Abstract
The ribosome is a large molecular complex that consists of at least three ribonucleic acid molecules and a large number of proteins. It translates genetic information from messenger ribonucleic acid and makes protein accordingly. To better understand ribosomal function and provide information for designing biochemical experiments require knowledge of the complete structure of the ribosome. For expanding the structural information of the ribosome, we took on the challenge of developing a detailed Thermus thermophilus ribosomal structure computationally. By combining information derived from the low-resolution x-ray structure of the 70S ribosome (providing the overall fold), high-resolution structures of the ribosomal subunits (providing the local structure), sequences, and secondary structures, we have developed an atomic model of the T. thermophilus ribosome using a homology modeling approach. Our model is stereochemically sound with a consistent single-species sequence. The overall folds of the three ribosomal ribonucleic acids in our model are consistent with those in the low-resolution crystal structure (root mean-square differences are all <1.9 Å). The large overall interface area (∼2500 Å2) of intersubunit bridges B2a, B3, and B5, and the inherent flexibility in regions connecting the contact residues are consistent with these bridges serving as anchoring patches for the ratcheting and rolling motions between the two subunits during translocation.
INTRODUCTION
The ribosome is a complex and dynamic cellular machine (Spirin, 2002) that is responsible for one of the most important cellular functions—protein synthesis. Structurally and functionally, the ribosome can be divided into the small (30S) and the large (50S) subunits. The 30S subunit is responsible for decoding genetic information residing in messenger ribonucleic acid (mRNA). The 50S subunit is responsible for catalyzing the peptidyl transferase reaction. Both subunits are composed of long RNA strands complexed with ribosomal proteins. Translocation, or the movement of the ribosome along the mRNA, is achieved by simultaneous motion of the small and large subunits, whose interdependent conformational changes act in concert to move the transfer RNAs (tRNAs) through the ribosome. This is an essential step of protein synthesis and remains poorly understood, even in light of recent cryoelectron microscopy (cryoEM) and x-ray crystal structures. This part of protein synthesis in particular demands knowledge of the atomic structure of the 70S ribosome complex—specifically, the intersubunit bridge regions. The pioneering study of Frank and Agrawal (2000) determined that the small subunit pivots with respect to the large subunit and identified the intersubunit contact regions using cryoEM. A candidate atomic structure of the intersubunit bridges (even in a single conformational state) is important for designing biochemical experiments to determine which intersubunit bridge regions are most critical for translocation (e.g., experiments which modify bases to test which groups are critical for translocation; Phelps et al., 2002). These critical regions represent obvious targets for antibiotics. Here, we present the first all-atom model of the 70S ribosome which has a consistent sequence for a single organism. This model contains the intersubunit bridges at atomic resolution.
Recently, a tremendous amount of x-ray crystallography data has been produced regarding the ribosome. After decades of concerted effort, high-resolution structures of the 30S ribosomal subunit (Schluenzen et al., 2000; Wimberly et al., 2000), and the 50S ribosomal subunit (Ban et al., 2000; Harms et al., 2001) have separately been solved. These high-resolution structures of the individual subunits provide a three-dimensional reference for interpreting existing biochemical and genetic data (Ramakrishnan, 2002). Recently, the structures of the Escherichia coli 70S ribosome were derived independently from low-resolution cryoEM (Gao et al., 2003) and low-resolution x-ray crystallography (Vila-Sanjurjo et al., 2003). Backbone structures based on low-resolution cryoEM were deposited in Protein Data Bank (accession codes 1P85, 1P86, 1P87) containing phosphorus atoms for rRNAs and Cα atoms for proteins. The large subunit proteins contain only Cα atoms in structures derived from low-resolution x-ray crystallography (PDB; accession codes 1PNX, 1PNY).
Although the low-resolution structure of the Thermus thermophilus 70S ribosome (Yusupov et al., 2001) provides accurate information of the backbone fold of the ribosome, it lacks atomic resolution information, which is not only important for planning biochemistry experiments where a particular chemical group on a nucleotide is modified, but also serves as an important test of the stereochemistry of the structure.
