Abstract
The orientation and pore-forming mechanisms of pandinin 2 (pin2), an antimicrobial peptide isolated from venom of the African scorpion Pandinus imperator, bound to magnetically oriented lipid bilayers were examined by 31P and 13C solid-state, and 15N liquid-state NMR spectroscopy. 31P NMR measurements at various temperatures, under neutral and acidic conditions, showed that membrane lysis occurred only under acidic conditions, and at temperatures below the liquid crystal-gel phase transition of the lipid bilayers, after incubation for two days in the magnet. Differential scanning calorimetry measurements showed that pin2 induced negative curvature strain in lipid bilayers. The 13C chemical shift values of synthetic pin2 labeled at Gly3, Gly8, Leu12, Phe17, or Ser18 under static or slow magic-angle spinning conditions, indicate that pin2 penetrates the membrane with its average helical axis perpendicular to the membrane surface. Furthermore, amide H-D exchange experiments of 15N-Ala4, Gly8, and Ala9 triply-labeled pin2 suggest that this peptide forms oligomers and confirms that the N-terminal region creates membrane pores.
INTRODUCTION
Antimicrobial peptides with α-helical conformations are commonly and broadly found in nature and often induce permeation of lipid membranes (Maloy and Kari, 1995; Andreu and Rivas, 1998). Many antimicrobial peptides have been classified as pore-forming peptides such alamethicin, cecropin, PGLa, magainin, mellitin, and mastoparan. The pore-forming mechanisms of antimicrobial peptides have been extensively studied and several mechanisms of pore formation have been proposed (Shai, 1999; Bechinger, 1999). Alamethicin (North et al., 1995; Bak et al., 2001) inserts into the membrane hydrophobic core and creates a pore by forming transmembrane helical bundles. PGLa (Bechinger et al., 1998), cecropin A (Marassi et al., 1999), and ovispirin (Yamaguchi et al., 2001) are situated at the membrane surface and self-associate, in a “carpet-like” manner, onto the acidic phospholipid-rich domains of lipid bilayers. When a threshold concentration of peptide is reached, it permeates into the membrane by increasing the positive curvature of the bilayer. Magainin 2 (Matsuzaki et al., 1998), mastoparan X (Matsuzaki et al., 1994, 1995a,b, 1996a,b), and LL-37 (Henzler Wildman et al., 2003) form toroidal pores in lipid bilayers, where the inner monolayer is associated with the outer one via a phospholipid lining, and pores are formed by the lipid polar headgroups and the helix bundles oriented perpendicular to the membrane surface.
Pandinin 2 (Pin2, FWGALAKGALKLIPSLFSSFSKKD) is a pore-forming peptide isolated from the crude venom of the African scorpion Pandinus imperator (Corzo et al., 2001). This peptide shows antimicrobial activity against Gram-positive and Gram-negative bacteria, as well as strong hemolytic activity against sheep erythrocytes. Circular dichroism studies reveal an unordered structure in aqueous solution, with a more ordered α-helical structure observed in a membrane-mimetic environment (DPC micelles). A high-resolution structure of pin2 in DPC micelles had been determined by solution NMR and showed a single α-helical structure up to residues 18–19, with no significant kink around the proline at position 14 (Corzo et al., 2001).
Since solid-state NMR methods do not need isotropic reorientation, they are frequently used to obtain structural information on insoluble, amorphous or fibrous molecules and membrane-bound peptides and proteins. When a sample is aligned with an applied magnetic field, the resulting signal shows an orientational dependence on the NMR frequencies, which can provide valuable information concerning the orientation of membrane-bound peptides in lipid bilayers, especially if the secondary structure already is known (Cross and Opella, 1994; Marassi and Opella, 1998). Many oriented systems have been examined by solid-state NMR, where mechanically oriented systems with glass plates have been used most frequently (Bechinger and Opella, 1991; Opella et al., 1999). Recently, a novel solid-state NMR method was reported (Naito et al., 2000; Toraya et al., 2004) which determines the conformation of peptides bound to highly hydrated, magnetically oriented lipid bilayers, and hence more closely resembles physiological conditions. Therefore, we chose this method to examine the pin2 binding and pore-formation mechanism in magnetically oriented phospholipid bilayers. We also investigated the pin2-lipid bilayer interactions as a function of peptide concentration and pH, using 31P solid-sate NMR in a magnetically oriented system. N-H amide solvent exchange in DPC micelles was also studied, to examine the aqueous and hydrophobic pin2 environments.
EXPERIMENTAL
Materials
Dimyristoyl-phosphatidylcholine (DMPC) and dimyristoyl-phosphatidylglycerol (DMPG) were purchased from Funakoshi (Tokyo, Japan). Dioleoyl-phosphatidylethanolamine (DOPE) and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Nonlabeled pin2, five kinds of site-specifically 13C=O labeled pin2; [1-13C]-Gly3, Gly8, Leu12, Phe17, or Ser18, and 15N-Ala4, Gly8, Ala9 triply-labeled pin2 were synthesized on solid-phase using fluoren-9-ylmethoxycarbonyl (Fmoc) methodology on an Applied Biosystems (Foster City, CA) 433A peptide synthesizer. Fmoc-Asp(ot-butyl)-Wang resin was used to provide a free carboxyl at the C-terminus of pin2. After synthesis, peptide deprotection, and cleavage from resin, the crude peptides were dissolved in 40% acetonitrile solution and purified by reverse-phase HPLC on a semipreparative C18 ODS column (10 × 250 mm, Nacalai Tesque, Kyoto, Japan). The mass and the purity of the peptides were confirmed by matrix-assisted laser-desorption ionization-time-flight-MS. Mastoparan was purchased from Peptide Institute (Osaka, Japan).
Preparation of multilamellar vesicles
The membrane system was made of DMPC/DMPG (4:1) and co-solubilized with varying quantities of peptide in chloroform/methanol (8:1). After solvent evaporation under vacuum for two days, the lipid film was hydrated with Tris buffer containing 100 mM NaCl (pH = 7.6) at 40°C, vortex-mixed, and the pH adjusted with Tris buffer. The suspension was freeze-thawed for 10 cycles and centrifuged. The supernatant was removed to adjust the water content to 85% (w/w), and the suspension transferred to NMR tubes sealed with glue to prevent dehydration. Five kinds of 13C=O labeled pin2 were incubated for two days at 40°C in the magnet, and then lyophilized, before 13C NMR experiments to determine the principal values of the carbonyl carbons, in the absence of any motion.
