen. 1999. Molecular identification of the ryanodine receptor pore-forming segment. J. Biol. Chem. 274:25971–25974. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]
- Zhou, M., J. H. Morais-Cabral, S. Mann, and R. MacKinnon. 2001. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 411:657–661. [DOI] [PubMed] [Google Scholar]