The large size of the complex (>105 atoms) presents a nontrivial task for standard modeling techniques. Yet, the major challenge in deriving an accurate all-atom ribosomal model lies in the delicate balance between the modeling of the overall fold and the modeling of the fine details in the local structure. Furthermore, the resultant model must be consistent with stereochemical (i.e., bond geometries and atomic contacts) and secondary structural (i.e., canonical A-type helices) constraints. The atomic structures of the large (Ban et al., 2000; Harms et al., 2001) and the small (Wimberly et al., 2000; Schluenzen et al., 2000) ribosomal subunits have revealed both the overall folds and detailed local structures of RNA and protein molecules constituting the ribosome. Comparison of these high-resolution crystal structures of the ribosomal subunits (Ban et al., 2000; Wimberly et al., 2000; Harms et al., 2001) with the low-resolution crystal structure of the ribosome (Yusupov et al., 2001) indicates that significant structural differences exist between the ribosomal RNAs (rRNAs) in the individual subunits and the 70S complex. These structural differences would produce inaccurate models of the rRNAs in the 70S ribosome if a straightforward homology modeling approach were used. To solve this problem, we have incorporated the low-resolution crystal structure (Yusupov et al., 2001) into our modeling process which was recently used to model the E. coli 30S ribosomal subunit (Tung et al., 2002). According to Yusupov et al. (2001), the phosphorus structures of the rRNAs can be traced with quite high accuracy from their crystal data of the ribosome at 5.5 Å resolution. Our all-atom model was constructed by using the backbone fold of the low-resolution 70S structure and the side-chain orientations of the high-resolution 30S and 50S structures via motif modeling. To be clear, we are in no way attempting to predict large-scale conformational changes of the ribosome. Rather, we are constructing a high resolution model of the same global conformation determined by low-resolution x-ray crystallography. Thus, using a knowledge-based approach, we have combined information derived from the low-resolution crystal structure of the 70S ribosome, high-resolution crystal structures of the large and small ribosomal subunits, sequences, and secondary structure, to develop an atomic model of the T. thermophilus 70S ribosome. In short, the “take home message” of this article is twofold: 1), we present the first all-atom model of the ribosome with a consistent single-species sequence that includes the intersubunit bridges, which are critical for understanding translocation at the molecular level; and 2), our modeling strategy serves as a proof-of-principle for using low-resolution structural data to construct all-atom candidate models for large biomolecular complexes, which are useful for designing biochemistry experiments where specific groups of RNA bases and protein residues are modified (Phelps et al., 2002). This model has lead to the hypothesis that three sets of intersubunit interactions (bridges) serve as possible anchoring patches for the ratchet/pivot motion (Frank and Agrawal, 2000) during translocation.
METHODS
Helix modeling
In general, RNA helices adopt a canonical A-form conformation whereas DNA helices adopt a canonical B-form conformation (Arnott et al., 1973). Using a set of reduced coordinates and a least-squares fitting procedure, a modeling method was developed that allows the prediction of DNA helical structures based on coordinates of the phosphorus atoms (Tung and Soumpasis, 1996). When used to predict various helical structures, this method has an accuracy of 0.5 Å overall root mean-square difference (RMSD). We have implemented the method to model structures of RNA helices from coordinates of phosphorus atoms. Due to the similarity in sizes, the structure of a purine-pyrimidine non-Watson-Crick basepair can be modeled directly, in a manner similar to that of a Watson-Crick basepair. To model the structure of a purine-purine basepair or a pyrimidine-pyrimidine basepair, due to differences in sizes when compared to a Watson-Crick basepair, adjustments are required in order for these basepairs to fit into a canonical A-type helix. This method was used to model 16S rRNA and was described in details previously (Tung et al., 2002).
Loop modeling
RNA loops can be divided into two groups, small and large, depending on their length and flanking structures. In this work, small loops are defined to be hairpin loops <15 nucleotides long and bulge loops <5 nucleotides long. For small loops, a motif modeling approach is applied. A motif library of small loops including both hairpin and bulge loops from known structures of 16S (PDB identification code 1FJF) and 23S (PDB identification codes 1JJ2 and 1KPJ) rRNAs has been developed. The structure of the target motif (phosphorus structure from the low-resolution crystal) is compared to those in the motif library. The structure of the motif in the library that has the smallest root-mean-square difference (in phosphorus structure) is chosen as a template to model the structure of the target motif. To model structures of large loops, a homology modeling approach similar to that described in our previous work (Tung et al., 2002) is used.
Structural optimization/constrained minimization
The purpose of subjecting the model to energy minimization is to refine reasonable stereochemistry produced by our motif modeling method. Both the overall folds and the local interactions for all the molecules constituting the ribosome are maintained through the use of our modeling approach and both the high-resolution and the low-resolution structures of the ribosome and subunits. The structure of the 70S ribosome was energy-minimized using the AMBER force-field (Cornell et al., 1995) and the NAMD2 (Kale et al., 1999) molecular dynamics simulation program on a 64 processor LINUX cluster. The phosphorus atoms in the model were targeted to those in the crystal structure (PDB identification codes 1GIX and 1GIY) using constraints of 5 kcal/mol Å2 (Fig. 1 a). To ensure the integrity of the helical structures in the model during the minimization, the bases involved in Watson-Crick pairing were constrained at 5kcal/mol Å2. Bases in the ultrastable tetraloops, including GNRA, CUYG, and UNCG (Batey et al., 1999), were constrained at 3 kcal/mol Å2. Where discontinuities in the rRNA existed, bases on the 5′ and 3′ ends were constrained to 5 kcal/mol Å2. Steepest descent minimization was performed for 5000 steps.
FIGURE 1.
Regions of ribosomal RNA in both the small and the large subunits. (a) Superposition of the modeled (pink) and the target (green) 5S rRNA is shown. (b) The original (cyan), the remodeled (pink), and the target (green) structures of the 16S spur region.