Solid-state NMR spectroscopy
All solid-state NMR spectra were acquired on a CMX Infinity 300 spectrometer (Chemagnetics, Varian, Palo Alto, CA) operating at a proton-resonance frequency of 300 MHz. 31P spectra were acquired using a 5-μs single excitation pulse with 30 kHz continuous wave (CW) 1H decoupling during acquisition. The dwell time was 50 μs and 256–512 transients were accumulated for each free induction decay with a 3-second delay. The 31P chemical shifts were referenced externally to 85% H3PO4 (0 ppm). For 13C NMR measurements, 20,000–66,000 transients were accumulated for each free induction decay with a 53-μs dwell time and a 3.7-second delay. For hydrated samples, spectra were acquired using 4-μs excitation pulses with 30 kHz CW 1H decoupling. The sample rotation speed was maintained at 100 ±15 Hz for slow MAS experiments. For lyophilized samples, a cross-polarization (CP) experiment was performed with a contact time of 2 ms, 40 kHz CP, and CW 1H decoupling radio frequency. The 13C chemical shifts were externally referenced to the methine carbon of adamantine (29.5 ppm). 13C spectra were processed using 50 Hz line broadening. Unless otherwise stated, samples were incubated for two days at 40°C before data acquisition.
Differential scanning calorimetry measurements
DOPE/POPC (6:1) and various amounts of pin2 were dissolved together in chloroform/methanol (2:1 v/v). The solvent was evaporated under a stream of dry nitrogen with last traces removed under high vacuum. The dried lipids were then dispersed in buffer, 20 mM Tris-HCl, 100 mM NaCl, and 2 mM EDTA (pH = 7.6), by vigorous vortexing to produce a 50 mM phospholipid concentration. Differential scanning calorimetry measurements were performed on a Microcal MCS differential scanning calorimeter (Microcal, Amherst, MA) at a heating rate of 1°C/min. All scans were made after freezing the dispersion in dry ice. The data were analyzed using software provided by the manufacturer.
H-D exchange
N-H amide exchange experiments were performed on a Bruker DMX-750 spectrometer (Karlsruhe, Germany) operating at a proton-resonance frequency of 750 MHz. A series of two-dimensional 1H-15N HSQC spectra at 40°C were taken at 5-min intervals after the addition of D2O, and were recorded with a recycle delay of 2 s with two scans. The data were acquired with 64 points in the 15N dimension corresponding to a 50-ppm spectral width, and 2048 points in the 1H dimension. A GARP decoupling sequence (Shaka et al., 1985) was applied during data acquisition. The spectra were acquired using TPPI for quadrature detection (Marion and Wüthrich, 1983) in the t1 dimension. The phase-shifted sine bell functions were applied for both t1 and t2 dimensions.
RESULTS
Concentration dependence of pin2–lipid bilayer interactions
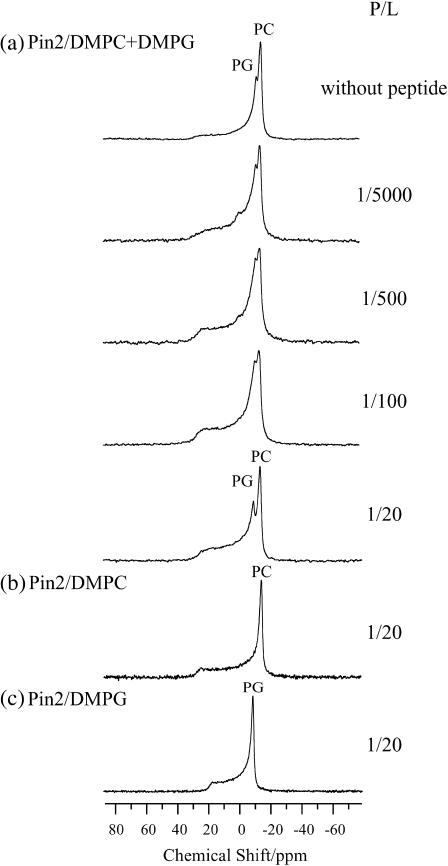
To examine the effect of pin2 on lipid bilayers, the 31P NMR spectra of lipid bilayers in the presence and absence of pin2 were measured under static conditions. Fig. 1 a shows the 31P NMR spectra of DMPC/DMPG (4:1) lipid bilayers at 40°C containing various amounts of pin2 at pH = 7.5. In the absence of pin2, an axially symmetric, motionally averaged powder pattern was observed where the δ⊥component is largest. This pattern indicates that the bilayers are magnetically aligned, with their normal perpendicular to the magnetic field (Dempsey and Watts, 1987) and the phospholipids are laterally diffused on the lipid bilayers. When pin2 was added to the bilayers in a peptide/lipid (P/L) molar ratio of 1:5000, there was an increase in the powder pattern-like nature of the 31P NMR spectrum. This indicated that the magnetic alignment of the lipid bilayers was disturbed by the interaction of pin2 with the lipid headgroups even in the presence of such small amounts of peptide. At P/L = 1:500–1:20, the lipid bilayer alignment showed greater disturbance, and at P/L = 1:200, the chemical shift of the DMPG signal was dramatically shifted downfield. Fig. 1, b and c, show the 31P NMR spectra of DMPC and DMPG lipid bilayers, respectively, at 40°C and pH = 7.5, with pin2 at P/L = 1:20. (Comparing Fig. 1, b and c, with Fig. 1 a, we can unequivocally assign the two DMPC and DMPG signals.) The DMPG shift at P/L = 1:20 could be due to the negatively charged DMPG headgroup interacting with the positively charged pin2 lysine, causing the phosphate group to tilt with respect to the axis of motional averaging, hence reducing the averaged chemical shift; i.e., the σ22(σ33) principal axis is parallel to the axis of motional averaging (Herzfeld et al., 1978; Scherer and Seelig, 1989).
FIGURE 1.
Effect of pin2, at various concentrations, on the 31P NMR spectra of phospholipids in lipid bilayers composed of (a) DMPC/DMPG (4:1), (b) DMPC, and (c) DMPG at 40°C under neutral pH conditions. PC and PG indicate the signals of DMPC and DMPG, respectively.
DSC measurement
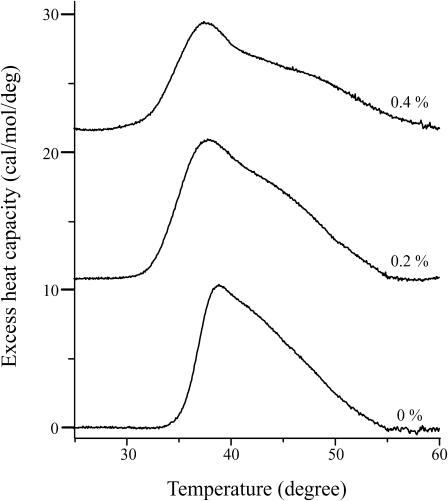
To elucidate the ability of pin2 to induce curvature strain on lipid bilayers, the effect of pin2 on the phase transition temperature TH from lamellar liquid-crystalline phase (Lα) to inverted hexagonal phase (HII) of DOPE was investigated. Fig. 2 shows a series of DSC scans in the absence and presence of pin2 in a DOPE/POPC (6:1) lipid mixture. TH in the absence of pin2 was 39°C and higher pin2 content resulted in lower TH values, implying that pin2 induces negative curvature strain in lipid bilayers (Hallock et al., 2002). All DSC scans showed line broadening as compared with experiments involving magainin2 (Matsuzaki et al., 1998) and MSI-78 (Hallock et al., 2003), probably because both Lα and HII phases create lipid systems that show isotropic motional averaging, as seen in other PC/PE lipid mixtures (Cullis et al., 1978; Tilcock et al., 1982).