Solvent accessible surface area and interface area
Solvent accessible surface areas (SASAs) were calculated using version 2.1 of NACCESS (Hubbard and Thornton, 1993). The interface area (IA) of the ith bridge region is given by IAi = SASAA,i + SASAB,i − SASAAB,i, where SASAA,i is the SASA of the 30S portion of the ith bridge region, SASAB,i is the SASA of the 50S portion of the ith bridge region, and SASAAB,i is the SASA of the entire ith bridge region.
RESULTS AND DISCUSSION
With the high-resolution structure of the T. thermophilus small-subunit ribosome known, modeling the structure of the small-subunit in T. thermophilus 70S ribosome is relatively straightforward. The main challenge of this work is to model the structure of the large-subunit and to combine the two subunits to form the T. thermophilus 70S ribosome.
The 30S ribosomal subunit
The high-resolution structures of the 16S rRNA in the 30S ribosomal subunit have been solved independently and are available in the PDB (Berman et al., 2000) with identification codes 1FJF and 1FKA. We have chosen to use 1FJF as our template because it has a slightly better resolution (3.05 Å) than that of 1FKA (3.3 Å). If the 16S rRNA in 1FJF is used as the 16S model and superimposed with the low-resolution 16S structure (Yusupov et al., 2001) solved in the 70S ribosome (PDB identification code 1GIX), the overall RMS is 2.69 Å. Several regions show large differences between the two structures. One such difference occurs at the spur region (nucleotides 73–99) with an RMSD of 6.93 Å (the cyan and the green structures in Fig. 1 b). We note that E. coli numbering of rRNA positions is used unless otherwise noted. This large RMSD in the spur region could be due to the involvement of the spur in the crystal packing of 30S subunits (Yusupov et al., 2001). Our remodeled spur region (using the low-resolution phosphorus structure as the target) produces a significantly smaller RMSD of 2.46 Å for this region (the pink and the green structures in Fig. 1 b). Two additional regions, helix 41 (residues 1241–1296) and helix 44 (residues 1400–1502), were subjected to additional remodeling. The remodeled 16S produced overall RMSDs of 2.36 Å and 1.81 Å before and after the constrained minimization respectively.
Although structures of several regions in the 16S rRNA required remodeling, these regions are not in the vicinity of the small subunit proteins. Since the structures of these proteins are dictated by their sequences as well as the interactions with the 16S rRNA, it is reasonable to assume that the folds of the small subunit proteins in the isolated 30S ribosomal subunit and those in the 70S ribosome are similar. When the structures of the small-subunit proteins in our model are compared to those in the 30S (Wimberly et al., 2000), the RMSDs for the Cα atoms are small (ranging from 0.17 to 0.42 Å with an average of 0.22 Å), consistent with this assumption.
The 50S ribosomal subunit
The sequence of the 5S rRNA from T. thermophilus is highly homologous to those of Haloarcula marismortui and Deinococcus radiodurans (63% and 73% sequence identities, respectively). The higher sequence similarity makes the D. radiodurans 5S structure the choice of template for modeling T. thermophilus 5S structure. Using the sequence of T. thermophilus and the high-resolution D. radiodurans 5S structure, we model the atomic structure of the T. thermophilus 5S rRNA (Fig. 1 a). RMSDs between the model and the target are 1.76 and 1.23 Å, respectively, for the initial model and the model after the constrained minimization for the 5S rRNA.
Although the sequence homologies are high between the 23S of T. thermophilus and those of D. radiodurans and H. marismortui (73% and 60% identities, respectively), there are significant structural differences between the 23S in the 50S subunit and in the 70S ribosome (Yusupov et al., 2001). The entire 23S rRNA structure is modeled using a hybrid method that combines the detailed local folds derived from high-resolution crystal structures (Ban et al., 2000; Harms et al., 2001) and the global fold from the low-resolution crystal structure (Yusupov et al., 2001). Based on conformational variability, our modeling approach is divided into two parts: 1), modeling of the double-stranded (helix) regions, and 2), modeling of the single-stranded (loop) regions. An algorithm developed previously for modeling structures of nucleic acid helices with known phosphorus structures is used to model the structures of helical regions. A motif library was developed and used to model structures of short single-stranded regions (up to 15 nucleotides for hairpin loops and 5 nucleotides for bulge loops). A homology modeling approach similar to that described in our previous work (Tung et al., 2002) is used to model structures of large single-stranded loop regions. The overall RMSDs are 2.49 and 1.72 Å, respectively, for the initial 23S rRNA model and the model after the constrained minimization from the target.
Since the atomic structures of the T. thermophilus large-subunit proteins are not known experimentally, significant effort is required to model these proteins. We use a conservative approach of modeling only those proteins which have a known target sequence, a known template Cα structure with high sequence identity to the target, and a known all-atom homolog. There are a total of fourteen large subunit proteins that have all three sets of information (i.e., Cα structures from D. radiodurans, all-atom structures from H. marismortui and sequences from T. thermophilus). The structures of a portion of the large subunit proteins from H. marismortui are determined to atomic resolution using x-ray crystallography (PDB accession code 1JJ2). The large-subunit proteins in the low-resolution crystal structure of the ribosome (PDB accession code 1GIY) only have Cα atoms. Several large subunit proteins (e.g., L3, L4, L5, L15, L18, L24, and L29) in 1GIY have the sequences of the corresponding proteins in H. marismortui. Likewise, large subunit proteins in D. radiodurans (1KPJ) only have Cα structures. Furthermore, the sequence homologies between large-subunit proteins from T. thermophilus and H. marismortui are significantly lower than between those from T. thermophilus and D. radiodurans. With these considerations, information from different sources has to be merged for our modeling purpose. We modeled the T. thermophilus large subunit protein main-chain structures using the Cα structures from D. radiodurans. The side chains are modeled using the approach of Tung (1999). In this approach, side-chain atoms are attached to the main chain with torsional angles maintained close to those in the template. This particular consideration has the advantage of allowing the target and the template residues to utilize the same, often limited, space available to the side chains in a folded protein as well as proteins that are parts of a large molecular complex. The proteins were energy-minimized before and after inserting back into the 50S subunit. The insertion was achieved by matching the surrounding 23S rRNA structures between those from T. thermophilus and D. radiodurans.
The mRNA, tRNAs
The structures of the six nucleotides in the mRNA that interact with the anticodons of the A- and P-site tRNAs as well as the A-, P-, and E-site tRNAs themselves are present in the low-resolution crystal structure (PDB identification code 1GIX). During the protein synthesis, the ribosome is occupied by A- and P-site tRNAs in the pretranslocational state and P- and E-site tRNAs in the posttranslocational state. We have included the six nucleotide mRNA and the A- and P-tRNAs in our model to represent the pretranslocational structure of the ribosome.
70S ribosome
Based on the modeled structures of the individual molecules, the 70S ribosomal complex is assembled and subjected to a constrained energy minimization using the AMBER force-field (Cornell et al., 1995). The energy-minimized structure of the T. thermophilus 70S ribosome is shown in Fig. 2. When compared to the target, our models have RMSDs of 1.12, 1.70, and 1.64 Å, respectively, for 5S, 16S, and 23S ribosomal RNAs. The overall RMSD for the three rRNAs is 1.65 Å.
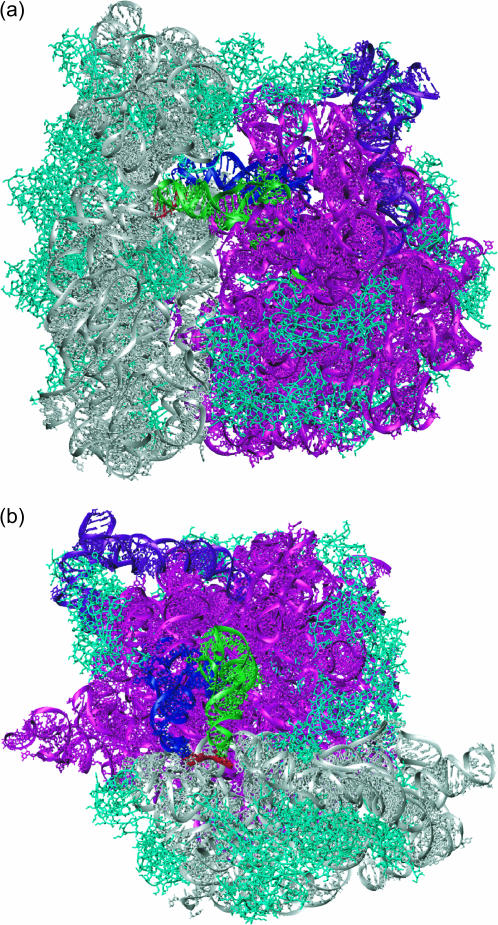
FIGURE 2.
(a) Structure of the T. thermophilus 70S ribosome. The 5S rRNA, 16S rRNA, 23S rRNA, A-site and P-site tRNAs, and mRNA are shown in purple, gray, magenta, green, blue, and red, respectively. Both the large subunit and the small subunit ribosomal proteins are shown in cyan. (b) Top view of the T. thermophilus 70S ribosome. The upper portion of the complex is not shown to view the tRNAs.
Peptidyl transferase center
The peptidyl transferase site on the 50S subunit is highly conserved across kingdoms and has a similar sequence composition in T. thermophilus, D. radiodurans, and H. marismortui. The T. thermophilus phosphorous peptidyl transferase center fold is consistent with both the D. radiodurans all-atom structure and the H. marismortui all-atom structure of the 50S subunit complexed with an acceptor stem analog. The local base conformation of the D. radiodurans all-atom structure and the H. marismortui differ. Thus, we have constructed two models of the 50S peptidyl transferase site. Both are models of the T. thermophilus peptidyl transferase center.