FIGURE 2.
Differential scanning calorimetry curves of pin2 in DOPE/POPC lipid bilayers. Lipid membranes were composed of DOPE/POPC (6:1) at the given concentrations of pin2. The position of the calorimetry scans is arbitrary.
pH dependence of lipid bilayer alignment in the presence of pin2
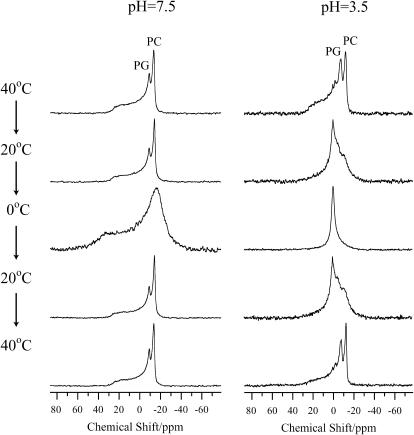
To determine the orientation between pin2 and lipid bilayers by NMR solid-state analysis in a fully hydrated system, we require a strongly magnetic oriented lipid bilayer system, prepared by lipid membrane lysis and re-fusion (Naito et al., 2000; Kimura et al., 2002; Toraya et al., 2003). Hence, elongated and magnetically orientated lipid bilayers were evaluated under different pH conditions. Fig. 3 shows the 31P NMR spectra of lipid bilayers at pH = 7.5 and 3.5 in the presence of pin2, at various temperatures. At pH = 7.5, as the temperature was lowered from 40°C to 0°C, the 31P signal becomes broadened, due to interference from averaging of the chemical shift anisotropy (CSA) by lateral diffusion at temperatures below the liquid crystal-gel transition temperature (Tm = 24°C) of DMPC and DMPG. When the temperature was raised back to 40°C, the same powder pattern was observed as before. At pH = 3.5 and 40°C, increased powder pattern-like character was observed. The small signal at −6 ppm indicates the presence of lysolipids in the lipid bilayers, generated by phospholipid hydrolysis (Naito et al., 2002) and the signal at 0 ppm is probably caused by lysolipid micellization (Naito et al., 2002). The formation of lysolipids was verified by TLC separation (after NMR measurements) using 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)] and 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine as a lysolipid standard. When the temperature was lowered to 0°C, a narrow signal was observed. This isotropic narrow signal in the 31P NMR spectrum has been reported in several other peptide-containing lipid dispersion systems, such as the melittin-DMPG system (Dufourcq et al., 1986b), the cyclic peptide gramicidin S-PC/PE system (Prenner et al., 1997), and the dinorphin A-DMPC system (Naito et al., 2002). This isotropic signal may be ascribable to fast isotopic tumbling of phospholipids, caused by a lipid bilayer phase inversion from multilamellar vesicles to another structure, such as micelles, small unilamellar vesicles, small discoidal bilayers (Dufourcq et al., 1986a), or cubic phases (Prenner et al., 1997). When the temperature was raised back to 20°C, a mixture of the isotropic components and the powder pattern components were observed, indicating the coexistence of an isotropic phase and a lamellar phase. At 40°C, the δ‖ component of the 31P NMR powder pattern disappeared suggesting that the small membrane bilayer particles were re-fused to make elongated lipid bilayers during the temperature increase from 0°C to 40°C (Naito et al., 2002). In summary, membrane lysis of lipid bilayers occurred under acidic conditions, in the presence of pin2.
FIGURE 3.
31P NMR spectra of phospholipids in lipid bilayers composed of DMPC/DMPG (4:1) in the presence of pin2 (P/L = 1/20) under neutral and acidic pH conditions, at various temperatures. PC and PG indicate the signals of DMPC and DMPG, respectively.
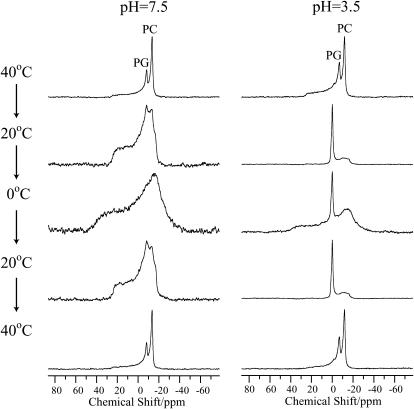
For comparison reasons, we also measured the 31P NMR spectra of the same DMPG/DMPC lipid bilayers under neutral and acidic conditions in the presence of mastoparan (INLKALAALAKKIL), a well-known pore-forming peptide from wasp venom, at various temperatures (Fig. 4). As with pin2, the isotropic signal was absent under neutral conditions, whereas under acidic conditions, the narrow isotropic signal appeared at temperatures below Tm. To date, no isotropic narrow signals indicating membrane lysis have been reported for fully hydrated lipid bilayers in the presence of mastoparan (Hori et al., 1999). It was therefore concluded that acidic conditions are important for membrane fusion and lysis in the presence of pin2 or mastoparan, under magnetic alignment of the peptide-lipid system. The membrane lytic behaviors of mastoparan and pin2 are different, probably due to the structural differences between the two peptides, such as length or hydrophobicity.
FIGURE 4.
31P NMR spectra of phospholipids in lipid bilayers composed of DMPC/DMPG (4:1) in the presence of mastoparan (P/L = 1/10) under neutral and acidic pH conditions, at various temperatures. PC and PG indicate the signals of DMPC and DMPG, respectively.