The first model is based on the D. radiodurans all-atom structure. The 2247–2257 hairpin of the T. thermophilus 23S rRNA is in close proximity of the CCA residues at the 3′ end of the acceptor arm of the P-site tRNA. The sequence of the T. thermophilus 2247–2257 hairpin is identical to that in D. radiodurans (residues 2226–2236). Using the structure of the D. radiodurans hairpin as a template, the structure of the T. thermophilus hairpin is modeled as shown in Fig. 3, a and b (pink), along with that from the T. thermophilus (PDB identification code 1GIX) phosphorus structure used as the target (green).
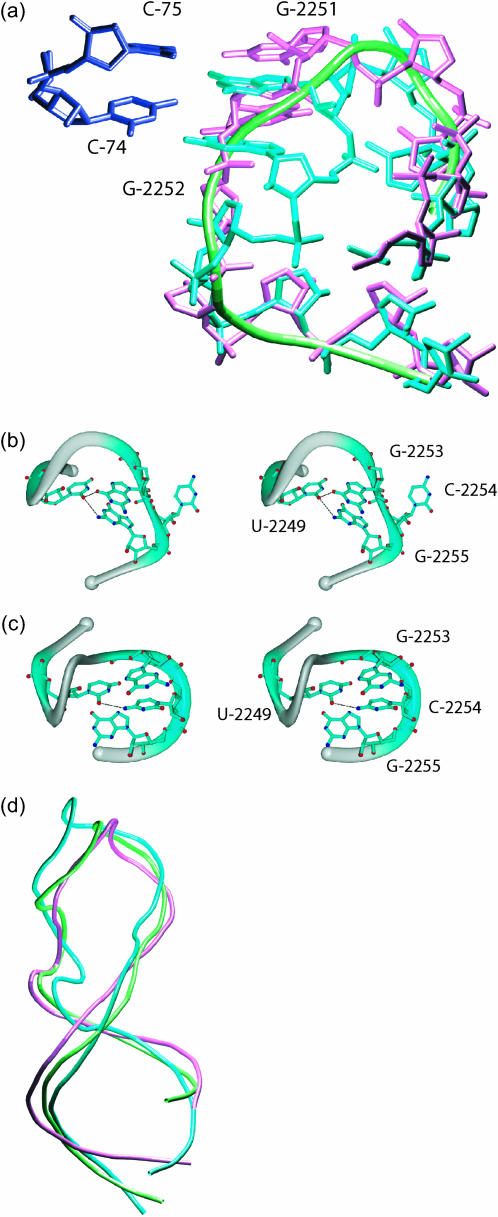
FIGURE 3.
Structures of a conformational switch in peptidyl transferase center and the sarcin/ricin loop. (a) Pretranslocation (cyan), modeled (pink), and targeted (green) structures of the 23S PTC regions are plotted alongside with C-74 and C-75 of the P-site tRNA. 23S rRNA G-2251 and G-2252 interact with C-74 and C-75 of P-site tRNA. (b) In the D. radiodurans-based model, U-2249 interacts with G-2253 and G-2255. In the H. marismortu-based model, U-2249 interacts with C-2254. (c) Both the NMR derived (cyan) and the x-ray derived (green) structures of the isolated SRL are more extended than the modeled SRL (pink) in the ribosome.
The second model is based on the H. marismortui all-atom structure. It has been previously demonstrated that in the case of H. marismortui, two guanines (residues 2251, 2252) in the above hairpin are basepaired with C-74 and C-75 of the P-site tRNA when the ribosome large subunit is in a pretranslocational intermediate state (Schmeing et al., 2002; PDB identification code 1KQS). However, these two guanines (residues 2251, 2252) in the D. radiodurans-based model (shown in pink in Fig. 3 a) do not make direct contacts with the CCA residues on the acceptor arm of the P-site tRNA. To model the structure of the hairpin in the pretranslocational intermediate state, the structure of the corresponding hairpin in H. marismortui (Schmeing et al., 2002) in the pretranslocational state is used as a template and the resultant structure (cyan) is shown in Fig. 3 a. A translation of ∼5 Å of the backbone is observed between structures of the hairpin in two different states. In the H. marismortui-based model, U-2249 interacts with C-2254. In the D. radiodurans-based model, C-2254 has flipped out and U-2249 interacts with G-2253 and G-2255 (Fig. 3 b). These two distinct structures of the 2247–2257 hairpin may serve as a localized, conformational switch during P-site tRNA binding or during translocation (Frank and Agrawal, 2000). We note that the overall fold of this local structure does not change; rather, the bases rearrange to establish basepairs with the acceptor stem. Insertion of E-tRNA does not result in steric conflicts with either model.
Sarcin/ricin loop
The sarcin/ricin loop (SRL) contains the longest (11 nucleotides) conserved rRNA sequence (Szewczak and Moore, 1995) that folds into a conformation required for ribotoxin (sarcin, ricin) recognition (Correll et al., 1998). In addition to ribotoxins, the SRL is also recognized by the pokeweed antiviral protein (Marchant and Hartley, 1995) and elongation factors (Szewczak et al., 1993; Correll et al., 1998). The SRL was previously defined as a 29-residue oligonucleotide, corresponding to C-2646 to G-2674 in E. coli 23S rRNA and U-4310 to U-4338 in rat 28S rRNA (Correll et al., 1998). The structure of the isolated SRL has been solved using NMR (Szewczak and Moore, 1995) and x-ray crystallography (Correll et al., 1998). Here, we compare the isolated SRL structures solved using two different experimental methods to that in the ribosome derived in our model. The RMSDs between the phosphorus atoms of our model and the six NMR-derived SRL structures (Szewczak and Moore, 1995) range from 4.28 Å to 6.18 Å, whereas the RMSD between our model and the x-ray structure (Correll et al., 1998) of the SRL is 2.52 Å. Fig. 3 c shows the superposition of our model (pink), the x-ray derived structure (green) and the NMR-derived structure (cyan) which has the smallest RMSD from our modeled SRL. In either case (NMR-derived or x-ray derived), the isolated SRL structure is more extended than the structure of SRL embedded in the ribosome. The structures of the universally conserved 11 nucleotide region of the SRL are under further study. The structure of this region is highly conserved with an overall RMSD (for all nonhydrogen atoms) between our model and the closest NMR-derived SRL of 1.12 Å (0.88, 1.03, and 1.69 Å, respectively, for base, sugar, and phosphates) and that between our model and the x-ray derived SRL of 1.04 Å (0.92, 0.98, and 1.38 Å, respectively, for base, sugar, and phosphates). The structural variations for the 11 nucleotide conserved region are significantly smaller than those for the whole SRL. This result is consistent with the notion that this universally conserved region can be treated as a structural motif, independent of its structural context (isolated or embedded in the ribosome) and environmental conditions (solution or crystal), as defined by Moore (1999).
Intersubunit bridge interactions
The 70S ribosome model allows us to inspect the intersubunit bridge interactions at atomic resolution. A bridge interaction is defined by regions on both subunits that are in direct contact (<4 Å). The list of intersubunit bridge interactions derived from the model (Table 1) is mostly consistent with those of Yusupov et al. (2001). Some regions are shifted by one nucleotide due to the differences in the structures used (all-atom versus phosphorus and Cα atoms only). This list of bridge interactions is also consistent with previous naming schemes (Frank et al., 1995; Yusupov et al., 2001).
TABLE 1.
Bridge interactions between 30S and 50S subunits in the 70S ribosome
| Bridge | 30S | 50S | IA [Å2] |
|---|---|---|---|
| B1a | S13: 78–79, 82–83, 93 | 886–888 | 382.0 |
| B1b | S13: 3, 6–7, 9, 68–69, 71 | L5: 113–116, 140–141 | 722.7 |
| B2a | 1406–1409, 1495–1496 | 1914–1915, 1917–1919 | 535.8 |
| B2b | 783–785, 791, 1516–1517 | 1835–1837, 1919, 1921, 1931 | 621.0 |
| B2c | 770–771, 773, 899, 900 | 1793, 1831–1832 | 322.1 |
| B3 | 1418–1419, 1421–1422, 1483–1486 | 1947–1950, 1959–1961 | 846.6 |
| L14: 46–49 | |||
| B4 | 762–763 | 713–717 | 575.5 |
| S15: 53, 56–57, 60,89; S17: 93, 104 | |||
| B5 | 1427–1430, 1472, 1474–1477 | 1686, 1688–1690, 1700–1704, 1988 | 1068.8 |
| L14: 17; L19: 108 | |||
| B6a | 1431–1432, 1464, 1467 | L19: 108–110 | 314.9 |
| B6b | 1446 | L19: 91 | 87.8 |
| B7a | 699, 702 | 1846, 1848, 1895–1897 | 435.8 |
| B8 | 345–346 | L19: 42, 44 | 270.8 |
| L14: 119 | |||
| Total | 6138.8 |
The A-minor motif and its variants (Doherty et al., 2001; Nissen et al., 2001), are types of bridge interactions commonly observed in the 70S ribosome. In B2a, the ribose of 23S:A1918 is in the minor groove of 16S helix 44 and is close to U1406. The 16S:C1496 and 23S:A1919 nucleotides are also in close contact. Similar to the A-minor motif, U-minor interactions occur at 16S:A1408-23S:U1914 and 16S:C1407-23S:U1917. Although bridge B5 has mainly O1P-O2′ interactions (16S:A1429-23S:G1704; 16S:U1428-23S:G1704), the A-minor-like interaction 16S:U1427-23S:G1703 also occurs. Finally, bridge B7a shows the flipped out 16S:A702 interacting with 23S:G1846 at the minor groove.