Orientation and dynamics of pandinin 2 in lipid bilayers
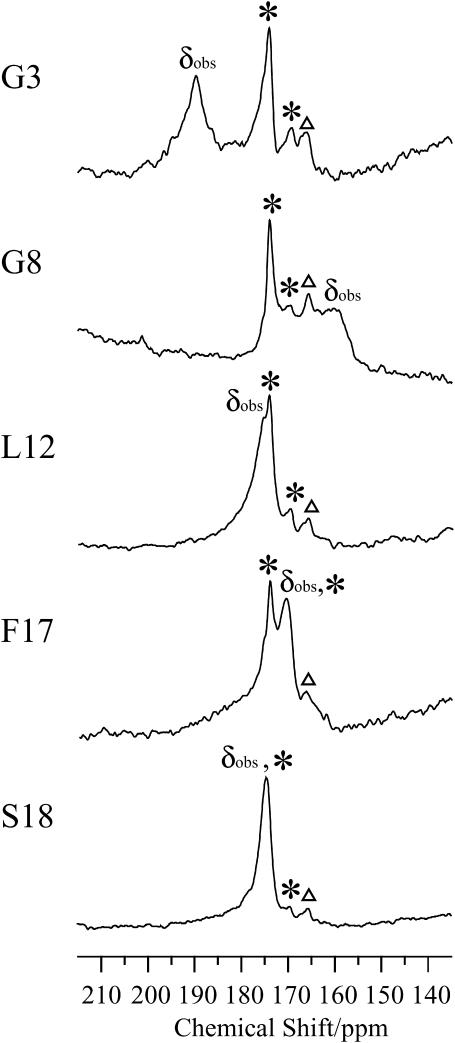
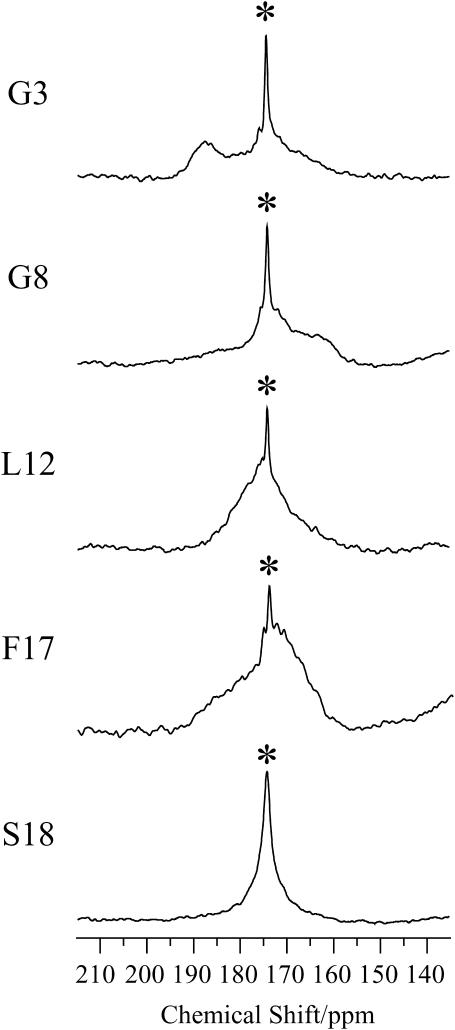
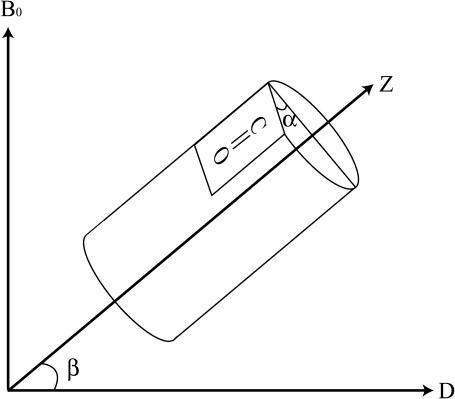
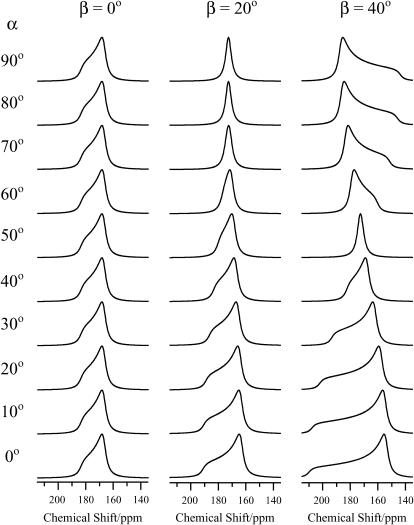
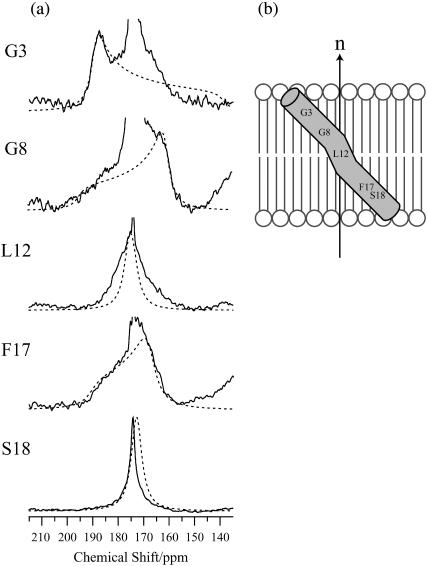
To determine the relative orientation between pin2 and the lipid bilayers, the 13C NMR spectra of five kinds of site-specifically 13C=O labeled pin2 (Gly3, Gly8, Leu12, Phe17, and Ser18) bound to lipid bilayers, were measured. Before each measurement, the 31P NMR spectra of the samples were acquired to confirm that the membrane surface was oriented parallel to the magnetic field. Figs. 5 and 6 show the 13C NMR spectra measured under static and slow MAS conditions, respectively. Table 1 lists the 13C chemical shift values (δobs) under static conditions, and the principal values of 13C=O CP spectra (not shown) of the lyophilized samples. The δ22 values show the greatest variation of the principal values of Gly3 and Gly8 13C=O chemical shift tensors. This confirms earlier observations, showing the δ22 value of the Gly13C=O CSA tensor is the most dependent on adjacent residues (Separovic et al., 1990). These principal values show the typical broad asymmetrical powder pattern of carbonyl carbons (∼150 ppm; Hartzell et al., 1987). The 13C NMR spectra obtained under slow MAS conditions (Fig. 6) also show broad signals, obtained by disturbing the magnetic ordering under slow-speed magic-angle spinning, but the CSA (δ‖–δ⊥) was ∼0–45 ppm, which is smaller than normal. All of these spectra show an axially symmetrical powder pattern, different to normal. Generally, the principal axis directions of the carbonyl carbon chemical shift anisotropy in peptides are reported as follows: the δ11 and the δ22 axes are in the peptide plane where the δ22 axis points roughly along the C=O bond, between 0° and 12° off the parallel to the C=O bond direction, and the δ11 axis is perpendicular to the δ22, whereas the δ33 axis is approximately perpendicular to the peptide plane (Hartzell et al., 1987; Separovic et al., 1990; Wei et al., 2001). From the slow MAS results, it is presumed that the α-helix rapidly rotates around the average helical axis assuming the C=O bond direction is nearly parallel to the helical axis (Naito et al., 2000). Further, the 13C=O chemical shift values, δobs obtained under static conditions (Fig. 5), are close to the δ⊥ values (Fig. 6), indicating that the rotating axis is parallel to the lipid long axis (Toraya et al., 2004). During the slow MAS study the values and signs of the CSA (δ‖–δ⊥) vary from residue to residue, suggesting that the C=O (δ22) direction deviates angle β (−90° ≤ β < 90°) from the membrane normal, perpendicular to the magnetic field. Therefore, the peptide plane makes an angle α (0° ≤ α < 360°) with the Z–D plane as shown in Fig. 7 (Toraya et al., 2004). Fig. 8 shows the simulated 13C NMR spectra using δ11, δ22, and δ33 values of 247, 184, and 88 ppm, respectively, on the basis of the CP spectra of lyophilized Gly3 C=O labeled pin2 at β = 0°, 20°, and 40°. The lineshapes of the spectrum are similar for any of the following orientations: (α, β), (−α, β), (π−α, β), (−π +α, β), (α, −β), (−α, −β), (π−α, −β), (−π +α, −β). The values and signs of the CSA vary depending on the α- and β-angles. Hence, by comparing the experimental and simulated lineshapes for the five kinds of 13C=O labeled pin2, and assigning the difference of α-angle between the neighboring peptide plane as 100° ±10°, the β-orientations could be determined. The carbonyl region of the 13C NMR spectra of a variety of 13C-labeled pin2, bound to lipid bilayers, obtained under slow MAS conditions with their best-fit simulated spectra superposed, are shown in Fig. 9 a. The best β-orientations are listed in Table 1. From this result, we can determine that the N- and C-terminal helical rods are inclined ∼45°, with the middle section inclined ∼25° to the average helical axis (lipid long axis). Based on these data, a structural model of pin2 in a lipid bilayer can be proposed as in Fig. 9 b.