The IA, which represents the surface area on the subunits inaccessible to the solvent due to the formation of the complex, is an indicator of the binding strength (Nadassy et al., 1999) for the bridge interactions. The IA for the 70S ribosomal bridge interaction has a wide range (from <100 Å2 to >1000 Å2) as listed in Table 1. All the individual IAs are smaller than the value (1600 ± 400 Å2) observed in stable complexes (Nadassy et al., 1999). However, the sum of individual IAs gives a total area of 6139 Å2, large enough to form a stable complex. This observation is consistent with the ribosome adopting different conformations during the translocational cycle (Frank and Agrawal, 2000). The weak individual bridge interactions allow the subunits to adopt different conformations relative to each other during the process. One particular set of bridges (B2a, B3, B5) involves interactions between helix 44 of the small subunit and domain IV (nucleotides 1650–2000) of the large subunit (Fig. 4, a and b). These bridges include helix-to-loop (B2a), helix-to-helix (B3), and helix-to-helix/loop (B5) interactions (Fig. 4, c–e) and give a total area of ∼2,500 Å2, large enough to constitute a stable complex. The large number of bulge loops and mismatched basepairs in helix 44, combined with the separations between the bridge residues on domain IV, provide flexibility' connecting these particular bridge areas to the rest of the complex. These observations are consistent with the three bridges (B2a, B3, and B5) serving as anchoring patches for the ratchet/pivot motions between the two subunits. This motion has been observed in cryoEM studies (Frank and Agrawal, 2000) and has been determined to be the lowest frequency motion by normal mode analysis (Chacon et al., 2003). Interestingly, the strong bridge interactions of B2a, B3, and B5 are also consistent with the second lowest frequency normal mode motion, which entails the rolling of the small subunit with respect to the large subunit about the H44 axis.
FIGURE 4.
Intersubunit bridge interactions. (a) Surface of domain IV (pink) of the 23S rRNA has a structural indentation that is complementary to helix 44. (b) Bridges B2a, B3, and B5 are highlighted in white and green, respectively, for residues from 16S helix 44 and 23S domain IV. (c) Stereo plot of the bridge B2a. (d) Stereo plot of the bridge B3. (e) Stereo plot of the bridge B5. In panels c–e, 16S rRNA is shown in white (nonbridging nucleotides) and cyan (bridging nucleotides), and the 23S rRNA is shown in magenta (nonbridging nucleotides) and green (bridging nucleotides).
CONCLUSIONS
Although the low-resolution structure of the T. thermophilus 70S ribosome provides accurate information of the overall folds for the components, it lacks the information of detailed local interactions. To put the information derived from biochemical studies of the ribosome into its appropriate structural context, as well as to aid in the design of new biochemical studies, an atomic structure of the 70S ribosome is necessary. We took on the challenge of developing an atomic model of the 70S ribosome. Although the basic concept as well as the overall goals of our modeling approach are similar to those used in x-ray or NMR refinement, the main target function is different (i.e., experimental data versus the low-resolution structure). To achieve our goal, we have used novel methods such as RNA motif modeling, RNA homology modeling, and protein homology modeling using information derived from different sources as templates. As a result, we have developed an atomic model of the T. thermophilus 70S ribosome by combining information derived from the low-resolution crystal structure of 70S ribosome, high-resolution crystal structures of the large and small ribosomal subunits, sequences, and secondary structure. Our modeled structures of the rRNAs are consistent with those in the low-resolution crystal structure (with RMSDs all <1.9 Å), and therefore, accurate in overall fold. A homology modeling approach is used to model the local structures based on high-resolution structures of the ribosomal subunits to preserve the detailed local interactions. The energy minimization procedure guarantees a good stereochemistry for the final model. This model represents a proof-of-principle for constructing stereochemically correct all-atom structural models from low-resolution structural data.
Our model allows the study of detailed structures as well as the interactions between ribosomal subunits by calculating solvent accessible surface areas. We have seen that the sequence universally conserved SRL region is also structurally conserved. The SRL region can be treated as a structural motif independent of its structural contexts and environmental conditions. We propose that the two distinct structures of the 2247–2257 hairpin may serve as a local conformational switch during the translocation or during P-site tRNA binding. The intersubunit bridge interactions were studied by the calculation of the IAs. None of the individual bridges have an IA large enough to form a stable complex. However, the sum of individual IAs gives a total area of >6000 Å2, large enough to form a stable complex. One particular set of bridges (B2a, B3, B5) involves the interactions between helix 44 of the small subunit and domain IV of the large subunit. These bridges have a total area of ∼2500 Å2, also large enough to form a stable complex. These bridges may serve as an anchoring patch for the ratchet/pivot motions between the two subunits during translocation.
Acknowledgments
The authors thank Drs. Angel Garcia, Byron Goldstein, Carla Wofsy, and Tom Terwilliger for their critical reading of this manuscript.
This study was performed under the auspices of the Department of Energy under contract to the University of California. C.S.T. and K.Y.S. are supported by Los Alamos National Laboratory/Laboratory Directed Research and Development funding.
The accession codes for the small and the large subunits are 1TWT, 1TWV, respectively, in PDB.
References
- Arnott, S., D. W. L. Hukins, S. D. Dover, S. D. D. Fuller, and A. R. Hodgson. 1973. Structures of synthetic polynucleotides in the A-RNA and A′-RNA conformations: X-ray diffraction analyses of the molecular conformations of polyadenylic acid-polyuridylic acid and polyinosinic acid·polycytidylic acid. J. Mol. Biol. 81:107–122. [DOI] [PubMed] [Google Scholar]
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. Steitz. 2000. Structure of the 30S ribosomal subunit. Science. 289:905–920. [DOI] [PubMed] [Google Scholar]
- Batey, R. T., R. P. Rambo, and J. Doudna. 1999. Tertiary motifs in RNA structure and folding. Angew Chem. Int. Ed. 38:2326–2343. [DOI] [PubMed] [Google Scholar]
- Berman, H., M. J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucl. Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon, P., F. Tama, and W. Wriggers. 2003. Mega-dalton biomolecular motion captured from electron microscopy reconstructions. J. Mol. Biol. 326:485–492. [DOI] [PubMed] [Google Scholar]
- Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Gerguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force-field for the simulation of proteins, nucleic-acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- Correll, C. C., A. Munishkin, Y. L. Chan, Z. Ren, I. G. Wool, and T. A. Steitz. 1998. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. USA. 95:13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, E. A., R. T. Batey, B. Masquida, and J. A. Doudna. 2001. A universal model of helix packing in RNA. Nat. Struct. Biol. 8:339–343. [DOI] [PubMed] [Google Scholar]
- Frank, J., and R. Agrawal. 2000. A Ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 406:318–322. [DOI] [PubMed] [Google Scholar]
- Frank, J., A. Li, Y. H. Verschoor, J. Zhu, R. K. Lata, M. Radermacher, P. Penczek, R. Grassucci, R. K. Agawal, and S. Srivastava. 1995. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem. Cell Biol. 73:757–765. [DOI] [PubMed] [Google Scholar]
- Gao, H., J. Sengupta, M. Valle, A. Korostelev, N. Eswar, S. M. Stagg, S. C. Harvey, A. Sali, M. S. Chapman, and J. Frank. 2003. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 113:789–801. [DOI] [PubMed] [Google Scholar]
- Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 107:679–688. [DOI] [PubMed] [Google Scholar]
- Hubbard, S. J., and J. M. Thornton. ‘NACCESS’, computer program. Department of Biochemistry and Molecular Biology, University College, London.1993.
- Kale, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Philips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: Greater scalability for parallel molecular dynamics. J. Comp. Phys. 151:283–312. [Google Scholar]
- Marchant, A., and M. R. Hartley. 1995. The action of pokeweed antiviral protein and ricin A-chain on mutants in the α-sarcin loop of Escherichia coli 23S ribosomal RNA. J. Mol. Biol. 254:848–855. [DOI] [PubMed] [Google Scholar]
- Moore, P. B. 1999. Structural motifs in RNA. Annu. Rev. Biochem. 68:287–300. [DOI] [PubMed] [Google Scholar]
- Nadassy, K., S. J. Wodak, and J. Janin. 1999. Structural features of protein-nucleic acid recognition sites. Biochemistry. 38:1999–2017. [DOI] [PubMed] [Google Scholar]
- Nissen, P., J. A. Ippolito, N. Ban, P. B. Moore, and T. A. Steitz. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA. 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps, S. S., O. Jerinic, and S. Joseph. 2002. Universally conserved interactions between the ribosome and the anticodon stem-loop of A-site tRNA important for translocation. Mol. Cell. 10:799–807. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell. 108:557–572. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 102:615–623. [DOI] [PubMed] [Google Scholar]
- Schmeing,T. M., A. C. Seila, J. L. Hansen, B. Freeborn, J. K. Soukup, S. A. Scaringe, S. A. Strobel, P. B. Moore, and T. A. Steitz. 2002. A pretranslocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9:225–230. [DOI] [PubMed] [Google Scholar]
- Spirin, A. S. 2002. Ribosome as a molecular machine. FEBS Lett. 514:2–10. [DOI] [PubMed] [Google Scholar]
- Szewczak, A. A., and P. B. Moore. 1995. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 247:81–98. [DOI] [PubMed] [Google Scholar]
- Szewczak, A. A., P. B. Moore, Y. L. Chan, and I. G. Wool. 1993. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. USA. 90:9581–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung, C.-S., and D. M. Soumpasis. 1996. Structural prediction of A- and B-DNA duplexes based on coordinates of the phosphorus atoms. Biophys. J. 70:917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung, C.-S. 1999. Structural study of homeodomain protein-DNA complexes using a homology modeling approach. J. Biomol. Str. & Dyn. 17:347–354. [DOI] [PubMed] [Google Scholar]
- Tung, C.-S., S. Joseph, and K. S. Sanbonmatsu. 2002. All-atom homology model of the Escherichia coli 30S ribosomal subunit. Nat. Struct. Biol. 9:750–755. [DOI] [PubMed] [Google Scholar]
- Vila-Sanjurjo, A., W. K. Ridgeway, V. Seymaner, W. Zhang, S. Santoso, K. Yu, and J. H. D. Cate. 2003. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl. Acad. Sci. USA. 100:8682–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly, B. T., D. E. Brodersen, W. M. Clemons Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature. 407:327–339. [DOI] [PubMed] [Google Scholar]
- Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. D. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science. 92:883–896. [DOI] [PubMed] [Google Scholar]