FIGURE 5.
Carbonyl region of the 13C NMR spectra of a variety of 13C-labeled pin2, bound to lipid bilayers (P/L = 1/20) under static conditions, at 40°. Asterisks denote the phospholipid C=O group. Triangles denote unknown impurities.
FIGURE 6.
Carbonyl region of the 13C NMR spectra of a variety of 13C-labeled pin2 bound to lipid bilayers (P/L = 1/20) under slow MAS conditions (ωR = 100 Hz) at 40°C. Asterisks denote the phospholipid C=O group.
TABLE 1.
13C=O chemical shift values and β-angles of 13C-labeled peptides
| δobs/ppm* | δ11/ppm† | δ22/ppm† | δ33/ppm† | β/degree | |
|---|---|---|---|---|---|
| Gly3 | 189.4 | 246.6 | 183.6 | 87.6 | 45° |
| Gly8 | 160.1 | 247.8 | 179.3 | 90.1 | 46° |
| Leu12 | 175.2 | 247.0 | 190.9 | 87.4 | 25° |
| Phe17 | 170.5 | 244.3 | 188.9 | 92.8 | 45° |
| Ser18 | 174.5 | 250.5 | 182.3 | 85.6 | 47° |
Chemical shift values are for hydrated samples under static conditions.
Principal values are for lyophilized powder samples.
FIGURE 7.
Schematic representation of the coordinate frames in which the orientation and dynamics of pin2 were defined. The bilayer normal D is perpendicular to the magnetic field Bo. D is identical to the average helix axis. The value β is the tilt angle of helix axis (Z) from D. Z is identical to the δ22 principal axis of C=O chemical shift tensor. The value α is the angle between a peptide plane and a Z–D plane.
FIGURE 8.
The α-angle dependence of the carbonyl region in simulated 13C NMR spectra. The principal values of carbonyl carbon of Gly3, δ11 = 247, δ22 = 184, and δ33 = 88 ppm. The β-angle is fixed at 0°, 20°, and 40°.
FIGURE 9.
(a) 13C NMR spectra showing the carbonyl regions for a variety of 13C-labeled pin2 bound to lipid bilayers (P/L = 1/20) under slow MAS conditions (solid line, same as Fig. 5), with the best-fit simulated spectra (dotted line) superimposed. (b) Proposed model for orientation of pin2 in lipid bilayers.
Amide H-D exchange
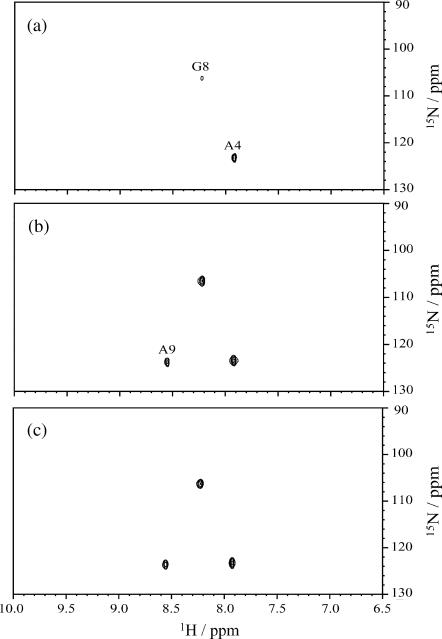
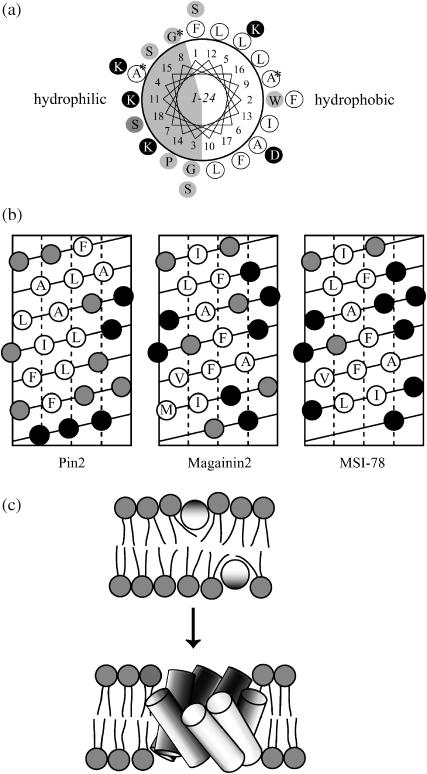
To understand how pin2 interacts with the lipid membrane, the amide H-D exchange of 15N-labeled pin2 was examined (Williams et al., 1996; Almeida and Opella, 1997). Fig. 10 shows the HSQC spectra measured at 5, 30, and 60 min after adding D2O to a mixture of 3 mM 15N-Ala4, Gly8, and Ala9 triply-labeled pin2 and 120 mM DPC micelles. The amide proton of residue Ala4 underwent >50% exchange after 5 min and the exchange was complete within 30 min; hence it was classified as a fast exchange. The amide proton of residue Ala9 showed little exchange after 5 min, and incomplete exchange after 60 min; hence it was classified as a slow exchange. Lastly, the amide proton of residue Gly8 underwent moderate exchange. The varying amounts of amide exchange would indicate that Ala4 is exposed to the aqueous environment whereas Ala9 is located in a hydrophobic micelle region, and Gly8 borders these regions. These results are in good agreement with the hydrophobicity of pin2 as shown in Fig. 11 a. According to the 13C NMR pin2 orientation and dynamics studies, the average helical axis of pin2 penetrates perpendicular to the bilayer membrane. Together, the amide H-D exchange and 13C NMR data would indicate that pin2 forms pores by monomer association, where Ala4 faces the aqueous environment of the pore and Ala9 faces the acyl chains of the membrane. Although we presume that the α-helix of pin2 laterally diffuses while rapidly rotating around the membrane normal, the amide H-D exchange results suggest that the helix bundle might rapidly rotate around the membrane normal. This rotation is essentially the same as the rotation of the α-helix around the membrane normal. If the helix bundle of pin2 laterally diffuses into the lipids of the lipid bilayer, then rotation around the membrane normal is essentially the same as the lateral diffusion of the helix bundle.
FIGURE 10.
15N-1H HSQC spectra of 15N-Ala4, Gly8, Ala9 triply labeled pin2 (3 mM) in DPC micelles (P/L = 1/40) at 40°C after amide H-D exchange at (a) 5 min, (b) 30 min, and (c) 60 min.
FIGURE 11.
(a) Helical-wheel projection of pin2 (the 15N-labeled residues are indicated by asterisks), (b) Helical net representations, and (c) model for mechanism of pin2 pore formation. Hydrophobicity of each amino acid is represented in shading with the most polar residues in rendered in solid symbols for a and b.
DISCUSSION
Pore-forming mechanisms that have been proposed to explain the mode of action of α-helical membrane peptides include the transmembrane-helical bundle, the carpet-like, and the toroidal models. However, α-helical membrane peptides can adopt more than one pore-forming mechanism, depending on experimental conditions. For example, the pore-forming mechanism of pardaxin depends on the type of membranes used. Solid-state NMR studies of pardaxin showed it to adopt a carpet-type orientation in lipid bilayers composed of 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC), but displayed a transmembrane orientation in lipid bilayers composed of DMPC (Hallock et al., 2002). LAH4, a pore-forming peptide, exhibited carpet-type orientation under acidic conditions, but a transmembrane orientation at neutral pH (Bechinger, 1996). Even under similar experimental conditions, mastoparan simultaneously exhibited two distinct orientations; 10% transmembrane and 90% carpet-type (Hori et al., 2001). Therefore, the study of peptide pore-forming mechanisms is highly dependent on experimental conditions, since the peptides can adopt different orientations in response to their environment.
The DSC measurements (Fig. 2) suggested that pin2 induces negative curvature strain in lipid bilayers, unlike other peptides such as magainin2 (Matsuzaki et al., 1998) and MSI-78 (Hallock et al., 2003), which induce a positive curvature. It has been proposed that peptide hydrophobicity dictates this behavior, and is dependent on the position of the hydrophobic amino acids in the peptide sequence (Tachi et al., 2002). When hydrophobic residues are clustered, the deepest penetrating portion of the peptide will significantly expand the hydrophobic core of the bilayer and induce negative curvature. Fig. 11 b compares helical net representations (Tachi et al., 2002), where hydrophobicity is referred to as hydropathy index (Kyte and Doolittle, 1982). Fig. 11, a and b, show the hydrophobic residues of pin2 are more ordered and more hydrophobic than those in magainin2 or MSI-78. Therefore, it is likely that this clustering of hydrophobic residues enables pin2 to penetrate further into the lipid bilayer and induce negative curvature strain on the bilayer.
Concerning the biological activity of pin2 on lipid membranes, we should clarify the difference between antimicrobial and hemolytic activities, and membrane lysis (Figs. 3 and 4). Membrane lysis was observed under acidic conditions and temperatures below Tm where the membranes were completely broken, with no traces of the original bilayer remaining. However, lysed or completely broken membranes are not a requisite for antimicrobial or hemolytic activities, which can occur when molecules leak through a membrane pore.
The 13C solid-state NMR data and subsequent analyses (Figs. 5–9) showed the orientation of pin2 within a lipid bilayer and the dynamics in the membrane mimetic environment. As a comparison, melittin in a mechanically oriented DTPC bilayer system with glass plates also shows a transmembrane orientation (Smith et al., 1994); i.e., melittin reorients around the bilayer normal illustrated by the 90°-oriented spectra showing large chemical shift anisotropy below Tm, observable from the carbonyl chemical shift values varying from different residues. In highly hydrated systems, melittin forms a transmembrane α-helix in the lipid bilayer and laterally diffuses via rapidly rotating around the membrane normal (Naito et al., 2000), where the local helical axis rotates around the membrane normal, making an angle of ±30° and ±10° in a DMPC bilayer system (Naito et al., 2000), or −36° and 25° in a DPPC bilayer system (Toraya et al., 2004) for the N- and C-terminal helical rods, respectively. Pin2 in the present DMPC/DMPG bilayer system also laterally diffuses by rapidly rotating around the membrane normal. Since a high-resolution structure of pin2, determined by solution NMR, showed no significant kink around proline, we suggest that the angles between the local helical axis and the average helical axis of pin2 have similar signs for the N- and C-terminals and around Leu12. Therefore, the local helical axis may proceed around the bilayer normal by making an angle of 45° for the N- and C-terminal helical rods and 25° around Leu12, and pin2 may be doubly kinked around Leu12 in DMPC/DMPG lipid bilayers as shown in Fig. 9 b. Other peptides have also shown steep tilt angles and similar rotational motions. For example, Protegrin-1, a β-sheet antimicrobial peptide, is tilted by 55° from the DLPC bilayer normal (Yamaguchi et al., 2002), and rotates uniaxially around the bilayer normal. The helix-break-helix fusogenic peptide B18 shows a tilt angle of 54° in DMPC/DMPG (4:1) lipid bilayers and binds to membranes in a boomerang-like fashion (Afonin et al., 2004). The steep tilt angles and rotation around the bilayer normal might be a requisite for certain types of pore-forming peptides.
We propose a pore-forming mechanism for the action of pin2 on DMPG/DMPC lipid bilayers, based on our orientation and dynamic NMR studies. The DSC measurements show that pin2 induces negative curvature strain in lipid bilayers, implying that deep penetration of pin2 significantly expands the hydrophobic core of the lipid bilayer, providing a driving force for further penetration into the membrane (Fig. 11 c). The pin2 average helical axis is oriented perpendicular to the membrane surface, with the N(Gly3)- and C(Ser18)-terminal helical rods inclined at ∼45° to the average helical axis (Table 1). Pin2 probably forms an open α-helical bundle or twisted-barrel type oligomer by self-association, where a pore is formed within the peptide complex. At temperatures below Tm the lateral diffusion rate slows down, suggesting that the pores associate with each other. The small area of lipid bilayer surrounded by peptides would then form a small discoidal bilayer which would disperse, as previously observed by Dufourcq et al. (1986a) and Naito et al. (2000). The static 31P NMR isotropic signal reveals the isotropic dispersion of the small discoidal bilayers. Moreover, the static 13C=O NMR spectrum of Gly3 also shows an isotropic signal below Tm (not shown), even though the lateral diffusion rate of both lipids and peptides becomes slow at temperatures below Tm.
In conclusion, this article demonstrates a novel pore-forming mechanism for α-helical peptides.
Acknowledgments
We thank Prof. Matsuzaki for helpful discussion. We are grateful to Prof. Aimoto for the usage of the Microcal MCS calorimeter. We also thank Dr. M. Horikawa for synthesizing the 13C=O labeled Fmoc amino acid, Dr. E. Villegas for help during peptide synthesis, and Drs. M. Hisada and K. Masuda for acquiring the mass spectra of synthetic peptides.
Abbreviations used: DLPC, 1,2-dilauroyl-sn-glycero-3-phosphatidylcholine; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine; DMPG, 2-dimyristoyl-sn-glycero-3-phosphatidylglycerol; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine; DPC, dodecylphosphocholine; DSC, differential scanning calorimetry; DTPC, 1,2-ditetradecyl-sn-glycero-3-phosphatidylcholine; HPLC, high-performance liquid chromatography; HSQC, heteronuclear single quantum coherence; MALDI-TOF, matrix-assisted laser-desorption ionization-time-flight; MAS, magic-angle spinning; ODS, octa decyl silica; PC, phosphatidylcholine; PE, phosphatidylethanolamine; pin2, pandinin 2; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine; TPPI, time-proportional phase-incrementation.
References
- Afonin, S., U. H. N. Durr, R. W. Glaser, and A. S. Ulrich. 2004. “Boomerang”-like insertion of a fusogenic peptide in a lipid membrane revealed by solid-state 19F NMR. Magn. Reson. Chem. 42:195–203. [DOI] [PubMed] [Google Scholar]
- Almeida, F. C. L., and S. J. Opella. 1997. Fd coat protein structure in membrane environments: structural dynamics of the loop between the hydrophobic trans-membrane helix and the amphipathic in-plane helix. Mol. Biol. 270:481–495. [DOI] [PubMed] [Google Scholar]
- Andreu, D., and L. Rivas. 1998. Animal antimicrobial peptides: an overview. Biopolymers. 47:415–433. [DOI] [PubMed] [Google Scholar]
- Bak, M., R. P. Bywater, M. Hohwy, J. K. Thomsen, K. Adelhorst, H. J. Jakobsen, O. W. Sørensen, and N. C. Nielsen. 2001. Conformation of alamethicin in oriented phospholipid bilayers determined by 15N solid-state nuclear magnetic resonance. Biophys. J. 81:1684–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger, B., and S. J. Opella. 1991. Flat-coil probe for NMR spectroscopy of oriented membrane samples. J. Magn. Reson. 95:585–588. [Google Scholar]
- Bechinger, B. 1996. Towards membrane protein design: pH-sensitive topology of histidine-containing polypeptides. J. Mol. Biol. 263:768–775. [DOI] [PubMed] [Google Scholar]
- Bechinger, B., M. Zasloff, and S. J. Opella. 1998. Structure and dynamics of the antibiotic peptide PGLa in membranes by solution and solid-state nuclear magnetic resonance spectroscopy. Biophys. J. 74:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1462:157–183. [DOI] [PubMed] [Google Scholar]
- Corzo, G., P. Escoubas, E. Villegas, K. J. Barnham, W. He, R. Norton, and T. Nakajima. 2001. Characterization of unique amphipathic antimicrobial peptides from venom of the scorpion Pandinus imperator. Biochem. J. 359:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, T. A., and S. J. Opella. 1994. Solid-state NMR structural studies of peptides and proteins in membranes. Curr. Opin. Struct. Biol. 4:574–581. [Google Scholar]
- Cullis, P. R., P. W. M. Van Dijck, B. De Kruijff, and J. De Gier. 1978. Effects of cholesterol on the properties of equimolar mixtures of synthetic phosphatidylethanolamine and phosphatidylcholine. A 31P NMR and differential scanning calorimetry study. Biochim. Biophys. Acta. 513:21–30. [DOI] [PubMed] [Google Scholar]
- Dempsey, C. E., and A. Watts. 1987. A deuterium and phosphorus-31 nuclear magnetic resonance study of the interaction of melittin with dimyristoylphosphatidylcholine bilayers and the effects of contaminating phospholipase A2. Biochemistry. 26:5803–5811. [DOI] [PubMed] [Google Scholar]
- Dufourcq, E. J., J. F. Faucon, G. Fourche, J. Dufourcq, T. G. Krzywicki, and M. le Marire. 1986a. Reversible disc-to-vesicle transition of melittin-DPPC complexes triggered by the phospholipid acyl chain melting. FEBS Lett. 201:205–209. [Google Scholar]
- Dufourcq, E. J., I. C. P. Smith, and J. Dufourcq. 1986b. Molecular details of melittin-induced lysis of phospholipid membranes as revealed by deuterium and phosphorus NMR. Biochemistry. 25:6448–6455. [DOI] [PubMed] [Google Scholar]
- Hallock, K. J., D. K. Lee, J. Omnaas, H. I. Mosberg, and A. Ramamoorthy. 2002. Membrane composition determines pardaxin's mechanism of lipid bilayer disruption. Biophys. J. 83:1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallock, K. J., D. K. Lee, and A. Ramamoorthy. 2003. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys. J. 84:3052–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler Wildman, K. A., D. K. Lee, and A. Ramamoorthy. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 42:6545–6558. [DOI] [PubMed] [Google Scholar]
- Hartzell, C. J., M. Whitfield, T. G. Oas, and G. P. Drobny. 1987. Determination of the 15N and 13C chemical shift tensors of l-[13C]alanyl-L-[15N]alanine from the dipole-coupled powder patterns. J. Am. Chem. Soc. 109:5966–5969. [Google Scholar]
- Herzfeld, J., R. G. Griffin, and R. A. Haberkorn. 1978. Phosphorus-31 chemical-shift tensors in barium diethyl phosphate and urea-phosphoric acid: model compounds for phospholipid head-group studies. Biochemistry. 14:2711–2718. [DOI] [PubMed] [Google Scholar]
- Hori, Y., M. Demura, T. Niidome, H. Aoyagi, and T. Asakura. 1999. Orientational behavior of phospholipid membranes with mastoparan studies by 31P solid state NMR. FEBS Lett. 455:228–232. [DOI] [PubMed] [Google Scholar]
- Hori, Y., M. Demura, M. Iwadate, A. S. Ulrich, T. Niidome, H. Aoyagi, and T. Asakura. 2001. Interaction of mastoparan with membranes studied by 1H-NMR spectroscopy in detergent micelles and by solid-state 2H-NMR and 15N-NMR spectroscopy in oriented lipid bilayers. Eur. J. Biochem. 268:302–309. [DOI] [PubMed] [Google Scholar]
- Kimura, S., A. Naito, S. Tuzi, and H. Saito. 2002. Dynamics and orientation of transmembrane peptide from bacteriorhodopsin incorporated into lipid bilayer as revealed by solid state 31P and 13C NMR spectroscopy. Biopolymers. 63:122–131. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. [DOI] [PubMed] [Google Scholar]
- Maloy, W. L., and U. P. Kari. 1995. Structure-activity studies on magainins and other host defense peptides. Biopolymers. 37:105–122. [DOI] [PubMed] [Google Scholar]
- Marassi, F. M., and S. J. Opella. 1998. NMR structural studies of membrane proteins. Curr. Opin. Struct. Biol. 8:640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi, F. M., S. J. Opella, P. Juvvadi, and R. B. Merrifield. 1999. Orientation of cecropin A helices in phospholipid bilayers determined by solid-state NMR spectroscopy. Biophys. J. 77:3152–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, D., and K. Wüthrich. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113:967–974. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., O. Murase, H. Tokuda, S. Funakoshi, N. Fujii, and K. Miyajima. 1994. Orientational and aggregational states of magainin 2 in phospholipid bilayers. Biochemistry. 33:3342–3349. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1995a. Translocation of a channel-forming antimicrobial peptide, magainin 2, across lipid bilayers by forming a pore. Biochemistry. 34:6521–6526. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., O. Murase, and K. Miyajima. 1995b. Kinetics of pore formation by an antimicrobial peptide, magainin 2, in phospholipid bilayers. Biochemistry. 34:12553–12559. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1996a. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 35:11361–11368. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., S. Yoneyma, O. Murase, and K. Miyajima. 1996b. Transbilayer transport of ions and lipids coupled with mastoparan X translocation. Biochemistry. 35:8450–8456. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K., K. Sgishita, N. Ishibe, M. Ueha, S. Nakata, K. Miyajima, and R. M. Epand. 1998. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 37:11856–11863. [DOI] [PubMed] [Google Scholar]
- Naito, A., T. Nagao, K. Norisada, T. Mizuno, S. Tuzi, and H. Saito. 2000. Conformation and dynamics of melittin bound to magnetically oriented lipid bilayers by solid-state 31P and 13C NMR spectroscopy. Biophys. J. 78:2405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito, A., T. Nagao, M. Obata, Y. Shindo, M. Okamoto, S. Yokoyama, S. Tuzi, and H. Saito. 2002. Dynorphin induced magnetic ordering in lipid bilayers as studied by 31P NMR spectroscopy. Biochim. Biophys. Acta. 1558:34–44. [DOI] [PubMed] [Google Scholar]
- North, C. L., M. B. Mathys, and D. S. Cafiso. 1995. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys. J. 69:2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opella, S. J., F. M. Marassi, J. J. Gesell, A. P. Valente, Y. Kim, M. O. Montal, and M. Montal. 1999. Structures of M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 6:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenner, E. J., R. N. A. H. Lewis, K. C. Neuman, S. M. Gruner, L. H. Kondejewski, R. S. Hodges, and R. N. McElhaney. 1997. Nonlamellar phases induced by the interaction of gramicidin S with lipid bilayers. A possible relationship to membrane-disrupting activity. Biochemistry. 36:7906–7916. [DOI] [PubMed] [Google Scholar]
- Scherer, P. G., and J. Seelig. 1989. Electric charge effects on phospholipid headgroups. Phosphatidylcholine in mixtures with cationic and anionic amphiphiles. Biochemistry. 28:7720–7728. [DOI] [PubMed] [Google Scholar]
- Separovic, F., R. Smith, C. S. Yannoni, and B. A. Cornell. 1990. Molecular sequence effect on the 13C carbonyl chemical shift shielding tensor. J. Am. Chem. Soc. 112:8324–8328. [Google Scholar]
- Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1462:55–70. [DOI] [PubMed] [Google Scholar]
- Shaka, A. J., P. B. Barker, and R. Freeman. 1985. Computer-optimized decoupling scheme for wideband applications and low-level operation. J. Magn. Reson. 64:547–552. [Google Scholar]
- Smith, R., F. Separovic, T. J. Milne, A. Whittaker, F. M. Bennett, B. A. Cornell, and A. Makriyannis. 1994. Structure and orientation of the pore-forming peptide, melittin, in lipid bilayers. J. Mol. Biol. 241:456–466. [DOI] [PubMed] [Google Scholar]
- Tachi, T., R. F. Epand, R. M. Epand, and K. Matsuzaki. 2002. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide-lipid interactions and selective toxicity. Biochemistry. 41:10723–10731. [DOI] [PubMed] [Google Scholar]
- Tilcock, C. P. S., M. B. Bally, S. B. Farren, and P. R. Cullis. 1982. Influence of cholesterol on the structural preferences of dioleoylphosphatidylethanolamine-dioleoylphosphatidylcholine systems: a phosphorus-31 and deuterium nuclear magnetic resonance study. Biochemistry. 21:4596–4601. [DOI] [PubMed] [Google Scholar]
- Toraya, S., K. Nishimura, and A. Naito. 2004. Dynamic structure of vesicle-bound melittin in a variety of lipid chain lengths by solid-state NMR. Biophys. J. In press. [DOI] [PMC free article] [PubMed]
- Wei, Y., D. K. Lee, and A. Ramamoorthy. 2001. Solid-state 13C NMR chemical shift anisotropy tensors of polypeptides. J. Am. Chem. Soc. 123:6118–6126. [DOI] [PubMed] [Google Scholar]
- Williams, K. A., N. A. Farow, C. M. Deber, and L. E. Kay. 1996. Structure and dynamic of bacteriophage IKe major coat protein in MPG micelles by solution NMR. Biochemistry. 35:5145–5157. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., D. Huster, A. Waring, R. I. Lehrer, W. Kearney, B. F. Tack, and M. Hong. 2001. Orientation and dynamics of an antimicrobial peptide in the lipid bilayer by solid-state NMR spectroscopy. Biophys. J. 81:2203–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., T. Hong, A. Waring, R. I. Lehrer, and M. Hong. 2002. Solid-state NMR investigations of peptide-lipid interaction and orientation of a β-sheet antimicrobial peptide, protegrin. Biochemistry. 41:9852–9862. [DOI] [PubMed] [Google Scholar]